Abstract
According to theory, present eukaryotic cells originated from a beneficial association between two free-living cells. Due to this endosymbiotic event the pre-eukaryotic cell gained access to oxidative phosphorylation (OXPHOS), which produces more than 15 times as much ATP as glycolysis. Because cellular ATP needs fluctuate and OXPHOS both requires and produces entities that can be toxic for eukaryotic cells such as ROS or NADH, we propose that the success of endosymbiosis has largely depended on the regulation of endosymbiont OXPHOS. Several studies have presented cytochrome c oxidase as a key regulator of OXPHOS; for example, COX is the only complex of mammalian OXPHOS with known tissue-specific isoforms of nuclear encoded subunits. We here discuss current knowledge about the origin of nuclear encoded subunits and the appearance of different isozymes promoted by tissue and cellular environments such as hypoxia. We also review evidence for recent selective pressure acting on COX among vertebrates, particularly in primate lineages, and discuss the unique pattern of co-evolution between the nuclear and mitochondrial genomes. Finally, even though the addition of nuclear encoded subunits was a major event in eukaryotic COX evolution, this does not lead to emergence of a more efficient COX, as might be expected from an anthropocentric point of view, for the “higher” organism possessing large brains and muscles. The main function of these subunits appears to be “only” to control the activity of the mitochondrial subunits. We propose that this control function is an as yet underappreciated key point of evolution. Moreover, the importance of regulating energy supply may have caused the addition of subunits encoded by the nucleus in a process comparable to a “domestication scenario” such that the host tends to control more and more tightly the ancestral activity of COX performed by the mtDNA encoded subunits. This article is part of a Special Issue entitled: Respiratory Oxidases.
Keywords: Evolution, Oxidative phosphorylation, Mitochondria, Endosymbiosis, Regulation
1. Introduction
Lynn Margulis’ theory about a prokaryotic origin for eukaryotic mitochondria is now broadly accepted [1–3]. According to this theory, present eukaryotic cells originated from a beneficial symbiosis between two free-living cells. Indeed, an α-proteobacterium was supposedly taken inside the pre-eukaryotic (host) cell and then formed an obligate endosymbiont.
Due to symbiotic association with the endosymbiont, i.e., the mitochondrial ancestor, the host cell gained access to oxidative phosphorylation (OXPHOS), which generates an ATP yield that is more than 15 times higher than glycolysis. With the exception of some parasitic organisms (such as Giardia or microsporidia), the conservation of OXPHOS across most eukaryotic lineages (e.g., plants, animals, fungi) suggests that this system is crucial for eukaryotic life. However, besides ATP, OXPHOS also produces reactive oxygen species (ROS, also sometimes referred to as ‘free radicals’) and heat, and requires oxygen and nutrients. Too large or too small an amount of these substrates and products can be toxic for eukaryotic cells. For example, an excess of substrate such as NADH can lead to lactic acidosis by driving lactate dehydrogenase to produce lactate [4] whereas an excess of products or by-product such as ROS can lead to apoptosis [5]. OXPHOS activity, therefore, has to be adjusted to take into account supply of nutrients and demand for energy. Furthermore, energy requirements differ among different cells from the same eukaryotic organism and from the same cell during its lifespan. All things considered, we propose that the success of endosymbiosis has largely depended on the regulation of OXPHOS activity, implying tight host-endosymbiont communication.
Several studies have presented complex IV (cytochrome c oxidase, COX, EC 1.9.3.1) as a key regulator of overall respiratory chain activity in intact mammalian cells: (i) COX has a high control coefficient in vivo on OXPHOS activity, meaning a decrease of COX activity decreases ATP production [6–8]; (ii) expression, assembly, and activity of COX were shown to be highly regulated [9,10]; and (iii) intrinsic biochemical parameters of COX were shown to be tissue specific [11] due to different isoform expression; for example, liver-type COX, which is expressed in tissues that rely fully on aerobic energy metabolism but cannot spare more room to increase the mitochondrial complement, has a higher basal activity compared to skeletal muscle/heart-type COX [11–16].
Mitochondrial encoded subunits carry out both electron transfer and proton-pumping functions, but it has been proposed that these enzymatic activities are mainly regulated through the nuclear encoded subunits [17]. Here, we discuss how evolutionary events that adapted OXPHOS activity to cellular requirements increased the fitness of the two genomes and were then positively selected and conserved. The importance of regulating energy supply may have caused a process comparable to a “domestication scenario” such that the host tends to control more and more tightly the ancestral activity of COX performed by the mtDNA encoded subunits through the addition of subunits encoded by the nucleus.
After a brief summary of our current understanding about the electron transfer and proton-pumping functions, we discuss the origin of nuclear encoded subunits and the appearance of different isozymes promoted by tissue and cellular environments such as hypoxia. Finally, we review evidence for recent selective pressure acting on COX among vertebrates, particularly in primate lineages, and discuss the unique pattern of co-evolution between the nuclear and mitochondrial genomes.
2. Ancestral function of cytochrome c oxidase
The mitochondrial respiratory chain couples the reduction of molecular oxygen to the translocation of protons across the inner mitochondrial membrane [18]. In mammals, the first step of the respiratory chain is the oxidation of NADH or FADH2 by, respectively, complexes I and II, followed by electron transfer to complex III via coenzyme Q, and finally transfer via cytochrome c to complex IV (COX), which reduces the final acceptor oxygen to water. Complexes I, III, and IV couple the redox reactions to the translocation of protons across the inner mitochondrial membrane. These translocations generate a proton gradient that permits ATP synthase to synthesize ATP from ADP and inorganic phosphate.
Mitochondrial COX, the terminal complex of the respiratory chain, belongs to a large family of heme-copper terminal oxidases, also containing prokaryotic aa3-type COX. Although mammalian mitochondrial COX has 13 subunits and prokaryotic aa3-type has only three to four subunits [19,20], the amino acid sequences of the core subunits I and II of both enzymes are more conserved than is typical between bacteria and mammalian homologs. For example, there is 52% identity between Bos taurus and Paracoccus denitrificans subunits I (69% similarity) and 34% identity between subunits II (59% similarity). The X-ray structures of P. denitrificans and bovine enzymes confirmed that the structure is also very similar between prokaryotic and mitochondrial COX [21–24]. Although the proton pumping mechanism is not yet fully understood, the different models proposed are consistent with both the mitochondrial and prokaryotic structures.
