Summary
Genomic and metagenomic sequencing efforts, including human microbiome projects, reveal that microbes often encode multiple systems that appear to accomplish the same task. Whether these predictions reflect actual functional redundancies is unclear. We report that the prominent human gut symbiont Bacteroides thetaiotaomicron employs three functional, homologous vitamin B12 transporters that in at least two cases confer a competitive advantage in the presence of distinct B12 analogs (corrinoids). In the mammalian gut, microbial fitness can be determined by the presence or absence of a single transporter. The total number of distinct corrinoid transporter families in the human gut microbiome likely exceeds those observed in B. thetaiotaomicron by an order of magnitude. These results demonstrate that human gut microbes use elaborate mechanisms to capture and differentiate corrinoids in vivo and that apparent redundancies observed in these genomes can instead reflect hidden specificities that determine whether a microbe will colonize its host.
Introduction
In genetically tractable microbes, deletion of multiple systems is often required to create a phenotype of interest (Epstein, 2003; Kehres and Maguire, 2003; Maguire, 2006; Miethke and Marahiel, 2007; Wood, 2006). With advances in genome sequencing, identification of these apparent functional redundancies is no longer restricted to model organisms and now extends widely to human-associated species that lack genetic tools (Temperton and Giovannoni, 2012). It has been proposed that such redundancies provide backup for the most important cellular features (Dean et al., 2008; Li et al., 2010). However, the observation that DNA polymerase and many other essential proteins are generally encoded in single copy is inconsistent with this hypothesis, suggesting that seemingly redundant proteins are maintained for other reasons including environmental variables that are not understood.
Human microbiome projects illustrate the importance of understanding this problem. Trillions of microbes live in and on the human body, with the greatest numbers found in the distal gut. These microbes belong primarily to two phyla (Bacteroidetes and Firmicutes) and are only distantly related to Escherichia coli or other model organisms. However, microbiome sequencing has produced an emergent picture of enormous species-level diversity but considerable functional overlap between individuals (Arumugam et al., 2011; Turnbaugh et al., 2009). While the apparently duplicated functions observed in these genomes could reflect true redundancies, they could also reveal hidden fitness determinants, biomarkers, or therapeutic targets.
Specific factors that determine microbial fitness and shape community composition in the gut remain largely obscure. Systems that mediate acquisition of essential cofactors likely play key roles in these processes. Notably, E. coli and other model organisms encode multiple transporters for several essential cofactors (iron, magnesium, potassium) (Epstein 2003; Maguire 2006; Miethke and Marahiel 2007). However, one of the most well-characterized cofactor transport systems in bacteria, the BtuBFCD transporter, exists in single copy in E. coli and other bacteria studied to date and is their exclusive route for B12 acquisition (Chimento et al., 2003). BtuB is a TonB-dependent outer membrane transporter found only in Gram-negative bacteria, while the periplasmic binding protein BtuF and ABC transporter BtuCD are found across bacterial taxa. Curiously, the human gut is replete with B12 analogs (corrinoids) produced by select members of the gut microbiota (Allen and Stabler, 2008; Brandt et al., 1977; Zhang et al., 2009), but how bacteria sense and respond to these compounds is unexplored.
Here we demonstrate that, unexpectedly, human gut microbes often encode multiple B12 transporters in their genomes. We establish that this apparent redundancy instead represents a vastly expanded repertoire of corrinoid transporters. In the prominent human gut symbiont Bacteroides thetaiotaomicron, individual transporters confer distinct corrinoid preferences in vitro and play distinct roles in determining microbial fitness in gnotobiotic mice. Further, the extent of functional redundancy of these transport systems can be directly controlled through the diet of the host. Our results also suggest that the number of functionally distinct corrinoid transporters in the human gut microbiome exceeds those found in E. coli and other previously studied model organisms by at least 30-fold.
Results
Corrinoid transporters represent a widespread redundancy in the human gut microbiome
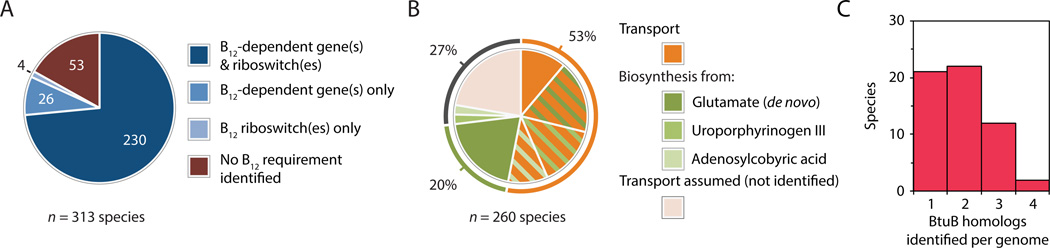
To identify genes and pathways involved in B12-related processes in the human gut microbiome, we first searched the genome sequences of 313 human gut bacterial species for B12-dependent genes and riboswitches (Table S1; Table S2). Based on these features, most (260/313; 83%) of these species involve B12 in their biology (Figure 1A; Figure S1A; Table S3). An elaborate pathway is required to produce this cofactor (Figure S1B) (Rodionov et al., 2003; Roth et al., 1996). However, the majority of the 260 B12-dependent human gut species identified above lack the capacity for de novo B12 biosynthesis and thus likely rely on transport to meet their B12 requirements (Figure 1B; Table S2; Table S3).
Figure 1. Widespread redundancies in B12 transport in human gut microbial genomes.
(A) Over 80% of sequenced human gut microbial species (260/313) encode B12-dependent genes or riboswitches. (B) Most of these 260 species lack the genes required to synthesize B12 de novo and rely on transport to meet their B12 requirements. Species that encode B12 transport are shown in orange and those that possess partial or complete B12 biosynthetic pathways are marked in green; species with both of these capabilities are designated with a hatched pattern. (C) The majority of human gut Bacteroidetes encode multiple B12 transporters within their genomes. See also Figure S1.
Consistent with this observation, we identified homologs of the gene encoding BtuB in over 70% of the Gram-negative species that involve B12 in their biology (100/139). BtuB homologs identified by protein sequence similarity can be cross-validated with an independent query for a conserved RNA element (the B12 riboswitch) that regulates gene expression (Nahvi et al., 2002; Vitreschak et al., 2003). Indeed, each of the BtuB homologs identified in our search is positioned downstream of a B12 riboswitch, and no proteins meeting these stringent search criteria are found in any of the 53 species predicted to lack B12-dependent enzymes (Table S2; Table S3). We investigated the utility of using the BtuFCD components as an alternate strategy to identify corrinoid transporters in human gut microbial genomes. We also identified BtuFCD homologs which are (1) present in an operon regulated by a predicted B12 riboswitch, and (2) a better match to a B12 transporter than a Co2+ transporter (Table S4) (Cheng et al., 2011). Using these criteria we identified 70 putative corrinoid ABC transporters encoded in the same operon as a previously identified btuB gene. This approach further identified 56 complete (BtuFCD) and one partial (BtuFC) B12 regulated ABC transporters, 33 of which were from Gram-positive bacteria and thus not detected by the BtuB searches (Table S3). This estimate remains conservative given the paucity of functionally characterized BtuFCD components from Gram-positive bacteria and the observation that in both B. thetaiotaomicron and E.coli, corrinoid ABC transporters are not always regulated by a B12 riboswitch (Figure 2A). However, for the purposes of our analysis we focused on the BtuB protein, which is best able to discriminate corrinoid transporters from systems known to transport other substrates. Notably, the 152 BtuB homologs we identified in the human gut microbiome are distributed not only between but also within genomes: in the phylum Bacteroidetes, which comprise over 80% of the microbiota of some individuals (Human Microbiome Project Consortium, 2012b), the majority of species possess duplicate, triplicate, or even quadruplicate copies of these proteins (Figure 1C).
