Abstract
Individual proteomes typically differ from the reference human proteome at ~10,000 single amino acid variants. When viewed at the population scale, this individual variation results in a wide variety of protein sequences. In targeted proteomics experiments, such variability can confound accurate protein quantification. To assist researchers in identifying target peptides with high variability within the human population we have created the Population Variation plug-in for Skyline, which provides easy access to the polymorphisms stored in dbSNP. Given a set of peptides, the tool reports minor allele frequency for common polymorphisms. We highlight the importance of considering genetic variation by applying the tool to public datasets.
Keywords: MRM/SRM, genetic variation, bioinformatics, dbSNP
Introduction
In the era of personalized genomics and precision medicine, tens of thousands of human genomes are being sequenced to elucidate the genetic basis for diversity and disease [1]. Compared to the reference human genome, individuals often differ at millions of nucleotides, including both small single nucleotide polymorphisms (SNPs) and larger variations. For SNPs, individual genomes typically show ~10,000 non-synonymous variants that change protein sequence [2], [3], [4]. A second category of SNPs, stop-gain or indels, have a more pervasive effect and alter all subsequent amino acids. Several large-scale sequencing efforts aim to categorize genomic diversity of the human population as a whole. The HapMap consortium initially obtained information for 1 million SNPs from 269 individuals [5]. More recently, the 1000 Genome Project performed whole genome sequencing to discover SNPs as well as larger sequence variants [6]. Such projects continue to expand their sampling and add to the knowledge of human genetic variation. One benefit of population studies is that they are able to estimate the frequency of variants for the entire human population or specific sub-populations.
Targeted proteomics measurements are a high throughput method to accurately quantify protein abundances. The reliability of the method lends it to use in biomarker development studies that require a large number of samples. For example, Whiteaker and colleagues utilized targeted proteomics to quantify proteins in 80 mouse plasma samples [7]. Targeted studies in humans often use cell lines, however recent work by the Carr group studied 13 human cardiac patients and 52 exercising controls to identify biomarkers for myocardial infarction [8].
The diversity in human protein sequences poses a computational challenge for targeted proteomics workflows. As peptide sequences are the quantified surrogate for protein abundance, studies need to account for possible sequence variation across the cohort. Individuals with a variant amino acid within the peptide region would have a null or noise value from a targeted assay. Selecting the best peptide to represent a protein, or assay design, is a crucial aspect of any targeted proteomics experiment [9]. Considerations for peptide selection typically include fragmentation intensity, potential for chemical modification, and interference from the background matrix; many software tools have been created to address these factors [10], [11], [12], [13], [14]. However, there is currently no tool to aiding researchers in identifying peptides that have high variability within the human population. We present the Population Variation tool which uses data from dbSNP to identify the minor allele frequency of peptide targets for MRM/SRM experiments. The tool is available as a plug-in from the Skyline store.
Methods
Database Setup
The human subset of dbSNP build 137 was downloaded in November 2013 from ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/. Our goal was to obtain a database containing SNPs of a known minor allele frequency. We limited our results using the following criteria: SNPs kept must have a minor allele frequency > 0.01; SNPs kept must have a non-null protein accession; SNPs kept must be of type missence, stop-gain, or frameshift. With these constraints, only three tables were relevant: SNPContigLocusID, Allele, and SNPAlleleFreq_TGP. We simultaneously filter, merge the tables and removed most columns, keeping: prot_acc, residue, aa_pos, snp_id, fxn_code, minorAlleleFreq. This produces a 9 MB database, whereas the original dbSNP download was >15 GB. The resulting data is stored in a SQLite database and distributed with the plug-in.
Database Access
PopulationVariation is programmed in C# using .NET 4.0 framework. It is available as an external tool for Skyline [13], or as a zip from http://omics.pnl.gov/software/PopulationVariation.php Protein accession and peptide sequences are obtained from Skyline via a custom data form. Protein objects in Skyline must contain proper accessions as this is the key into the SNP database. Accepted accessions are NCBI RefSeq (ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/protein/protein.fa.gz), or Uniprot (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/proteomes/HUMAN.fasta.gz). dbSNP uses RefSeq accessions as the key, therefore Uniprot accessions are mapped using ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/.
Results
In the context of clinical or population studies using targeted proteomics, a peptide with a high natural variation is problematic. Subjects with the variant allele have a different amino acid sequence. Therefore, targeted MRM/SRM approaches which isolate a specific m/z would register a null or noise value for the peptide target, confounding downstream analysis. To assist researchers in determining whether their target peptides have such variability we have created the Population Variation plug-in for the Skyline software program [13]. Three kinds of mutations that alter protein coding sequences are reported: non-synonymous variants that change a single amino acid, and frame-shift and stop-gain mutations that alter all downstream amino acids.
