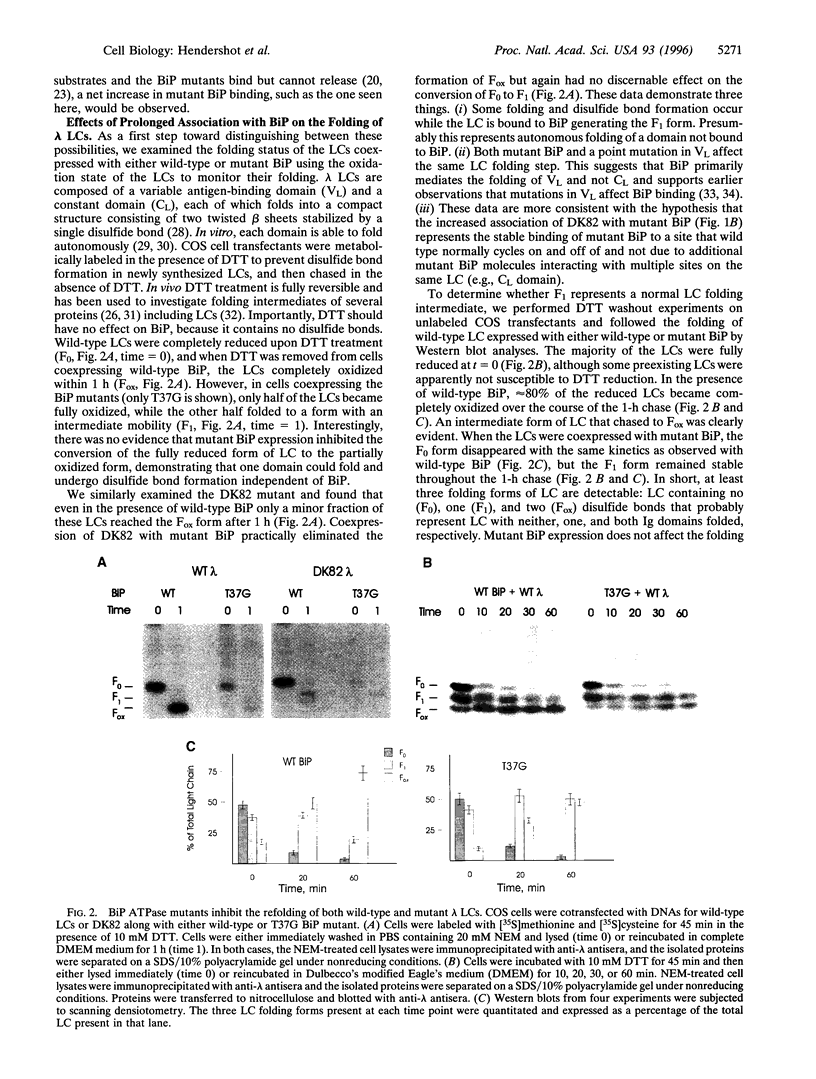
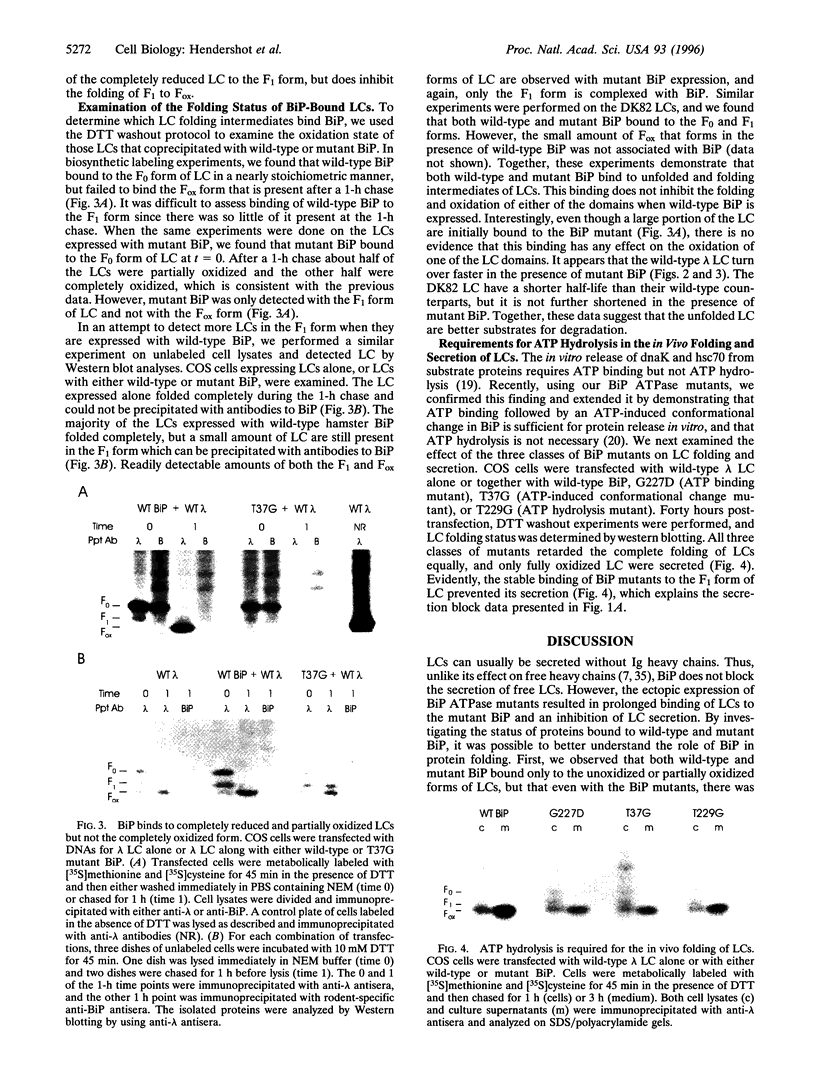
Abstract
A group of resident ER proteins have been identified that are proposed to function as molecular chaperones. The best characterized of these is BiP/GRP78, an hsp70 homologue that binds peptides containing hydrophobic residues in vitro and unfolded or unassembled proteins in vivo. However, evidence that mammalian BiP plays a direct role in protein folding remains circumstantial. In this study, we examine how BiP interacts with a particular substrate, immunoglobulin light chain (lambda LC), during its folding. Wild-type hamster BiP and several well-characterized BiP ATPase mutants were used in transient expression experiments. We demonstrate that wild-type lambda LCs showed prolonged association with mutant BiP which inhibited their secretion. Both wild-type and mutant BiP bound only to unfolded and partially folded LCs. The wild-type BiP was released from the incompletely folded LCs, allowing them to fold and be secreted, whereas the mutant BiP was not released. As a result, the LCs that were bound to BiP mutants were unable to undergo complete disulfide bond formation and were retained in the ER. Our experiments suggest that LCs undergo both BiP-dependent and BiP-independent folding steps, demonstrating that both ATP binding and hydrolysis activities of BiP are essential for the completion of LC folding in vivo and reveal that BiP must release before disulfide bond formation can occur in that domain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amzel L. M., Poljak R. J. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- Bernard O., Hozumi N., Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978 Dec;15(4):1133–1144. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993 Nov 19;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bole D. G., Hendershot L. M., Kearney J. F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986 May;102(5):1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Helenius J., Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992 May;11(5):1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Helenius J., Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992 Mar 19;356(6366):260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- Bulleid N. J., Freedman R. B. Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 1988 Oct 13;335(6191):649–651. doi: 10.1038/335649a0. [DOI] [PubMed] [Google Scholar]