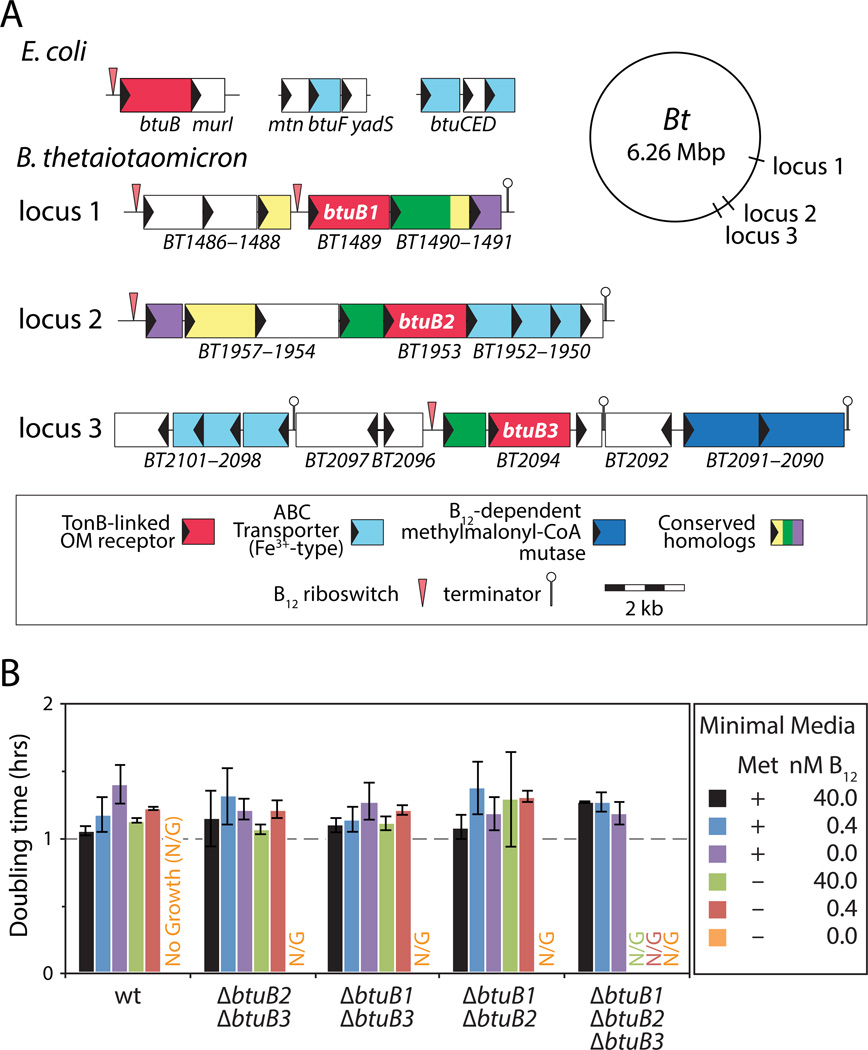
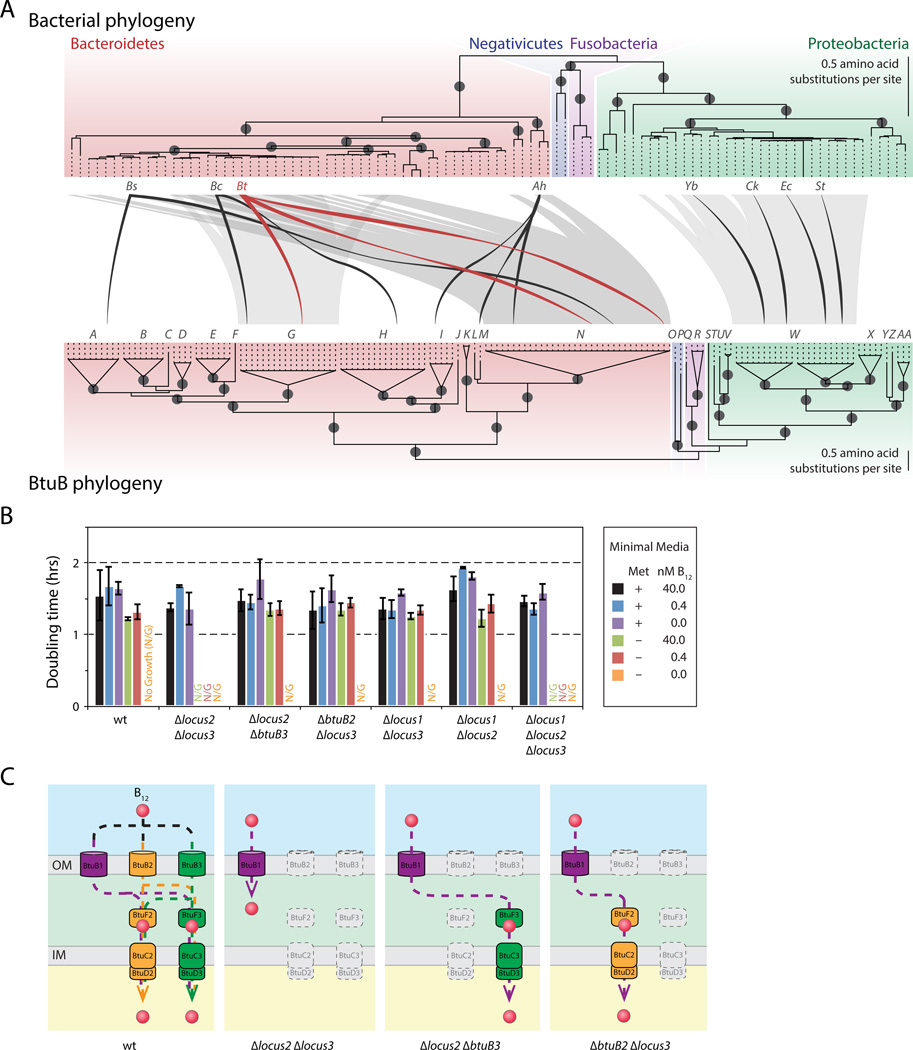
Figure 2. The prominent human gut symbiont B. thetaiotaomicron encodes three functional B12 acquisition systems.
(A) Genetic organization of the canonical btuBFCD B12 transporter in E. coli and the three B12 transport systems in B. thetaiotaomicron. (B) Each B. thetaiotaomicron BtuB homolog supports doubling times equivalent to the wildtype strain in conditions that require B12 transport. Mean doubling times and standard deviations of triplicate cultures are shown. Wt, wildtype; Met, methionine. See also Figure S2.
Experimental dissection of corrinoid transport redundancy in a prominent human gut symbiont
Bacteroides thetaiotaomicron typifies these observations from genome analysis. This prominent human gut microbe, like many other species, lacks the machinery for B12 biosynthesis but possesses multiple B12-dependent enzymes. The B. thetaiotaomicron genome contains three predicted B12 transport systems that are each located adjacent to a B12 riboswitch and each contain a BtuB homolog (designated BtuB1-BtuB3; Figure 2A; Table S3). B. thetaiotaomicron synthesizes methionine via the B12-dependent enzyme MetH (Table S3); as a result, B12 transport is required for growth in medium that lacks methionine (Varel and Bryant, 1974). To determine whether each BtuB homolog in B. thetaiotaomicron is capable of B12 transport, we constructed a panel of seven mutant strains carrying nonpolar, unmarked deletions of each btuB gene individually, in each pairwise combination, and in triplicate. Each single and double ΔbtuB mutant grows at rates indistinguishable from the wildtype strain in medium lacking methionine and containing B12, while deletion of all three btuB genes identified in our computational search prevents B12-dependent growth unless external B12 concentrations are increased by four orders of magnitude (Figure 2B; Figure S2A; Figure S2B). Liquid chromatography/mass spectrometry-based quantification of B12 levels further confirms that these systems are together responsible for the entirety of B12 transport in this organism (Figure S2C) and restoration of any of the three btuB homologs in single copy in the chromosome of the ΔbtuB1 ΔbtuB2 ΔbtuB3 mutant fully complements these phenotypes (Figure S2D; Figure S2E). Together, these experimental results establish that the computational approaches used to identify redundant B12 transporters across the human gut microbiome delineate these systems with the necessary sensitivity and specificity. Further, these results suggest that B. thetaiotaomicron encodes three functional BtuB proteins that allow equivalent growth rates in B12-dependent conditions.
