Abstract
We have previously shown that CK2 associates with the human high-mobility group protein SSRP1 and that this association increases in response to UV irradiation. CK2 also phosphorylates SSRP1 in vitro. Here we extend this work by investigating CK2 regulation of SSRP1 function through phosphorylation. Phosphorylation of SSRP1 by CK2 inhibited the nonspecific DNA-binding activity of SSRP1 and FACT (facilitating chromatin-mediated transcription) complex in vitro. Using a serine/threonine-scanning Auto-spot peptide array coupled with a filter-based kinase assay with synthetic peptides as substrates, we identified serines 510, 657, and 688 as phosphorylation targets of CK2 in vitro. Mutagenesis of the three serines revealed that serine 510 was more important for the regulation of SSRP1 DNA-binding activity. Furthermore, we found that SSRP1 was phosphorylated in cells in response to UV (but not γ) irradiation. These results suggest that CK2 regulates the DNA-binding ability of SSRP1 and that this regulation may be responsive to specific cell stresses.
CK2 is a ubiquitous and evolutionarily conserved kinase. This enzyme is a heterotetrameric protein complex consisting of two regulatory β subunits (28 kDa) and two catalytic α (42 kDa) or α′ (38 kDa) subunits with the stoichiometry of either α2β2, α′2β2, or αα′β2 as the holoenzyme (1). Genetic studies in yeast (2) and in mice (3) demonstrate that this enzyme is essential for viability and animal embryogenesis. Biochemical and functional analyses of this enzyme reveal that it can phosphorylate a broad spectrum of protein substrates in vitro and regulate a variety of cellular functions including transcription in the nucleus. Consistent with its regulatory role in the nucleus, CK2 associates with a number of chromatin and nuclear matrix proteins in the cell (4–6).
One of the mammalian CK2-interacting nuclear proteins is the previously identified DNA-binding protein called SSRP1 (structure-specific recognition protein 1) (7). SSRP1 (named T160 in mice) (8) is a member of the high-mobility group (HMG)1 family of proteins (9), which represent the most abundant chromatin-associated nonhistone proteins. SSRP1 is essential for cell (10) and animal (11) viability. Sequence analysis reveals that this 80-kDa phosphoprotein possesses several interesting and evolutionarily conserved domains: a large N-terminal region (aa 1–440) (>80% identity between Drosophila and human; 95% identity between Xenopus and human), an acidic domain (aa 440–496) with a limited homology to nucleolin, an HMG box domain (aa 539–614, 60% identity between Drosophila and human) with two flanking basic domains (aa 512–534 and aa 623–640), and a mixed charge domain at the extreme C-terminal region (aa 661–709). In yeast, Pob3 and Nhp6 form a bipartite SSRP1 analog (12). Pob3 is highly homologous with the N terminus of SSRP1, whereas Nhp6 is an HMG protein resembling the C terminus of SSRP1 (12). These domains are crucial for the functions of SSRP1. For example, the highly conserved N-terminal region of SSRP1 has been shown to directly interact with its partner, Spt16 (7), forming a tight heterodimer complex (13). This complex was initially identified from human cells as a facilitating chromatin-mediated transcription (FACT) complex to regulate transcriptional elongation with chromatin as a template (14). This complex was also shown to probably regulate DNA replication in yeast and Xenopus (10, 15). Furthermore, the C-terminal HMG box is able to bind to DNA nonspecifically as well as to specific structures of DNA, such as DNA modified by the anti-tumor drug cisplatin (9) or cruciform DNA (16). Finally, SSRP1 functions as a co-regulator for transcription, and this regulation is executed by interacting with other transcriptional activators such as SRF (17), Drosophila GATA factor (18), and p63 through its middle domain (19). Thus, SSRP1 appears to possess multiple functions in the nucleus. Whether these functions of SSRP1 are regulated by CK2 remains unclear despite the fact that CK2 associates with SSRP1 in a protein kinase complex (20).
As an initial step to address this question, we started to explore potential regulation of SSRP1 function by CK2 through phosphorylation. This idea originated from our previous findings that a UV irradiation-responsive p53 serine 392 kinase complex contains SSRP1, Spt16, and CK2 (20), and CK2 efficiently phosphorylates SSRP1 (but not Spt16) in vitro (20). Also, we found that the assembly of this ternary complex appears to be induced after UV irradiation of cells (7). Consistent with our findings is that CK2 was also shown to phosphorylate maize SSRP1 at the HMG region in vitro, and this phosphorylation seemed to induce the recognition of UV irradiation-damaged DNA by SSRP1 in vitro (21). Although SSRP1 possesses a number of CK2 consensus sites, it is still unclear which amino acids CK2 specifically phosphorylates and whether phosphorylation of these potential residues regulates the ability of SSRP1 to bind to DNA. Also, it is unknown whether SSRP1 phosphorylation is responsive to UV irradiation in cells. Hence, we performed a series of biochemical analyses to address these specific questions. As detailed below, we show that SSRP1 is phosphorylated at serine and threonine residues in cells, and that one or more target sites are indeed induced by UV (but not γ) irradiation. Also, CK2 phosphorylation of SSRP1 that is either alone or in the FACT complex inhibits the ability of SSRP1 to bind to nonmodified linear DNA in vitro. Employing a serine/threonine-scanning Auto-spot peptide array technique coupled with a CK2 kinase assay, we identified serines 510, 657, and 688 of SSRP1 as CK2 targets in vitro. Mutagenesis of the three serines revealed that serine 510 might be critical for regulation of SSRP1·DNA binding in vitro. These results indicate that CK2 can regulate the ability of SSRP1 to bind to nonmodified DNA by specifically phosphorylating serine 510 and suggest that these amino acids, although not residing in the DNA-binding HMG domain, may play a critical role in regulating SSRP1 function.
