Abstract
Infection by human rhinovirus (HRV) is a major cause of upper and lower respiratory tract disease worldwide and displays considerable phenotypic variation. We examined diversity by completing the genome sequences for all known serotypes (n = 99). Superimposition of capsid crystal structure and optimal-energy RNA configurations established alignments and phylogeny. These revealed conserved motifs; clade-specific diversity, including a potential newly identified species (HRV-D); mutations in field isolates; and recombination. In analogy with poliovirus, a hypervariable 5′ untranslated region tract may affect virulence. A configuration consistent with nonscanning internal ribosome entry was found in all HRVs and may account for rapid translation. The data density from complete sequences of the reference HRVs provided high resolution for this degree of modeling and serves as a platform for full genome-based epidemiologic studies and antiviral or vaccine development.
Human rhinovirus (HRV), the disease agent for the common cold, is responsible for ~50% of asthma exacerbations and is one of the factors that can direct the infant immune system toward an asthmatic phenotype (1–4). Direct and indirect costs from the common cold and related complications in asthmatics amount to an estimated ~$60 billion per year in the United States (5, 6). HRVs are single-stranded, positive-sense RNA enteroviruses in the Picornaviridae family and have been cataloged primarily by capsid serotyping relative to a historical repository of 99 strains, obtained from clinical specimens. HRVs are classified by their use of either intercellular adhesion molecule–1 (ICAM-1) (88 major viruses) or low-density lipoprotein receptor (LDLR) (11 minor viruses) as their receptor for cell entry (7). They have also been characterized by composite sensitivities across a panel of potential therapeutics (8) that have been used to parse the strains into two related drug-reactivity groups. The partial sequences of viral capsid-coding regions, noncoding regions, and a limited number of complete genomes have resulted in a division of the original 99 strains into two species: HRV-A (containing 74 serotypes) and HRV-B (containing 25 serotypes).
Recently, a number of previously unknown HRV-like sequences were detected in patients with influenza-like illnesses associated with severe respiratory compromise (9–11). The newly identified viruses have not been cultured, but their sequences indicate that they likely represent a third (HRV-C) species. The lack of whole-genome sequence data for the full cohort of HRVs has made it difficult to understand basic molecular and evolutionary characteristics of the viruses and has hampered investigations of the epidemiology of upper respiratory tract infections and asthma epidemics. To define the extent and nature of HRV diversity and their evolution, we sequenced the genomes for every previously undetermined HRV in the reference repository, completing the full set of 99 serotypes, as well as 10 additional field samples.
Genome sequences and alignments
Modifications (12) were made to the sequence-independent, single-primer amplification (SISPA) method (13) to determine the complete genomes of 70 HRVs from the reference repository, as well as 10 nasal-wash samples from patients with HRV upper respiratory tract infections. We sequenced these viruses to an average of sixfold coverage for each of the ~7-kb genomes. To provide phylogenetic accuracy for these organisms with relatively small genomes and (often) high degrees of sequence similarity, a stringent approach was taken for aligning the sequences. The initial sequence fits for the polyproteins were performed on the basis of superimposition of the amino acid sequences within virion crystal structure maps (14) and supplemented with additional structure data from other viral proteins. In a stepwise manner, profile hidden Markov models (HMM) augmented the founder set with the remaining sequences (13). The published sequences (including redundant determinations) for the remaining serotypes were added so that the final collection consisted of 138 full-length HRV genomes, including at least one representative for each of the 99 original strains, 10 field samples, and 7 HRV-C strains (table S1). The genome-length RNA and polyprotein alignments for all considered sequences are provided in tables S2 and S3. Regardless of species, all HRVs were found to have similar average base compositions. They are rich in A (31 to 34%) and U (25 to 30%) but low in G (19 to 22%) and C (18 to 22%). The third codon positions have the highest composition skew. An identity matrix for the polyproteins (fig. S1) shows that the average amino acid identity between pairs of HRV-C strains is slightly more diverse (78% identity, range 68% to 95%) than among the HRV-A (80%, range 64% to 99%) or HRV-B (83%, range 75% to 97%). The lack of broader diversity suggests that all HRVs are in a stable status for maintaining selection for certain traits, yet still have mutational flexibility for escape from immune responses.