In E. coli, it has been estimated that B12-dependent methionine synthesis requires very low levels of this cofactor (20 molecules/cell, or approximately 30 nanomolar intracellular concentration) (Di Girolamo et al., 1971). Thus, growth of bacterial mutants in isolation may provide insufficient resolution to evaluate transporter function in the highly competitive environment of the mammalian gut. To distinguish whether the three B12 transport systems in B. thetaiotaomicron also substitute for each other under more stringent competitive conditions, we quantified the contribution of each BtuB homolog to competitive growth in the presence of the wildtype strain. Serial passaging of mixed cultures of wildtype, ΔbtuB mutant, and complemented strains in medium lacking methionine reveals that each BtuB homolog makes a distinct contribution to bacterial fitness that varies by over three orders of magnitude when B12 is present in the culture medium (Figure 3A). Strains that transport B12 via BtuB1, for example, are drastically outcompeted by isogenic wildtype (and complemented mutant) bacteria, while mutants encoding only BtuB2 or BtuB3 show negligible or intermediate fitness defects, respectively, under these conditions and timescales. The minor competitive disadvantage in methionine-replete medium observed in strains encoding only BtuB1 or BtuB3 is fully complemented by restoration of the missing genes (Figure 3A).
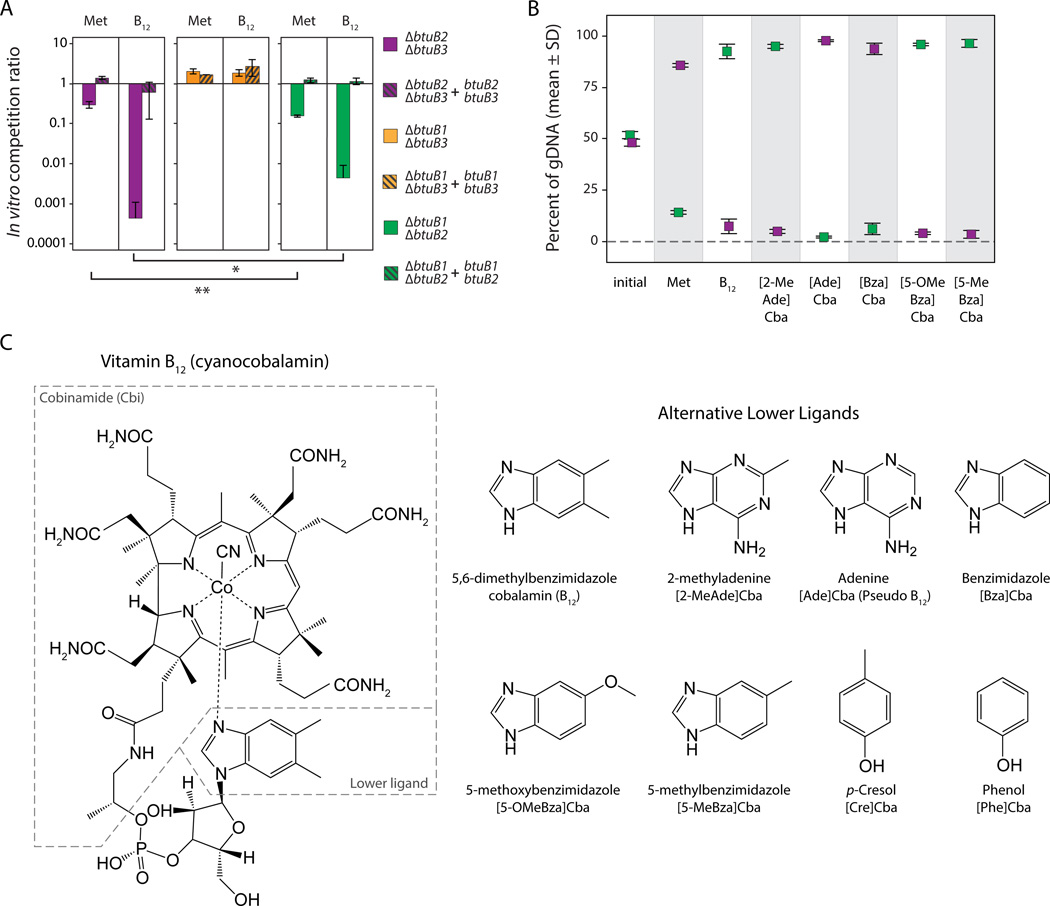
Figure 3. Competition assays reveal functional specificity for predicted redundant transporters in vitro.
(A) A B. thetaiotaomicron strain encoding only BtuB1 is dramatically outcompeted in competition with the wildtype parent in conditions in which B12 transport is required for growth, while a strain encoding only BtuB2 shows no competitive defect in these conditions and BtuB3 provides an intermediate phenotype. The slight competitive defect observed in methionine-replete conditions is complemented by expression of the deleted genes in trans. Wildtype, mutant and complemented strains were inoculated at an initial ratio of 1:8:1, passaged by daily 1:1,000 dilution for five days, and strain abundances determined by qPCR. Normalized mean ratios and standard deviations of mutant and complemented strains to the wildtype from triplicate cultures are shown. Asterisks indicate significant differences (t-test, * p < 0.05 of Log10 transformed data). (B) Direct competition of B. thetaiotaomicron strains encoding only BtuB1 or BtuB3 reproduces functional preferences observed in wildtype-mutant competition assays in media with methionine or B12. Corrinoid structure also determines the relative fitness of these strains. Strains were inoculated at an initial ratio of 1:1, passaged, and quantified as above and mean percentages and standard deviations of each strain across triplicate cultures are shown. (C) Molecular structure of vitamin B12 (cyanocobalamin) and the lower ligands of other corrinoids purified and examined in this study; abbreviations describe the corresponding cobamides (Cba) (Allen and Stabler, 2008; Renz, 1999). Dashed lines outline the structural precursor cobinamide (Cbi) and the lower ligand, which varies between corrinoids. See also Figure S3.
Because a strain encoding only BtuB2 is indistinguishable from the wildtype strain in these in vitro culture conditions, we next focused on the other two BtuB homologs. Based on the results above, direct competition of strains encoding only BtuB1 or BtuB3 should reveal that BtuB3 provides a competitive advantage in the presence of B12. In methionine-replete medium (where BtuB1 provides a slight, but reproducible and complementable, benefit compared to BtuB3) the relative fitness of these strains should be reversed. This is indeed the case (Figure 3B), confirming that these systems show consistent differences in their ability to transport vitamin B12. It is unlikely that differential expression is sufficient to explain these results (see Discussion).
