Abstract
Calcineurin-inhibitors CI are immunosuppressive agents prescribed to patients after solid organ transplant to prevent rejection. Although these drugs have been transformative for allograft survival, long-term use is complicated by side effects including nephrotoxicity. Given the narrow therapeutic index of CI, therapeutic drug monitoring is used to prevent acute rejection from underdosing and acute toxicity from overdosing, but drug monitoring does not alleviate long-term side effects. Patients on calcineurin-inhibitors for long periods almost universally experience declines in renal function, and a subpopulation of transplant recipients ultimately develop chronic kidney disease that may progress to end stage renal disease attributable to calcineurin inhibitor toxicity (CNIT). Pharmacogenomics has the potential to identify patients who are at high risk for developing advanced chronic kidney disease caused by CNIT and providing them with existing alternate immunosuppressive therapy. In this study we utilized BioVU, Vanderbilt University Medical Center’s DNA biorepository linked to de-identified electronic medical records to identify a cohort of 115 heart transplant recipients prescribed calcineurin-inhibitors to identify genetic risk factors for CNIT We identified 37 cases of nephrotoxicity in our cohort, defining nephrotoxicity as a monthly median estimated glomerular filtration rate (eGFR) < 30 mL/min/1.73m2 at least six months post-transplant for at least three consecutive months. All heart transplant patients were genotyped on the Illumina ADME Core Panel, a pharmacogenomic genotyping platform that assays 184 variants across 34 genes. In Cox regression analysis adjusting for age at transplant, pre-transplant chronic kidney disease, pre-transplant diabetes, and the three most significant principal components (PCAs), we did not identify any markers that met our multiple-testing threshold. As a secondary analysis we also modeled post-transplant eGFR directly with linear mixed models adjusted for age at transplant, cyclosporine use, median BMI, and the three most significant principal components. While no SNPs met our threshold for significance, a SNP previously identified in genetic studies of the dosing of tacrolimus CYP3A5 rs776746, replicated in an adjusted analysis at an uncorrected p-value of 0.02 (coeff(S.E.) = 14.60(6.41)). While larger independent studies will be required to further validate this finding, this study underscores the EMRs usefulness as a resource for longitudinal pharmacogenetic study designs.
1. Introduction
Calcineurin-inhibitors (CI), such as tacrolimus and cyclosporine, are immunosuppressants prescribed to recipients of allographs to reduce the risk of rejection by the immune system. These drugs function by dampening IL-2 signaling pathway in T-cells and avoid the vigorous inflammation and tissue damage typical of an alloresponse. While these drugs have led to dramatically improved survival among heart transplant recipients, the nephrotoxic side-effects of these drugs continue to diminish the long-term survival rates among patients [1; 2]. CI are dosed in a narrow therapeutic window requiring close monitoring of serum drug levels to prevent allograft rejection while minimizing the risk of adverse events.
Post-transplant, patients undergo continuous monitoring of their serum creatinine and glomerular filtration rates (GFR) to determine impact of the immunosuppressants on kidney function. A decline in kidney function is nearly universal among heart transplant recipients with significant variability in the development of severe kidney disease. Patients are frequently faced with the development of chronic kidney disease (CKD) which is classified into 5 stages of increasing severity, each defined by the estimated GFR. In a retrospective study 352 heart transplant recipients, 3% developed end-stage renal disease or CKD Stage 5 by 5 years and 12% by 10 years [3]. Clinical risk factors for developing post-transplant CKD include pre-transplant GFR, pre-transplant diabetes mellitus, a female cardiac donor, gender of the recipient, and post-operative renal replacement therapy [3].
Despite vast structural differences, the pharmacokinetics of cyclosporine and tacrolimus are surprisingly similar, and both agents are targets of the P-gp efflux pump ABCB1 and the cytochrome p450 CYP3A family of enzymes [4]. These genes are polymorphic for functional alleles, and variants have been examined in several pharmacogenetic studies of calcineurin-inhibitor dosing and nephrotoxicity in renal transplants [5–8]. Despite a large number of candidate gene studies on the effects of these variants on immunosuppression therapy, many of these analyses are narrow in their scope of genes tested. In this study, we explored the roles of other pharmacokinetic genes outside the CYP3A family and ABCB1 on the development of calcineurin inhibitor nephrotoxicity CNIT. For our study, we identified 127 heart transplant recipients in BioVU, Vanderbilt University Medical Center’s DNA biorepository linked to de-identified electronic medical records. From data collected in this patient population, we developed a longitudinal pharmacogenetic study to test the impact of ADME Core variants on the development of CNIT [9].