Picornaviruses encode a single open reading frame (ORF) representing about 90% of the RNA length. Translation produces a polyprotein (~215 kD for HRV), subsequently cleaved in a viral protease–dependent cascade to form the 11 to 12 mature viral proteins required to initiate and sustain an infection (fig. S2A). Local and global RNA structures play established roles in HRV biology; however, the extent, character, and relatedness of serotype-specific 5′- and 3′UTR (untranslated region) variation are unknown. The role of this variability has thus not been related to function by modeling techniques or in vivo approaches. Our alignment methods included optimal energy RNA structure considerations (15) and were therefore sensitive to potential differences among the HRVs in the 5′- and 3′UTRs.
HRV RNA structures
All enteroviruses encode 5′-terminal cloverleaf-like motifs (CLs) that bind viral and cellular proteins for the initiation of RNA synthesis and also help convert infecting genomes from translation to replication templates. The HRV CLs [80 to 84 bases (b)] were predicted in every sequence with minimal structural variation among the species (representative structures are shown in Fig. 1A and additional structures in fig. S3). Immediately 3′ to the CL, all HRVs were found to share an unusual pyrimidine-rich spacer segment with short oligo(C) and oligo(U) units interspersed with As (blue boxes, Fig. 1A and fig. S3A). The HRV-A have the shortest tracts (11 to 22 b) and HRV-B the longest (22 to 50 b). Nearly every HRV displayed a unique sequence in this region, and we identified unexpected variation even among isolates of the same serotype (Fig. 1A and fig. S3A). The equivalent genome location in poliovirus (10 b) interacts with poly(C)-binding protein 2 and is involved in the determination of the polio neurovirulent potential. The analogous regions in aphthoviruses or cardioviruses have homopolymeric poly(C) or poly(UC) tracts, the deletion of which markedly attenuates the virus through an RNA-activated protein kinase activation–dependent mechanism (16). If the HRV 5′ spacer tracts are functional analogs to those of these other picornaviruses, then it is possible that the pathogenic potential of an individual HRV may also be encoded, in part, by this region.
Fig. 1.
Genome-wide optimal energy RNA configurations for select motifs representative of each HRV species. (A) All HRVs display characteristic 5′UTR CL elements with minimal predicted structural variation (blue boxes). (B) The alignments and predicted structures of the IRES of HRVs reveal a bait-and-switch arrangement for initiation of translation. A stem-pairing AUG (light green box) is found for each species, but a highly variable region of intervening sequence near the initiator AUG (dark green box) was noted. (C) HRV 3′UTRs have a unique unbranched stem motif before the poly-A tail. The left-most codon (red box, white text) is the ORF terminator. Other boxes highlight additional terminators (tan boxes), including a characteristic codon (tan box, red text) found in the apical loop.
Picornaviruses use internal ribosome entry sites (IRESs) to mediate translation initiation of their polyprotein ORFs. The IRESs of all enteroviruses (termed type 1 IRESs) are thought to bind 40S ribosomal subunits internally within their 5′UTR and to then scan additional nucleotides to find the proper initiator AUG (17). Our modeling of the known serotypes confirms that all HRV IRESs start just 3′ to the pyrimidine-rich spacer tract. We found that the internal IRES sequences are highly conserved, with an average nucleotide identity of 82%. Indeed, this region of the genome has the greatest degree of identity among all HRV (fig. S2B), exceeding 95% for regional motifs within the IRES. However, we also observed that the dominant IRES sequence conservation did not extend completely to the initiator AUG and that for the 18 to 40 bases 5′- to this codon, the region scanned by ribosomes, there was little species-specific conservation (<60% nucleotide identity). Despite this, our folding predictions configured every one of these regions into virtually the same RNA motif, which suggests that this structure is conserved even when the underlying nucleotides are not (Fig. 1B and fig. S3B).