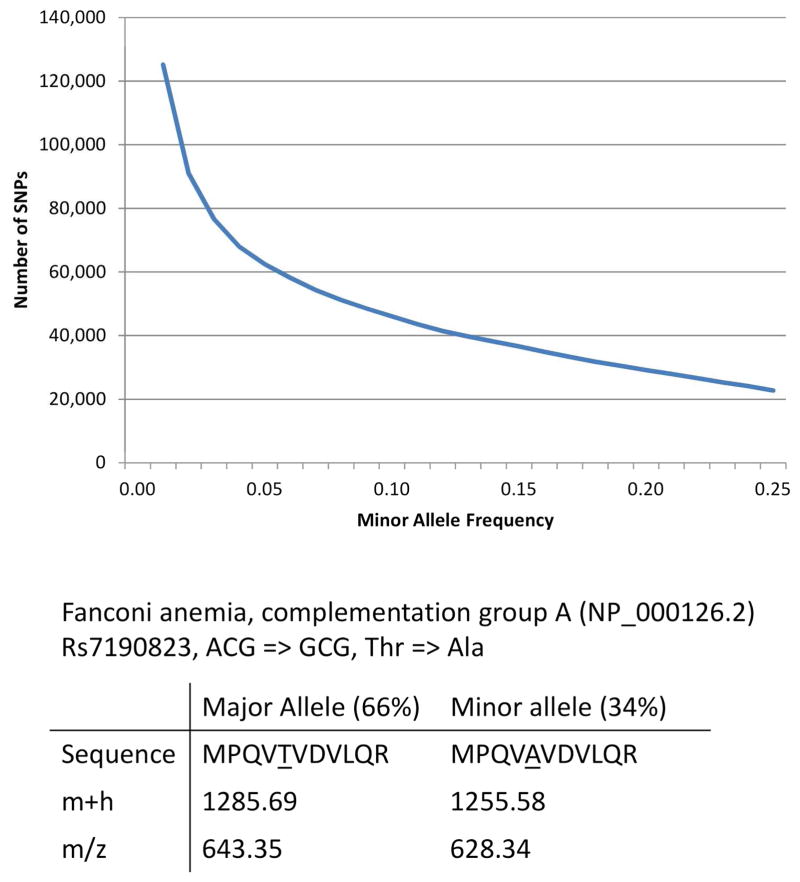
The Population Variation tool draws upon dbSNP as the source of information for genetic variation [15]. Among the many projects that contribute to dbSNP, the 1000 Genome Project uses its broad sampling to report not only the location and category of the polymorphism, but also a global population estimate for minor allele frequency (MAF) [6]. In its first phase, the 1000 Genome project reported 125,204 variants in 20,283 proteins with a minor allele frequency greater than 1%, and 62,418 variants in 13,792 proteins with greater than 5% MAF. There are also a large number of highly common variants; 22,740 variants in 6811 proteins are present with a MAF > 25% (Figure 1).
Figure 1.
Effects of SNPs on MRM assays. In the top graph, the number of SNPs is plotted by their minor allele frequency. For example, 46,018 SNPs have a minor allele frequency greater than or equal to 0.10 (10%). The bottom panel show dbSNP entry rs7190823, a missense mutation to the franconi anemia complementation group A protein. This SNP changes the sequence and therefore the mass of the tryptic peptide.
To highlight the utility of the Population Variation plug-in, we have chosen to re-analyze publicly available SRM studies to show peptides that are impacted by these variations. The plug-in accesses a local version of dbSNP using the protein accession listed in the Skyline file (see Methods). Using the 96 human transcription factors studied by Stergachis et al [16], we searched for common variants which affect protein coding sequences. Twelve peptides were discovered to harbor variants with greater than 5% MAF. In zinc finger protein 530 (NP_065931), peptide DILQMIELQASPCGQK has a 24% minor allele frequency and was one of only three high quality peptides for the protein. This peptide also highlights a subtle problem when using cell lines or other banked biomaterials. The original assay design by Stergachis derived protein sequences from the cDNA clone library used for protein overexpression. Curiously, for zinc finger protein 530 (HsCD00301131) this clone harbored the minor allele. Thus, the peptide identified in the original paper is not present in the majority of the population.
In identifying targets for >1000 prominent cancer proteins, the Aebersold group designed ~5500 SRM assays [17]. We further applied the plug-in on this large-scale dataset. Our result showed that 72 peptides contain variants with MAF > 5% and 30 peptides with a variant allele >20% MAF. We further checked the status of 34 potential candidate biomarkers mentioned in Aebersold’s paper. Five proteins out of 34 were detected with MAF > 1%, with one peptide from complement protein 7 harboring a with 8% minor allele frequency.
Conclusions
As proteomics research moves towards more clinical applications, accounting for natural genetic variation will become increasingly important. Unlike research in model organisms where individual subjects are typically inbred, human studies contain patients with diverse genetic backgrounds. In targeted proteomics, if the desire is to consistently measure the abundance of a single protein, peptide targets should be universal within the cohort. An alternate way of using the tool is to mimic a SNP microarray, and design peptide targets for both alleles. In this manner, researchers could examine the relative abundance of alleles. The Population Variation plug-in for Skyline assists researchers by exposing to them human sequence variation within the peptides and proteins in their experiment. This tool will be regularly updated as new data from 1000 Genome Project are stored in dbSNP, or other projects which estimate population level variation.
Acknowledgments
The authors thank Jia Guo, and Jintang He for early testing of the software. This work was supported by grant U24-CA-160019 from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), by the Department of Energy Science Undergraduate Laboratory Internships (SULI) program, and by the National Institute of General Medical Sciences (Grant 8 P41 GM103493-10). Work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
References
- 1.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. Genomes Project C. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, et al. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mead JA, Bianco L, Ottone V, Barton C, Kay RG, et al. MRMaid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol Cell Proteomics. 2009;8:696–705. doi: 10.1074/mcp.M800192-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 15.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stergachis AB, MacLean B, Lee K, Stamatoyannopoulos JA, MacCoss MJ. Rapid empirical discovery of optimal peptides for targeted proteomics. Nat Methods. 2011;8:1041–1043. doi: 10.1038/nmeth.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra194. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]