To test the hypothesis that the various B12 transporters in B. thetaiotaomicron exhibit distinct corrinoid preferences, we next extracted and purified seven distinct corrinoids representing each of the three known structural families (Figure 3C). Five of these (containing adenine, 2-methyladenine, benzimidazole, 5-methylbenzimidazole, or 5-methoxybenzimidazole as the lower ligand) allow growth of wildtype B. thetaiotaomicron in defined minimal medium with doubling times equivalent to those measured in the presence of vitamin B12, suggesting that this species readily transports these compounds and can utilize them as cofactors for methionine synthase (Figure S3A). Further, this transport requires at least one of the three BtuB-containing systems identified in our computational search of the B. thetaiotaomicron genome: a ΔbtuB1 ΔbtuB2 ΔbtuB3 mutant is rapidly and completely outcompeted by the wildtype strain in minimal medium containing any of these five corrinoids (Figure S3B). We next determined whether the differential efficiency of BtuB1 and BtuB3 observed in vitamin B12 transport applies to these alternate corrinoids. To this end, we repeated the competition assays between strains encoding only BtuB1 or BtuB3 in minimal media containing each of the five corrinoids that are transported and utilized by wildtype B. thetaiotaomicron. Strikingly, the relative fitness of strains encoding single BtuB homologs is determined by which corrinoid is present: BtuB1 exhibits a preference for corrinoids containing adenine or benzimidazole as the lower ligand, while BtuB3 favors corrinoids containing 2-methyladenine, 5-methoxybenzimidazole, 5-methylbenzimidazole, or 5,6-dimethylbenzimidazole (B12 itself) as the lower ligand (Figure 3B). Competition assays between wildtype, mutant, and complemented strains are consistent with these observations (Figure S3C). Together, these results suggest sampling a small fraction of known corrinoid diversity identifies B12 transport systems encoded in the genome of B. thetaiotaomicron that are not redundant but instead provide competitive advantages to this species in the presence of distinct corrinoids. It is also apparent that BtuB2, which was previously highlighted in an in vivo mutagenesis screen (Goodman et al., 2009), plays a distinct role in the presence of diverse corrinoids in vitro.
Functional differences between apparently redundant transporters determine microbial fitness in gnotobiotic mice
To measure the impact of transporter specificity in vivo, we established a simplified system in gnotobiotic mice in which the impacts of single microbial genes and single small molecules on microbial dynamics can be measured directly. We first colonized germfree Swiss Webster mice (n=10) consuming an irradiated, synthetic, B12-replete diet with three isogenic B. thetaiotaomicron strains (the wildtype parent, an isogenic ΔbtuB2 mutant, and the complemented ΔbtuB2 mutant) and assessed the relative abundance of each strain in fecal samples over time and along the length of the gut after 15d. Although the ΔbtuB2 mutant encodes two intact BtuB transporters that each allow doubling times equivalent to the wildtype strain in B12-dependent conditions in vitro (Figure 2A; Figure S2A), loss of this single gene impacts competitive fitness over 4,000- fold in the mammalian gut environment (t-test p < 0.0001; Figure 4A). Complementation of the ΔbtuB2 mutant with btuB2 under the control of the BT1311 (rpoD) promoter restores mRNA abundance to within 2-fold of native levels and abrogates the fitness defect in gnotobiotic mice (Figure 4A; Figure S4A). Similar trends were observed in gnotobiotic Swiss Webster mice consuming a standard, undefined diet (Figure S4B) and in gnotobiotic C57BL6/J mice (Goodman et al., 2009).
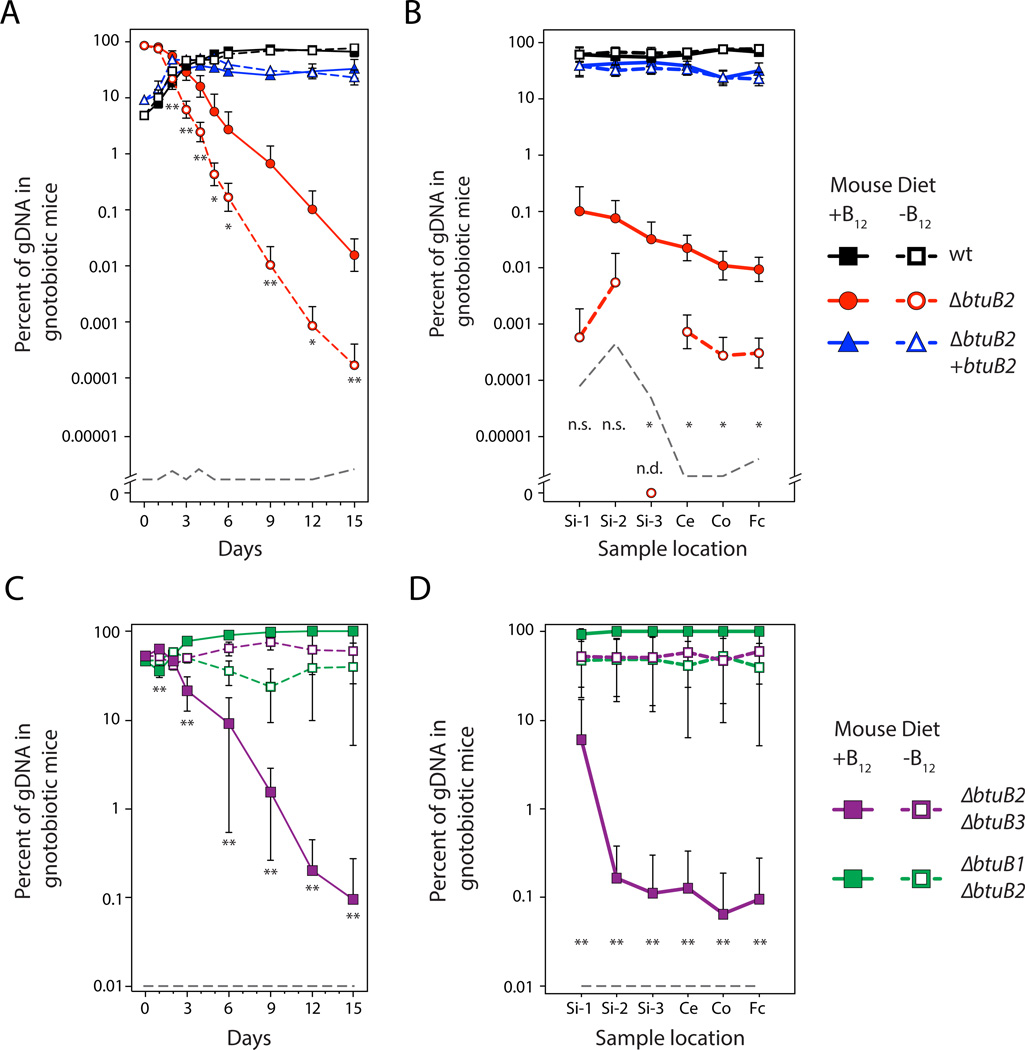
Figure 4. Functional specificity of homologous transporters determines microbial fitness in the mammalian gut.
Population dynamics of B. thetaiotaomicron wild type (black squares), ΔbtuB2 mutant (red circles), and complemented mutant strains (blue triangles) in gnotobiotic Swiss Webster mice are shown (A) in fecal samples over time (n=10/group) and (B) along the gastrointestinal tract (n=5/group). Strain abundances during in vivo competition between B. thetaiotaomicron strains encoding only BtuB1 (purple squares) or BtuB3 (green squares) in gnotobiotic Swiss Webster mice are shown over time (C) and along the length of the gut (D) (n=9/group). Asterisks indicate significant differences in ΔbtuB2 (A and B) or ΔbtuB2 ΔbtuB3 (C and D) mutant abundances in mice consuming a B12-replete diet (closed symbols and solid lines) compared to mice consuming a B12-deplete diet (open symbols and dashed lines; t-test * p < 0.05, ** p < 0.01). The limit of detection is shown with a dashed grey line; n.s., not significant; n.d., not detected. Genomic DNA was extracted and qPCR performed on the luminal contents of the proximal (Si-1), medial (Si-2) and distal (Si-3) small intestine, the cecum (Ce), colon (Co) and feces (Fc). See also Figure S4.