Near the bottom of a long [15 to 20 base pair (bp)] minimum energy unbranched stem, the ORF AUG was invariably paired with a conserved upstream noncoding AUG (green boxes, Fig. 1B), marking the 3′ boundary of the IRES, and the normal launch point for 40S scanning. Every HRV genome fold maintains this pairing. We predict that HRVs use the proximity of these AUGs to orient the 40S for direct transfer to the proper codon, without the need for scanning through the intervening nucleotides. The sequence and length variation between the AUGs was consistent with the idea that ribosomes would bypass this region entirely if they jump from one AUG to the other. This IRES folding is essentially a bait-and-switch mechanism, which we predict may enhance HRV translation competitiveness. This paired AUG motif is unique to HRV and is not found in other enteroviruses (e.g., poliovirus).
The HRV 3′UTRs (40 to 60 b) begin with the ORF termination codon and extend to the genetically encoded poly(A) tail. The ORF terminators themselves (solid red boxes in Fig. 1C and fig. S3D) included UAG, UAA, and UGA codons. The codon selection often differed among isolates from the same serotype. Multiple additional terminators (tan boxes), in and out of the ORF, were identified that punctuated each segment (3 to 9 per UTR). Despite large differences (>40%) in nucleotide identity (Fig. 1C), it was noted that all HRVs maintain a 13- to 16-bp unbranched stem, covering 67 to 88% of the 3′UTR, immediately abutting the poly(A) tail (see also fig. S3D). Some 3′ stems have small interior loops, but nearly all, except for the unusual sequences of clade-D, present 5-base terminal loops, anchored with apical U-G or U-A pairs. Inevitably, the 3′ sides of these terminal loops display UAG or UGA terminator codons, which may or may not synchronize with the local ORF. The function of these 3′ stems is unknown, but such conservation is usually indicative of a putative protein recognition motif (such as translation termination factors) or, alternatively, an RNA:RNA tertiary interaction between the terminal-loop segment and a different (unknown) region of the genome.
Phylogenetic relationships of the HRV
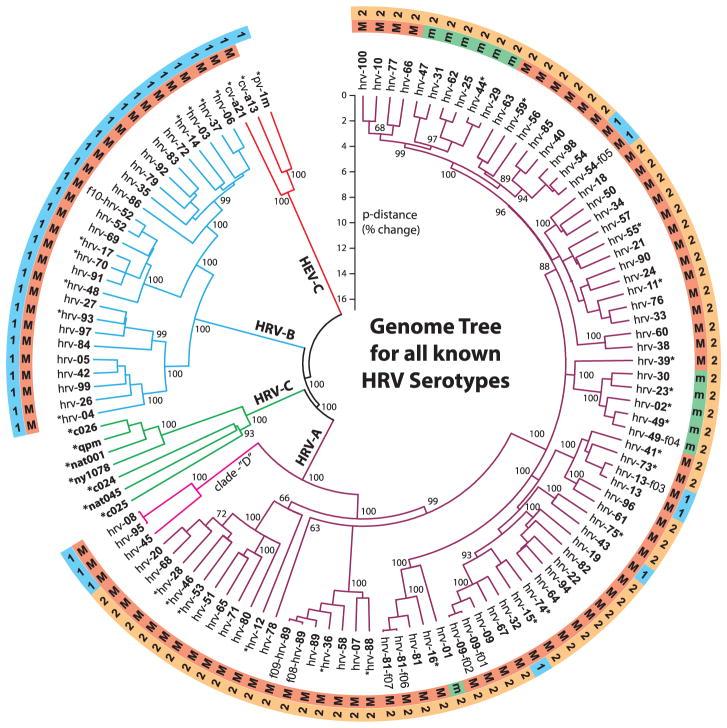
Multiple methods were used to compute and compare phylogenetic trees for the aligned RNA and protein data. Figure 2 is a neighbor-joining consensus tree (18), which considered the 5′- and 3′UTRs and the first and second codon positions for the RNA genomes. All major nodes of the tree topology were stable over a range of calculation parameters, regardless of whether they invoked minimum evolution, parsimony, unweighted pair group method with arithmetic mean, or maximum likelihood (ML) methods. (See fig. S6A for ML tree with bootstrap, likelihood ratio tests, and comparison to the neighbor-joining tree.) Statistical comparisons were performed between the ML tree generated from the full genomes and the ML constrained-topology trees generated from capsid sequences that have been used as a surrogate for serotypes (fig. S6, A to C) (13), with the null hypothesis that topologies of these limited-sequence trees were the same as that of the full genome-based tree. The approximately unbiased (AU) test calculated from the multiscale bootstrap P values for the capsid VP1 and VP0 trees were <10−69, strongly rejecting the null hypothesis. In a