EXPERIMENTAL PROCEDURES
Reagents and Buffers
Buffer C 100 (BC100) contains 20 mm Tris/HCl (pH 7.9), 0.1 mm EDTA, 15% glycerol, 100 mm KCl, 1 mm dithiothreitol, and protease inhibitors including 0.2 mm phenylmethylsulfonyl fluoride, 4 mm pepstatin A, 1 mg/ml leupeptin, and 1 mg/ml aprotinin.
Kinase buffer (1×) is 20 mm Tris/HCl (pH 7.5), 10 mm MgCl2, 50 mm KCl, and 1 mm dithiothreitol. The buffer for circular dichroism (CD) analysis is 10 mm Tris/H3PO4 (pH 7.9) and 10% glycerol.
Plasmids and Antibodies
pET24a-CK2α′, pET24a-CK2β, pET24a-SSRP1, and pGEX-KG-C-SSRP1 (aa 471–709) plasmids were as described previously (7).s The pGEX-KG-C-SSRP1 (aa 471–709) serine3alanine mutants were generated by site-directed mutagenesis using the QuikChange™ kit (Stratagene). Polyclonal anti-SSRP1 and anti-Spt16 antibodies were generated as described previously (7).
Purification of Recombinant Proteins
Histidine-tagged proteins were expressed and purified from bacteria using nickel-nitrilotriacetic acid-agarose (Qiagen), as described previously (7). GST fusion proteins were bound to glutathione-agarose beads (Sigma) and eluted with glutathione, followed by dialysis against BC100 buffer.
Purification of the FLAG·FACT Complex
FLAG-SSRP1 HEK293 cells (1 × 108 cells) were cultured in suspension and harvested for nuclear extracts as described previously (22). The extracts were loaded onto a phosphocellulose (P11) column and step-eluted at 0.1, 0.35, 0.5, and 1.0 m KCl. The 0.5 m KCl fraction was dialyzed against BC100 buffer and incubated with anti-FLAG M2-agarose affinity gel (Sigma). After washing rigorously, the immunoprecipitate was eluted from the beads with FLAG peptide, subjected to SDS-PAGE, and stained with colloidal blue. The FACT complex purified from the 0.5 m fraction was used for electrophoresis gel mobility shift assays.
Purification of the CK2 Kinase Complex
An equal molecular ratio of highly purified His-CK2α′ and His-CK2β (7) was incubated on ice for 1 h and run on a Superdex 200 (3.2/30) column (Smart HPLC, Pharmacia Corporation). The kinase-containing fractions were pooled and used for in vitro kinase assays.
In Vivo 32P-Labeled Inorganic Phosphate and Phosphoamino Acid Analysis—HeLa cells were either untreated, γ-irradiated (14 gray), or UVB-irradiated (1200 J/m2). Three hours after irradiation, 32P inorganic phosphate was added to 375 µCi/ml, along with 500 nm phosphatase inhibitor okadaic acid, and the cells were allowed to grow for an additional 3 h. The cells were harvested for immunoprecipitation. After extensive washing, the immunoprecipitates were subjected to SDSPAGE, and proteins were detected by autoradiography. The radioactive SSRP1 bands were sliced from the gel and subjected to phosphoamino acid analysis as described previously (23). After extraction from the gel, the samples, normalized by counts/min, were loaded onto a cellulose thin-layer chromatography plate (Fisher Scientific), which was subjected to two-dimensional electrophoresis. Signals were detected by autoradiography.
Electrophoresis Gel Mobility Shift Assay (EMSA)
EMSA was performed as described previously (20). Proteins, as indicated in each figure legend, were incubated with a 3′ end-labeled 87-bp DNA fragment (5000 cycles/min, 0.1–1.0 ng of DNA/assay) derived from the p21waf1/cip1 promoter (24). After incubation at room temperature for 30 min, the reaction mixtures were loaded onto a 4.5% nondenatured gel. Protein·DNA complexes were detected by autoradiography. EMSA for SSRP1 was also carried out with different sizes and sequences of DNA. The results were similar to what are shown in this study (data not shown).
In Vitro Kinase Assay
Radioactive in vitro kinase assays were performed with [γ-32P]ATP as described previously (7). Substrates were either 50 ng of His-SSRP1 or 100 ng of GST-C-SSRP1. For the kinase reaction that was coupled with EMSA, the reaction volume was 10 µl, and 1 mm unlabeled ATP, instead of [γ-32P]ATP, was used.
In Vitro Kinase Assay on Peptide Arrays
Peptide arrays were synthesized on cellulose membranes with an Auto-Spot Robot ASP 222 (AbiMed, Langenfeld, Germany) as described previously (25). In vitro kinase assays on the peptide arrays with our purified recombinant CK2 complexes were also performed as described previously (26).
Circular Dichroism
CD analyses were performed on an automated AVIV 215 spectrometer at 25 °C as described previously (27). All the protein concentrations were maintained at 0.1 mg/ml. Spectra were taken between 190–260 nm using a 0.5-mm path-length cuvette.