- Doms R. W., Lamb R. A., Rose J. K., Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993 Apr;193(2):545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- Dul J. L., Argon Y. A single amino acid substitution in the variable region of the light chain specifically blocks immunoglobulin secretion. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8135–8139. doi: 10.1073/pnas.87.20.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul J. L., Burrone O. R., Argon Y. A conditional secretory mutant in an Ig L chain is caused by replacement of tyrosine/phenylalanine 87 with histidine. J Immunol. 1992 Sep 15;149(6):1927–1933. [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Flynn G. C., Pohl J., Flocco M. T., Rothman J. E. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991 Oct 24;353(6346):726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Gaut J. R., Hendershot L. M. Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J Biol Chem. 1993 Apr 5;268(10):7248–7255. [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986 Sep 12;46(6):939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Goto Y., Hamaguchi K. Unfolding and refolding of the reduced constant fragment of the immunoglobulin light chain. Kinetic role of the intrachain disulfide bond. J Mol Biol. 1982 Apr 25;156(4):911–926. doi: 10.1016/0022-2836(82)90147-4. [DOI] [PubMed] [Google Scholar]
- Haas I. G., Meo T. cDNA cloning of the immunoglobulin heavy chain binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2250–2254. doi: 10.1073/pnas.85.7.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas I. G., Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983 Nov 24;306(5941):387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Hammond C., Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994 Oct 21;266(5184):456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Hendershot L. M., Ting J., Lee A. S. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol. 1988 Oct;8(10):4250–4256. doi: 10.1128/mcb.8.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L. M., Wei J. Y., Gaut J. R., Lawson B., Freiden P. J., Murti K. G. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995 Mar;6(3):283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L., Bole D., Köhler G., Kearney J. F. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol. 1987 Mar;104(3):761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman J., Inbar D., Givol D. An active antibody fragment (Fv) composed of the variable portions of heavy and light chains. Biochemistry. 1973 Mar 13;12(6):1130–1135. doi: 10.1021/bi00730a018. [DOI] [PubMed] [Google Scholar]