similar manner, ML constrained-topology trees generated from IRES and 3D-polymerase sequences were not consistent with the full genome tree (fig. S6, A and B). Additional statistical tests also confirmed these results from the AU (fig. S6, B and C) for all four of the constrained-topology trees. Taken together, these results suggest that trees based on sequences from small, albeit biologically important, regions of the HRV genome have a limited capacity to reflect the evolutionary relationships and underlying diversity of HRVs. The tree shown in Fig. 2 also accurately represents parallel calculations on the aligned polyprotein dataset (p-distances differed by <2%). All aligned full-length sequences contributed to the tree calculations, but published sequences (noted with asterisks on Fig. 2) are shown only as needed, to complete the cohort of reference serotypes. (Were the redundant published sequences illustrated, without exception they would lie on the same-serotype branches, with p-distances <0.5%.) The outer rings of this figure designate major (M: ICAM-1) or minor (m: LDLR) receptor preferences, and also each strain’s small-molecule drug reactivity group (“1” or “2”) (8). The tree is shown rooted with three outgroup sequences from the Human Enterovirus C species (poliovirus 1m, coxsackieviruses a13 and a21), but the use of five additional outgroups had no effect on overall tree topology or HRV p-distances (fig. S6). The field samples (i.e., f01 to f10) were assigned tentative serotype names on the basis of capsid similarity relative to the known reference strains. Although some of the full genomes from these samples proved close to their repository cognates, a divergence in sequence for several strains was noted.
Fig. 2.
A neighbor-joining (NJ) phylogenetic tree showing relationships between all known HRV serotypes created on the basis of full genome sequences. The HEV-C sequences (poliovirus 1M, coxsackievirus a13, and coxsackievirus a21) were used as outgroups. Branch lengths are proportional to similarity (p-distance). Key nodes on this tree are annotated with NJ bootstrap values (percentage of 2000 sampled trees). Asterisks in the strain names identify sequences obtained from published data (table S1). All other taxa are from this study. Letters in the outer rings designate whether that virus uses the major (M) ICAM-1 receptor or the minor (m) LDLR receptor (7) and whether its relative reactivity was more like group “1” or group “2” (if known) toward a panel of small-molecule antiviral compounds (8).
According to our tree(s), HRV-A and HRV-C share a common ancestor, which is a sister group to the HRV-B. Although the HRV-C clade currently has only seven full sequences, its genetic origin is clearly different from the reference set, and our phylogeny indicates that these represent a third HRV species, as has been recommended to the International Committee on Taxonomy of Viruses (19). The HRV-C have yet to be cultured or assessed for immunological cross-reactivity, but the sequence space occupied by the available samples suggests that there may be many additional HRV-C strains awaiting discovery. Distance extrapolations relative to the new full reference cohort predict that HRV-C may have an even broader range of serotypes than the original 99, of which each confers only limited immunologic cross-protection to another.
