Abstract
Proteins with LytM (Peptidase_M23) domains are broadly distributed in bacteria and have been implicated in a variety of important processes, including cell division and cell-shape determination. Most LytM-like proteins that have been structurally and/or biochemically characterized are metallo-endopeptidases that cleave crosslinks in the peptidoglycan (PG) cell wall matrix. Notable exceptions are the Escherichia coli cell division proteins EnvC and NlpD. These LytM factors are not hydrolases themselves, but instead serve as activators that stimulate PG cleavage by target enzymes called amidases to promote cell separation. Here we report the structure of the LytM domain from EnvC, the first structure of a LytM factor implicated in the regulation of PG hydrolysis. As expected, the fold is highly similar to that of other LytM proteins. However, consistent with its role as a regulator, the active site region is degenerate and lacks a catalytic metal ion. Importantly, genetic analysis indicates that residues in and around this degenerate active site are critical for amidase activation in vivo and in vitro. Thus, in the regulatory LytM factors, the apparent substrate binding pocket conserved in active metallo-endopeptidases has been adapted to control PG hydrolysis by another set of enzymes.
Keywords: autolysin, cytokinesis, morphogenesis, murein, sacculus, peptidoglycan
INTRODUCTION
Most bacteria surround themselves with a polysaccharide cell wall matrix called peptidoglycan (PG). This meshwork is essential for cellular integrity and is composed of glycan strands connected to one another by crosslinks between attached peptides (Typas et al., 2012). New PG material is synthesized by the high molecular weight penicillin-binding proteins (HMW-PBPs) (Typas et al., 2012). However, because the PG layer is a continuous molecule, enzymes that cleave bonds in the PG network, referred to as PG hydrolases or autolysins, are also required for the expansion and division of the matrix (Uehara and Bernhardt, 2011; Hashimoto et al., 2012; Typas et al., 2012; Singh et al., 2012).
Over the years, an incredibly diverse collection of enzymes with PG hydrolase activity have been identified. Every glycosidic or amide linkage in PG is a substrate for cleavage, and at least one, but often several distinct families of enzymes, are known to target individual bonds (Firczuk and Bochtler, 2007; Vollmer et al., 2008; van Heijenoort, 2011). Moreover, most bacterial genomes appear to encode a large number of different PG hydrolases. For example, the model gram-negative bacterium Escherichia coli encodes over 35 known or predicted PG hydrolases belonging to 12 different families (van Heijenoort, 2011). Importantly, however, our studies of the LytM-domain proteins in this organism indicate that at least some of the predicted PG hydrolases in bacterial genomes are likely to have developed alternative functions in addition to, or at the expense of, catalytic activity (Uehara et al., 2010).
Proteins with LytM (Pfam: Peptidase_M23) domains are broadly distributed in bacteria (Finn et al., 2008). They are homologs of lysostaphin, a well characterized zinc metallo-endopeptidase that cleaves the unique pentaglycine crosslinks found in Staphylococcus aureus PG (Browder et al., 1965). The protein family is named after LytM from S. aureus (SaLytM), which was the first member to be structurally characterized (Firczuk et al., 2005; Firczuk and Bochtler, 2007). Since the structure of LytM was described, members of the protein family from a diverse array of bacterial species have been implicated in a variety of important processes, including cell division, cell elongation, cell-shape determination, sporulation, and bacteriophage infection (Cohen et al., 2009; Uehara et al., 2009; Sycuro et al., 2010; Goley et al., 2010; Möll et al., 2010; Poggio et al., 2010; Bonis et al., 2010; Meisner and Moran, 2011; Singh et al., 2012). Several LytM-like proteins have also been structurally and/or biochemically characterized, the majority of which also appear to be metallo-endopeptidases that cleave the peptide moiety of PG to break crosslinks in the matrix (Lu et al., 2006; Cohen et al., 2009; Singh et al., 2012). The current exceptions are SpoIIQ from Bacillus subtilis (Meisner and Moran, 2011; Levdikov et al., 2012; Meisner et al., 2012) and the E. coli cell division proteins EnvC and NlpD (Uehara et al., 2010), for which no hydrolytic activity has been detected.
During sporulation, B. subtilis divides asymmetrically to form a larger mother cell compartment and a smaller (forespore) compartment. The mother cell then engulfs the forespore compartment and nurtures the developing spore within its cytoplasm. SpoIIQ interacts with SpoIIAH to form the core of a channel that connects the two compartments (Camp and Losick, 2008; Meisner et al., 2008; Camp and Losick, 2009; Doan et al., 2009; Meisner and Moran, 2011; Meisner et al., 2012). The channel is thought to supply the forespore with essential nutrients needed for the completion of sporulation. The structure of the SpoIIQ-SpoIIAH complex has been solved independently by two groups (Levdikov et al., 2012; Meisner et al., 2012). As expected, the LytM-like domain of SpoIIQ has a fold that resembles other members of the family. Thus far, however, SpoIIQ has only been implicated as a structural component of the inter-compartmental transport conduit. While other activities remain possible, it is unlikely to function as a metallopeptidase because it is missing residues implicated in metal-binding and catalysis in other LytM-like proteins, and no metal was observed in the region of the structure corresponding to the degenerate active site (Levdikov et al., 2012; Meisner et al., 2012).
The E. coli cell division proteins EnvC and NlpD are LytM-like proteins that also appear to have lost metallopeptidase activity (Uehara et al., 2010). These factors, along with the PG amidases AmiA, AmiB, and AmiC are required for processing the newly synthesized PG material that connects developing daughter cells at the division site (Heidrich et al., 2001; Priyadarshini et al., 2007; Uehara et al., 2009). Mutants defective for all three amidases or both LytM factors fail to cleave the connecting (septal) PG and form long chains of cells with distinct cytoplasmic compartments connected by shared layers of PG and a partially constricted outer membrane (Heidrich et al., 2001; Priyadarshini et al., 2007; Uehara et al., 2009). Our results indicate that rather than functioning as PG hydrolases themselves, EnvC and NlpD are recruited to the division site to directly activate septal PG hydrolysis by the PG amidases (Uehara et al., 2010). EnvC specifically activates AmiA and AmiB, while NlpD specifically activates AmiC. Structural and genetic analysis of the amidases has recently revealed that the activation mechanism involves the removal of an autoinhibitory alpha-helix from the amidase active site (Yang et al., 2012). Release of this helix is unlikely to involve proteolytic processing by the LytM factors because no change in the molecular weight of the amidases was observed following incubation with their cognate LytM activator (Uehara et al., 2010). Furthermore, like SpoIIQ, EnvC and NlpD are missing conserved metal-binding and catalytic residues conserved in active LytM-like metallopeptidases (Uehara et al., 2010). We have thus proposed that the LytM factors promote a reversible conformational change in the amidases that stimulates the release of the inhibitory helix from the active site (Yang et al., 2012). Interestingly, the ability of EnvC to activate AmiA and AmiB in vivo is itself subject to the control of the ABC transporter-like complex composed of the divisome proteins FtsE and FtsX (Yang et al., 2011). Although the mechanism of this regulation remains to be elucidated, the available data suggest that FtsEX may use conformational changes initiated by ATP hydrolysis in the cytoplasm to modulate EnvC activity on the external face of the cytoplasmic membrane (Yang et al., 2011). FtsEX is likely playing a similar regulatory role in controlling PG hydrolase activity throughout the bacterial domain (Crawford et al., 2011; Sham et al., 2011; Yang et al., 2011).
