Abstract
Oncogenic Ras mutations are widely considered to be locked in a permanent ‘On’ state and ‘constitutively active’. Yet, many healthy people have cells possessing mutant Ras without apparent harm, and in animal models mutant Ras causes transformation only after upregulation of Ras activity. Here, we demonstrate that oncogenic K-Ras is not constitutively active but can be readily activated by upstream stimulants to lead to prolonged strong Ras activity. These data indicate that in addition to targeting K-Ras downstream effectors, interventions to reduce K-Ras activation may have important cancer-preventive value, especially in patients with oncogenic Ras mutations. As other small G proteins are regulated in a similar manner, this concept is likely to apply broadly to the entire Ras family of molecules.
Keywords: cancer, Ras-GTP
INTRODUCTION
Ras proteins function as binary molecular switches that, when turned ‘on’ by occupancy with GTP, interact with downstream signaling molecules to activate a wide variety of intracellular signaling networks including those regulating proliferation, differentiation, apoptosis and cell migration.1 Specific point mutations in Ras disable the GTP hydrolysis process and thus impair its inactivation.2 These mutations are oncogenic and are observed in ~30% of all cancers. In particular, oncogenic K-Ras is found in nearly every pancreatic cancer—the fourth leading cause of cancer death in USA. Despite much evidence to the contrary existing in the literature, a persistent dogma exists that oncogenic Ras is locked in a permanent ‘On’ state and thus ‘constitutively active’. This idea has been included in a large number of papers and reviews in top journals.3–5
Surprisingly though, several studies have indicated that normal healthy people possess cells bearing mutant oncogenic Ras in different organs including the pancreas,6–8 colon7 and lung9 at rates far exceeding the rates of cancer development. Similarly, mouse models expressing oncogenic Ras in the whole body or in specific organs develop cancer, but the number of transformed cells is a very small fraction of those expressing oncogenic Ras.10 These findings indicate that the effects of expression of oncogenic Ras at physiological levels are relatively small. However, it has been shown in animal models that in the presence of oncogenic Ras, upstream stimuli that cause only transient effects in wild-type cells generate prolonged activation of Ras and accelerate the development of cancer.11,12 It is difficult to explain the importance of upstream stimuli on cancer development if oncogenic Ras is constitutively active.
In the current study, we found that oncogenic K-Ras is not constitutively active as has been widely believed. Rather, oncogenic K-Ras can be readily activated by upstream stimulants leading to prolonged strong K-Ras activity, which has been shown to be critical for pathological response.12 As a large population of normal healthy humans unknowingly harbor Ras mutations,9,13 we propose that in addition to targeting Ras downstream effectors, interventions to reduce Ras activation may have important cancer-preventive value especially in people with oncogenic Ras mutations.
RESULTS AND DISCUSSION
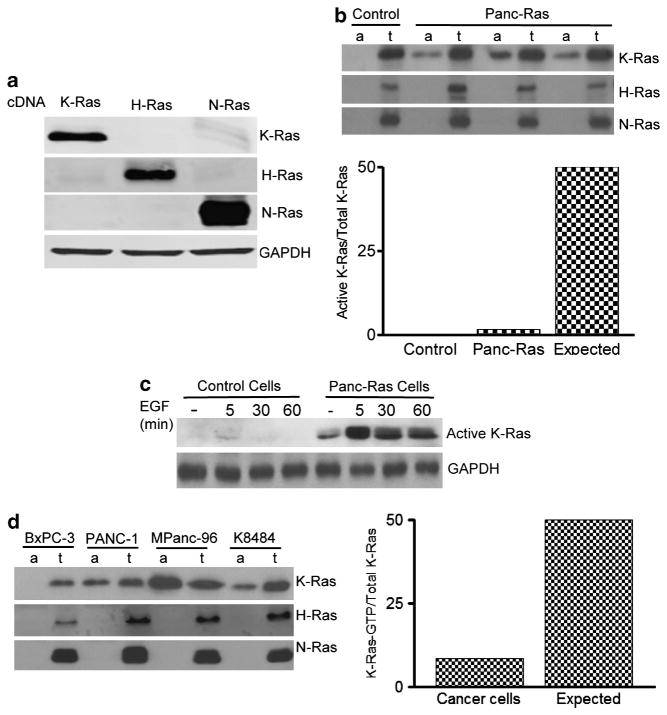
To better understand the role of oncogenic K-Ras in cancer initiation, we directly examined the activity of oncogenic K-Ras expressed at physiological levels using a model in which one of the two copies of the K-Ras gene expresses oncogenic K-RasG12D from its native promoter after Cre-mediated recombination specifically in the pancreas (hereafter referred as Panc-Ras mice).14 We used this model to test the prediction that if oncogenic Ras is always ‘on’, then in these cells 50% of total K-Ras should be active under basal conditions. We utilized a K-Ras isoform-specific antibody that did not cross react with either N-Ras or H-Ras (Figure 1a) to examine the fraction of active K-Ras in lysates prepared from pancreases of resting control and Panc-Ras mice. ‘Active’ K-Ras was defined as that which could be captured in a Raf pull-down assay, as Raf only binds to GTP-loaded Ras. The results indicated that the level of K-Ras that bound Raf in cells from Panc-Ras mice was not at all close to 50%, but actually <2% of total K-Ras (Figure 1b). This is very far from the expected fraction of active K-Ras if mutant K-Ras were constitutively active. This large difference from expected results is unlikely to be explained by inefficient recovery of Ras GTP in the Raf pull-down assay. Some pdx-Cre strains used to activate the LSL-KrasG12D allele develop a mosaic expression pattern in the pancreas.14 To determine the recombination efficiency of the pdx-Cre used in the current study, the mice were crossed with ROSA26-LacZ reporter mice and recombination efficiency was evaluated by X-gal staining. The resulting animals showed that nearly 100% of cells underwent recombination (Supplementary Figure 1). In addition, we also crossed the LSL-KRasG12D mice with Ela-CreERT mice, which also gave nearly 100% recombination in pancreatic acinar cells.15 The same effects on Ras activity were observed in all of these models.