A separate phylogenic finding was the unexpected basal divergence within HRV-A of a small (n = 3) group of distinct strains, denoted clade D (20). Although the major basis for discriminating clade D from other HRV-A lies in their general, genome-length sequence divergence, these particular isolates have RNA elements—such as the cis-acting replication element, the 3′UTR terminal loop feature (see above and fig. S3), and local insertions/deletions and sequence motifs—that are somewhat atypical of other HRV-A strains. Some of the distinguishing characteristics are highlighted in fig. S4. Among all other major A clades, and major B clades, none have p-distances (>10%) that segregate them so distinctly. We are cautious in proposing clade D as a fourth species (HRV-D), but the phylogenetic evidence (Fig. 2) and sequence characteristics (figs. S3 and S4) are highly suggestive. Other early topological divisions within HRV-A separate a major clade composed of 10 serotypes (counterclockwise, hrv-20 through hrv-12) from a second grouping composed of ~12 miniclades representing 61 serotypes (counterclockwise, hrv-89-f 09 through hrv-100). These particular relationships were not readily apparent when only partial genome sequences were examined (21, 22). Fig. S6 shows a comparison of results from trees constructed with whole genomes, VP4/VP2, and VP1 sequence. Neither of the trees derived from these shorter sequences revealed the miniclades, clade D, or multiple other features. We thus contend that comparison of full-genome data, the context wherein evolutionary events occur, most likely provides the defining relationships among the HRVs, allows a more comprehensive assessment of strain diversity, and allows for more accurate historical extrapolations. The phylogenetic diversity we describe at the whole-genome level is consistent with the clinical heterogeneity of HRV infections in humans (1, 3, 4), although mapping specific clinical characteristics (i.e., incubation period, severity, respiratory compromise, and pro-asthmatic phenotypes) to responsible genomic regions will require additional field isolates from a large number of patients with multiple traits. Given the genome-wide diversity we have documented (e.g., 5′ spacer elements, ORF start, protease, 3′UTRs), clinically relevant relationships may well depend on comparisons from multiple genome regions.
Recombination in HRV
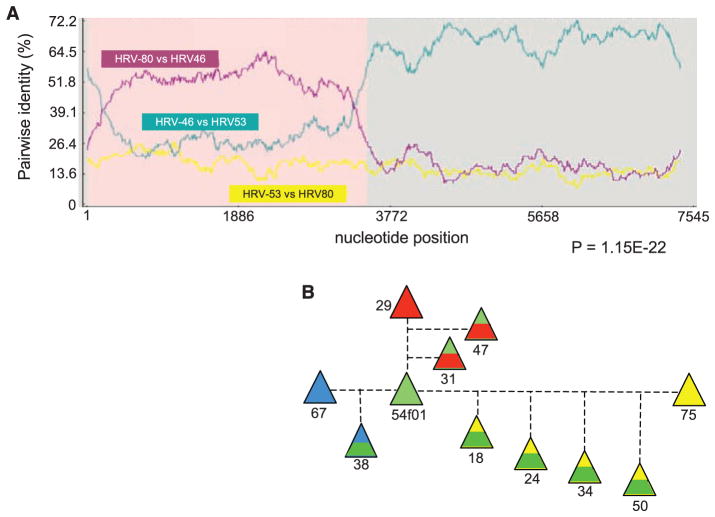
Results from earlier sequencing of a subset of HRV reference genomes concluded that RNA recombination was not a major mechanism for HRV diversity (23, 24) and asserted that known isolates were independently segregating entities. We have reevaluated the potential for recombination by scanning the full reference set and the new field strains with a suite of recombination detection programs (25) relying on phylogenetic distance and sequence similarity. Stringent criteria (P < 0.00001 from two or more analyses modes) identified 23 genomes with probable origins resulting from at least 12 independent recombination events. Figure 3A shows representative data indicating that hrv-46 arose by recombination between hrv-53 (major parent) and hrv-80 (minor parent). Within the hrv-46 genome, nucleotides 32 to 3222 are most similar to hrv-80, whereas the rest of the genome (nucleotides 3223 to 7200) is common to hrv-53. The result is consistent with this trio’s computed phylogenetic relationship (Fig. 2), placing the major parent (hrv-53) and the daughter (hrv-46) in the same clade and the minor parent (hrv-80) in a different, nearby clade. Results for all 23 identified recombination