- Hochstenbach F., David V., Watkins S., Brenner M. B. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock C. K., Kelly R. B. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989 May;8(5):1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. S., Arvan P. Calnexin and BiP act as sequential molecular chaperones during thyroglobulin folding in the endoplasmic reticulum. J Cell Biol. 1995 Jan;128(1-2):29–38. doi: 10.1083/jcb.128.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler M. R., Dirks S., Haas I. G. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler M. R., Haas I. G. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992 Apr;11(4):1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S. J., Jordan R., McMacken R., Gierasch L. M. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992 Jan 30;355(6359):455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- Machamer C. E., Doms R. W., Bole D. G., Helenius A., Rose J. K. Heavy chain binding protein recognizes incompletely disulfide-bonded forms of vesicular stomatitis virus G protein. J Biol Chem. 1990 Apr 25;265(12):6879–6883. [PubMed] [Google Scholar]
- Melnick J., Aviel S., Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem. 1992 Oct 25;267(30):21303–21306. [PubMed] [Google Scholar]
- Melnick J., Dul J. L., Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994 Aug 4;370(6488):373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986 Jul 18;46(2):291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Palleros D. R., Reid K. L., Shi L., Welch W. J., Fink A. L. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993 Oct 14;365(6447):664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- Schumacher R. J., Hurst R., Sullivan W. P., McMahon N. J., Toft D. O., Matts R. L. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994 Apr 1;269(13):9493–9499. [PubMed] [Google Scholar]
- Simons J. F., Ferro-Novick S., Rose M. D., Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995 Jul;130(1):41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C., Sitia R. The differential effects of dithiothreitol and 2-mercaptoethanol on the secretion of partially and completely assembled immunoglobulins suggest that thiol-mediated retention does not take place in or beyond the Golgi. Mol Biol Cell. 1994 Dec;5(12):1311–1324. doi: 10.1091/mbc.5.12.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Gaut J. R., Hendershot L. M. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995 Nov 3;270(44):26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Wild J., Kamath-Loeb A., Ziegelhoffer E., Lonetto M., Kawasaki Y., Gross C. A. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]