Here we report the structure of the LytM domain from EnvC, the first structure of a LytM factor implicated in the regulation of PG hydrolysis. As expected from sequence analysis, the domain has a fold that is highly similar to that of other LytM proteins. Importantly, the structure also shows that the region of EnvC corresponding to the metallopeptidase active site in other structures is indeed degenerate and lacks a catalytic metal ion. This observation strongly supports the model that EnvC is functioning as a regulator or PG hydrolysis and is not itself capable of cleaving PG. Residues in the EnvC structure required for amidase activation were identified using a genetic selection and site-directed mutagenesis. Our results indicate that residues in and around the degenerate EnvC active site are critical for amidase activation in vivo and in vitro. Thus, although the active site of EnvC is degenerate, the protein still employs the apparent substrate binding pocket conserved in active LytM-like metallopeptidases to promote cell wall hydrolysis by the amidases.
RESULTS
Structure of the LytM domain of EnvC
The primary sequence of EnvC contains three identifiable domains: a signal peptide (residues 1–34), a predicted coiled coil (CC) domain (residues 35–271), and a LytM domain (residues 318–413) (Ichimura et al., 2002; Hara et al., 2002; Bernhardt and de Boer, 2004). We crystallized a C-terminal fragment of EnvC calledLytEnvC (residues 278–419) that has previously been shown to be sufficient for amidase activation in vivo and in vitro (Uehara et al., 2010). The structure of this domain was solved by molecular replacement using the atomic coordinates of active LytM from S. aureus (Table S1) (Firczuk et al., 2005). Diffraction data obtained with the native crystals ofLytEnvC were of excellent quality and generated a density map in which we could trace the main chain from residue 290 to residue 419 for all four molecules in the asymmetric unit.
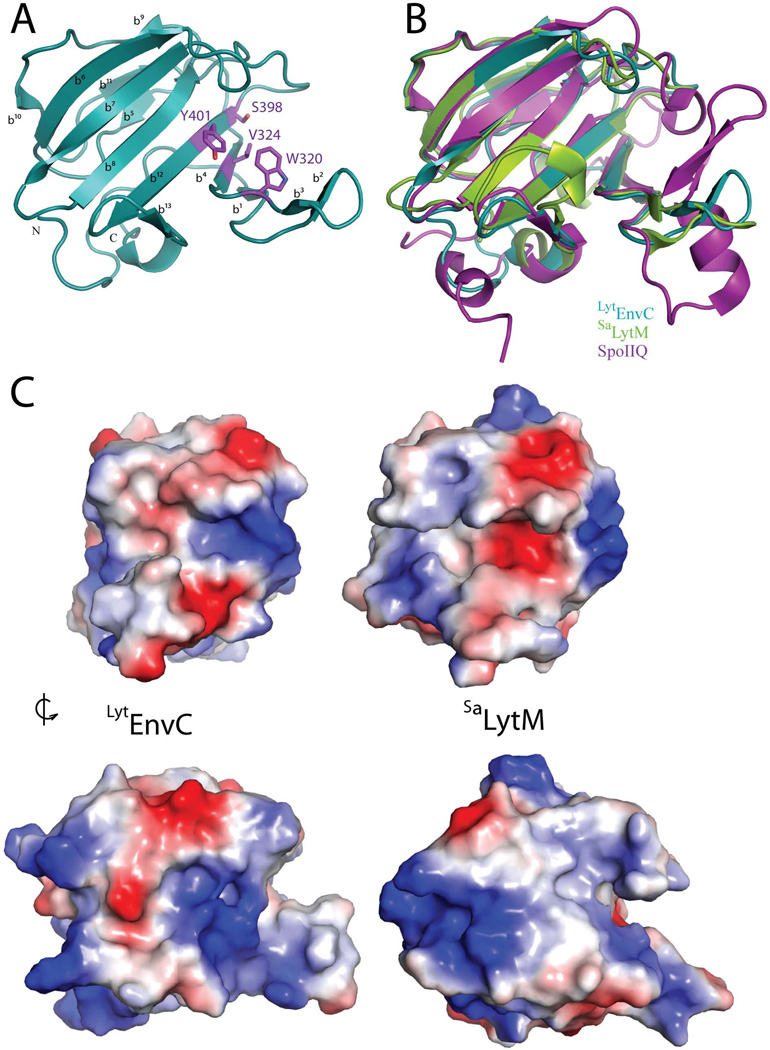
TheLytEnvC structure (Figure 1A) is a two-layered sandwich of anti-parallel β-sheets formed by a large sheet of 7 strands (β1-β4-β12-β8-β7-β6-β10) and a smaller sheet of 3 strands (β9-β11-β5) (Figure 2). Extending from the core β-sheets is a large β-turn (β2-β3) that forms the bottom portion of a prominent cleft in the structure (see below). Superposition of the Cα traces show that the four models in the asymmetric unit are nearly identical (rmsd of 0.05 Å between chains A and B; rmsd of 0.22 Å between chains A and C or D).
Figure 1. Structure ofLytEnvC and comparison with other LytM domains.
A. Ribbon diagram ofLytEnvC illustrated with N and C termini labelled. Residues corresponding to those required for metalloendopeptidase activity in other LytM domains are shown as purple colored sticks and labeled. β-sheets are numbered b1–b13. B. Superimposition ofLytEnvC (blue), truncatedSaLytM (green) (PDB ID: 2b13) (Firczuk et al., 2005) and B. subtilis SpoIIQ (violet) (PDB ID: 3TUF) (Levdikov et al., 2012; Meisner et al., 2012). C. Molecular surface ofLytEnvC (left) andSaLytM (right). Positive and negative charges are colored blue and red, respectively. Note that the cleft present on theLytEnvC structure is much wider than that ofSaLytM. Lower structures are rotated roughly 90 degrees counterclockwise relative to the upper structures.
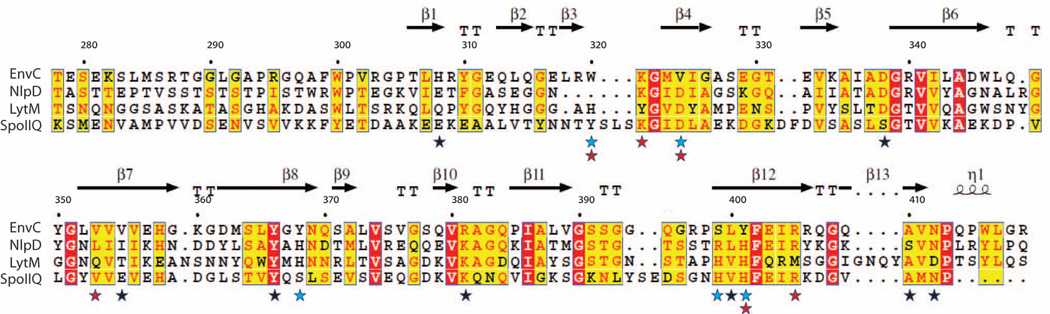
Figure 2. LytEnvC secondary structure map and sequence alignment with other LytM domains.