RESULTS
SSRP1 Phosphorylation Is Induced by UV (but Not by γ) Irradiation
Our previous findings that the CK2-SSRP1 interaction increased after UV irradiation and that CK2 efficiently phosphorylated SSRP1 in vitro prompted us to examine whether SSRP1 phosphorylation is induced by UV irradiation in cells. To this end, human cervical carcinoma HeLa cells were irradiated with either UVB or γ rays for 6 h, including 3 h of labeling with [32P]orthophosphate. The cells were harvested for co-immunoprecipitation and 32P-labeled proteins were detected by autoradiography. As shown in Fig. 1A, irradiation of the cells with either UV or γ rays did not change the steady-state level of SSRP1 (upper panel). Interestingly, and consistent with our previous work (7), 32P-labeled SSRP1 increased 2.4-fold specifically in response to UV (but not γ) irradiation (Fig. 1A, lower panel), suggesting that the UV irradiation-inducible CK2·SSRP1 complex might also lead to inducible SSRP1 phosphorylation by CK2. To determine which amino acids are phosphorylated after UV irradiation, the 32P-labeled SSRP1 bands in Fig. 1Awere gel-purified for phosphoamino acid analysis. Greater than 95% of phosphorylated amino acids in SSRP1 were serines, ~5% were threonines, and none was tyrosine (Fig. 1B). These results indicate that SSRP1 is indeed phosphorylated mainly at serines in cells and that this phosphorylation is UV irradiation-responsive.
Fig. 1. UV irradiation induction of SSRP1 phosphorylation at Ser and Thr in vivo.
A, phosphorylation of SSRP1 in cells was induced by UV irradiation. Hela cells were labeled in vivo by 32P-labeled inorganic phosphate 3 h after irradiation by UVB or γ rays, followed by an additional 3 h of incubation before being harvested for immunoprecipitation (IP). 200 µg of protein was used for the immunoprecipitation. Immunoprecipitates were subjected to SDS-PAGE and labeled SSRP1 was detected by autoradiography. B, phosphoamino acid analysis was carried out as described under “Experimental Procedures,” revealing that both serines and threonines are phosphorylated on SSRP1. Samples (1720 cycles/min) were loaded onto a cellulose thin-layer chromatography plate, which was subjected to two-dimensional electrophoresis and autoradiography. Only the UVB irradiation-treated sample is shown in the figure, although nontreated or γ-treated samples show the same pattern. WB, Western blot.
CK2 Phosphorylation of SSRP1 Inhibits the Ability of SSRP1 and the FACT Complex to Bind to Nonmodified DNA in Vitro
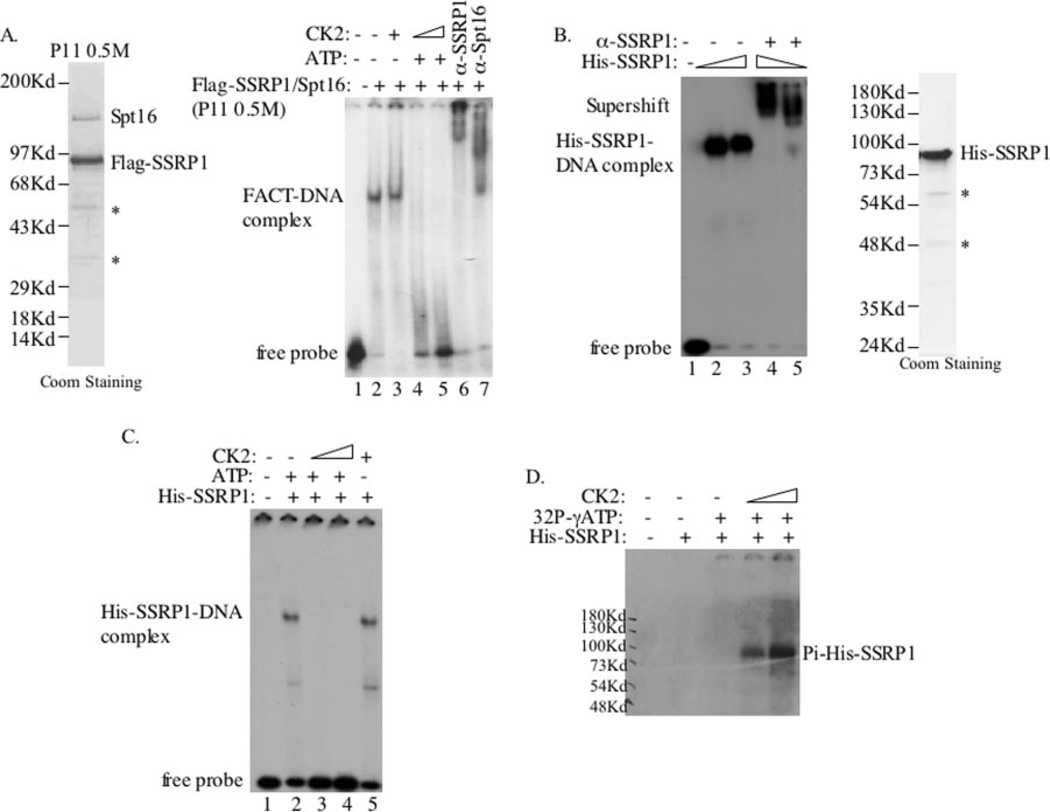
To determine whether CK2-mediated phosphorylation of SSRP1 affects its DNA-binding activity, we compared a purified FACT complex containing both FLAG-SSRP1 and Spt16 from the FLAG-SSRP1 expressing HEK293 cells versus recombinant histidine-tagged SSRP1. The purity of FACT and His-SSRP1 is shown in Fig. 2A (left panel) and Fig. 2B (right panel), respectively. Recombinant CK2 α and α′ subunits were either purified to homogeneity and reconstituted into a heterotetrameric complex as described previously (7) or purchased from Promega. With these purified proteins, we carried out a set of electrophoresis gel mobility shift assays using 32P-labeled 87-bp oligomers digested from the pWaf-1-Δ50 plasmid (24) as probes. As shown in Fig. 2A (right panel), FACT bound to DNA efficiently in vitro as expected (28). This FACT·DNA complex was indeed formed with SSRP1 and Spt16, as both anti-SSRP1 and anti-Spt16 antibodies super-shifted the complex (Fig. 2A, lanes 6 and 7). Interestingly, CK2 reduced the FACT·DNA complex in the presence (but not in the absence) of 1 mM ATP, suggesting that this inhibition may be phosphorylation-dependent. Supporting this assumption is the fact that CK2 phosphorylated SSRP1 (but not Spt16) in FACT in vitro (20).