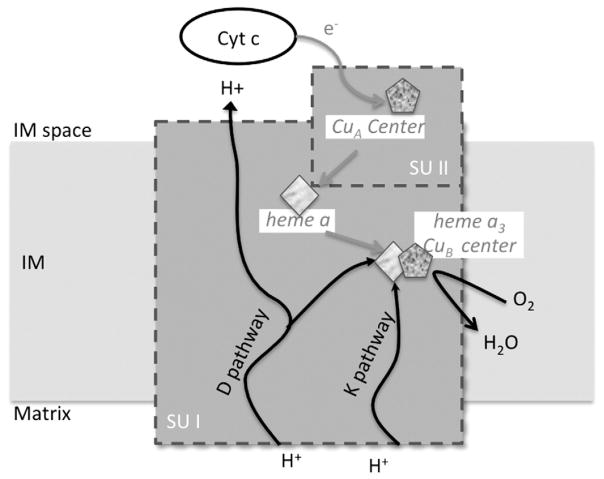
In mammalian mitochondria COX is embedded in the inner membrane and reduced cytochrome c binds to COX on the inter-membrane space side [17]. Electrons from cytochrome c are passed through the CuA center located in subunit II (Fig. 1), then go to subunit I, from heme a to the heme a3/CuB center. This is the catalytic center of the enzyme, which reduces oxygen to water, by consuming electrons that come initially from cytochrome c and four chemical protons that are taken up from the matrix. This reaction is exergonic and is coupled to the translocation of four additional protons from the matrix to the intermembrane space. The proton gradient generated powers the conversion of ADP and phosphate to ATP by ATP synthase [25]. At least two pathways are currently thought to conduct protons inside subunit I [26–29]: (i) the K pathway (via Lys319, according to bovine numbering), which leads to the heme a3/CuB (catalytic center), and (ii) the D pathway (via Asp91 to Glu242, according to bovine numbering), where protons can be directed to the heme a3/CuB center and/or to the intermembrane space.
Fig. 1.
Mechanism of cytochrome c oxidase, squares symbolize heme and pentagons symbolize coppers. IM space, intermembrane space; IM, inner membrane; gray arrows represent electron pathway and black arrows represent proton transfer pathway and oxygen reduction.
As prokaryotic subunits I and II are sufficient to carry out the electron transfer and proton-pumping functions, COX presumably has been functional since the initiation of endosymbiosis. However, because the energetic needs of eukaryotic cells undoubtedly changed over time, selective pressures would have rapidly appeared to favor synchronization of energy production to those cellular needs.
3. Addition of nuclear genes
Due to the presence of ten nuclear in addition to the three mitochondrial encoded COX subunits, their origin and evolution is likely to be insightful about the regulation of ATP production by OXPHOS [9,10].
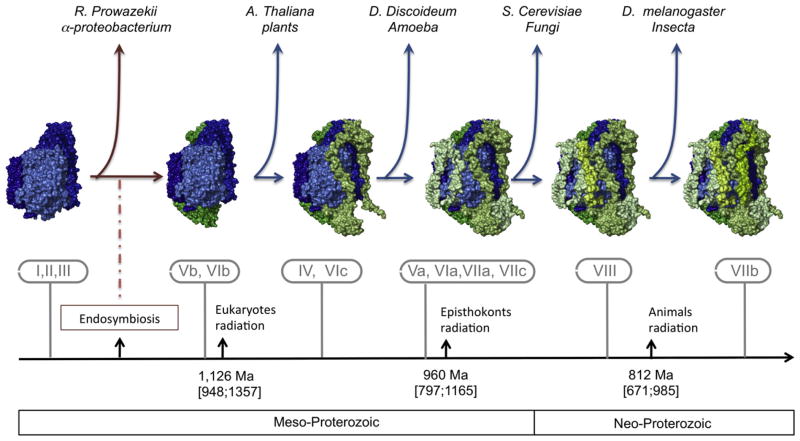
Because many ancestral endosymbiont genes have been transferred to the nuclear genome [30], the current location of the subunits is not informative about their possible endosymbiont or host origin. Indeed, even the three mammalian mitochondrial encoded subunits have been reported to be completely (COX II, III) or partially (COX I) encoded by the nucleus in some organism [31–33]. To address this question about genome origin, Das et al. have looked for homologous sequences of the nuclear-encoded COX subunits in the Rickettsia prowazekii genome [34], which is the α-proteobacterium thought to be the most closely related descendant of the species that became the ancestral mitochondria [35]. They also searched for the presence of homologs in the mitochondrial genome of the protozoan Reclinomonas americana, which contains the largest number of genes (97) of any mitochondrion studied [36]. Computational searches failed to find any homolog, which likely suggests that these proteins were not present in the ancestral endosymbiont even if it does not exclude some secondary loss.
In contrast, homologs of several human nuclear-encoded COX subunits have been found in invertebrates (Drosophila melanogaster), fungi (Saccharomyces cerevisiae), mycetazoan amoeba (Dictyostelium discoideum), and plants (Arabidopsis thaliana) [34]. These results suggest that the emergence of nuclear-encoded subunits started before the radiation of the major eukaryote lineages. However, all subunits are not present in all organisms [34]; this could be due to a secondary loss as mentioned above or also be an indicator of the time of appearance (Fig. 2). Both hypotheses are likely; further complete genome sequencing will refine our knowledge about the timing of their appearance. It is worth noting that the largest human subunits [37] are the most shared across phyla: subunits IV (about 17 kDa in human), Vb (about 11 kDa), VIb (about 10 kDa), and VIc (about 8 kDa) are also present in plants or in amoeba, suggesting an old origin; in contrast, according to Das et al., two of the smallest subunits, VIIb (about 6 kDa) and VIII (about 5 kDa), seem only present in animals [34,37].
Fig. 2.
Hypothetical genesis of vertebrate cytochrome c oxidase. Based on Das et al. [34], mitochondrial encoded subunits are represented in blue and nuclear encoded subunits are represented in green.
The import of nuclear encoded subunits requires both the development of a protein import machinery in the endosymbiont membranes and specific targeting information in nuclear encoded subunits. How this complex system has emerged is not clear; however, the presence of a protein import machinery, translocase of the outer membrane (TOM complex) and translocases of the inner membrane (TIM complexes) in plants, fungi, and animals, suggests that the import of host polypeptides was one of the first evolution steps after the endosymbiotic event [38,39]. The emergence of an import system has raised the questions of the timing of its emergence and of whether those systems were derived from the host or from the endosymbiont genome [40–43].
Regarding the emergence of the “targeting information,” several studies have proposed a concept of “preadaptations” where pre-existent proteins with positively charged amino acid sequences would have been imported “by chance” into the mitochondria by TIM-TOM complexes [44,45]. This could have been the case for the oldest nuclear COX subunits.
Once in mitochondria, the most parsimonious scenario would be a random association due to physicochemical characteristics between the nuclear and endosymbiont subunits. Then advantageous association would have been selected for, fixed, and conserved and implemented as part of the assembly process. Since the ancestral endosymbiont COX was already a complex formed by several subunits, an assembly process was already in existence that the nuclear encoded subunits could adapt to form their association. Even if bacterial and human COX assembly are very different, the presence of homologous assembly genes between human and α-proteobacteria suggests that the eukaryotic assembly process is an evolution of the prokaryotic assembly process rather than a de novo process [46]. Despite a high conservation of nuclear encoded subunits and of the copper insertion pathway between yeast and humans, only a few COX assembly factors are common [47], which suggests that the assembly process was improved over a long time scale rather than just after the endosymbiont event.