The secondary structure elements found in theLytEnvC structure are indicated above the sequence alignment. Residue numbering is for EnvC.LytEnvC residues important for function are highlighted with a red star.LytEnvC residues selected for mutagenesis but resulting in unstable mutant proteins are highlighted with a black star. Residues required forSaLytM catalysis are indicated by a blue star. Residues that are conserved or similar in at least three proteins in the alignment are boxed in yellow. Positions that are absolutely conserved are boxed in red. β, β-strand; T, turn
Comparison with other LytM domains
A search of the Dali database (Holm and Rosenström, 2010) confirmed the close similarity ofLytEnvC with the M23 family of metalloproteases, and in particular with theSaLytM glycyl-glycine endopeptidase (PDB 2b13; rmsd of 1.7 Å over 123 residues, Z-score = 19.4) (Firczuk et al., 2005; Firczuk and Bochtler, 2007) (Figure 1B and S1).LytEnvC also aligned well with the degenerate LytM protein SpoIIQ (PDB 3TUF; rmsd of 1.9 Å over 120 residues, Z-score = 17.6) (Figure 1B). The most striking difference between the structure ofLytEnvC and that of activeSaLytM is in the putative substrate binding cleft (Figure 1C and S1). The active site zinc ofSaLytM is located within a long, relatively narrow cleft in the structure formed by loops and the β-turn that extend from the core β-sheets. The corresponding cleft on theLytEnvC structure has a wider mouth. The cleft also does not extend all the way across the face of theLytEnvC protein as it does inSaLytM and therefore looks more like a three-finger pincer than the long narrow groove ofSaLytM (Figure 1C). Importantly, none of the Zn-coordinating residues inSaLytM (H210, D214 and H293) are conserved inLytEnvC (W320, V324, and Y401) (Figure 2 and S1) and accordingly, no Zn2+ ligand is found in the structure (Figure 1A). This observation is consistent with the fact that no hydrolase activity has been detected for EnvC (Uehara et al., 2010).
Genetic selection for the identification of defectiveLytEnvC variants
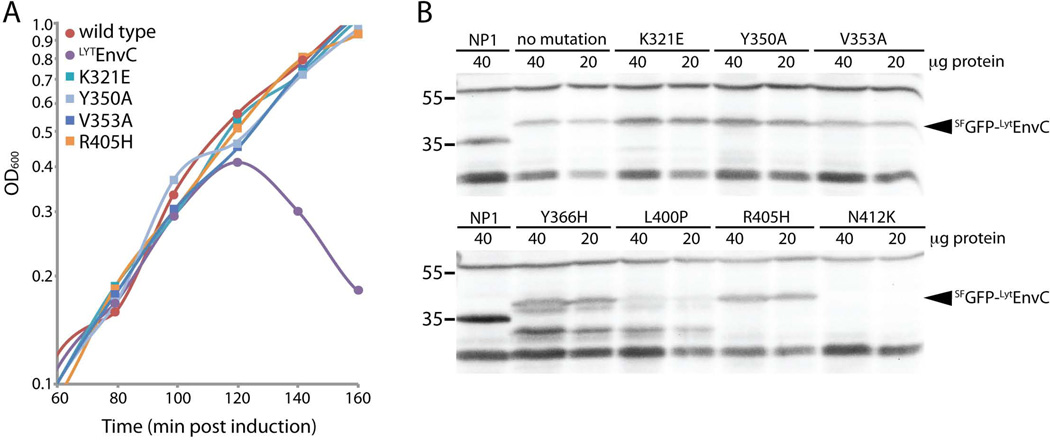
We employed both an unbiased genetic selection and site-directed mutagenesis to identify residues on theLytEnvC structure important for its amidase activation function. The unbiased selection for defective mutants was based on the lethal activity ofLytEnvC. We previously demonstrated that the production of the superfolding green fluorescent protein (SFGFP) fusionSFGFP-LytEnvC exported by the signal sequence of DsbA (ssDsbA) induces cell lysis and death in an amidase dependent manner (Uehara et al., 2010). We therefore reasoned that we should be able to identify residues inLytEnvC critical for amidase activation by selecting for survivors following induction of a mutagenized copy of ssdsbA-sfgfp -lytenvC. However, with such a selection, many of the resulting mutants would likely encode uninteresting variants ofLytEnvC that are either truncated, unstable, or defective for transport to the periplasm. In order to more easily eliminate the background of these mutants, we engineered an in-frame fusion of the bla reporter to the 3‘end ofssdsbA-sfgfp -lytenvC. The addition of β-lactamase to the C-terminus ofSFGFP-LytEnvC fusion did not significantly affect its lytic activity (data not shown). Importantly, when the fusion was produced in a resistant mutant strain lacking AmiA and AmiB, it conferred resistance to ampicillin, indicating that the β-lactamase portion of the fusion also remained functional (data not shown). Thus, ampicillin resistance could be used as a simple diagnostic for the production of full-length, exported variants ofSFGFP-LytEnvC-Bla. For the selection,lytenvC was mutagenized using error-prone PCR and inserted into an integration vector to generatessdsbA-sfgfp -lytenvC-bla under control of the arabinose promoter (Para). The resulting plasmid library was integrated at the phage λ att site of strain TB28 [WT]. Survivors were then selected on LB agar containing 0.2% arabinose and 50 µg/ml ampicillin. ThelytenvC portion of the fusion was amplified from the chromosome of the survivors and sequenced. Mutant genes encoding variant proteins with the single amino acid substitutions V353A, L400P, R405H, or N412K were identified (Table 1). These mutatedlytenvC genes were used to construct new integration vectors (pNP33-derivatives) encodingssDsbA-SFGFP fusions to theLytEnvC variants without Bla and with production of the fusion being controlled by the lactose promoter (Plac). The resulting constructs were integrated at the phage HK022 att site of strain TB28 [WT] and tested for lytic activity following induction with IPTG. As expected, all of the identified variants failed to induce cell lysis (Figure 3A, Table 1). Immunoblot analysis using anti-GFP antisera revealed thatSFGFP-LytEnvC(V353A) andSFGFP-LytEnvC(R405H) accumulated to near wild-type levels (Figure 3B). On the other hand,SFGFP-LytEnvC(L400P) appeared to be cleaved because only a lower molecular weight product was detectable in the blot. Accumulation ofSFGFP-LytEnvC(N412K) was not detected. We conclude thatLytEnvC(V353A) andLytEnvC(R405H) are likely to be functionally defective for amidase activation in vivo, while the variantsLytEnvC(L400P) andLytEnvC(N412K) are most likely poorly folded or otherwise unstable. Thus, while the bla fusion construct likely helped us reduce the background of misfolded or otherwise uninterestingLytEnvC variants identified in the selection, the system was obviously not perfect.
Table 1.