Fig. 2. Phosphorylation of SSRP1 by CK2 inhibits its DNA-binding activity.
A, phosphorylation of FACT by CK2 inhibited its DNA-binding ability. The FACT complex was purified from FLAG-SSRP1-expressing HEK293 cells, as described under “Experimental Procedures.” The 0.5 m fraction, which contains the FACT complex, is shown in the left panel. 4 µl of this fraction was used in an in vitro kinase assay followed by EMSA. 10-µl CK2 kinase reactions were carried out with the FACT complex for 1 h at 30 °C and then incubated with 3′ end-labeled DNA (87 bp) probes for 20–25 min. Also, antibodies specific against SSRP1 (lane 6) and Spt16 (lane 7) were used. B, the purified recombinant His-SSRP1 protein bound to DNA. The purity of the recombinant His-SSRP1 is shown in the right panel. The EMSA was done using 250 and 500 ng of purified His-SSRP1. The His·SSRP1·DNA complex was supershifted by an anti-SSRP1 antibody (lanes 4 and 5). C, phosphorylation of SSRP1 by CK2 inhibited its DNA-binding activity. The same kinase/EMSA assay was conducted as in A, except that 50 ng of His-SSRP1 was used. D, the purified recombinant His-SSRP1 was phosphorylated by CK2. In vitro kinase reactions were conducted for 30 min using [γ-32P]ATP and 50 ng of His-SSRP1 as substrates. Coom, Coomassie.
To verify whether the reduction of the FACT·DNA complex formation is through CK2 phosphorylation of SSRP1, we conducted EMSA using purified His-SSRP1. As expected (9), recombinant SSRP1 efficiently bound to DNA (Fig. 2B) under the same conditions as those used for FACT (Fig. 2A), and the anti-SSRP1 antibody also supershifted this complex (Fig. 2B). Again, CK2 completely inhibited the formation of SSRP1·DNA complex in the presence (but not in the absence) of ATP (Fig. 2C). This inhibition was not because of the change of SSRP1 level, as its level stayed the same after incubation (data not shown). The elimination of SSRP1·DNA complexes by CK2 was closely related to SSRP1 phosphorylation by this kinase, as 32P labeling of SSRP1 was detected in a CK2 dose-dependent manner (Fig. 2D). These results suggest that CK2 can phosphorylate SSRP1 and thus prevent SSRP1, either alone or in the FACT complex, from binding to nonmodified DNA in vitro.
SSRP1 Phosphorylates the C-terminal Domain of SSRP1 and Inhibits Its Ability to Bind to DNA in Vitro
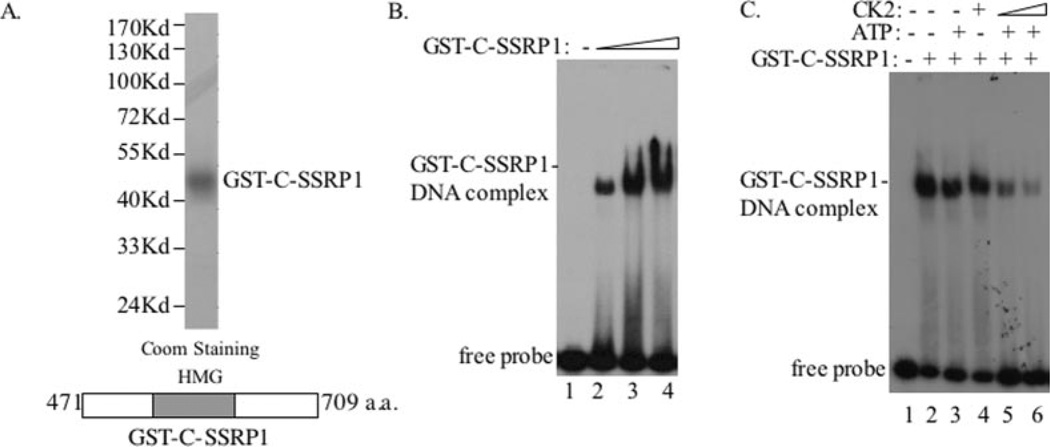
It was shown previously (28) that SSRP1 binds to DNA through its C-terminal HMG-containing domain. Next we wanted to determine whether CK2 inhibits the DNA-binding activity of SSRP1 by phosphorylating the C terminus of SSRP1, although the middle domain of SSRP1 was also phosphorylated by CK2 (7). GST C-terminal SSRP1 (aa 471–709), encompassing the HMG box, was purified as described previously (7) to homogeneity (Fig. 3A) for EMSA. As anticipated (28), this GST-C-SSRP1 fragment bound to DNA in a dose-dependent fashion (Fig. 3B). Again, this DNA-binding ability was inhibited by CK2 when only ATP was present (Fig. 3C). This inhibition was dose-dependent (Fig. 3C, lanes 5 and 6) and associated with phosphorylation of the C terminus of SSRP1 by CK2, as detected when [γ-32P]ATP, instead of unlabeled ATP, was used (see Fig. 6D and data not shown). Taken together, these results demonstrated that CK2 inhibits the DNA-binding activity of SSRP1 by phosphorylating its C-terminal domain.