In mice and humans, B12 is stored in the liver and efficiently recycled through the gut via enterohepatic circulation. As a result, short-term removal of vitamin B12 from the diet reduces, but does not eliminate, this vitamin from the host (Nielsen et al., 2012). To establish whether the role of BtuB2 in determining B. thetaiotaomicron fitness can be modulated via diet, we maintained germfree mice (n=10) on a synthetic diet lacking B12 for 7d prior to colonization with the three isogenic B. thetaiotaomicron strains described above and measured strain dynamics as above. In mice consuming the B12-depleted diet, the contribution of BtuB2 to microbial fitness nearly doubles as compared to the B12-replete diet (ANCOVA, d.f.=1, F=25.6, p < 0.0001), leading to a difference in relative abundance of nearly two orders of magnitude by d15 (Figure 4A). This difference is maintained along the entire length of the gut (Figure 4B).
To determine whether the in vitro phenotypes for BtuB1 and BtuB3 also occur in vivo, we prepared simplified populations consisting of equal proportions of strains expressing only BtuB1 or BtuB3, colonized gnotobiotic mice consuming B12-replete or B12-deplete diets, and monitored strain dynamics over time and along the length of the gut as above. In agreement with the in vitro studies (Figure 3B), strains encoding only BtuB3 outcompete strains encoding only BtuB1 in mice consuming a B12-replete diet, both over time (Figure 4C) and along the length of the gut (Figure 4D). This fitness benefit is abrogated when mice are fed diets lacking B12, conditions in which BtuB1 and BtuB3 make little (or no) contribution to fitness (Figure 4). The reduced contribution of BtuB1 and BtuB3 in low B12 concentrations is also observed in vitro (Figure S4C). Together, these results suggest that the functional specificities between corrinoid transporters demonstrated in vitro also determine colonization and persistence in vivo.
Characterization of corrinoid transporters across 313 members of the human gut microbiota
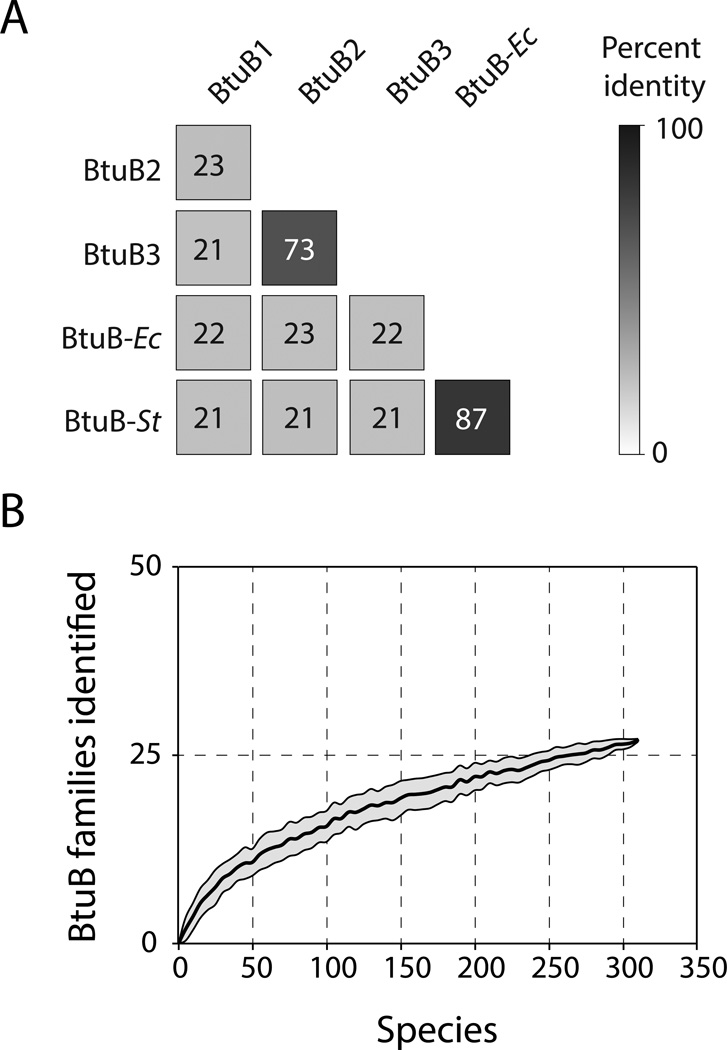
Given that few members of the human gut microbiota are genetically tractable, these studies of B. thetaiotaomicron provide an experimental framework for interpreting the diversity of corrinoid transport systems across the human gut microbiome. Pairwise protein identity calculations between each BtuB homolog in B. thetaiotaomicron and the corresponding E. coli and S. typhimurium proteins reveal that B. thetaiotaomicron BtuB1 and BtuB2 share no greater protein identity with each other than either protein has with the homologs in the distantly related Proteobacteria (Figure 5A). Notably, although strains encoding BtuB2 and BtuB3 exhibit distinct phenotypes, these proteins share over 70% identity. Based on these observations, we used a conservative 50% protein identity cutoff to cluster the B12-riboswitch regulated BtuB homologs from 313 human gut microbial genomes into families (Table S1; Table S3). This analysis suggests that these genomes encode at least 27 corrinoid transporter families that share less than 50% protein sequence identity with each other and thus are predicted to exhibit distinct corrinoid preferences (Figure 5B). Phylogenetic reconstruction of these BtuB homologs shows evidence of both duplication and divergence of these systems within closely related bacterial lineages, as well as horizontal transfer of these genes between species (Figure 6A; Figure S5A; Figure S5B; Figure S5C).
Figure 5. Human gut microbes likely encode at least 27 distinct corrinoid transporter families.
(A) Amino acid sequence identity from >80% of the length aligned between BtuB orthologs from B. thetaiotaomicron (BtuB1-BtuB3), E. coli K-12 (BtuB-Ec), and S. typhimurium LT2 (BtuB-St). (B) Rarefaction analysis on BtuB protein families as defined by a 50% amino acid identity cutoff. The mean number of BtuB families observed as increasing numbers of genomes are sampled is shown as a bold line; standard deviations from 100 permutations are shown with shading. See also Figure S5.
Figure 6. Duplication, diversification and horizontal transfer of corrinoid transporters among human gut microbes.
(A) Phylogenetic relationships between human gut microbial species and between BtuB sequences were reconstructed by maximum likelihood. Terminal branches on the BtuB phylogeny have been collapsed and labeled according to their placement into the 27 families (A-AA) predicted by rarefaction analysis. Species distributions of representative BtuB families with evidence of recent horizontal gene transfer are indicated with shaded lines. BtuB sequences present in representative species are shown in black (Bs, B. stercoris; Bc, B. caccae; Ah, Alistipes sp. HGB5; Yb, Yersinia bercovieri; Ck, Citrobacter koseri; Ec, E. coli; St, S. typhimurium) and red (Bt, B. thetaiotaomicron). Internal nodes with bootstrap support ≥ 75% are shown as dark gray circles and colored regions correspond to the indicated bacterial phylum or class. (B) The corrinoid transport system encoded in locus 1 requires proteins encoded in the other corrinoid transport systems for function. B. thetaiotaomicron strains encoding only locus 1 are not viable in conditions requiring B12 transport for growth, while strains encoding locus 1 and btuFCD genes from locus 2 or locus 3 grow at wildtype rates in conditions that require B12 transport. Colors correspond to the key and mean doubling times and standard deviations from triplicate cultures are shown. (C) Predicted routes of corrinoid transport in mutant strains described in (B). In the wildtype strain, B12 can be transported across the outer membrane by any of the three BtuB homologs and cross the inner membrane through either of the two BtuFCD systems. Because locus 1 does not encode its own btuFCD genes, a strain lacking locus 2 and locus 3 is not viable in B12 dependent conditions. The btuFCD genes from locus 2 or locus 3 restore B12-dependent growth to this strain, suggesting that B12 transported through the outer membrane via BtuB1 can enter the cytoplasm via BtuFCD homologs encoded in the other two corrinoid transporter loci. See also Figure S6.