Summary of mutational analysisa
| EnvC variant |
LytEnvC lytic activity |
LytEnvC protein stability |
FLEnvC function |
FLEnvC septal localization |
LytEnvC amidase activationd (%) |
|---|---|---|---|---|---|
| wild type | + | + | + | + | 100.00 |
| H308Ac | + | ND | ND | ND | ND |
| W320Ac | + | ND | ND | ND | ND |
| K321Ac | + | ND | ND | ND | ND |
| V324Ac | + | ND | ND | ND | ND |
| R381Ec | + | ND | ND | ND | ND |
| Y401Fc | + | ND | ND | ND | ND |
| K321Ec | − | + | − | + | 7.12 |
| V324Ec | − | + | − | + | ND |
| Y350Ac | − | + | − | + | 22.31 |
| V353Ab | − | + | − | + | 10.04 |
| R405Hb | − | + | − | + | 4.73 |
| Y366Hc | − | +/− | ND | ND | ND |
| Y401Ec | − | +/− | − | + | ND |
| W320Ec | − | − | ND | ND | ND |
| D338Kc | − | − | ND | ND | ND |
| V355Ec | − | − | ND | ND | ND |
| L400Pb | − | − | ND | ND | ND |
| A410Ec | − | − | ND | ND | ND |
| N412Kb | − | − | ND | ND | ND |
Rows highlighted in green, pink, blue, or grey denote variants that are functional, non-functional/stable, non-functional/partially stable, or unstable, respctively.
Indicates a variant obtained by random selection.
Indicates a variant obtained by site directed mutagenesis.
Values represent the percentage of AmiB activity observed in reactions containingLytEnvC variants relative to wild-type after 30 minutes (see Figure 6)
Figure 3. Lytic activity and steady state accumulation of defectiveSFGFP-LytEnvC variants.
A. Cells from single colonies of strain TB28 [WT] or TB28 with plasmid pNP33 [Plac::ssdsbA-sfgfp-lytenvC] or pNP33 derivatives encoding the indicatedSFGFP-LytEnvC variants were grown in LB broth without IPTG at 30°C to an OD600 of 0.4–0.5. Cells were then diluted to an OD600 of 0.0125 in LB supplemented with 100 µM IPTG and growth was continued at 30°C. After one hour, OD600 measurements were taken every 20 mins.SFGFP-LytEnvC variants with the substitutions W320E, V324E, D338K, V355E, Y366H, L400P, Y401E, A410E, and N412K also failed to induce lysis (Table 1). B. Samples of cultures producingSFGFP-LytEnvC variants were harvested at 100 minutes post-induction for the preparation of protein extracts. Equivalent amounts of total protein for each sample were separated by SDS-PAGE and blotted to membranes.SFGFP-LytEnvC variants were detected with affinity purified anti-GFP antibodies (Rockland). Strain NP1 [zapA-gfp] was used as a positive control for the detection of GFP. Positions of molecular weight markers are indicated by the numbers (kDa) to the left of the blots. Non-specific cross-reacting bands are observed above 55 kDa and below 35 kDa. Results for a subset of theSFGFP-LytEnvC variants are presented in the figure. See Figure S2 for the remaining set of immunoblots.
Site-directed mutagenesis to identify additional defectiveLytEnvC variants
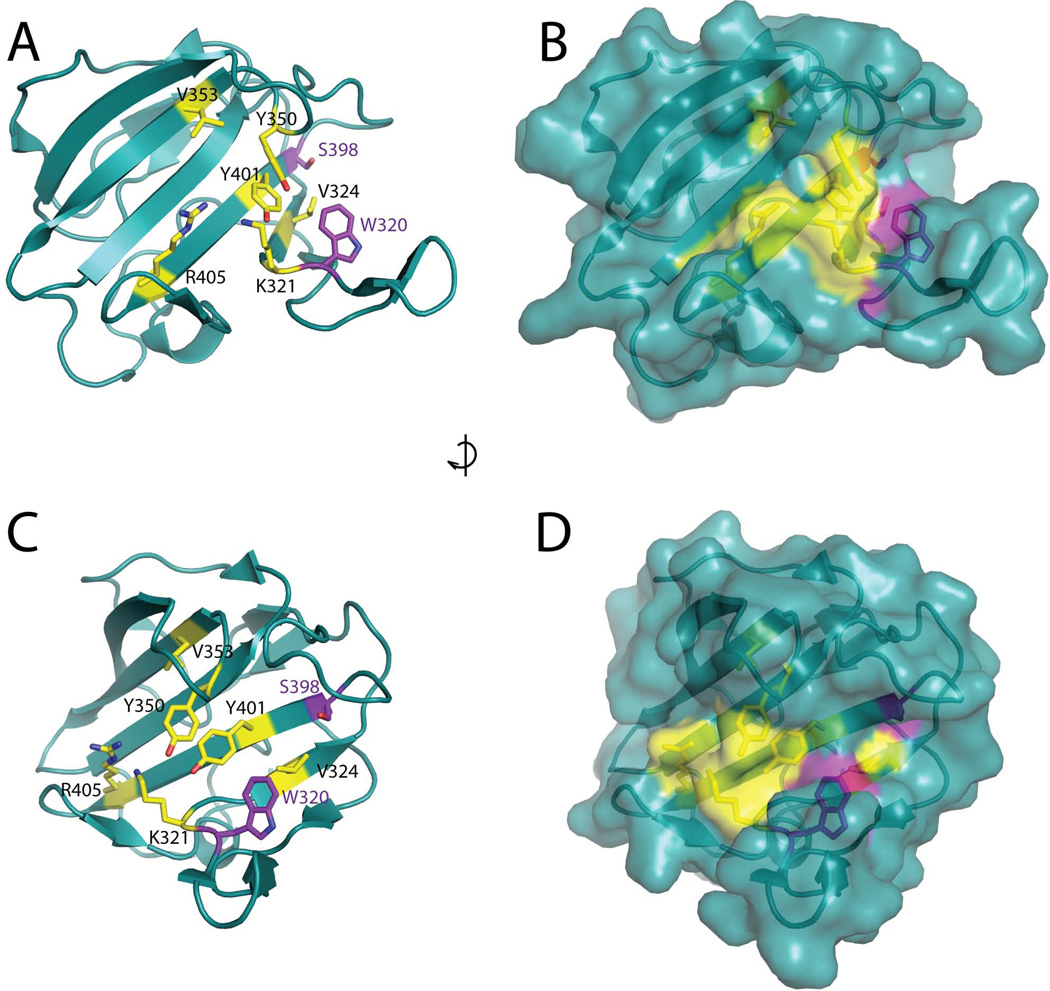
Residues V353 and R405 map to a region of theLytEnvC structure that is adjacent to the potential binding cleft where the degenerate metallopeptidase active site is located (Figure 4). To identify additional residues important for EnvC function, we used site-directed mutagenesis to create constructs producing variants ofSFGFP-LytEnvC with other amino acid substitutions in the degenerate active site region. We also generated variants with substitutions in conserved, surface-exposed residues in other areas of the structure and a variantLytEnvC(Y366H) corresponding to that encoded by the original envC mutant allele, envC 61, which was identified in a screen for temperature sensitive cell division mutants by Starka and co-workers in 1973 and mapped by Hara and colleagues many years later (Rodolakis et al., 1973; Hara et al., 2002). The vector pNP33 (Plac:: ssdsbA-sfgfp -lytenvC) was used as the template for site-directed mutagenesis. The resulting derivatives were then integrated at the phage HK022 att site of strain TB28 [WT] and tested for their ability to induce lysis upon induction with IPTG. The results are summarized in Table 1. With the exception ofSFGFP-LytEnvC(Y350A) andSFGFP-LytEnvC(Y366H), only substitutions that introduced a negative charge into the degenerate active site region (W320E, K321E, V324E, V355E, Y401E, and A410E) resulted in a non-lyticSFGFP-LytEnvC derivative (Figure 3A, Table 1). Interestingly, in many cases, variants where the same residues were substituted with alanine (W320A, K321A, V324A) retained lytic function (Table 1). Immunoblot analysis indicated thatSFGFP-LytEnvC(K321E),SFGFP-LytEnvC(V324E), andSFGFP-LytEnvC(Y350A) accumulate to near wild-type levels, whileSFGFP-LytEnvC(Y401E) andSFGFP-LytEnvC(Y366H) were partially degraded withSFGFP-LytEnvC(Y401E) appearing to accumulate more full length protein thanSFGFP-LytEnvC(Y366H). In the case of these partially degraded variants, however, they both accumulated comparable levels of full-length protein relative to the wild-type control such that one would expect them to have been lytic if they were functional (Figure 3B, S2, and Table 1). The remaining non-functional variants were apparently unstable and almost completely processed to lower molecular weight forms (Figure 3B, S2, and Table 1). Our results therefore indicate that in addition to V353 and R405 identified above, residues K321, V324, Y350, and Y401 are likely to be important for EnvC function. The functional importance of Y366 is less clear given the greater degree of degradation observed for this variant. Significantly, all of the functionally important residues are clustered in and around the degenerate LytM active site (Figure 4), suggesting that although this region has likely lost catalytic activity, it remains functional as an “active site” for amidase activation. Furthermore, because the addition of negative charges into the putative binding cleft was often required to disrupt activity, we infer that the highly positive electrostatic potential of this region is likely to be a key functional feature (Figure 1C).