2. Methods
2.1. Study Population
As stated above, our study population of heart transplant recipients was obtained from BioVU. A full description of BioVU as a resource, including its ethical, privacy and other protections has been described in detail elsewhere [10]. In brief, BioVU extracts and stores DNA from blood collected from routine clinical testing that is scheduled to be discarded after a three-day waiting period at the Vanderbilt University Medical Center (VUMC) in Nashville, TN. DNA samples are linked to a de-identified version of the patient’s electronic medical record, known as the Synthetic Derivative (SD), which can be accessed by investigators for research purposes after approval by the local internal review board and BioVU Review Committee. Patients eligible for possible inclusion into BioVU are those with an out-patient laboratory blood draw, have signed the consent to treatment form, and have not made a formal indication to opt-out [11].
Using the SD, we identified initial candidates for our study by screening for patients who met the following criteria: a) a heart transplant documented with three or more ICD9 code V42.1 (heart replaced by transplant) and/or one CPT code 33945, b) one or more mention of an immunosuppressant, c) DNA available in the biorepository and genotyped on the Illumina ADME Core Panel, and d) the patient was over the age of 15 at the date of the transplant operation. This initial screen identified 152 potential candidates. We then manually extracted the date of the transplant operation from each record. We excluded 10 patients with an ambiguous transplant operation date in the record or miscoded with a kidney, lung, liver, or multiple heart transplants during his/her lifespan. We extracted immunosuppressant data from the de-identified records of this heart transplant sample population with MedEx. MedEx extracts medications and their signature mentions from free-text entries in the EMRs. We used only medications with at least one mention of a dose, route, frequency or strength to limit the medications to those the patient was actually prescribed. A more detailed description of the software has been published elsewhere [12].
We also extracted additional clinical information from the SD. For quantitative measurements such as body mass index (BMI, kg/m2), serum creatinine (mg/dl), and systolic and diastolic blood pressure (mmHg), monthly medians were calculated. Prior to transplant chronic kidney disease and diabetes mellitus were defined by ICD9 codes before the transplant date. Chronic kidney disease was defined by three or more mentions of the following ICD9-codes: 403, 585.1, 585.2, 585.3, 585.4, 585.5, 585.6, and 585.9. Patients were considered to have diabetes mellitus pre-transplant if they had three or more mentions of the following ICD-9 codes: 250.3, 250.32, 250.2, 250.22, 250.9, 250.92, 250.8, 250.82, 250.7, 250.72, 250.6, 250.62, 250.5, 250.52, 250.4, 250.42, 250, and 250.02. Pre-transplant hypertension was defined as median systolic blood pressure > 140 mmHg, systolic and /or > 90 mmHg diastolic, or prescribed one of the following hypertension medications: hydralazine, minoxidil, renin antagonist, central alpha agonists, ACE inhibitors (ACEI)/angiotensin receptor blockers (ARB), aldosterone antagonists, diuretics, K-sparing diuretics, loop diuretics, alpha antagonists, calcium channel blockers (CCB), beta blockers (BB), thiazide/BB, thiazide/ACEI/ARB, thiazide/aldosterone antagonist, thiazide/renin antagonist, and diuretic combinations, all before the transplant date.
2.2. Phenotype Definition
The outcome of interest was time to develop severe nephrotoxicity clinically attributed to CNIT, which we defined in our patient population as the development of CKD stage 4 or 5 in the setting of CI use. To assess kidney function over the course of immunosuppression therapy, we estimated the glomerular filtration rate from the “four variable” Modification of Diet in Renal Disease formula [13]:
| (1) |
All patients who entered into the SD by the time of their transplant date were included in this study. Patients who entered the SD post-transplant were included if their initial eGFR measurement upon entering the SD was > 30 mL/min/1.73m2this included patients with CKD stages 1, 2, and 3. These patients were assumed not to have CKD 4 or 5 in the setting of CI prior to their entry into BioVU and were entered into the analysis at their heart transplant date. Patients who entered the SD after their heart transplant date with an eGFR < 30 were excluded from the analysis. Our definition of severe chronic kidney disease 4 was a monthly median eGFR of < 30 mL/min/1.73m2 for three consecutive months. This threshold is adapted from the National Kidney Foundation’s definition for CKD stage 4: GFR of 15–29 and CKD Stage 5: GFR < 15 or dialysis [14].