Four lines of evidence suggest that this estimate of 27 functionally distinct systems is conservative. Most simply, additional genome sequencing will reveal additional corrinoid transporters: rarefaction analysis suggests that a 2-fold increase in genome sequences will add 13 additional transporter families. Second, although the outer-membrane protein BtuB provides the most informative sequence for excluding false positives (i.e., differentiating systems that transport corrinoids from those that target other small molecules), this specificity is gained at a large cost in sensitivity. Corrinoid transport systems in Gram-positive species that lack an outer membrane, including the majority of the Firmicutes phylum, are not represented in a BtuB-based search. Among the 154 Gram-positive species in our dataset, nearly half (69) encode B12-dependent genes and/or riboswitches but lack a complete B12 biosynthetic pathway and thus likely possess one or more transporters not included in the results of our search. Notably, this is a lower bound as many species that are fully capable of de novo corrinoid biosynthesis, including S. typhimurium, also encode functional transport systems. Third, clustering BtuB proteins at 50% identity lacks sufficient resolution to separate B. thetaiotaomicron BtuB2 and BtuB3, which have distinct functions, into different families (Figure 6A; Figure S5B). A 75% identity cutoff is sufficient to separate these proteins into separate groups and adds over 30 additional families to the estimate (Figure S5D). Finally, in genomes that encode multiple corrinoid transport systems, proteins encoded in separate loci could cross-assemble to form additional variants. Consistent with this possibility, B12 transported through the B. thetaiotaomicron outer membrane via locus 1 (which lacks its own inner membrane transporter BtuFCD; Figure 2A) reaches the cytoplasm via proteins encoded in locus 2 or locus 3 (Figure 6B; Figure 6C; Figure S6A). Together, these results suggest that the three transporters identified in B. thetaiotaomicron represent a small fraction of the diversity encoded in the human gut microbiome.
Discussion
Do microbes possess multiple systems to accomplish the same task? In the pre-genomics era, functional redundancies in bacteria emerged primarily from genetic studies revealing that deletion of multiple genes is necessary to induce a phenotype of interest. These studies require both genetic tools for the target species and a phenotype that can be readily measured in the laboratory: as a result, examples of functional redundancies remained limited to a select number of well-known, though enigmatic, cases. Genome sequencing, however, allows direct prediction of functional redundancies without genetic manipulation of the organism or experimental tractability of the phenotype. The thousands of bacterial genome sequences that are currently available (largely from species that lack genetic tools and experimental models of their environment) amplify the scope of these predictions.
Recent advances in human gut microbiome research offer a unique combination of resources for delineating shared and unique functions and determining the extent to which microbial communities share a core metagenome. In this study, we utilize the large number of completed human gut microbial genome sequences, microbial genetics, and germfree mouse models of the gut environment to dissect a broadly distributed example of apparent functional redundancy in the human gut microbiome. We establish that the human gut Bacteroidetes tend to encode multiple copies of the BtuBFCD transporter. B. thetaiotaomicron, a genetically tractable representative of this phylum, contains three B12 transport systems and thus is typical in this regard. We demonstrate that although all of these systems are capable of transporting this substrate, each exhibits a unique relative preference for five additional corrinoids. Notably, the corrinoids we tested represent less than half of the known corrinoid structural diversity present in the human gut from dietary sources and microbial biosynthesis; it is likely that additional corrinoids will reveal further preference differences among BtuB homologs. Experiments in gnotobiotic mice reveal that this functional specificity observed in vitro also occurs in the mammalian gut and that a single transporter can determine microbial fitness in vivo. Further, the role of these systems in determining microbial fitness can be modulated by dietary intervention of the host.
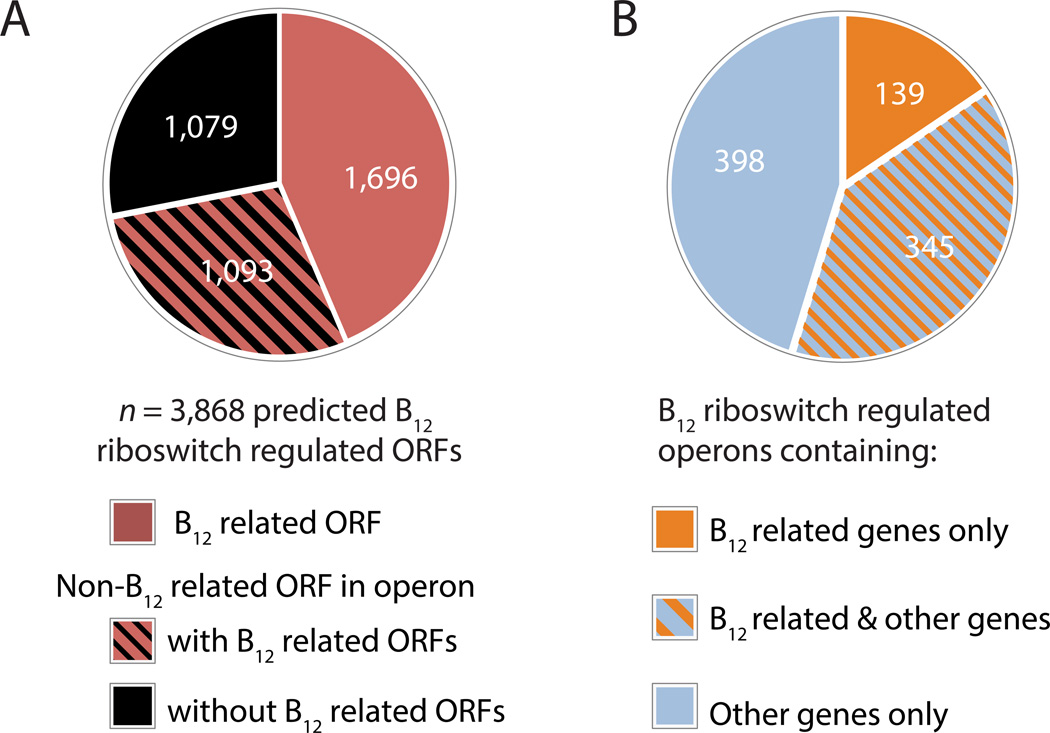
Why does B12 transport have such a dramatic effect on B. thetaiotaomicron strain dynamics in vivo? Given that methionine synthase is not required for B. thetaiotaomicron fitness in gnotobiotic mice (Goodman et al., 2009), the importance of BtuB2-dependent B12 transport in vivo suggests that B. thetaiotaomicron fitness depends on additional corrinoid-dependent enzymes. It is also likely that other members of the human gut microbiota require corrinoids for functions absent from B. thetaiotaomicron entirely (Table S3). The 313 genomes examined in this study collectively encode 809 B12 riboswitches predicted to regulate 3,868 genes, half of which have not previously been associated with B12 (Figure 7A; Figure 7B). Together, these results suggest that the in vivo requirement for corrinoid transport may be due to a wide range of B12-dependent and/or -regulated proteins across the human gut microbiota.
Figure 7. Many genes not previously associated with vitamin B12 are encoded downstream of predicted B12 riboswitches.
(A) B12 riboswitches were identified across 313 human gut microbial genomes; B12 related ORFs are listed in Table S3 and include transport, biosynthesis, and dependent proteins and their B12-independent isoenzymes. (B) A large fraction of predicted B12 riboswitch-regulated operons in sequenced human gut microbial species do not contain any genes directly (B12 biosynthesis, transport, or dependency) or indirectly (B12-independent isozymes) related to corrinoid utilization. 882 predicted B12 riboswitch-regulated operons from 234 species are shown.