Figure 4. EnvC residues important for function cluster in and around the degenerate active site cleft.
Residues important for EnvC function are shown as yellow colored sticks on the structure ofLytEnvC including two corresponding to those required for metallo-endopeptidase activity in other LytM domains. The two remaining “degenerate” active site residues are shown as purple colored sticks. Both ribbon (A, C) and surface (B, D) representations of the structures are shown. Side views (A, B) and front views (C, D) of the cleft are shown. Structures in C and D are rotated roughly 90 degrees clockwise relative to the structures in A and B.
Activity of full length EnvC variants with amino acid substitutions in the LytM domain
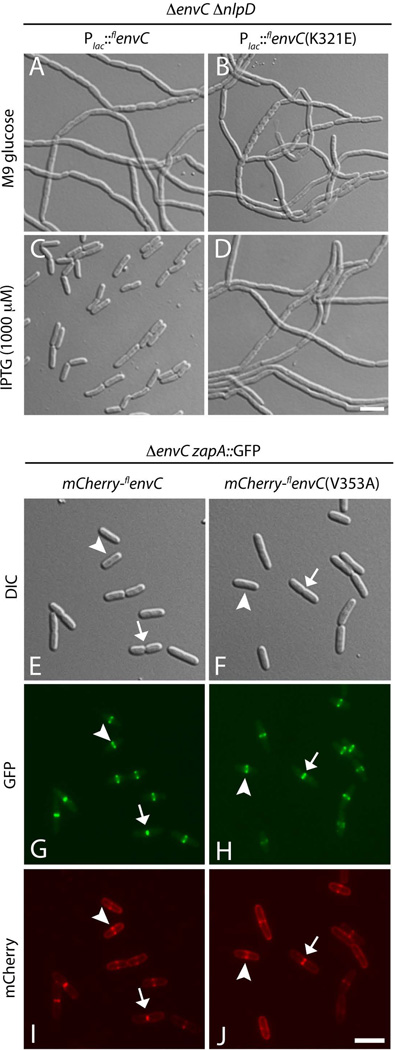
TheSFGFP-LytEnvC fusions that have been studied thus far are not subject to the normal regulatory controls governing amidase activation by full-length EnvC (FLEnvC) (Uehara et al., 2010). We therefore wondered whether the residues we identified as important for lysis induction bySFGFP-LytEnvC are also important for regulated amidase activation during cell division byFLEnvC. To investigate this, we generated constructs encodingFLEnvC derivatives with the identical amino acid substitutions found to abolish the lytic activity ofSFGFP-LytEnvC. We then assayed the ability of these constructs to correct the severe chaining phenotype of a strain inactivated for both EnvC and NlpD. We chose to monitor correction of the EnvC− NlpD− phenotype over that of a single envC deletion because the phenotype of the double mutant is more dramatic and correction is easier to assess. All of the amino acid substitutions that were found to inactivateSFGFP-LytEnvC also disrupted the ability of the correspondingFLEnvC variants to promote normal cell separation in the EnvC− NlpD−background (Figure 5A–D and S3). Additionally, consistent with our previous results demonstrating that the CC domain of EnvC is necessary and sufficient for its recruitment to the division site (Uehara et al., 2010), mCherry fusions to the defectiveFLEnvC variants all displayed normal septal localization patterns (Figure 5G–H and S4). Importantly, the mCherry-FLEnvC fusions were found to localize even in cells without visible constrictions (Figure 5G–H, arrowheads), indicating that the observed septal recruitment is unlikely to be the result of increased periplasmic volume at the division site due to delayed septal PG splitting (Bernhardt and de Boer, 2004). Thus, although theFLEnvC variants localize normally, they fail to promote normal cell separation.
Figure 5. Defective EnvC variants are impaired for amidase activation in vivo but still localize to the septal ring.
A–D. Cells of strain NP130(att HKNP62) [Δ envC ΔnlpD (Plac::flenvC (WT)] (A and C) or NP130(attHKNP61) [ΔenvC ΔnlpD (Plac::flenvC (K321E)] (B and D) were plated on M9 media containing either 0.2% glucose (A and B) or 0.2% maltose plus 1000 µM IPTG (C and D), and grown overnight at 37°C. Cells from single colonies were resuspended in medium and imaged on agarose pads using DIC optics. E–J. Cells of strain NP32(attHKNP62) [ΔenvC zapA- GFP (Plac::ssdsbA -mCherry-flenvC(WT)] (E, G, I) or NP32(attHKNP61) [ΔenvC zapA- GFP (Plac::ssdsbA-mCherry-flenvC (V353A)] (F, H, J) were grown overnight in LB at 37°C, diluted 1:100 in M9 glucose and grown at 30°C to an OD600 of 0.4–0.6. Cells were then imaged on agarose pads using DIC (E and F), GFP (G anf H), and mCherry (I and J) optics. Similar results were obtained with the entire set of defective EnvC variants (See Figures S3 and S4).