Interestingly, some nuclear subunits seem to have specific functions in assembly. For example, in yeast subunit VIb is needed for assembly but can be removed from the mature enzyme without any effect on activity [48]. Other mammalian subunits (Vb, VIa, and VIb) might be involved in the stability of the dimer state [23]. By increasing or limiting the enzyme amount, the regulation of assembly and enzyme stability are an important part of the enzyme activity control [9].
Some nuclear encoded subunits play a more direct role in COX regulation. Indeed, functional work has shown that nuclear subunits are central in the complex regulation of enzyme activity. For example, COX IV binds ATP, leading to an allosteric inhibition of enzyme activity [49], and this inhibition can be reversed by binding the thyroid hormone T2 to subunit Va [50]. This example shows nuclear encoded subunits’ ability to regulate COX activity based on cellular energy demand (ATP level) and to integrate external stimuli like thyroid hormone. These new proprieties allow endosymbiont OXPHOS to respond to intracellular and extracellular stimuli to adapt host energetics.
4. Addition of tissue specific subunits
In multicellular organisms, the same extracellular stimulus can require different OXPHOS responses depending of the type of cell. In mammals, it has been shown that biochemical characteristics of COX differ regarding the tissue of origin [11,12]. However, given that mitochondrial encoded subunits, which are responsible for the catalytic functions, are the same in all tissues, one possible scenario would be that the observed differences in COX activity can be related to the emergence of nuclear subunit isoforms. Indeed, isoforms for at least 5 subunits have been described in mammals: COX subunit IV, VIa, VIb, VIIa and VIII [15,51–54]. Based on phylogenetic evidence, it has been proposed that the emergence of these subunit isoforms is caused by duplications of ancestral subunit coding genes [55]. These duplications lead to two gene homologues co-existing in the same genome. Two genes presumed to be derived from a single duplication event and potentially coding for two isoforms are called paralogous. Interestingly, among the OXPHOS gene family, setting aside numts [56], COX has more paralogs than any other OXPHOS complex, emphasizing the key role of COX in OXPHOS adaptation and regulation in different tissues [8,55]. In fact, COX and cytochrome c are the only components of mammalian OXPHOS with known tissue-specific isoforms (reviewed in [57]).
Despite high conservation of nuclear subunits between yeast and mammals, the isoforms of homologous subunits between the two taxa do not share a common origin. Indeed, a recent study has shown that vertebrate and invertebrate COX subunit isoforms are due to distinct duplication events [55]. These observations could suggest poor conservation of paralogous genes once duplicated or recent duplications. The second hypothesis is supported by the fact that the common explanation for isoform diversity in vertebrates is the suggested two rounds of whole-genome duplication [58]. This duplication of complete genomes was suggested based on the HOX gene cluster and would have occurred at the stem of the vertebrate lineage [59]. For COX, this old global duplication seems consistent with at least three subunits, COX IV, COXVIa, and COX VIIa, which have two paralogs in fish and tetrapods [55]. However, some duplication events appear to be more recent; for example; COX VIb has only two paralogs in fish and most tetrapods but three isoforms are present in the primate lineage [55].
After duplication, when the presence of a double copy of the same gene is not deleterious, selective pressures decrease for at least one copy. Indeed, one copy can accumulate mutations with only a small fitness decrease due to the presence of the other “back up” gene copy. Since constraints are relaxed, the divergence rate between the two copies is accelerated and new functional properties are allowed to appear (neofunctionalization). A high divergence rate can also specialize two copies for two or more different functions previously done by the ancestral gene (subfunctionalization) [60]. One form of subfunctionalization is a differential expression pattern depending on the cellular environment, e.g., tissue-specific isoform expression. Once this pathway is engaged, selection is reinstated. Indeed, mutations allowing an optimization of the copy to the new cellular environment or function are positively selected and deleterious mutations are negatively selected (due to the absence of the “backup copy” doing the new function) [61].
It is unclear how the paralog of each nuclear COX subunit developed its expression pattern. However, three pairs of subunits, VIa, VIIa, and VIII, have what was originally called H (heart/skeletal muscle) type and L (liver) type isoforms. For each subunit the L isoform is actually ubiquitously expressed whereas H shows developmental and tissue specificity, being primarily expressed postnatally in mature contractile muscles [62].
The evolution of this tissue-specific expression has led to a dichotomy between these two types of tissue: (i) In the first type, called liver-type tissue based on the COX isozyme expression in tissues like liver and kidney, the COX enzyme shows a higher basal activity [12] but lower quantity of enzyme due to a relatively low number of mitochondria [11]. This is likely so because those tissues have other specialized functions, which do not allow a further increase in mitochondrial mass. (ii) The second type, called heart-type based on the COX isozyme expressed in contractile tissues (e.g., heart and skeletal muscle), is characterized by a high capacity of COX due to a high amount of mitochondria [11] coupled with a lower basal COX activity compared to liver-type COX [12].
This dichotomy allows for different regulatory systems. For example, in muscle, exercise increases COX complex biogenesis and then OXPHOS activity [63]. Energy regulation based on complex amount seems well adapted for muscle cells. Indeed, skeletal muscle is less size constrained than other tissues, e.g., muscle amount increases and decreases during lifespan depending on stimulation. Interestingly, the mammalian brain COX isozyme belongs to the liver-type [64] and is characterized by an intermediary amount and intermediary basal COX activity [11]. This last pattern may have been selected in response to specific-brain constraints like high energy need despite limited space for mitochondria in neurons.
5. COX adaptation to oxygen
The emergence of nuclear encoded subunit isoforms after the whole genome duplication became crucial for vertebrate cells to adapt OXPHOS activity to the cellular oxygen environment. Indeed, vertebrate cells do not possess the prokaryotic cell’s ability to switch between different respiratory chains in response to the extracellular environment. In contrast to only one terminal oxidase present in vertebrates, P. denitrificans or Bacillus subtilis each possess three different terminal oxidases. The switch between different terminal oxidases has been shown to be a key regulatory element; for example, cytochrome bd, present in B. subtilis and Escherichia coli, is involved in oxygen regulation [65,66]. E. coli possesses two quinol oxidases and expresses predominantly cytochrome bo3 in normal aerobic conditions. However, E. coli expresses cytochrome bd at low oxygen tension, which has a high affinity for oxygen but does not pump protons [67]. This hypoxic system has a lower efficiency but allows OXPHOS to produce some energy and to regenerate NAD+ from NADH to maintain metabolic flux even at low oxygen tensions.
An alternative terminal oxidase (AOX) that couples ubiquinol oxidation with oxygen reduction to water without proton pumping has also been described in plants. In contrast to cytochrome bd of E. coli, AOX has a lower affinity for oxygen than COX but it is also regulated by oxygen concentration [68]. By following transcription, protein amount, and activity of AOX during and after hypoxia, Szal and collaborators have shown (i) an absence of AOX amount and activity during hypoxia, and (ii) a similar AOX amount and activity before and just after hypoxia [68]. AOX transcript levels showed no significant differences between control, hypoxic, and post-hypoxic tissue indicating that the regulation of AOX expression by oxygen availability may take place at the translational level. It has been suggested that, in plants, AOX could be a part of the post-hypoxic tissue response characterized by Biermelt and coworkers, who observed higher respiration rates and an activation of antioxidant defense systems due to the increase of oxygen concentration [69]. The physiological role of AOX is still unclear; however, a main function would be to prevent an over-reduction of ubiquinone and thus limit ROS production [70]. This function seems to be particularly important when oxygen concentration changes in hyperoxia and post-hypoxia.