Fig. 3. Phosphorylation of the C terminus of SSRP1 by CK2 inhibits its DNA-binding ability.
A, a schematic shows the GST fusion C terminus of SSRP1 (bottom of the panel). GST-C-SSRP1 was subjected to SDS-PAGE and stained with Coomassie Brilliant Blue (Coom). B, the purified GST-C-SSRP1 fragment bound to DNA. The EMSA was conducted using increasing amounts of GST-C-SSRP1 (50, 100, and 200 ng). C, phosphorylation of GST-C-SSRP1 by CK2 inhibited its DNA-binding activity. The same kinase/EMSA assay was conducted as described in the legend to Fig. 2 using 100 ng of GST-C-SSRP1.
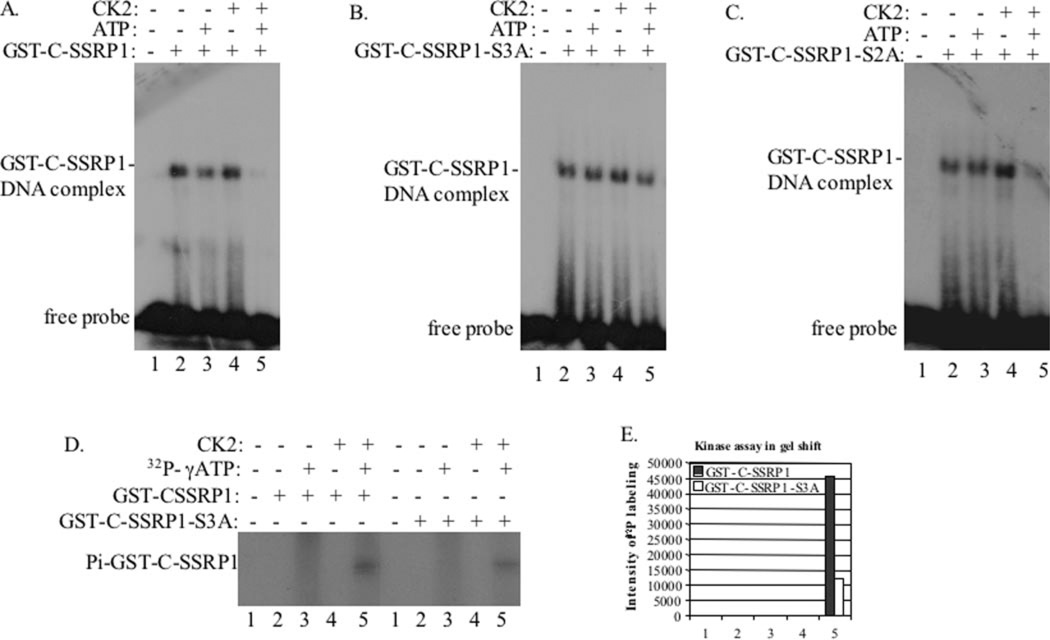
Fig. 6. Phosphorylation of GST-C-SSRP1-S3A (but not GST-C-SSRP1-S2A) by CK2 fails to inhibit the DNA-binding activity of this protein.
A, phosphorylation of GST-C-SSRP1 by CK2 inhibits its DNA-binding activity. The same kinase/EMSA assay was conducted as described in the legend to Fig. 3, and 50 ng of GST-C-SSRP1 was used. B, phosphorylation of GST-C-SSRP1-S3A by CK2 failed to inhibit its DNA-binding ability. The kinase/EMSA assay on the triple mutant of GST-C-SSRP1 was conducted under the same conditions as those for wild type GST-C-SSRP1 in A. C, phosphorylation of GST-C-SSRP1-S2A by CK2 still inhibited its DNA-binding ability. The kinase/EMSA assay on the double mutant of GST-C-SSRP1 was conducted under the same conditions as those for wild type GST-C-SSRP1 in A. D, GST-C-SSRP1-S3A is partially phosphorylated by CK2. In vitro radioactive kinase assays were carried out under the same conditions as used for the EMSA reactions in A and B. The 32P-labeled SSRP1 fragment signals were quantified and plotted in a graph shown in E using imaging software.
CK2 Phosphorylates Serines 510, 657, and 688 of SSRP1 in Vitro
Although the C terminus of SSRP1 was phosphorylated by CK2 in vitro (Fig. 6 and data not shown), it was challenging to determine which amino acids are CK2 target sites. There are 46 serines and four threonines in this region of SSRP1, seven of which exist in the HMG box (Fig. 4). Also, 20 of these potential phosphorylation sites display a similarity to the CK2 consensus sequence ((S/T)XX(D/E) or (S/T)(D/E)). Moreover, mass spectrometry of phosphorylated SSRP1 failed to map CK2 sites (data not shown) mainly because of the relatively low efficiency of SSRP1 phosphorylation (<30%) in vitro and dense distribution of phosphorylation target residues in the middle and Cterminal regions of SSRP1 (Fig. 4 and data not shown). To surmount these obstacles, we employed an Auto-spot peptide array technique coupled with a filter-based kinase assay (25, 26). Using this approach with pairs of synthetic wild type and corresponding serine→alanine mutant 10mer peptides spotted on a nitrocellulose filter as substrates, we identified three serines as CK2 sites. As shown in Fig. 4, peptide pair 10 (note, each pair of peptides has a wild type and a serine→alanine mutant peptide), encompassing serine 510, peptide pair 15 with serines 657 and 659, and peptide pair 19 with serines 685 and 688, was phosphorylated by CK2. By contrast, phosphorylation of the peptides by CK2 was markedly reduced when their corresponding serine→alanine mutants were used as substrates. Although peptides 15 and 19 harbored two serines in each case, only the S657A mutation of peptide 15 (in comparison with peptide 16 that contains S657A and S659A) or the S688A of peptide 19 had reduced phosphorylation (Fig. 4, right column), suggesting that serines 657 and 688 are the CK2 sites (of note, the residual signals of the mutant S510A-, S657A-, or S657A/S659A-containing peptides 10, 15, and 16 might be due to less stringent washing, as these peptides should not be phosphorylated). Consistently, the three sites also display a similarity to the CK2 consensus sequence (Fig. 4). Surprisingly, all of these three serines reside outside of the HMG domain of SSRP1 (Fig. 4, lower panel). Although some of the synthetic peptides might not serve as ideal substrates for CK2 in this assay, perhaps because of improper folding (thereby giving false negative results), this study at least identifies serines 510, 657, and 688 as CK2 phosphorylation sites for further analysis.