Figure 1.
Oncogenic Ras requires activation for enhanced activity. (a) Lysates from HEK293 cells transfected with K-, N- and H-Ras cDNA were used to determine the specificity of corresponding antibodies by western blotting. (b) Active K-, N- and H-Ras were measured by a Raf pull-down assay from pancreas lysates of 2-month-old control and Panc-Ras mice (lanes a). For comparison with total corresponding Ras isoform proteins, 5% of total lysates used for pull-down assay were loaded to adjacent wells (lanes t) and probed with isoform-specific antibodies. Densitometry was performed using ImageJ software. (c) Active K-Ras was measured in isolated pancreatic acinar cells from control and Panc-Ras mice after EGF (50 ng/ml) stimulation. (d) Oncogenic Ras is partially active in cancer cells. Active K-, N- and H-Ras (lanes a) were compared with 10% of the corresponding Ras isoform levels used for pull-down assay (lanes t) in human pancreatic cancer cells (Bxpc3, Panc1 and Mpanc96) and pancreatic cancer cells derived from Panc-Ras mouse (K8484). Note all cells except BxPC-3 possess oncogenic K-Ras mutations.
Another prediction made based on oncogenic Ras being ‘always on’ would be that the maximal stimulation of K-Ras activity by upstream activators would be ≤2-fold, as only the wild-type 50% of K-Ras could be activated. To test this prediction, we treated isolated pancreatic acinar cells prepared from control and resting Panc-Ras mice with a Ras activator, epidermal growth factor (EGF). We found that EGF stimulation increased the level of active K-Ras by ~10-fold compared with unstimulated cells (Figure 1c, Supplementary Figure 2). Moreover, the difference between theoretical and actual levels is quite large. Taken together, these results are inconsistent with the hypothesis that oncogenic K-Ras is ‘always on’. However, in the presence of oncogenic K-Ras EGF stimulated prolonged elevation of Ras activity, which has been shown to cause accelerated development of cancer.11,12 In contrast, EGF caused only transient effects in cells with wild-type K-Ras (Figure 1c). These data indicate that even though oncogenic K-Ras is not constitutively active, it can be readily activated by upstream stimulants.
To determine whether this observation were also true in transformed cells, we compared the level of active K-Ras to that of total K-Ras in lysates from human cancer cells and cancer cells developed from tumors in Panc-Ras mice. Surprisingly, even in the transformed cells active K-Ras was only ~9% of total K-Ras (Figure 1d). Ras signaling was also significantly upregulated by EGF in these cells (Supplementary Figure 3). Similar results were observed in human colon cancer cells (data not shown). Therefore, in neither normal nor transformed cells did the percentage of active K-Ras match that predicted if oncogenic K-Ras was ‘constitutively active.’