AOX has been well studied in plants and is known to be present in a few other eukaryotes like parazoa and fungi, but had been supposed to be absent in animals. However, recent evidence has shown the presence AOX in metazoans [71]. Surprisingly, AOX appears to have a broad distribution across all the main metazoan phyla, e.g., Cnidaria, Annelida, Mollusca, Nematoda and Chordata [72]. Based on this broad distribution, it is possible to conclude that AOX was present at an early stage of eukaryotic history and was an element of the original plant, animal, and fungal respiratory chain (Fig. 3). Despite missing evidence about the existence of AOX in other eukaryote lineages, e.g. Excavates and Chromoalveolates, Atteia et al. have found a gene encoding an AOX homolog in one α-proteobacterium, suggesting an endosymbiont origin of AOX [73,74]. However, a recent gain of AOX due to a horizontal gene transfer cannot yet be fully excluded.
Fig. 3.
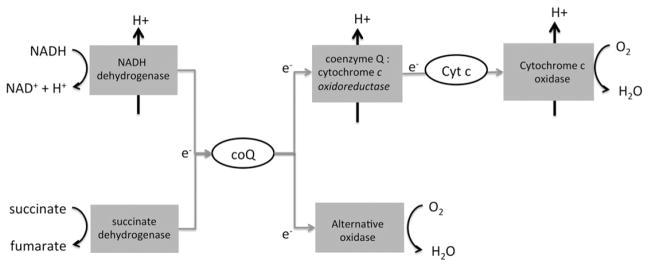
Branched respiratory chain. Respiratory complexes are represented as gray squares, and gray arrows represent electron transfer. Other reactions are represented with black arrows. This is presumably the ancestral respiratory chain for plants, fungi, and animals, containing in addition to cytochrome c oxidase of vertebrates an alternative terminal oxidase (AOX).
Despite a great number of genomes analyzed, McDonald and colleagues failed to show any evidence of AOX in vertebrates [72]. This result suggests that AOX was lost at the vertebrate stem. Interestingly, this event is concomitant with the whole genome duplication and the proposed origin of isoform pairs for three COX nuclear encoded subunits [55]. Remarkably, one of these new isoform pairs is COX IV isoform 1 and COX IV isoform 2 [15,75]. The expression pattern of these two isoforms is well known to be oxygen dependent, although the precise mechanism remains controversial [76,77]. The two COX IV isoforms also have a tissue expression pattern related to the oxygen use of the tissue [57], i.e., in mammals COX IV isoform 2 is specifically expressed in the well-oxygenated tissues lung and trachea in addition to placenta [78], the latter being the site for embryonic gas exchange, which is sometimes referred to as the lung of the embryo. A similar expression pattern has been observed in zebrafish, with the mRNA for COX IV isoform 2 more abundant in the gills [55]. This common expression pattern in oxygen exchange tissues between mammals and fish suggests an old subfunctionalization for the two isoforms probably since the stem of vertebrates and concomitant with the loss of AOX. The pattern of expression of isoform 2 is odd. Indeed, this isoform is mainly present in tissues with high oxygen like lung, but is also induced during hypoxia [76,77]. Interestingly, cells expressing this second isoform produce fewer free radicals, which is similar to plant cells expressing AOX [76].
We suggest that the ability to change COX IV isoform expression would become an alternative way to regulate OXPHOS as a function of oxygen concentration in the absence of AOX in the vertebrate lineage. Thus, AOX studies could help us to better understand the expression pattern of COX IV isoforms. For example, in plants, the level of AOX mRNA has been found high during hypoxia but this does not influence the protein level, which suggests post-transcriptional regulation [68]. The high amount of mRNA during hypoxia accelerates protein synthesis during the post-hypoxia response. Despite the fact that this pattern has been observed in plant cells, which are able to support much greater variation of oxygen concentration than mammalian cells, it would be interesting to carry out similar studies for COX IV isoforms in mammals.
AOX has been lost independently in numerous taxa, as for example in Saccharomyces cerevisiae [72]. Interestingly, S. cerevisiae possesses only one pair of COX subunit isoforms. Indeed, only the proposed homolog of vertebrate COX IV (named COX5 in yeast) is present in two isoform copies. As in vertebrates, isoform expression is also regulated by oxygen availability and the hypoxic isoform enhances the catalytic constant relative to the ‘aerobic’ isoform [79]. This proposed convergent evolution of oxygen regulation between yeast and vertebrate ancestors separated by one billion years of evolution could appear as astonishing. However, with a closer look several points should limit the enthusiasm about a functional homology between the mammalian COX IV and the yeast COX V isoform pairs [77]. First, the promoter sequences do not share any homology. Secondly, the amino-acid sequence itself is proportionately the least conserved of the nuclear-encoded subunits, e.g., 9% between mammals and yeast (39% for mammalian subunit Va, 22% for VIa, 24% for VIb, or 16% for VIIc compared to the corresponding subunits in yeast), and the homology hypothesis is mostly supported by the presence of conserved domains [34]. Finally, the yeast hypoxic Vb gene is induced under strong hypoxia whereas mammals have COX IV-2 always expressed in lung (a highly oxygenated tissue). Therefore, COX IV-2 appears to be a primary adaptation to a high oxygen environment.
Additionally, a direct link between subunit IV duplication, AOX loss, and hypoxia is not universal. For example, the amoeba Dictyostelium discoideum presents a different pattern of hypoxia response at the level of COX. Despite this organism’s still having AOX, hypoxia leads to an expression switch between two isoforms homologous to mammalian subunit VIc (called VIIe and VIIs in D. discoideum) [80,81].
The responses to varying oxygen environments across different species and different kingdoms underline the importance of the nuclear encoded subunits as modulators of COX activity and as modulators of global endosymbiont OXPHOS activity.