Activity ofLytEnvC variants in vitro
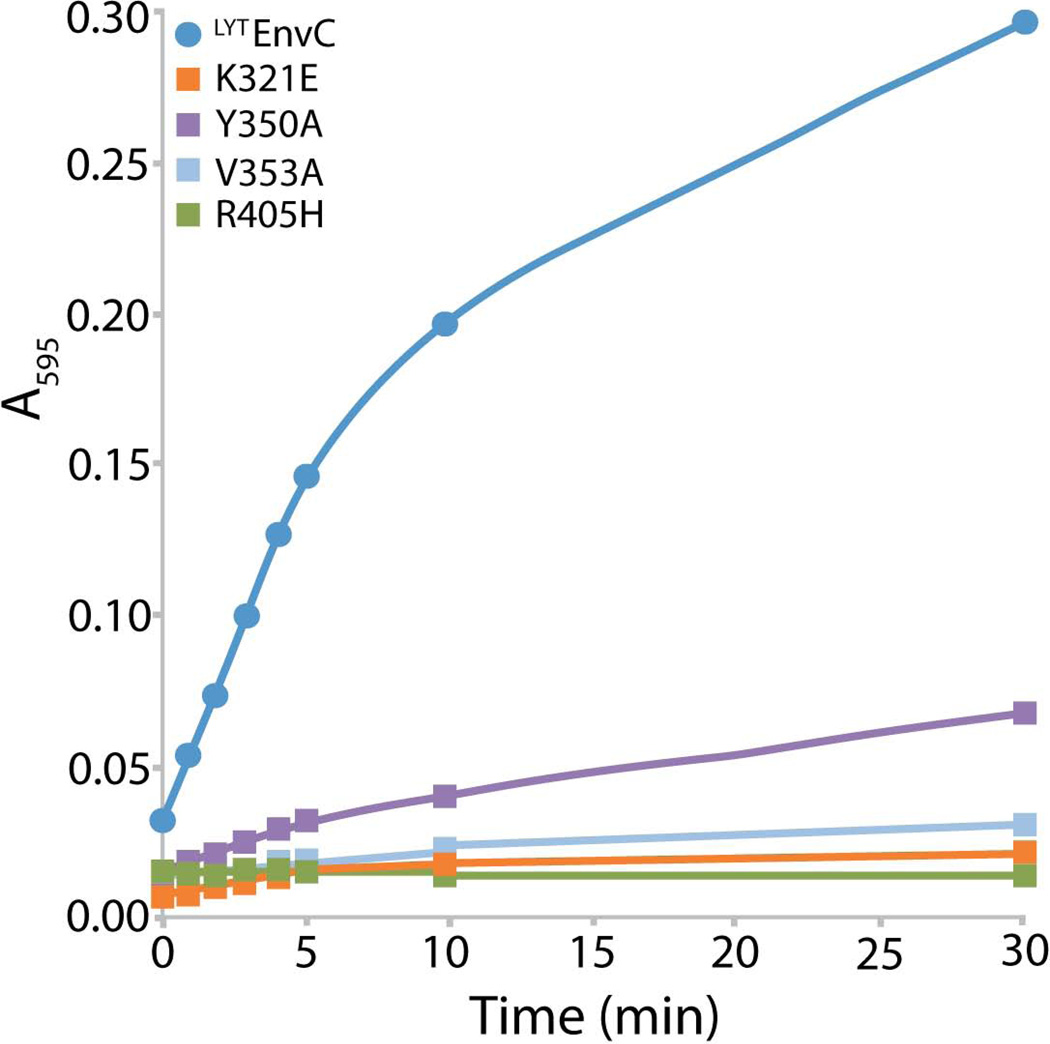
The simplest interpretation of the mutational analysis is that the defective EnvC variants described above have lost the ability to activate PG hydrolysis by the amidases. To test this, we purified untagged versions ofLytEnvC(WT) and the variantsLytEnvC(K321E),LytEnvC(Y350A),LytEnvC(V353A), andLytEnvC(R405H) and monitored their ability to activate AmiB in vitro. PG hydrolysis was monitored using PG sacculi covalently labeled with the dye Remazol Brilliant Blue (RBB). The RBB-labeled sacculi were added to reactions containing AmiB andLytEnvC or its derivatives (1 µM each). At the indicated time points following sacculi addition, a portion of the reaction was removed and terminated with the addition of an equal volume of ethanol. Once the time course was completed, undigested sacculi were pelleted and the absorbance of the supernatant in each sample was measured (Figure 6). As expected based on the in vivo analysis, all of theLytEnvC variants tested were defective for AmiB activation in vitro. OnlyLytEnvC(Y350A) retained a low but measurable rate of activation. We therefore conclude that the degenerate substrate binding cleft observed in theLytEnvC structure is the “active site” of EnvC required for it to stimulate amidase activity during cell division.
Figure 6. DefectiveLytEnvC variants fail to activate AmiB in vitro.
Reactions containing RBB-labeled PG, AmiB (1 µM), and the indicatedLytEnvC derivative (1 µM) were incubated at 37°C for the indicated times when they were terminated by the addition of ethanol. Undigested PG was pelleted by centrifugation and the absorbance of the supernatants at 595 nm were determined.
DISCUSSION
This study reports the first structure of a LytM domain protein functioning as an activator of PG hydrolases. We have previously inferred that mixtures of EnvC with either AmiA or AmiB possess robust PG hydrolysis activity because EnvC stimulates catalysis by the amidases as opposed to promoting a PG cleavage reaction itself that synergizes with amidase activity (Uehara et al., 2010). In support of the activation model, we failed to detect PG hydrolytic activity for EnvC alone, and sequence analysis suggested that EnvC lacks metal-binding residues conserved in other LytM peptidases (Uehara et al., 2010). The structure ofLytEnvC provides important confirmation of the activation model. While its fold is highly similar to that ofSaLytM and other proteins in the LytM family that have been structurally characterized, the region of theLytEnvC structure corresponding to the metallo-peptidase active site is devoid of Zn2+ and an obvious metal-chelating motif. Thus, the protein has almost certainly lost metallo-peptidase activity and is unlikely to function as a PG hydrolase itself. The lack of a catalytic metallo-peptidase center in theLytEnvC structure also supports the idea that amidase activation is proceeding via the induction of a reversible conformational change rather than through a proteolytic processing event.