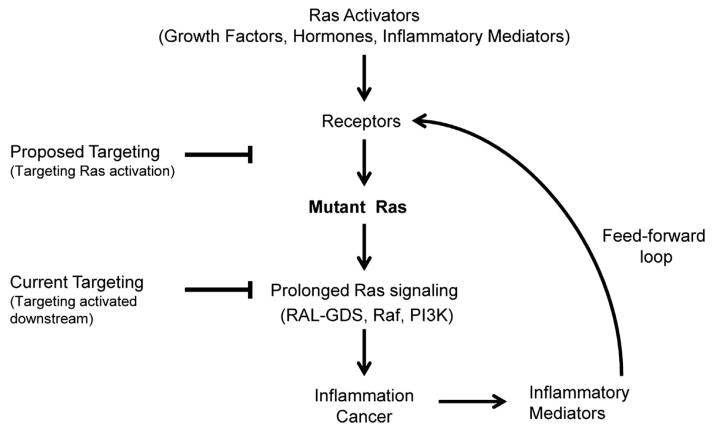
In summary, our results indicate that oncogenic K-Ras is not constitutively active as has been widely believed. Rather, oncogenic K-Ras can be readily activated by upstream stimulants. However, as previously reported, once activated oncogenic K-Ras has delayed kinetics of returning to the basal state and can participate in a pathological feed-forward loop with inflammation.12 As other small G proteins are regulated in a similar manner, this concept is likely to apply broadly to the entire Ras family of molecules. On the basis of this new understanding, it could be predicted that any stimulus that activates Ras or that promotes the development of inflammation would facilitate the initiation of cancer in cells bearing oncogenic Ras. This prediction has already been upheld for a variety of stimuli including EGF, tumor growth factor-α, CCK, PGE2 and lipopolysaccharide.12,16–18 These concepts take on an increased clinical relevance when coupled with the knowledge that there is a large population of normal healthy humans that unknowingly harbor Ras mutations.9,13 A reasonable conclusion would be that a reduction of factors that can activate Ras signaling may have important cancer-preventive values in patients with oncogenic Ras mutations (Figure 2).
Figure 2.
Targeting both upstream and downstream of Ras signaling for cancer prevention. Multiple factors are able to act as Ras activators through interactions with upstream receptors (e.g., EGF receptor). The effect on wild-type Ras is a transient elevation of downsteam signaling pathways including those regulated by RALGDS, RAF1 and PI3K (not shown). However, when oncogenic Ras is present, the downstream signaling after stimulation is prolonged at a high level. Elevated Ras activity generates inflammatory mediators that further activate Ras, and thus sustain Ras activity at high levels through a feed-forward loop. Previously emphasis has been focused on targeting downstream pathways activated by Ras, as oncogenic Ras was considered to be constitutively active. However, endogenous expression of mutant Ras gives rise only low level of Ras activity, which does not cause pathological effects unless Ras activity is upregulated. On the basis of the evidence that oncogenic Ras is not active without input from upstream activators, we propose that targeting the upstream mechanisms of Ras activation would be an alternative approach for cancer prevention, especially in patients with Ras mutations.
MATERIALS AND METHODS
Genetically engineered mice
LSL-KRasG12D mice, which possess the conditional knock-in mutant K-RasG12D, and pdx1-Cre mice were obtained from the Mouse Models for Human Cancer Consortium Repository (Rockville, MD, USA).19 For targeted expression of K-RasG12D in the pancreas, LSL-K-RasG12D mice were bred with pdx1-Cre mice to generate LSL-K-RAS/pdx1-Cre double-transgenic mice (Panc-Ras mice).
Acini preparation
Pancreatic acini were prepared as described previously20 with modifications. Briefly, pancreata from mice were digested with purified collagenase (100 unit/ml, Worthington Biochemicals, Lakewood, NJ, USA) and incubated at 37 °C for 50 min with shaking at 120 r.p.m. Digested tissue was than mechanically dispersed via pipette and passed through a 150-μm mesh nylon cloth. Acini were then purified three times with gradient separation in Dulbecco’s modified Eagle’s medium (DMEM) containing 4% bovine serum albumin. Purified acini were resuspended in DMEM containing 0.1 mg/ml soybean trypsin inhibitor and 1% bovine serum albumin for 1 h in a tissue culture incubator. After stimulation with EGF 50 ng/ml (Sigma-Aldrich, St Louis, MO, USA) for different time periods, acini were assayed for Ras activity.
Ras activity assay
Ras activity refers to the level of guanosine triphosphate-bound Ras, which is able to bind Raf as measured using a Raf pull-down assay kit as recommended by the manufacturer (Millipore, Billerica, MA, USA).21 Briefly, pancreatic samples were homogenized on ice in lysis buffer containing 25 mM 4-(2–hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.5), 1% Igepal CA-630, 150 mM NaCl, 0.25% sodium deoxycholate, 10% glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM ethylenediaminetetraacetic acid, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 mM sodium orthovanadate. These samples were sonicated and centrifuged at 15 000 g for 10 min at 4 °C to remove cellular debris. Aliquots of lysates were set aside to allow quantification of total Ras and protein concentrations. Equal amounts of lysate were incubated for 30 min at 4 °C with agarose beads coated with Raf-Ras binding domain. The beads were then washed three times with ice-cold lysis buffer, boiled for 5 min at 95 °C, and active Ras was analyzed by immunoblotting following standard western blot analysis protocols using K-, N- and H-Ras-specific antibodies. For comparison with total corresponding Ras protein, 5–10% of total lysates used for pulldown were loaded to adjacent wells.