scenarios are summarized in Table 1. [See also (13) and table S4.] Of the recombination locales suggested by these events, the majority (10 of 12) involve the 5′UTR or the adjacent capsid genes, which seemingly have been collectively rearranged to produce at least 20 separate progeny strains. Among the 138 full-length sequences, hrv-54 (or its ancestor) was apparently the most active in recombination. Field strain hrv-54-f05 links closely to three separate events (see Fig. 3B), contributing to at least seven different serotype progeny. In confirmation studies, we used a progressive alignment method for the HRV genomes (13), then repeated the suite of recombination detection programs. Of the 23 recombination events from the HMM alignment (Table 1), 19 were also found after the progressive alignment, although in some cases different major and/or minor parents were selected from within the same closely related clade (table S4).
Fig. 3.
Recombination of HRVs creates additional serotypes. (A) Representative results showing that hrv-46 arose from a recombination of hrv-53 (major parent) and hrv-80 (minor parent). Shown are normalized pairwise identities between each parent and the daughter hrv (purple and green) and the two parents (yellow). As indicated, hrv-46 nucleotides 32 to 3222 are from hrv-80, and nucleotides 3223 to 7200 are from hrv-53. (B) Recombination with an ancestor of hrv-54-f05 has resulted in seven serotype progeny. Each parental hrv is shown as a solid color. The contribution of each parent is proportional to the area of its color in the offspring. See Table 1 and table S4 for nucleotide boundaries and results from other recombination events.
Table 1.
Recombination events in HRV serotypes. The P value listed is the lowest obtained. See table S4 for full data and P values.
| Major parent | Minor parent | Recombinant | Genome region | P |
|---|---|---|---|---|
| hrv-45 | hrv-21 | hrv-8 | 5′UTR | 1.094 × 10−21 |
| hrv-45 | hrv-21 | hrv-95 | 5′UTR | 1.094 × 10−21 |
| hrv-65 | hrv-21 | hrv-80 | 5′UTR, VP4 | 6.129 × 10−23 |
| hrv-51 | hrv-11 | hrv-20 | 5′UTR, VP4 | 8.402 × 10−26 |
| hrv-51 | hrv-11 | hrv-68 | 5′UTR, VP4 | 8.402 × 10−26 |
| hrv-28 | hrv-62 | hrv-71 | 5′UTR | 8.484 × 10−15 |
| hrv-54-f05 | hrv-75 | hrv-18 | 5′UTR, VP4, VP2, VP3, VP1 | 3.737 × 10−7 |
| hrv-54-f05 | hrv-75 | hrv-24 | 5UTR, VP4, VP2, VP3, VP1 | 3.737 × 10−7 |
| hrv-54-f05 | hrv-75 | hrv-50 | 5′UTR, VP4, VP2, VP3, VP1, P2A | 3.737 × 10−7 |
| hrv-54-f05 | hrv-75 | hrv-34 | VP4, VP2, VP3, VP1 | 3.737 × 10−7 |
| hrv-54-f05 | hrv-67 | hrv-38 | VP2, VP3, VP1, P2A, P2B, P2C, P3A | 1.561 × 10−29 |
| hrv-54 | hrv-67 | hrv-60 | VP2, VP3, VP1, P2A, P2B, P2C, P3A | 1.561 × 10−29 |
| hrv-53 | hrv-80 | hrv-46 | 5′UTR, VP4, VP2, VP3, VP1 | 1.291 × 10−62 |
| hrv-13-f03 | HRV41 | hrv-73 | 5′UTR, VP4, VP2, VP3, VP1 | 9.419 × 10−12 |
| hrv-30 | hrv-59 | hrv-39 | 5′UTR, VP4, VP2, VP3, VP1 | 2.963 × 10−12 |
| hrv-68 | hrv-20 | hrv-28 | VP4, VP2, VP3, VP1, P2A, P2B, P2C | 6.839 × 10−12 |
| hrv-100 | hrv-10 | hrv-56 | P2A, P2B, P2C, P3A, P3B, P3C, P3D | 4.230 × 10−17 |
| hrv-29 | hrv-54-f05 | hrv-31 | P3C, P3D | 7.934 × 10−13 |
| hrv-29 | hrv-54-f05 | hrv-47 | P3C, P3D | 1.232 × 10−10 |
| hrv-42 | hrv-97 | hrv-4 | 5′UTR, VP4 | 8.233 × 10−35 |
| hrv-84 | hrv-37 | hrv-27 | 5′UTR, VP4 | 1.500 × 10−10 |
| hrv-84 | hrv-37 | hrv-93 | 5′UTR, VP4 | 1.500 × 10−10 |
| hrv-84 | hrv-37 | hrv-97 | 5′UTR, VP4 | 1.500 × 10−10 |