The LytC-type amidases are also metallo-enzymes with a catalytic Zn2+ ion present in their active site cleft (Shida et al., 2001; Korndörfer et al., 2006; Mayer et al., 2011; Yang et al., 2012). We recently showed that cell separation enzymes belonging to this family are regulated by an autoinhibitory alpha-helix that occludes the active site and blocks access to substrate (Yang et al., 2012). Mutations predicted to destabilize the interaction of the autoinhibitory helix with the amidase active site led to elevated basal amidase activity in the absence of an activator (Yang et al., 2012). We therefore proposed that the amidases are likely capable of spontaneously interconverting between an active “open” and inactive “closed” conformation with the closed form predominating at equilibrium due to the strength of the interaction between the inhibitory helix and the active site (Yang et al., 2012). To activate the amidases, EnvC and NlpD most likely stabilize the open conformation of their cognate amidases, thus biasing the equilibrium to favor the active state. An attractive mechanism by which the open form of the amidase might be stabilized is through the direct binding of the autoinhibitory helix by the LytM activator. Furthermore, our structure-function analysis ofLytEnvC suggests that the amidase activation factors may use their degenerate metallo-endopeptidase active site cleft to mediate this binding event. UnlikeSaLytM, which has a narrow groove in its structure containing the catalytic Zn2+ ion that can likely only accept extended peptide substrates (Firczuk et al., 2005), the corresponding cleft in theLytEnvC structure is wide enough to easily accept the amidase regulatory alpha-helix. Consistent with this hypothesis, our mutational analysis indicates that residues in and around the degenerate active site cleft are critical for the amidase activation function of EnvC. Moreover, when molecular modeling was used to investigate the potential interaction of the regulatory helix of AmiB (residues 293–309) withLytEnvC, seven of the ten best rigid-body docking models identified a potential binding site for the helix in theLytEnvC cleft (Figure S5). Interestingly, in the structure of latent (inactive)SaLytM, the N-terminal regulatory domain of the proenzyme makes extensive contacts with the active site and the adjacent face of the endopeptidase domain (Odintsov et al., 2004). The analogous region of theLytEnvC structure was also implicated in amidase activation function by the mutational analysis, suggesting that it may provide a platform for an even larger protein-protein interaction interface with AmiA and AmiB that is needed to stimulate PG hydrolysis.
Our model for amidase activation predicts that the inactiveLytEnvC variants identified in this study should be defective for an interaction with the amidases. Unfortunately, we have been unable to test this directly because our attempts at detecting EnvC-AmiA or EnvC-AmiB complexes using gel-filtration chromatography or in vitro “pull-down” experiments have so far been unsuccessful. We suspect that this is due a relatively low affinity between EnvC and its cognate amidases, which is not unexpected given the regulatory nature of the interaction. Further experimentation is therefore needed to test the activation model proposed here. To this end, we are currently pursuing site-specific crosslinking and co-crystalization studies to better understand the nature of the EnvC-amidase complex, how the interaction stimulates PG cleavage by the amidases, and ultimately how this activation reaction is controlled by the FtsEX ABC-system to properly coordinate septal PG splitting with PG biogenesis at the cytokinetic ring. Continued structural and genetic analysis of this PG remodeling system should also reveal opportunities to subvert its regulation for the development of new classes of lysis-inducing antibiotics.
EXPERIMENTAL PROCEDURES
Media, bacterial strains, and plasmids
Cells were grown in LB [1% tryptone, 0.5% yeast extract, 0.5% NaCl] or minimal M9 medium (Miller, 1972) supplemented with 0.2% casamino acids and 0.2% sugar (glucose, arabinose or maltose as indicated). Unless otherwise indicated, antibiotics were used at 10 (chloramphenicol; Cm), 15 (ampicillin; Amp) or 20 (kanamycin; Kan) µg/ml. Bacterial strains used are listed in Table S2. All E. coli strains used in the reported experiments are derivatives of MG1655. Plasmids used in this study are listed in Table S3.
Selection for defectivelytenvC mutants
To mutagenizelytenvC, the corresponding fragment of envC was amplified with Taq DNA polymerase and inserted into pTD70 [Para::ssdsbA-sfgfp-stop-bla], generating pTU206 [Para::ssdsbA-sfgfp-lytenvC-bla], and transformed into DH5α(λpir). The resulting transformants were pooled and plasmid DNA was purified. Plasmid from the pool were integrated at the λ att site of MG1655 as described previously (Haldimann and Wanner, 2001). Pooled integrants were plated on LB containing 0.2% arabinose and 50 µg/ml ampicillin to select for constructs encoding defectiveSFGFP-LytEnvC-Bla fusions that fail to induce cell lysis. Note that the pTD70 parent plasmid has a stop codon in the linker betweenssdsbA-sfgfp and bla, so that plasmids lacking an insertedlytenvC gene are eliminated by the ampicillin selection. Eight survivors were isolated and the mutation sites in the pTU206 derivatives were mapped by sequencing. Six isolates encodedSFGFP-LytEnvC-Bla with an N412K substitution (base changes: AAT to AAA) with or without additional substitutions. The other two isolates encoded variants with either an L400P or V353A substitution (base changes: CTC to CCC or GTG to GCG, respectively). A second round of selections was performed to identify additional non-functionallytenvC alleles. An identical procedure was followed except thatlytenvC was amplified under conditions that enhance mutagenesis by Taq polymerase (high MgCl2, dCTP, and dTTP concentrations). In this round, 17/18 isolates encodedSFGFP-LytEnvC-Bla with an N412K substitution, while the remaining isolate encoded a fusion with an R405H substitution (base change: CGC to CAC).
Protein purification
All proteins were over-expressed and purified with a 6xHis-SUMO (HSUMO) tag fused to their N-termini (Mossessova and Lima, 2000; Marblestone et al., 2006). The sequence of the affinity tag in all cases was MRGSHHHHHHMASG. The SUMO sequence was amplified from the Saccharomyces cerevisiae genome (gene Smt3) as described earlier (Bendezú et al. 2009). After purification of the H-SUMO fusion protein by metal-affinity chromatography, the H-SUMO tag was removed using 6xHis-tagged SUMO protease (H-SP) (Bendezú et al., 2009). Cleavage reactions were passed through Ni-NTA resin to remove free H-SUMO and H-SP, yielding a pure preparation of the desired protein. As a result of fusion construction, all proteins have an additional Ser residue at their N-termini.
LytEnvC(WT),LytEnvC(K321E),LytEnvC(R405H),LytEnvC(Y350A), andLytEnvC(V353A) were purified from BL21(λDE3) containing pDY280, pDY281, pDY282, pDY283, and pDY286, respectively. Overnight cultures were diluted 1:100 into 1 L of LB supplemented with ampicillin (50 µg/ml) and glucose (0.04%) and cells were grown at 30°C to an OD600 of 0.47–0.71. IPTG was then added to 1 mM and growth was continued for an additional 3.0–3.5 hours when cells were harvested by centrifugation. Cell pellets were resuspended in 20 ml buffer A [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10% glycerol] with 20 mM imidazole, split into two 10 ml aliquots, and stored at −80°C prior to use for protein purification. For purification, a 10 ml aliquot (representing 0.5 L of induced culture) was thawed, and the cells were disrupted by sonication. Cell debris and membranes were pelleted by centrifugation at 105,000×g for one hour at 4°C. H-SUMO fusions were purified using 2.0 ml Ni-NTA agarose resin (Qiagen) according to the manufacturer’s instructions. Resin was equilibrated in buffer A with 20 mM imidazole, washed with buffer A containing 20 mM imidazole once and then buffer A containing 50 mM once. The fusion proteins were then eluted with buffer A containing 300 mM imidazole. The purified H-SUMO fusions were incubated with a 1:1000 dilution of an H-SP preparation (Uehara et al., 2010) and dialyzed overnight in buffer A containing no imidazole at 4°C. The following morning the protein preparation was then passed over 2.0 ml Ni-NTA agarose resin (Qiagen). Untagged proteins were collected in the flowthrough, aliquoted and stored at −80°C in buffer A containing no imidazole. Protein concentrations were determined using the NI (Non-Interfering) Protein Assay (G-Biosciences). AmiB was purified as previously described (Uehara et al., 2010).