The observations that most species of human gut Bacteroidetes have multiple corrinoid transporters and that these systems cluster into dozens of functionally distinct families is surprising in light of the long history of the Btu transporter as a classical model for small molecule transport in bacteria. It is unlikely that the Bacteroidetes BtuB proteins are by themselves fully responsible for the functional differences observed between corrinoid transporters. The structure of E. coli BtuB bound to B12 suggests that the protein does not interact extensively with the lower ligand of the corrinoid (Chimento et al., 2003). Further, E. coli BtuB binds B12 and cobinamide (which lacks the lower ligand) with equal affinity (Kenley et al., 1978). We note, however, that E. coli does not encode multiple corrinoid transporters within its genome and E. coli BtuB shares less than 25% of its amino acid sequence with the BtuB homologs in the human gut Bacteroidetes. It is also unlikely that the periplasmic and inner-membrane components BtuF, BtuC, and BtuD are sufficient to determine corrinoid preferences because these proteins seem to be involved in corrinoid transport via multiple systems (Figure 6B; Figure 6C).
Although all three corrinoid transport systems are encoded downstream of predicted B12 riboswitches, it is possible that individual riboswitches allow distinct, corrinoid-specific regulatory responses that contribute to the observed phenotypes (and, potentially, to corrinoid-specific gene expression programs across the genome). Because different microbial species produce distinct corrinoids, an intriguing possibility is that these small molecules could thus serve as molecular signals that would allow B. thetaiotaomicron and other species to sense and respond to other species in their community. However, regulatory differences alone do not appear to be sufficient to explain corrinoid transporter preferences: for example, btuB1 is expressed at significantly higher levels than btuB3 in B12-limiting conditions (t-test, p < 0.001, Figure S6B) (Martens et al., 2008; Sonnenburg et al., 2005), yet strains expressing only btuB1 are outcompeted by strains expressing only btuB3 in these conditions in vitro (Figure 3B) and in vivo (Figure 4C; Figure 4D). Even though BtuB2 appears to function as a primary transporter in B. thetaiotaomicron for the limited number of corrinoids tested in these studies, we note that over half of the genome-sequenced human gut Bacteroidetes do not encode a BtuB2 homolog (family "N" in Figure 6A; Figure S5) and instead rely on other transporters. Additionally, all but one of the species that have a BtuB2 homolog also encode additional corrinoid transporters.
The evolutionary advantage for different microbial species to produce different corrinoids is unknown. Notable parallels exist, however, between corrinoid transport and iron transport. Some microbes express siderophore scaffolds to capture ferrous iron and cognate receptors that recognize the siderophore-iron complex (Miethke and Marahiel, 2007). Other species, however, express only the receptor and capture iron bound to siderophores produced by other species (Smith et al., 2005). This “siderophore piracy” is reminiscent of the corrinoid acquisition strategy observed in B. thetaiotaomicron and many other human gut microbes, which do not produce corrinoids but have extensive machinery to capture these compounds from other species. Many bacteria produce species- or strain-specific siderophores and cognate receptors, perhaps in response to siderophore piracy (Meyer et al., 2002). It is tempting to speculate that microbes able to synthesize, transport, and utilize a corrinoid with a novel lower ligand would gain a competitive advantage in the context of other species less able to transport this modified molecule. Notably, studies in humans show that gut microbes efficiently remove the lower ligand from 80% of consumed B12 and replace it with a wide range of alternate lower ligands (Allen and Stabler, 2008). This is consistent with the observation that the first 5–20 genes in the corrinoid biosynthetic pathway are absent in over 40% of gut microbes that encode the later steps required for lower ligand attachment (Figure S1B; Table S3). While microbial modification and capture of corrinoids may exhibit some features shared with iron acquisition strategies, there are at least two fundamental differences. First, while siderophore-receptor pairs can exhibit extreme specificity, corrinoid preferences are relative rather than absolute (i.e., each system is capable of corrinoid transport). Second, siderophores serve as a temporary decoration of iron that is shed upon internalization. In contrast, because microbes vary the structure of the corrinoid cofactor itself, corrinoid transport preferences must be coupled with the requirements of the cell’s corrinoid-dependent enzymes and regulatory elements. Notably, corrinoids with different lower ligands are not functionally equivalent as cofactors, as some corrinoid-dependent metabolic processes function only with specific corrinoids (Yi et al., 2012).
Together, these results suggest that apparent functional redundancies in human gut (and other) microbial communities can instead represent unappreciated complexities in microbial interactions with their environment and each other. Lessons learned from genetic redundancies in individual human gut microbial species thus provide a critical reference for interpreting genomes and metagenomes. Because the human gut microbiota is implicated in diverse aspects of health and disease, it is likely that understanding functional variation in these communities will unlock new diagnostic and therapeutic avenues.
Experimental Procedures
Computational definition of B12-associated genes in the human gut microbiome
We subsampled 495 publicly available human gut microbial genomes (Human Microbiome Project Consortium, 2012a) into a custom database that includes a single representative of each species (313 genomes predicted to encode 1,076,251 genes; Table S1). Experimentally validated BtuBFCD protein sequences were compiled from E. coli str. K-12 substr. MG1655, S. typhimurium LT2, B. thetaiotaomicron VPI-5482 and Dehalococcoides ethenogenes 195 and used as Blastp search queries against these 313 genomes (Table S1; Table S2). Similar searches were performed with the amino acid sequences of experimentally validated B12 dependent, and biosynthetic proteins and a collection of B12 independent isozymes (Table S2). The number of available queries for each ortholog group varies due to the presence of enzymes with multiple subunits, split genes, fused genes, and differing numbers of species with experimentally validated genes identified from the literature. Putative hits were filtered based on a conservative E-value cutoff (1e-45) and positive identification of ≥1 of the expected TIGRFAM or PFAM domains within the candidate gene. For enzyme families with both B12-dependent and B12-independent representatives, candidates with greater similarity to the B12-independent homolog were placed in the latter group. Genomes were scanned for vitamin B12 riboswitches (RFAM family RF00174) using the default parameters of Infernal v1.0.2 (Nawrocki et al., 2009). Each of the genes identified above were also evaluated for genomic context (presence in an operon regulated by a B12 riboswitch). B12-regulated operons were defined as co-directional transcribed genes, ≤ 125 nucleotides apart, encoding a predicted B12 riboswitch located ≤ 405 nucleotides upstream of the operon or within the operon itself. Operons containing incomplete subsets of the BtuFCD proteins as defined by a stringent e-value cutoff were further queried for sequences with correct TIGRFAM or PFAM domains corresponding to missing transporter subunits. Any predicted btuFCD B12 transport operon where the majority of the subunits had a better match to the CbtJKL Co2+ transporter from Sinorhizobium meliloti was excluded. Any predicted btuB candidates that were not regulated by a B12 riboswitch or that were located within a B12 biosynthetic operon were also excluded.
Candidate BtuB protein sequences were aligned using Muscle (Edgar, 2004) and clustered with Usearch (Edgar, 2010). To predict the effect of sampling more species, the data were rarified using a custom Perl script and power law regressions were fit to the resulting rarefaction curves (50% clusters: y=1.17x0.56, R2=0.979; 75% clusters: y=0.923x0.740, R2=0.993). The same alignment was then subjected to maximum likelihood phylogenetic reconstruction using RAxML with a general time reversible (GTR) model and default bootstrap settings (Stamatakis, 2006). The phylogeny of the species encoding BtuB homologs was reconstructed based on 13 orthologous core proteins conserved across all three domains of life (Brown et al., 2001; Stamatakis, 2006). Further details of the computational methods are provided in Supplemental Experimental Procedures.
Bacterial culture and manipulation
Bacteroides thetaiotaomicron VPI-5482 (ATCC 29184) strains were cultured anaerobically at 37ºC in liquid TYG medium (Holdeman, 1977) or on brain heart infusion (BHI; Beckton Dickinson) agar amended with 10% horse blood (Colorado Serum Co. or Quad Five). A flexible anaerobic chamber (Coy Laboratory Products) containing 20% CO2, 10% H2, and 70% N2 was used for all anaerobic microbiology procedures. E. coli S17-1 λ pir strains were grown in LB medium at 37ºC aerobically. Antibiotics were added when appropriate at the following final concentrations: ampicillin 100 µg/mL, gentamicin 200 µg/mL, erythromycin 25 µg/mL, and tetracycline 2 µg/mL. In-frame, unmarked deletions were generated using a counter-selectable allelic exchange procedure and confirmed by sequencing (Martens et al., 2008). Complementation constructs and oligonucleotide barcodes in pNBU2 vectors were introduced into the genome in single copy as described (Martens et al., 2008). All vectors and primers used in this study are provided in Table S5.