Fig. 4. Identification of serine 510, 657, and 688 of SSRP1 as CK2 phosphorylation sites in vitro.
A panel of designed SSRP1 10mer peptides was spotted on a nitrocellulose membrane. In vitro CK2 kinase reactions were conducted on the membrane using [γ-32P]ATP. After extensive washing, radioactive signals were detected by autoradiography. The asterisks denote potential phosphorylation sites, and the underlined asterisks indicate the identified CK2 sites. Only serines or threonines and their corresponding mutant alanines are shown here. The corresponding positions of these amino acids in the C terminus of SSRP1 are approximately marked on the schematic at the bottom. The sequences for peptide pairs 10, 15, and 19 are shown in corresponding positions, and CK2 target sites are underlined.
Serine 510 (but Not Serines 657 and 688) Plays a Role in CK2 Regulation of SSRP1 DNA-binding Activity
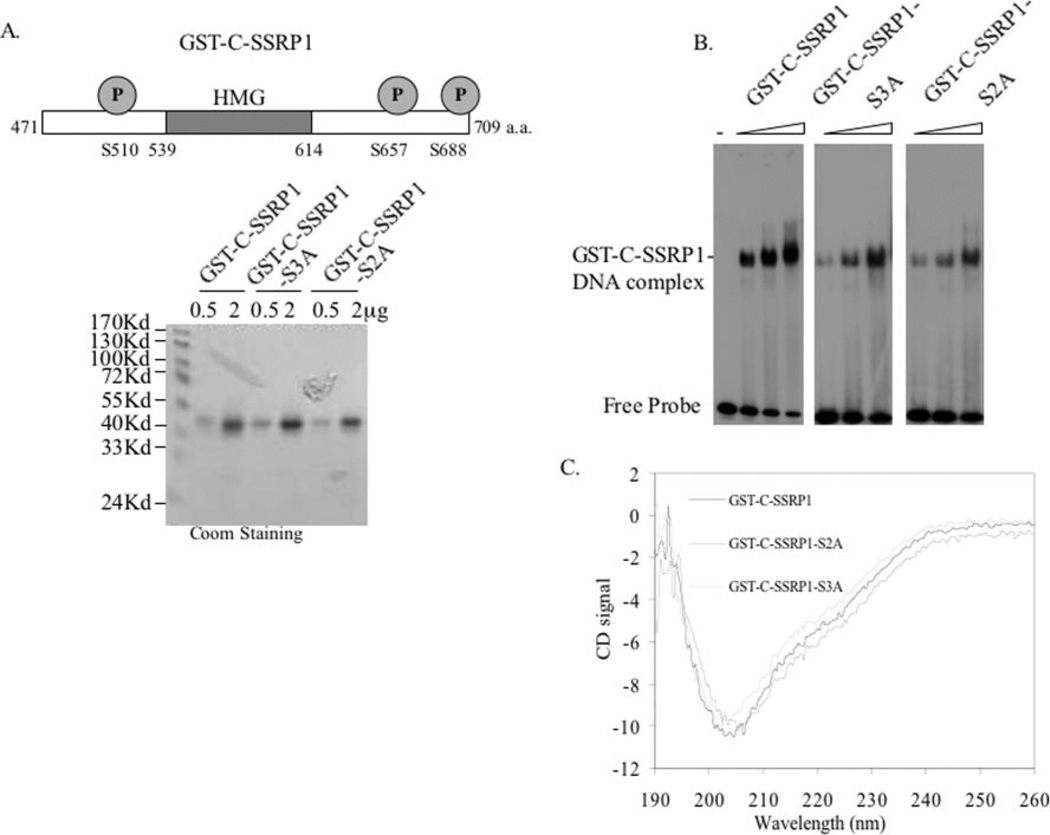
To investigate whether phosphorylation of SSRP1 at serines 510, 657, and 688 by CK2 is important for regulating the DNA-binding activity of SSRP1, we generated an SSRP1 C-terminal triple mutant S3A (denoting S510A/S657A/S688A) and a double mutant S2A (denoting S657A/S688A) in the GST-C-SSRP1 fusion protein using a mutagenesis kit from Stratagene. These mutants were confirmed by DNA sequencing (data not shown). These GST fusion proteins and its wild type counterpart were purified to homogeneity as shown in Fig. 5A. To ensure that these mutant proteins are still able to bind to DNA, we carried out EMSA with the purified proteins, as described in Fig. 2. As shown in Fig. 5B, at the same concentrations, the wild type and two mutant GST-C-SSRP1 proteins were able to bind to DNA, although the DNA-binding ability of the mutant proteins was a bit weaker. CD analysis of the wild type and mutant proteins showed that these proteins displayed a similar secondary structure (Fig. 5C).