Western blot analysis
Cell or tissue lysates were prepared, separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes. The membranes were blocked for 1 h at room temperature with 3% nonfat milk in Tris-buffered saline containing 0.05% Tween 20 and incubated overnight at 4 °C with one of the following primary antibodies: K-Ras (Santa Cruz sc-30, Santa Cruz, CA, USA); H-Ras (Santa Cruz sc-520) and N-Ras (Santa Cruz sc-519) (1:200; Santa Cruz), GAPDH (1:10000; Sigma-Aldrich), phospho-Erk and total Erk (1:1000; Cell Signaling Technology, Danvers, MA, USA). Immunodetection was performed with the corresponding Alexa Fluor 680-conjugated secondary antibodies to allow detection with the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). All images were converted to grayscale. Densitometry was measured using ImageJ software (http://rsbweb.nih.gov/ij/).
Study approval
All animal experiments were approved by The University of Texas MD Anderson Cancer Center’s Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Dr David Tuveson (UK Cambridge Research Institute) for providing the mouse pancreatic cancer cell line K8484. This work was supported by NIH DK052067, CA016672, P20 CA101936, CA155165, P50 CA102701 and the Lockton Endowment. The work was supported in part by grant 30910103911 (to Z Li) from the National Natural Science Foundation of China.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Spaargaren M, Bischoff JR, McCormick F. Signal transduction by Ras-like GTPases: a potential target for anticancer drugs. Gene Expr. 1995;4:345–356. [PMC free article] [PubMed] [Google Scholar]
- 2.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 3.Koeffler HP, McCormick F, Denny C. Molecular mechanisms of cancer. West J Med. 1991;155:505–514. [PMC free article] [PubMed] [Google Scholar]
- 4.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan L, McFaul C, Howes N, Leslie J, Lancaster G, Wong T, et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology. 2005;128:2124–2130. doi: 10.1053/j.gastro.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Xu T, Qian J, Wen X, Wu D. Detecting K-ras and p53 gene mutation from stool and pancreatic juice for diagnosis of early pancreatic cancer. Chin Med J. 2002;115:1632–1636. [PubMed] [Google Scholar]
- 8.Parsons BL, Meng F. K-RAS mutation in the screening, prognosis and treatment of cancer. Biomark Med. 2009;3:757–769. doi: 10.2217/bmm.09.95. [DOI] [PubMed] [Google Scholar]
- 9.Yakubovskaya MS, Spiegelman V, Luo FC, Malaev S, Salnev A, Zborovskaya I, et al. High frequency of K-ras mutations in normal appearing lung tissues and sputum of patients with lung cancer. Int J Cancer. 1995;63:810–814. doi: 10.1002/ijc.2910630611. [DOI] [PubMed] [Google Scholar]
- 10.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 11.Carriere C, Young AL, Gunn JR, Longnecker DS, Korc M. Acute pancreatitis markedly accelerates pancreatic cancer progression in mice expressing oncogenic Kras. Biochem Biophys Res Commun. 2009;382:561–565. doi: 10.1016/j.bbrc.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniluk J, Liu Y, Deng D, Chu J, Huang H, Gaiser S, et al. An NF-kappaB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J Clin Invest. 2012;122:1519–1528. doi: 10.1172/JCI59743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tada M, Ohashi M, Shiratori Y, Okudaira T, Komatsu Y, Kawabe T, et al. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology (Research Support, Non-US Gov’t) 1996;110:227–231. doi: 10.1053/gast.1996.v110.pm8536861. [DOI] [PubMed] [Google Scholar]
- 14.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 15.Ji B, Song J, Tsou L, Bi Y, Gaiser S, Mortensen R, et al. Robust acinar cell transgene expression of CreErT via BAC recombineering. Genesis. 2008;46:390–395. doi: 10.1002/dvg.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, et al. EGF Receptor Is Required for KRAS-Induced Pancreatic Tumorigenesis. Cancer cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siveke JT, Einwachter H, Sipos B, Lubeseder-Martellato C, Kloppel G, Schmid RM. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer cell. 2007;12:266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams JA, Korc M, Dormer RL. Action of secretagogues on a new preparation of functionally intact, isolated pancreatic acini. Am J Physiol. 1978;235:517–524. doi: 10.1152/ajpendo.1978.235.5.E517. [DOI] [PubMed] [Google Scholar]
- 21.de Rooij J, Bos JL. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene. 1997;14:623–625. doi: 10.1038/sj.onc.1201005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.