Detecting B12 dependent in vitro growth phenotypes
Cells were grown for 16h in TYG medium, washed, and resuspended in triplicate at OD600 0.0005 in minimal medium (Martens et al., 2008) supplemented with 536 µM DL-methionine (Sigma-Aldrich Co.) and/or vitamin B12 (Sigma-Aldrich Co.) or other corrinoids. Doubling times were calculated from OD600 measurements collected with a UV spectrophotometer (Thermo Scientific) during early exponential phase.
Guided biosynthesis and purification of corrinoids
Adeninylcobamide ([Ade]Cba) and 2-methyladeninylcobamide ([2-MeAde]Cba) were purified from S. typhimurium, and p-cresolylcobamide ([Cre]Cba) was purified from Sporomusa ovata, as previously described (Gray and Escalante-Semerena, 2009; Mok and Taga, 2013; Yi et al., 2012). Benzimidazolylcobamide ([Bza]Cba), 5-methylbenzimidazolylcobamide ([5-MeBza]Cba), and 5-methoxybenzimidazolylcobamide ([5-OMeBza]Cba) were produced in S. typhimurium upon supplementation of cultures with 1 µM each of dicyanocobinamide ((CN)2Cbi, Sigma-Aldrich Co.) and the respective lower ligand. Phenolylcobamide ([Phe]Cba) was produced by adding 1 mM phenol to S. ovata grown in media adapted from betaine standard medium (Möller et al., 1984) with 124 mM methanol as previously described (Mok and Taga, 2013).
Purification was performed on an Agilent Series 1200 high-performance liquid chromatography (HPLC) system (Agilent Technologies) equipped with a diode array detector. Samples were injected onto an Agilent Eclipse plus C18 column (5 µm, 9.4 by 250 mm) at 45ºC and separated using a solvent system consisting of solvent A, 0.1% formic acid in water, and solvent B, 0.1% formic acid in methanol. Benzimidazolyl and purinylcobamides were separated at a flowrate of 1.8 ml/min with a linear gradient of 10 to 40% solvent B over 17 min; [Cre]Cba and [Phe]Cba were separated at a flowrate of 2.0 mL/min with a linear gradient of 18 to 60% solvent B over 28.5 min.
In vitro competition experiments
B. thetaiotaomicron strains carrying unique oligonucleotide barcode sequences (Martens et al., 2008) were grown individually for 16h in TYG, washed, and combined in minimal medium containing methionine, B12, or other corrinoids. Cultures were incubated anaerobically at 37ºC and passaged every 24h by 1:1,000 dilution into pre-reduced culture medium. After 5d, total gDNA was recovered from cultures (Truett et al., 2000) and the relative abundance of each strain determined by qPCR as described (Martens et al., 2008) using a CFX96 instrument (BioRad) and SYBR FAST universal mastermix (KAPA Biosystems). Mean strain quantities were calculated using a standard curve and relative fold-changes were calculated with the efficiency-corrected ΔCq method (Bookout et al., 2006). All statistical comparisons were performed in JMP 9 (SAS Institute).
Gnotobiotic animal experiments
All experiments using mice were performed using protocols approved by the Yale University Institutional Animal Care and Use Committee. Germfree Swiss Webster mice were maintained in flexible plastic gnotobiotic isolators with a 12-hr light/dark cycle. Individually caged animals (n=9–10/group) were provided with either standard, autoclaved mouse chow (5K67 LabDiet, Purina) or irradiated, defined synthetic diets with either 86 µg/kg (TD.09511, Harlan Laboratories) or 0 µg/kg vitamin B12 (TD.09512, Harlan Laboratories) ad libitum 7d prior to gavage. Barcoded bacterial strains were grown individually for 16h in TYG medium, combined, and ~109 CFUs were administered to each animal by oral gavage. Fecal samples were collected and frozen at −80ºC every day for the first three or six days and every three days thereafter. Animals were sacrificed on d15, samples collected along the length of the gut and stored as above. Total gDNA was recovered from each initial gavage, fecal and luminal content samples collected. DNA was extracted by bead-beating and phenol-chloroform extraction (Ley et al., 2005) and stored at −20ºC. Strain abundances were determined using the barcode qPCR assay described above.
Supplementary Material
Highlights.
Human gut microbes often encode multiple transporters for vitamin B12
The B12 transporters preferentially capture distinct B12-like small molecules (corrinoids)
Despite apparent redundancy, single transporters determine microbial fitness in mice.
The human gut microbiome contains dozens of corrinoid transporter families.
Acknowledgements
We thank E. Groisman, E. Seth, N. Moran, H. Ochman, W. Schofield, A. Hansen, and T. Cullen for useful discussion and L. Valle for laboratory assistance. We would also like to thank E. Martens for unpublished barcode vectors. This work was supported by National Institutes of Health grants DK089121 and GM103574 (to A.L.G.) and GM083303 (to M.E.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am. J. Clin. Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr. Protoc. Mol. Biol. 2006;15:15.8.1–15.8.28. doi: 10.1002/0471142727.mb1508s73. [DOI] [PubMed] [Google Scholar]
- Brandt LJ, Bernstein LH, Wagle A. Production of vitamin B12 analogues in patients with small-bowel bacterial overgrowth. Ann. Intern. Med. 1977;87:546–551. doi: 10.7326/0003-4819-87-5-546. [DOI] [PubMed] [Google Scholar]
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat. Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genetics. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo PM, Kadner RJ, Bradbeer C. Isolation of vitamin B12 transport mutants of Escherichia coli. J. Bacteriol. 1971;106:751–757. doi: 10.1128/jb.106.3.751-757.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Epstein W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid. Res. Mol. Biol. 2003:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides. Mol. Microbiol. 2009;74:1198–1210. doi: 10.1111/j.1365-2958.2009.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012a;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012b;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- Kenley JS, Leighton M, Bradbeer C. Transport of vitamin B12 in Escherichia coli. Corrinoid specificity of the outer membrane receptor. J. Biol. Chem. 1978;253:1341–1346. [PubMed] [Google Scholar]
- Li J, Yuan Z, Zhang Z. The cellular robustness by genetic redundancy in budding yeast. PLoS Genetics. 2010;6:e1001187. doi: 10.1371/journal.pgen.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire ME. Magnesium transporters: Properties, regulation and structure. Front. Biosci. 2006;11:3149–3163. doi: 10.2741/2039. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM, Geoffroy VA, Baida N, Gardan L, Izard D, Lemanceau P, Achouak W, Palleroni NJ. Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads. Appl. Environ. Microbiol. 2002;68:2745–2753. doi: 10.1128/AEM.68.6.2745-2753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KC, Taga ME. Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J. Bacteriol. 2013;195:1902–1911. doi: 10.1128/JB.01282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nielsen MJ, Rasmussen MR, Andersen CB, Nexo E, Moestrup SK. Vitamin B12 transport from food to the body's cells--a sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol. 2012;9:345–354. doi: 10.1038/nrgastro.2012.76. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Temperton B, Giovannoni SJ. Metagenomics: microbial diversity through a scratched lens. Curr. Opin. Microbiol. 2012;15:605–612. doi: 10.1016/j.mib.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) BioTechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varel VH, Bryant MP. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 1974;28:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JM. Osmosensing by bacteria. Sci STKE. 2006;2006:pe43. doi: 10.1126/stke.3572006pe43. [DOI] [PubMed] [Google Scholar]
- Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 2012;78:7745–7752. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.