6. Evolution
Environment and way of life generate selective pressures that act not only on regulatory processes but also on intrinsic enzyme activity. Due to mitochondrial DNA’s high mutation rate, mitochondrial encoded COX subunits are particularly sensitive to new selective pressures. For example, selection and adaptation of mitochondrial-encoded subunits has been shown in vertebrate species living in low oxygen conditions [82,83]. By sequencing the mitochondrial DNA of pika (Ochotona curzoniae) living at a high altitude Tibetian plateau, Luo et al. have identified three very specific mutations affecting COX I and COX II amino acid sequences. Because two amino acid replacements of COX substitute non-polar to polar residues they propose that the mutations are functionally significant. Using a functional approach, Scott et al. have shown that cardiac myocytes of high altitude geese have reduced COX Vmax and a higher cytochrome c affinity than geese flying at moderate altitude [82]. The authors propose that because less reduced cytochrome c is needed to sustain maximal rates of respiration by COX, “Bar-headed geese may therefore be capable of sustaining cardiac output during hypoxia without experiencing large shifts in mitochondrial redox toward a more reduced state, thus minimizing cellular damage induced by ROS.” They failed to observe any difference in COX mRNA and protein amount, suggesting a structural evolution rather than a regulatory evolution. They then suggested that the replacement of a neutral residue (tryptophan) at position 116 of COX III by a positively charged amino-acid (arginine) could change the structure of COX and explain their results. However, the authors did not rule out other explanations such as the possibility of different phosphorylation patterns between the different species.
During the past decades our group and others have shown accelerated evolution of the COX enzyme during primate evolution [84–92]. To date, nine of the thirteen COX subunits have been shown to have an accelerated amino acid replacement rate in anthropoid primates [93]. We have also demonstrated that the targeting pre-sequence does not show accelerated evolution [94]. Because pre-sequences are subject to the same selection-independent factors, such as population size and mutation rate, it is possible to conclude that the evolution rate differences between sequence and pre-sequence are due to selective pressure. In parallel, functional work has shown a very specific binding interaction between COX and anthropoid cytochrome c compared to other primates and mammals [95–97]. Based on structural data regarding the cytochrome c binding site, it was proposed that accelerated COX evolution particularly affected the positions binding cytochrome c (27 of the 57 residues that bind cytochrome c were replaced in the anthropoid lineage). Because 11 charge-bearing residues involved in binding cytochrome c have been replaced with uncharged residues, it appears that there was a drastic reduction of the electrostatic interaction between COX and cytochrome c in the anthropoid lineage [98].
Anthropoid primates include New World and Old World monkeys and apes (including humans). Because the main phenotypic evolution that occurred on the anthropoid lineage (e.g., longer lifespan, larger neocortex, and prolonged fetal development) is related to aerobic energy metabolism, it is tempting to speculate that there is a link between molecular and phenotypic evolution. Interestingly, we have also shown evolution of the brain expression pattern of at least one subunit across primates: COX Va is expressed in cerebral cortical tissue at a higher level in human than in chimpanzee or gorilla [92].
This co-adaptation of both mitochondrial and nuclear subunits makes it unlikely that this evolution is due to a random increase of mutation rate at a few points in the genome. This also emphasizes the co-evolution between nuclear and mitochondrial subunits. Since mitochondrial and nuclear DNAs evolve at considerably different rates [99] the importance of this co-evolution between genomes was previously demonstrated by studying cybrid cells containing nDNA and mtDNA from different species. For example, oxidative phosphorylation of mtDNA-depleted human cells can be restored by mtDNA from common chimpanzee, bonobo, and gorilla but not by mtDNA from orangutan and representative species of Old World monkeys, New World monkeys, and strepsirrhines [100]. This incompatibility of mtDNA from primate species that diverged from humans as recently as 8–18 mya has specifically been attributed to a COX deficiency. Indeed, Barrientos et al. have shown that human COX nuclear subunits and human orangutan mitochondrial subunits are not able to be assembled together correctly [101]. COX assembly appears to be very sensitive to small amino acid variation.
By studying the evolution rate of interacting nDNA-encoded and mtDNA-encoded residues across vertebrates, our group has shown that nDNA-encoded residues in close physical proximity to mtDNA-encoded residues evolve more slowly than the other nuclear-encoded residues [102]. This is due to a greater purifying selection at the interface between subunits and confirms the greater structural–functional constraints suggested by xeno-cybrid studies. However, mtDNA-encoded residues in close physical proximity to nDNA-encoded residues evolve more rapidly than the other mitochondrial-encoded residues, suggesting that adaptive evolution has occurred at these residues. It appears that only residues encoded by fast mutating DNA (mtDNA) have undergone adaptive selection. Thus, we propose that selective pressures appear and disappear often but each window is only open during a short period of time (i.e., episodic positive selection). A fast mitochondrial mutation rate could actually be a major adaptive advantage when a population changes its way of life or environment and when this change requires a different ROS, heat, and energy balance. This is one of many hypotheses that could explain why the original endosymbiont subunits are still mitochondrially encoded [103].
7. Conclusion
The addition of nuclear encoded subunits was a major event in eukaryotic COX evolution, and some of these subunits are very conserved across eukaryotes. However, they are not directly involved in the COX catalytic process, and prokaryotic COX is very efficient without these sub-units. The main function of these subunits appears to be “only” to control the activity of the mitochondrial subunits. This control function is perhaps an underappreciated key point of evolution, especially in multicellular organisms. Indeed, once the respiratory chain was integrated into the eukaryotic cell the challenge was no longer to produce more energy but to produce it exactly when needed, and to conserve resources when less or no energy was needed. The evolutionary studies point out that selective pressures do not promote an increase of COX activity but promote a COX activity more adapted to the needed trade-off between energy, heat, and ROS production. An important control function that has not been sufficiently studied as yet is cell signaling. Evidence does exist, however, that subunit amino-acid evolution affects cell signaling in a species-specific manner [104]. Several amino acids identified as regulatory phosphorylation sites in non-human mammals cannot be phosphorylated in human [8]. For example, Tyr 11 of COX subunit IV-1 is phosphorylated in cow liver but in humans the corresponding amino acid is a phenylalanine and thus cannot be phosphorylated. Inversely, Thr 35 and Thr 38 of COX subunit Va have been shown phosphorylated in human but are glycines in other species. Unfortunately, studies about evolution of cell signaling pathways targeting OXPHOS, and especially COX, are still quite rare [105]. Finally, eukaryotic evolution is not characterized by the emergence of a more efficient COX, as might be expected from an anthropocentric point of view, for the “higher” organism possesses a big brain and big muscles. This failure to find a more efficient COX is perhaps due to the fact that the basic three subunit prokaryotic COX constitutes a system that was already fully optimized before the endosymbiotic event and not much could have been done to improve this system even by adding other subunits. “Perfection is achieved, not when there is nothing more to add, but when there is nothing left to take away.” Antoine de Saint-Exupery, French writer (1900–1944).
Acknowledgments
Supported by NIH GM65580, and by the National Science Foundation (grants BCS-0550209, BCS0827546 and BCS 9910679) and the Wayne State University Research Excellence fund.
Footnotes
This article is part of a Special Issue entitled: Respiratory Oxidases.