Fig. 5. GST-C-SSRP1-S3A triple and S2A double mutants can bind to DNA but with relatively lower affinity.
A, a schematic shows the positions of S510A, S657A, and S688A on GST-C-SSRP1 (upper panel). GST-C-SSRP1, GST-C-SSRP1-S3A, and GST-C-SSRP1-S2A (S657A/ S688A) were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue (Coom, lower panel). The amounts of proteins used are indicated on top. B, comparison between GST-C-SSRP1, GST-C-SSRP1-S3A, and GST-C-SSRP1-S2A in EMSA. 25, 50, and 100 ng of GST-C-SSRP1, GST-C-SSRP1-S3A, or GST-C-SSRP1-S2A were used in the EMSA. C, CD analysis of GST-C-SSRP1, GST-C-SSRP1-S3A, and GST-C-SSRP1-S2A. CD spectra of the wild type, triple, and double mutant GST-C-SSRP1 proteins were recorded in the wavelength range of 190–260 nm.
Next, we performed a set of EMSA experiments to determine whether these three serines are crucial for CK2 phosphorylation of the SSRP1 C terminus and thus its regulation of SSRP1 DNA-binding activity. Again phosphorylation of the GST-SSRP1-C terminus by CK2 markedly inhibited the formation of SSRP1·DNA complexes (Fig. 6A). By striking contrast, the triple mutant GST-C-SSRP1-S3A was still able to bind to DNA in the presence of CK2 and ATP (Fig. 6B), suggesting that substitution of the three serines by alanines may prevent CK2 phosphorylation of the C terminus of SSRP1 at these residues and thus rescue CK2-mediated inhibition of SSRP1 DNA binding. Consistent with this assumption was that phosphorylation of the triple mutant GST-C-SSRP1-S3A by CK2 was reduced to ~30% of that of the wild type protein (Fig. 6, D and E). Although the triple mutant was still phosphorylated by CK2 (Fig. 6D), the remaining phosphorylation did not appear to efficiently block the DNA-binding activity of the C terminus of SSRP1 (Fig. 6B). To further determine which serine is more critical for the inhibitory effect of CK2 on SSRP1 DNA-binding activity, we conducted the same EMSA assay with the SSRP1 C-terminal double mutant S2A (S657A/S688A). We found that this double mutant was unable to prevent the inhibition of CK2-mediated phosphorylation on SSRP1 DNA-binding activity (Fig. 6C). Taken together, these results suggest that serine 510 may play a crucial role in mediating CK2 regulation of SSRP1 DNA-binding function.
DISCUSSION
It has been shown that CK2 forms a complex with SSRP1 and Spt16 in cells and phosphorylates SSRP1 (but not Spt16) in vitro (20). Little is known about the regulation of SSRP1 function by CK2. As an initial step to understand the role of CK2 in human SSRP1 regulation, we have performed a series of biochemical experiments using purified proteins. We found that CK2-mediated phosphorylation of SSRP1 at its HMG-containing C-terminal region led to a reduction of SSRP1·DNA binding (Fig. 3). This inhibition occurred when either FACT or recombinant SSRP1 was used as the substrate of CK2 (Figs. 2). Moreover, by means of the Auto-spot peptide array/filter kinase approach, we identified serines 510, 657, and 688 as CK2 target sites in vitro (Fig. 4). Replacing all three serines (but not merely the serine 657 and 688 residues together) with alanines alleviated the inhibition of SSRP1 DNA-binding activity by CK2 (Figs. 5 and 6), indicating that serine 510 is more critical for regulating SSRP1 DNA-binding activity. Consistent with these results is that CK2 was also previously reported to inhibit the DNA-binding activity of maize HMGB proteins through phosphorylation (29). Hence, our results demonstrate that CK2 can phosphorylate the serines outside of the HMG box domain and thereby inhibit its ability to interact with nonmodified DNA.
Intriguingly, phosphorylation of maize SSRP1 by CK2 was recently shown to enhance the ability of the SSRP1 protein to bind to UV irradiation-damaged DNA in vitro (21), although specific phosphorylation amino acids were not identified and mutated. This finding, along with our current study, suggests that inhibiting the nonspecific DNA-binding activity of SSRP1 by CK2 would, in effect, increase the specificity of SSRP1 for UV irradiation-damaged DNA. In this scenario, DNA-damaging agents, such as UV irradiation, would signal through these molecules. Because SSRP1, together with Spt16, often associates with chromatin for its regulatory role in transcription and replication (10, 14, 15, 30–32), it is also likely that by dissociating SSRP1 from nonspecific DNA sequences, CK2 may enhance its ability to bind to specific structures of chromatin, where transcription, replication, or DNA repair may take place. Alternatively and conversely, by phosphorylating certain residues of SSRP1, CK2 may negatively regulate transcription or replication in response to UV irradiation-caused DNA damage. It is known that UV irradiation often causes global inhibition of transcription or replication (33). Yet, detailed mechanisms underlying this effect remain elusive, although one general thought is that intrastrand cross-linking of neighboring bases in DNA caused by UV irradiation may block elongation of transcription or replication. Given that SSRP1/Spt16 has been proposed to be functional in transcriptional elongation and replication (10, 14, 15, 30–32), one mechanism would be that CK2 may inhibit this function by phosphorylating SSRP1 and thus preventing it from associating with chromatin, consequently leading to transcriptional stall or replication pause in response to UV irradiation. Consistent with this speculation is that the assembly of the CK2·SSRP1·Spt16 complex, as well as phosphorylation of SSRP1, is induced in response to UV irradiation (Fig. 1) (20). Another possibility is that by dissociating SSRP1 or FACT from chromatin, CK2 would form a kinase complex with these proteins, which in turn assist CK2 in selectively phosphorylating and activating p53 in response to UV irradiation (7, 20). In sum, our finding, as described here, suggests a potential mechanism underlying the regulation of SSRP1 function by CK2 in response to UV irradiation.