Structure determination
LytEnvC(WT) purified as described above except that the flowthrough fractions from the final NiNTA column were concentrated with Amicon Ultra centrifugation filter units (molecular weight cut-off of 3,000; Millipore) and injected onto a 10/300 GL Superdex 200 gel filtration column (GE Healthcare).LytEnvC was eluted with 25 mM Tris, pH 8.0, 150 mM NaCl and again concentrated with Amicon Ultra centrifugation filter units. Protein concentrations were measured by using a Bradford assay (Bio-Rad).
Crystallization trials were performed in 96-well Greiner Crystal Quick plates with 100 nL protein (30 mg/mL) and 100 nL reservoir solution. NativeLytEnvC crystallized in 0.2 M sodium iodide, 17% PEG 3350, pH 6.9 at 20 °C in 24 to 48h.LytEnvC crystals were cryoprotected by exchanging the mother liquor with crystallization solution containing 20% glycerol, and then flash-frozen in liquid nitrogen.
LytEnvC crystallized in the space group P31 with the following unit cell parameters: a = 57.404 Å, b = 57.404 Å, c = 129.320 Å, α = β = 90°C and γ = 120° (Table S1). Native diffraction data were collected at ID14-1 of the European Synchrotron Facility (Grenoble, France). Phase determination was carried out by the molecular replacement method (CCP) by using the structure of S. aureus truncated LytM (PDB accession no. 2b13) (Firczuk et al., 2005). The diffraction data were reduced by using XDS (Kabsch, 2010), and the crystallographic model was refined by using CCP4 programs iterated with manual rebuilding. Water and ion molecules were added manually in COOT (Emsley et al., 2010). Model quality was determined by using Molprobity (Chen et al., 2010), with all residues within most favorable or allowed regions of the Ramachandran plot. A summary of the data collection and refinement statistics is given in Table S1. The coordinates and structure factors have been deposited in the PDB, with accession no. 4bh5.
Preparation of sacculi labelled with RBB
Sacculi were prepared from strain TU163 (Δlpp) as previously described (Uehara et al., 2009). Isolated sacculi were treated with 200 µg/ml amylase (Sigma, St Louis, MO) at 37°C for 2 hours in 1× PBS and washed with water. The amylase-treated sacculi were then incubated with 20 mM RBB (Sigma) in 0.25 M NaOH overnight at 37°C. The preparation was neutralized with HCl, and RBB-labelled sacculi were pelleted by centrifugation (21000×g, 20 min, room temperature). The sacculi were then repeatedly resuspended in water and pelleted by centrifugation until the supernatant was clear. The final pellet was resuspended in water containing 0.02% azide and stored at 4°C.
Dye-release assay for PG hydrolysis
RBB-labelled sacculi (12 µl) were incubated with AmiB (1 µM) and indicatedLytEnvC derivatives (1 µM) at 37°C in 60 µl of PBS buffer (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl and 2.7 mM KCl, pH 7.4) for the indicated time points when they were terminated by the addition of 30 µl of ethanol. Following termination, all reactions were centrifuged at 21000×g for 20 min at room temperature. The absorbance of the supernatants at 595 nm was then measured.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Thuy Dinh for plasmid construction and summer students Ben Dyer and Francesca Romana Cianfanelli for help with site-directed mutagenesis. We would also like to thank J. Marquez and the HTX Lab team (Partnership for Structural Biology, Grenoble) for access to high throughput crystallization instrumentation, and the European Synchrotron Radiation Facility (ESRF) for access to beamlines. Special thanks to Carlos Contreras-Martel and Andrea Dessen for support and access to data processing and refinement software. This work was supported by the National Institutes of Health (R01 AI083365-01). NTP is supported by a Ruth L. Kirschstein F32 fellowship (GM100609-02).
Abbreviations
- PG
peptidoglycan
- RBB
remazol brilliant blue
REFERENCES
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonis M, Ecobichon C, Guadagnini S, Prévost M-C, Boneca IG. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol. 2010;78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- Browder H, Zygmunt W, Young J, Tavormina P. Lysostaphin: Enzymatic Mode of Action. Biochem Biophys Res Commun. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DN, Sham YY, Haugstad GD, Xiang Y, Rossmann MG, Anderson DL, Popham DL. Shared catalysis in virus entry and bacterial cell wall depolymerization. J Mol Biol. 2009;387:607–618. doi: 10.1016/j.jmb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford MA, Lowe DE, Fisher DJ, Stibitz S, Plaut RD, Beaber JW, et al. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc Natl Acad Sci USA. 2011;108:17159–17164. doi: 10.1073/pnas.1108495108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk M, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev. 2007;31:676–691. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- Firczuk M, Mucha A, Bochtler M. Crystal structures of active LytM. J Mol Biol. 2005;354:578–590. doi: 10.1016/j.jmb.2005.09.082. [DOI] [PubMed] [Google Scholar]
- Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol Microbiol. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y, Nishimura Y. Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett. 2002;212:229–236. doi: 10.1111/j.1574-6968.2002.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Ooiwa S, Sekiguchi J. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall. J Bacteriol. 2012;194:796–803. doi: 10.1128/JB.05569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S. Proteolytic activity of YibP protein in Escherichia coli. J Bacteriol. 2002;184:2595–2602. doi: 10.1128/JB.184.10.2595-2602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korndörfer IP, Danzer J, Schmelcher M, Zimmer M, Skerra A, Loessner MJ. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J Mol Biol. 2006;364:678–689. doi: 10.1016/j.jmb.2006.08.069. [DOI] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci USA. 2012;109:5441–5445. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J Biol Chem. 2006;281:549–558. doi: 10.1074/jbc.M509691200. [DOI] [PubMed] [Google Scholar]
- Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol. 2011;193:5477–5486. doi: 10.1128/JB.00439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Moran CP. A LytM domain dictates the localization of proteins to the mother cell-forespore interface during bacterial endospore formation. J Bacteriol. 2011;193:591–598. doi: 10.1128/JB.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Maehigashi T, André I, Dunham CM, Moran CP. Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci USA. 2012;109:5446–5451. doi: 10.1073/pnas.1120113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci USA. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Möll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol Microbiol. 2010;77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–785. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Mol Microbiol. 2010;77:74–89. doi: 10.1111/j.1365-2958.2010.07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis A, Thomas P, Starka J. Morphological mutants of Escherichia coli. Isolation and ultrastructure of a chain-forming envC mutant. J Gen Microbiol. 1973;75:409–416. doi: 10.1099/00221287-75-2-409. [DOI] [PubMed] [Google Scholar]
- Sham L-T, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci USA. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Hattori H, Ise F, Sekiguchi J. Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. J Biol Chem. 2001;276:28140–28146. doi: 10.1074/jbc.M103903200. [DOI] [PubMed] [Google Scholar]
- Singh SK, L S, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyze an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Bernhardt TG. More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr Opin Microbiol. 2011;14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol Mol Biol Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol Microbiol. 2012;85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.