References
- 1.Richards TA, Archibald JM. Cell evolution: gene transfer agents and the origin of mitochondria. Curr Biol. 2011;21:R112–R114. doi: 10.1016/j.cub.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer C. Origins. On the origin of eukaryotes. Science. 2009;325:666–668. doi: 10.1126/science.325_666. [DOI] [PubMed] [Google Scholar]
- 3.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 4.Veech RL. The metabolism of lactate. NMR Biomed. 1991;4:53–58. doi: 10.1002/nbm.1940040204. [DOI] [PubMed] [Google Scholar]
- 5.Avery SV. Molecular targets of oxidative stress. Biochem J. 2011;434:201–210. doi: 10.1042/BJ20101695. [DOI] [PubMed] [Google Scholar]
- 6.Pacelli C, Latorre D, Cocco T, Capuano F, Kukat C, Seibel P, Villani G. Tight control of mitochondrial membrane potential by cytochrome c oxidase. Mitochondrion. 2011;11:334–341. doi: 10.1016/j.mito.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in wild-type and mitochondrial DNA mutation-carrying human cells. Proc Natl Acad Sci U S A. 1997;94:1166–1171. doi: 10.1073/pnas.94.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hüttemann M, Helling S, Sanderson TH, Sinkler C, Samavati L, Mahapatra G, Varughese A, Lu G, Liu J, Grossman LI, Doan JW, Marcus K, Lee I. Regulation of mitochondrial respiration and apoptosis through cell signaling: cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim Biophys Acta. 2012;1817:598–609. doi: 10.1016/j.bbabio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 10.Follmann K, Arnold S, Ferguson-Miller S, Kadenbach B. Cytochrome c oxidase from eucaryotes but not from procaryotes is allosterically inhibited by ATP. Biochem Mol Biol Int. 1998;45:1047–1055. doi: 10.1002/iub.7510450522. [DOI] [PubMed] [Google Scholar]
- 11.Benard G, Faustin B, Passerieux E, Galinier A, Rocher C, Bellance N, Delage JP, Casteilla L, Letellier T, Rossignol R. Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol. 2006;291:C1172–C1182. doi: 10.1152/ajpcell.00195.2006. [DOI] [PubMed] [Google Scholar]
- 12.Vijayasarathy C, Biunno I, Lenka N, Yang M, Basu A, Hall IP, Avadhani NG. Variations in the subunit content and catalytic activity of the cytochrome c oxidase complex from different tissues and different cardiac compartments. Biochim Biophys Acta. 1998;1371:71–82. doi: 10.1016/s0005-2736(97)00278-2. [DOI] [PubMed] [Google Scholar]
- 13.Seelan RS, Grossman LI. Cytochrome c oxidase subunit VIIa isoforms. Characterization and expression of bovine cDNAs. J Biol Chem. 1991;266:19752–19757. [PubMed] [Google Scholar]
- 14.Lomax MI, Grossman LI. Tissue-specific genes for respiratory proteins. Trends Biochem Sci. 1989;14:501–503. doi: 10.1016/0968-0004(89)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Huttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- 16.Jaradat SA, Ko MS, Grossman LI. Tissue-specific expression and mapping of the Cox7ah gene in mouse. Genomics. 1998;49:363–370. doi: 10.1006/geno.1998.5279. [DOI] [PubMed] [Google Scholar]
- 17.Ludwig B, Bender E, Arnold S, Huttemann M, Lee I, Kadenbach B. Cytochrome c oxidase and the regulation of oxidative phosphorylation. Chembiochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Wikstrom MK. Proton pump coupled to cytochrome c oxidase in mitochondria. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 19.Steinrucke P, Steffens GC, Panskus G, Buse G, Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987;167:431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- 20.Raitio M, Jalli T, Saraste M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987;6:2825–2833. doi: 10.1002/j.1460-2075.1987.tb02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 22.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 23.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 24.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 26.Hosler JP, Ferguson-Miller S, Calhoun MW, Thomas JW, Hill J, Lemieux L, Ma J, Georgiou C, Fetter J, Shapleigh J, et al. Insight into the active-site structure and function of cytochrome oxidase by analysis of site-directed mutants of bacterial cytochrome aa3 and cytochrome bo. J Bioenerg Biomembr. 1993;25:121–136. doi: 10.1007/BF00762854. [DOI] [PubMed] [Google Scholar]
- 27.Thomas JW, Lemieux LJ, Alben JO, Gennis RB. Site-directed mutagenesis of highly conserved residues in helix VIII of subunit I of the cytochrome bo ubiquinol oxidase from Escherichia coli: an amphipathic transmembrane helix that may be important in conveying protons to the binuclear center. Biochemistry. 1993;32:11173–11180. doi: 10.1021/bi00092a029. [DOI] [PubMed] [Google Scholar]
- 28.Thomas JW, Puustinen A, Alben JO, Gennis RB, Wikstrom M. Substitution of asparagine for aspartate-135 in subunit I of the cytochrome bo ubiquinol oxidase of Escherichia coli eliminates proton-pumping activity. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 29.Fetter JR, Qian J, Shapleigh J, Thomas JW, Garcia-Horsman A, Schmidt E, Hosler J, Babcock GT, Gennis RB, Ferguson-Miller S. Possible proton relay pathways in cytochrome c oxidase. Proc Natl Acad Sci U S A. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk R, Huynen MA. Mosaic origin of the mitochondrial proteome. Proteomics. 2010;10:4012–4024. doi: 10.1002/pmic.201000329. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Martinez X, Vazquez-Acevedo M, Tolkunova E, Funes S, Claros MG, Davidson E, King MP, Gonzalez-Halphen D. Unusual location of a mitochondrial gene. Subunit III of cytochrome c oxidase is encoded in the nucleus of Chlamydomonad algae. J Biol Chem. 2000;275:30144–30152. doi: 10.1074/jbc.M003940200. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Martinez X, Antaramian A, Vazquez-Acevedo M, Funes S, Tolkunova E, d’Alayer J, Claros MG, Davidson E, King MP, Gonzalez-Halphen D. Subunit II of cytochrome c oxidase in chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J Biol Chem. 2001;276:11302–11309. doi: 10.1074/jbc.M010244200. [DOI] [PubMed] [Google Scholar]
- 33.Gawryluk RMR, Gray MW. An ancient fission of mitochondrial cox1. Mol Biol Evol. 2010;27:7–10. doi: 10.1093/molbev/msp223. [DOI] [PubMed] [Google Scholar]
- 34.Das J, Miller ST, Stern DL. Comparison of diverse protein sequences of the nuclear-encoded subunits of cytochrome c oxidase suggests conservation of structure underlies evolving functional sites. Mol Biol Evol. 2004;21:1572–1582. doi: 10.1093/molbev/msh161. [DOI] [PubMed] [Google Scholar]