Acknowledgments
We thank Mary MacPartlin for proofreading this manuscript, Shelya Zeng and Hunjoo Lee for technique assistance, and Ezhilkani Subbian and Ujwal Shinde for their assistance in CD analysis.
Footnotes
This work was supported in part by NCI, National Institutes of Health Grants CA93614, CA095441, and CA079721 (to H. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Supported by National Institutes of Health Grant GM48231.
The abbreviations used are: HMG, high-mobility group; aa, amino acid; FACT, facilitating chromatin-mediated transcription; CD, circular dichroism; GST, glutathione S-transferase; EMSA, electrophoresis gel mobility shift assay.
REFERENCES
- 1.Gietz RD, Graham KC, Litchfield DW. J. Biol. Chem. 1995;270:13017–13021. doi: 10.1074/jbc.270.22.13017. [DOI] [PubMed] [Google Scholar]
- 2.Padmanabha R, Chen-Wu JL, Hanna DE, Glover CV. Mol. Cell. Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchou T, Vernet M, Blond O, Jensen HH, Pointu H, Olsen BB, Cochet C, Issinger OG, Boldyreff B. Mol. Cell. Biol. 2003;23:908–915. doi: 10.1128/MCB.23.3.908-915.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tawfic S, Davis AT, Faust RA, Gapany M, Ahmed K. J. Cell. Biochem. 1997;64:499–504. [PubMed] [Google Scholar]
- 5.Guo C, Davis AT, Ahmed K. J. Biol. Chem. 1998;273:13675–13680. doi: 10.1074/jbc.273.22.13675. [DOI] [PubMed] [Google Scholar]
- 6.Guo C, Yu S, Davis AT, Ahmed K. Cancer Res. 1999;59:1146–1151. [PubMed] [Google Scholar]
- 7.Keller DM, Lu H. J. Biol. Chem. 2002;277:50206–50213. doi: 10.1074/jbc.M209820200. [DOI] [PubMed] [Google Scholar]
- 8.Shirakata M, Huppi K, Usuda S, Okazaki K, Yoshida K, Sakano H. Mol. Cell. Biol. 1991;11:4528–4536. doi: 10.1128/mcb.11.9.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruhn SL, Pil PM, Essigmann JM, Housman DE, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2307–2311. doi: 10.1073/pnas.89.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlesinger MB, Formosa T. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, Gariglio M, Oltz EM, Ruley HE. Mol. Cell. Biol. 2003;23:5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewster NK, Johnston GC, Singer RA. Mol. Cell. Biol. 2001;21:3491–3502. doi: 10.1128/MCB.21.10.3491-3502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 14.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 15.Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. Curr. Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- 16.Gariglio M, Ying GG, Hertel L, Gaboli M, Clerc RG, Landolfo S. Exp. Cell Res. 1997;236:472–481. doi: 10.1006/excr.1997.3742. [DOI] [PubMed] [Google Scholar]
- 17.Spencer JA, Baron MH, Olson EN. J. Biol. Chem. 1999;274:15686–15693. doi: 10.1074/jbc.274.22.15686. [DOI] [PubMed] [Google Scholar]
- 18.Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng SX, Dai MS, Keller DM, Lu H. EMBO J. 2002;21:5487–5497. doi: 10.1093/emboj/cdf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H. Mol. Cell. 2001;7:283–292. doi: 10.1016/s1097-2765(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 21.Krohn NM, Stemmer C, Fojan P, Grimm R, Grasser KD. J. Biol. Chem. 2003;278:12710–12715. doi: 10.1074/jbc.M300250200. [DOI] [PubMed] [Google Scholar]
- 22.Flores O, Lu H, Reinberg D. J. Biol. Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 23.van der Geer P, Hunter T. Electrophoresis. 1994;15:544–554. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- 24.Zeng X, Levine AJ, Lu H. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6681–6686. doi: 10.1073/pnas.95.12.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tegge WJ, Frank R. Methods Mol. Biol. 1998;87:99–106. doi: 10.1385/0-89603-392-9:99. [DOI] [PubMed] [Google Scholar]
- 26.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. Mol. Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Yabuta Y, Subbian E, Oiry C, Shinde U. J. Biol. Chem. 2003;278:15246–15251. doi: 10.1074/jbc.M212003200. [DOI] [PubMed] [Google Scholar]
- 28.Yarnell AT, Oh S, Reinberg D, Lippard SJ. J. Biol. Chem. 2001;276:25736–25741. doi: 10.1074/jbc.M101208200. [DOI] [PubMed] [Google Scholar]
- 29.Stemmer C, Schwander A, Bauw G, Fojan P, Grasser KD. J. Biol. Chem. 2002;277:1092–1098. doi: 10.1074/jbc.M109503200. [DOI] [PubMed] [Google Scholar]
- 30.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 31.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 32.Mason PB, Struhl K. Mol. Cell. Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda S, Naruse S, Yatani R. Nature. 1967;213:696–697. doi: 10.1038/213696a0. [DOI] [PubMed] [Google Scholar]