Although the sequence fingerprints clearly trace these ancestral patterns, nevertheless all extant progeny also showed evidence of subsequent sequence divergence within the exchanged regions. Recombination was not identified between isolates from different species (i.e., HRV-A and HRV-B), but receptor binding preferences between ICAM and LDLR apparently presented no barriers to exchange. Major group hrv-54 and minor group hrv-29, for example, both contributed to the common ancestor of the minor group viruses, hrv-31 and hrv-47. These results, particularly for HRVs with different receptor preferences and those from distant clades, such as the parents hrv-21 and hrv-65, suggest that coinfection of the host with multiple parental strains is not uncommon and may lead to variant progeny with different biological properties. Our field isolates also deviated from the reference sequences in a manner that was not confined to any specific portion of the genome. In fact, field strain deviation relative to the reference isolates (for example, hrv-52-f01 versus hrv-52 differ by 838 nucleotides) was frequently greater than that observed between pairs of characterized serotypes (hrv-44 and hrv-29 differ by 385 nucleotides). Indeed, field samples of the same serotype collected from the same geographical region within 1 year showed marked variability (tables S2, S3, and S5). The propensity for such variation could underlie the marginal efficacy, or lack of efficacy, of anti-HRV therapeutics in clinical trials (26, 27). Given our reference set of full genomes, along with the means to rapidly sequence full genomes from field samples, new strategies may become apparent to engineer clade-specific agents by targeting their commonalities.
Conclusions
Our data complete and define the full set of genome-length sequences in the canonical reference repository of 99 HRV-A and HRV-B serotypes. Alignment and examination of these genomes confirmed species-specific sequence and RNA structural elements that differentiate the HRV-A and HRV-B from newly described HRV-C and further suggest that the HRV-A serotypes harbor a distinct, uncharacteristic clade, which may represent a fourth species (HRV-D). Local sequence variation, particularly in the 5′UTR, characterized each isolate within regions associated with the pathogenic potential for other picornaviruses. Parallel RNA structure comparisons defined several 5′ and 3′ elements as common to all isolates and unique to the HRV. Motifs like the AUG-presenting 5′ ORF initiation stem, or the UAG-presenting 3′ stem, may contribute to HRV-specific IRES translation mechanisms, ORF termination, or polymerase recognition. We also found embedded within multiple sequences, including recent field isolates, clear evidence for repeated, historical genome recombination. Coinfection with multiple HRVs is known to occur (28), and we now know that this can lead to strains that may have distinct biologic properties and clinical characteristics. The required host environment for HRV recombination is not known, but with complete genome sequences from additional patient isolates such factors may become apparent. Our repository data set provides a baseline framework for the analysis of additional HRVs that may be in communities, including the HRV-Cs, and will enable larger-scale studies of basic molecular and evolutionary characteristics and assignment of disease phenotypes to specific genome regions. The clustering of small clades, the recombinations, and the mutations found in all regions of these genomes suggest that future HRV epidemiologic studies might benefit from full genome sequencing rather than the more limited serotyping. With such an approach, correlations may be more informative in inferring pathogenic potential and in designing antiviral agents and vaccines.