- 35.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 36.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 37.Capaldi RA. Structure and function of cytochrome c oxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- 38.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 39.Perry AJ, Rimmer KA, Mertens HDT, Waller RF, Mulhern TD, Lithgow T, Gooley PR. Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol Biochem. 2008;46:265–274. doi: 10.1016/j.plaphy.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Alcock F, Clements A, Webb C, Lithgow T. Evolution. Tinkering inside the organelle. Science. 2010;327:649–650. doi: 10.1126/science.1182129. [DOI] [PubMed] [Google Scholar]
- 41.Clements A, Bursac D, Gatsos X, Perry AJ, Civciristov S, Celik N, Likic VA, Poggio S, Jacobs-Wagner C, Strugnell RA, Lithgow T. The reducible complexity of a mitochondrial molecular machine. Proc Natl Acad Sci U S A. 2009;106:15791–15795. doi: 10.1073/pnas.0908264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross J, Bhattacharya D. Endosymbiont or host: who drove mitochondrial and plastid evolution? Biol Direct. 2011;6:12. doi: 10.1186/1745-6150-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong J, Dolezal P, Selkrig J, Crawford S, Simpson AG, Noinaj N, Buchanan SK, Gabriel K, Lithgow T. Ancestral and derived protein import pathways in the mitochondrion of Reclinomonas americana. Mol Biol Evol. 2011;28:1581–1591. doi: 10.1093/molbev/msq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucattini R, Likic VA, Lithgow T. Bacterial proteins predisposed for targeting to mitochondria. Mol Biol Evol. 2004;21:652–658. doi: 10.1093/molbev/msh058. [DOI] [PubMed] [Google Scholar]
- 45.Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnesano F, Banci L, Bertini I, Martinelli M. Ortholog search of proteins involved in copper delivery to cytochrome c oxidase and functional analysis of paralogs and gene neighbors by genomic context. J Proteome Res. 2005;4:63–70. doi: 10.1021/pr049862f. [DOI] [PubMed] [Google Scholar]
- 47.Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim Biophys Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaMarche AEP, Abate MI, Chan SHP, Trumpower BL. Isolation and characterization of COX12, the nuclear gene for a previously unrecognized subunit of Saccharomyces cerevisiae cytochrome coxidase. J Biol Chem. 1992;267:22473–22480. [PubMed] [Google Scholar]
- 49.Arnold S, Kadenbach B. The intramitochondrial ATP/ADP-ratio controls cytochrome c oxidase activity allosterically. FEBS Lett. 1999;443:105–108. doi: 10.1016/s0014-5793(98)01694-9. [DOI] [PubMed] [Google Scholar]
- 50.Arnold S, Goglia F, Kadenbach B. 3,5-Diiodothyronine binds to subunit Va of cytochrome-c oxidase and abolishes the allosteric inhibition of respiration by ATP. Eur J Biochem. 1998;252:325–330. doi: 10.1046/j.1432-1327.1998.2520325.x. [DOI] [PubMed] [Google Scholar]
- 51.Grossman LI, Lomax MI. Nuclear genes for cytochrome c oxidase. Biochim Biophys Acta. 1997;1352:174–192. doi: 10.1016/s0167-4781(97)00025-0. [DOI] [PubMed] [Google Scholar]
- 52.Huttemann M, Schmidt TR, Grossman LI. A third isoform of cytochrome c oxidase subunit VIII is present in mammals. Gene. 2003;312:95–102. doi: 10.1016/s0378-1119(03)00604-8. [DOI] [PubMed] [Google Scholar]
- 53.Smeenk R, Hylkema M. Detection of antibodies to DNA: a technical assessment. Mol Biol Rep. 1992;17:71–79. doi: 10.1007/BF01006401. [DOI] [PubMed] [Google Scholar]
- 54.Huttemann M, Jaradat S, Grossman LI. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb—the counterpart to testes-specific cytochrome c? Mol Reprod Dev. 2003;66:8–16. doi: 10.1002/mrd.10327. [DOI] [PubMed] [Google Scholar]
- 55.Little AG, Kocha KM, Lougheed SC, Moyes CD. Evolution of the nuclear-encoded cytochrome oxidase subunits in vertebrates. Physiol Genomics. 2010;42:76–84. doi: 10.1152/physiolgenomics.00015.2010. [DOI] [PubMed] [Google Scholar]
- 56.Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat. 2004;23:125–133. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- 57.Huttemann M, Lee I, Pecinova A, Pecina P, Przyklenk K, Doan JW. Regulation of oxidative phosphorylation, the mitochondrial membrane potential, and their role in human disease. J Bioenerg Biomembr. 2008;40:445–456. doi: 10.1007/s10863-008-9169-3. [DOI] [PubMed] [Google Scholar]
- 58.Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- 60.Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonne G, Seibel P, Possekel S, Marsac C, Kadenbach B. Expression of human cytochrome c oxidase subunits during fetal development. Eur J Biochem. 1993;217:1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- 63.Hood DA, Zak R, Pette D. Chronic stimulation of rat skeletal muscle induces coordinate increases in mitochondrial and nuclear mRNAs of cytochrome-c-oxidase subunits. Eur J Biochem. 1989;179:275–280. doi: 10.1111/j.1432-1033.1989.tb14551.x. [DOI] [PubMed] [Google Scholar]
- 64.Kuhn-Nentwig L, Kadenbach B. Isolation and properties of cytochrome c oxidase from rat liver and quantification of immunological differences between isozymes from various rat tissues with subunit-specific antisera. Eur J Biochem. 1985;149:147–158. doi: 10.1111/j.1432-1033.1985.tb08905.x. [DOI] [PubMed] [Google Scholar]
- 65.Kelly MJ, Poole RK, Yates MG, Kennedy C. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J Bacteriol. 1990;172:6010–6019. doi: 10.1128/jb.172.10.6010-6019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill S, Viollet S, Smith AT, Anthony C. Roles for enteric D-type cytochrome-oxidase in N-2 fixation and microaerobiosis. J Bacteriol. 1990;172:2071–2078. doi: 10.1128/jb.172.4.2071-2078.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belevich I, Borisov VB, Konstantinov AA, Verkhovsky MI. Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability. FEBS Lett. 2005;579:4567–4570. doi: 10.1016/j.febslet.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Szal B, Jolivet Y, Hasenfratz-Sauder MP, Dizengremel P, Rychter AM. Oxygen concentration regulates alternative oxidase expression in barley roots during hypoxia and post-hypoxia. Physiol Plantarum. 2003;119:494–502. [Google Scholar]
- 69.Biemelt S, Keetman U, Albrecht G. Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiol. 1998;116:651–658. doi: 10.1104/pp.116.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta KJ, Zabalza A, van Dongen JT. Regulation of respiration when the oxygen availability changes. Physiol Plant. 2009;137:383–391. doi: 10.1111/j.1399-3054.2009.01253.x. [DOI] [PubMed] [Google Scholar]
- 71.McDonald A, Vanlerberghe G. Branched mitochondrial electron transport in the Animalia: presence of alternative oxidase in several animal phyla. IUBMB Life. 2004;56:333–341. doi: 10.1080/1521-6540400000876. [DOI] [PubMed] [Google Scholar]
- 72.McDonald AE, Vanlerberghe GC, Staples JF. Alternative oxidase in animals: unique characteristics and taxonomic distribution. J Exp Biol. 2009;212:2627–2634. doi: 10.1242/jeb.032151. [DOI] [PubMed] [Google Scholar]
- 73.Atteia A, van Lis R, van Hellemond JJ, Tielens AG, Martin W, Henze K. Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX) Gene. 2004;330:143–148. doi: 10.1016/j.gene.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 74.McDonald AE, Vanlerberghe GC. Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:357–364. doi: 10.1016/j.cbd.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Wotton KR, Shimeld SM. Comparative genomics of vertebrate Fox cluster loci. BMC Genomics. 2006;7:271. doi: 10.1186/1471-2164-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 77.Huttemann M, Lee I, Liu J, Grossman LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007;274:5737–5748. doi: 10.1111/j.1742-4658.2007.06093.x. [DOI] [PubMed] [Google Scholar]
- 78.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, III, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burke PV, Poyton RO. Structure/function of oxygen-regulated isoforms in cytochrome c oxidase. J Exp Biol. 1998;201:1163–1175. doi: 10.1242/jeb.201.8.1163. [DOI] [PubMed] [Google Scholar]
- 80.Sandona D, Gastaldello S, Rizzuto R, Bisson R. Expression of cytochrome c oxidase during growth and development of Dictyostelium. J Biol Chem. 1995;270:5587–5593. doi: 10.1074/jbc.270.10.5587. [DOI] [PubMed] [Google Scholar]
- 81.Schiavo G, Bisson R. Oxygen influences the subunit structure of cytochrome c oxidase in the slime mold Dictyostelium discoideum. J Biol Chem. 1989;264:7129–7134. [PubMed] [Google Scholar]
- 82.Scott GR, Schulte PM, Egginton S, Scott AL, Richards JG, Milsom WK. Molecular evolution of cytochrome c oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 2011;28:351–363. doi: 10.1093/molbev/msq205. [DOI] [PubMed] [Google Scholar]
- 83.Luo Y, Gao W, Gao Y, Tang S, Huang Q, Tan X, Chen J, Huang T. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion. 2008;8:352–357. doi: 10.1016/j.mito.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Ramharack R, Deeley RG. Structure and evolution of primate cytochrome c oxidase subunit II gene. J Biol Chem. 1987;262:14014–14021. [PubMed] [Google Scholar]
- 85.Adkins RM, Honeycutt RL. Evolution of the primate cytochrome c oxidase subunit II gene. J Mol Evol. 1994;38:215–231. doi: 10.1007/BF00176084. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt TR, Jaradat SA, Goodman M, Lomax MI, Grossman LI. Molecular evolution of cytochrome c oxidase: rate variation among subunit VIa isoforms. Mol Biol Evol. 1997;14:595–601. doi: 10.1093/oxfordjournals.molbev.a025798. [DOI] [PubMed] [Google Scholar]
- 87.Wu W, Goodman M, Lomax MI, Grossman LI. Molecular evolution of cytochrome c oxidase subunit IV: evidence for positive selection in simian primates. J Mol Evol. 1997;44:477–491. doi: 10.1007/pl00006172. [DOI] [PubMed] [Google Scholar]
- 88.Schmidt TR, Goodman M, Grossman LI. Molecular evolution of the COX7A gene family in primates. Mol Biol Evol. 1999;16:619–626. doi: 10.1093/oxfordjournals.molbev.a026144. [DOI] [PubMed] [Google Scholar]
- 89.Andrews TD, Easteal S. Evolutionary rate acceleration of cytochrome c oxidase subunit I in simian primates. J Mol Evol. 2000;50:562–568. doi: 10.1007/s002390010059. [DOI] [PubMed] [Google Scholar]
- 90.Wu W, Schmidt TR, Goodman M, Grossman LI. Molecular evolution of cytochrome c oxidase subunit I in primates: is there coevolution between mitochondrial and nuclear genomes? Mol Phylogenet Evol. 2000;17:294–304. doi: 10.1006/mpev.2000.0833. [DOI] [PubMed] [Google Scholar]
- 91.Goldberg A, Wildman DE, Schmidt TR, Hüttemann M, Goodman M, Weiss ML, Grossman LI. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci U S A. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uddin M, Opazo JC, Wildman DE, Sherwood CC, Hof PR, Goodman M, Grossman LI. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doan JW, Schmidt TR, Wildman DE, Uddin M, Goldberg A, Huttemann M, Goodman M, Weiss ML, Grossman LI. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 2004;33:944–950. doi: 10.1016/j.ympev.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 94.Schmidt TR, Goodman M, Grossman LI. Amino acid replacement is rapid in primates for the mature polypeptides of COX subunits, but not for their targeting presequences. Gene. 2002;286:13–19. doi: 10.1016/s0378-1119(01)00800-9. [DOI] [PubMed] [Google Scholar]
- 95.Osheroff N, Speck SH, Margoliash E, Veerman EC, Wilms J, Konig BW, Muijsers AO. The reaction of primate cytochromes c with cytochrome c oxidase. Analysis of the polarographic assay. J Biol Chem. 1983;258:5731–5738. [PubMed] [Google Scholar]
- 96.Van Kuilenburg AB, Gorren AC, Dekker HL, Nieboer P, Van Gelder BF, Muijsers AO. Presteady-state and steady-state kinetic properties of human cytochrome c oxidase. Identification of rate-limiting steps in mammalian cytochrome c oxidase. Eur J Biochem. 1992;205:1145–1154. doi: 10.1111/j.1432-1033.1992.tb16884.x. [DOI] [PubMed] [Google Scholar]
- 97.Rodriguez-Roldan V, Garcia-Heredia JM, Navarro JA, Hervas M, De la Cerda B, Molina-Heredia FP, De la Rosa MA. A comparative kinetic analysis of the reactivity of plant, horse, and human respiratory cytochrome c towards cytochrome c oxidase. Biochem Biophys Res Commun. 2006;346:1108–1113. doi: 10.1016/j.bbrc.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 98.Schmidt TR, Wildman DE, Uddin M, Opazo JC, Goodman M, Grossman LI. Rapid electrostatic evolution at the binding site for cytochrome c on cytochrome c oxidase in anthropoid primates. Proc Natl Acad Sci U S A. 2005;102:6379–6384. doi: 10.1073/pnas.0409714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc Natl Acad Sci U S A. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barrientos A, Muller S, Dey R, Wienberg J, Moraes CT. Cytochrome c oxidase assembly in primates is sensitive to small evolutionary variations in amino acid sequence. Mol Biol Evol. 2000;17:1508–1519. doi: 10.1093/oxfordjournals.molbev.a026250. [DOI] [PubMed] [Google Scholar]
- 102.Schmidt TR, Wu W, Goodman M, Grossman LI. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol Biol Evol. 2001;18:563–569. doi: 10.1093/oxfordjournals.molbev.a003836. [DOI] [PubMed] [Google Scholar]
- 103.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 104.Lee I, Salomon AR, Yu K, Samavati L, Pecina P, Pecinova A, Hüttemann M. Isolation of regulatory-competent, phosphorylated cytochrome c oxidase. Methods Enzymol. 2009;457:193–210. doi: 10.1016/S0076-6879(09)05011-3. [DOI] [PubMed] [Google Scholar]
- 105.Gnad F, Forner F, Zielinska DF, Birney E, Gunawardena J, Mann M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol Cell Proteomics. 2010;9:2642–2653. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]