Supplementary Material
Acknowledgments
Funded by the University of Maryland School of Medicine internal funds and by NIH grant U19-AI070503. We thank A. Wolf and J.-Y. Sgro for implementation of the tree topology tests. The accession numbers for the hrv strains are FJ445111 to FJ445190 (see table S1).
Footnotes
References and Notes
- 1.Gwaltney JM, Jr, Hendley JO, Simon G, Jordan WS., Jr N Engl J Med. 1966;275:1261. doi: 10.1056/NEJM196612082752301. [DOI] [PubMed] [Google Scholar]
- 2.Johnston SL, et al. Am J Respir Crit Care Med. 1996;154:654. doi: 10.1164/ajrccm.154.3.8810601. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson KG, Kent J, Ireland DC. Br Med J. 1993;307:982. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson DJ, et al. Am J Respir Crit Care Med. 2008;178:667. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fendrick AM, Monto AS, Nightengale B, Sarnes M. Arch Intern Med. 2003;163:487. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- 6.Weiss KB, Sullivan SD. J Allergy Clin Immunol. 2001;107:3. doi: 10.1067/mai.2001.112262. [DOI] [PubMed] [Google Scholar]
- 7.Vlasak M, et al. J Virol. 2005;79:7389. doi: 10.1128/JVI.79.12.7389-7395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andries K, et al. J Virol. 1990;64:1117. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez SR, et al. J Clin Virol. 2008;43:219. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McErlean P, et al. PLoS ONE. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SK, et al. J Clin Microbiol. 2007;45:3655. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djikeng A, et al. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supporting material on Science Online.
- 14.Palmenberg AC, Sgro J-Y. In: Molecular Biology of Picornaviruses. Semler BL, Wimmer E, editors. ASM Press; New York: 2001. [Google Scholar]
- 15.Palmenberg AC, Sgro JY. Semin Virol. 1997;8:231. [Google Scholar]
- 16.Martin LR, Duke GM, Osorio JE, Hall DJ, Palmenberg AC. J Virol. 1996;70:2027. doi: 10.1128/jvi.70.3.2027-2031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson RJ. In: Translational Control. Hershey WB, Mathews MB, Sonenberg N, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.On the basis of these data, the International Committee on Taxonomy of Viruses Picornavirus Study Group recently proposed to recognize them as a new species (29).
- 20.hrv-08 and hrv-95 are designated as two different serotypes by ATCC but differ by 67 nucleotides, perhaps indicating a misidentification in the repository.
- 21.Ledford RM, et al. J Virol. 2004;78:3663. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savolainen C, Blomqvist S, Mulders MN, Hovi T. J Gen Virol. 2002;83:333. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 23.Kistler AL, et al. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmonds P. J Virol. 2006;80:11124. doi: 10.1128/JVI.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D, Rybicki E. Bioinformatics. 2000;16:562. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 26.Fleischer R, Laessig K. Clin Infect Dis. 2003;37:1722. doi: 10.1086/379830. [DOI] [PubMed] [Google Scholar]
- 27.Barnard DL. Curr Pharm Des. 2006;12:1379. doi: 10.2174/138161206776361129. [DOI] [PubMed] [Google Scholar]
- 28.Savolainen C, Mulders MN, Hovi T. Virus Res. 2002;85:41. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 29.Knowles N. personal communication.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.