2.3. Genotyping
DNA samples from a total of 115 heart transplant recipients were genotyped on Illumina’s ADME Core Panel as part of Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA). In short, Illumina’s pharmacogenetic-targeted ADME Core panel is designed for the genotyping of 184 markers in 34 genes. A full description of the panel’s content and performance has been published elsewhere [9]. Genotyping for this study was conducted at the Center for Human Genetics Research DNA Resources Core at Vanderbilt University. Genotype calling was performed with ADME Module Version 1.0.0.3. Formatting of the ADME Core Panel data set and quality control of the markers was performed with PLATO and PLINK [15; 16]. SNPs were filtered from the analysis if the allele frequency was below 5%, genotyping efficiency < 95%, or a statistically significant deviation from Hardy Weinberg expectations (p < 0.001) in the European American population. After filtering, 49 SNPs remained in our analysis. A principal components analysis (PCA) was performed with the Eigensoft software using available genome-wide data in the full dataset and in the subset of European Americans [17]. We tested for relatedness of individuals in subsets of samples stratified by race/ethnicity. One sample from a related pair of European Americans was removed. The genome-wide inflation factor for this study was 1. We extracted 333,804 overlapping markers from the samples’ genotype data from the following platforms: 18 individuals on Illumina’s HumanOmni5-Quad, 109 on the HumanOmni1-Quad, and four on Illumina’s 1M-Duo BeadChip.
2.4. Statistical Analysis
Cox proportional hazard models were calculated using the date of the heart transplant as the starting time in a time-to-event analysis. Genotypes were modeled additively against development of CKD stage 4. Factors that were associated with renal function in univariate analyses (p < 0.05) were included in the final multivariable model. Patients who did not develop CKD stage 4 were censored from the analysis at their final eGFR measurement. For the linear mixed effects modeling of post-transplant eGFR, we used the R package, nlme. SNPs and covariates that met a 0.05 threshold in univariate analyses were included as fixed effects and the subject identifier was included as a random effect. The within subject correlation was 0.70 and we chose to account for it in our models with an autoregressive-moving average model with one autoregressive and one moving average parameters. Plots were generated with STATA 11 and RStudio Version 0.97.551
3. Results
3.1. Demographics
Table 1 presents the clinical characteristics of our study population identified in BioVU. Overall, this is an ancestrally cosmopolitan cohort where 80.8% of the patients were administratively assigned [18] as of European descent, while the remainder was reported as African American with the exception of one sample reported as Hispanic. The median age at transplant was 52.5 years of age. This is a slightly overweight population with the median body mass index of 27.4 kg/m2. Prior to transplant, 10.4% and 60.6% patients had evidence of diabetes mellitus and hypertension, respectively. A majority of patients (52.7%) had their heart transplant at VUMC. Twenty-five patients died during post-transplant follow up. All patients were prescribed a calcineurin-inhibitor: 35.7% were prescribed cyclosporine alone, 25.2% tacrolimus alone, and 39.1% were prescribed a combination of the two (at different times).
Table 1.
Clinical Characteristics of heart transplant samples.
| Patients | 115 |
| European Descent (%) | 86.0 |
| Female (%) | 33.9 |
| Transplant Operation at VUMC (%) | 80.8 |
| Pre-transplant Diabetes Mellitus (%) | 10.4 |
| Median Systolic (mmHg) | 100.2, IQR: 94.3–107.0 |
| Median Diastolic (mmHg) | 64.0 IQR: 59.9–66.9 |
| Pre-transplant Hypertension (%) | 66.0 |
| Pre-transplant Chronic Kidney Disease | 9.56 |
| Median Age at Tx (years) | 52.5, IQR: 40.5–58.1 |
| Required Dialysis Post-Transplant (%) | 18.2 |
| Median Post Tx Follow up Time (years) | 8.8, IQR: 4.8 – 12.2 |
| Median Pre-eGFR (mL/min/1.73m2) | 68.0, IQR: 57.4–87.2 |
| Median Body Mass Index (kg/m2) | 27.4, IQR:24.6–31.1 |
| Died (%) | 21.7 |
| Cyclosporine Only (%) | 35.7 |
| Tacrolimus Only (%) | 25.2 |
| Cyclosporine and Tacrolimus (%) | 39.1 |
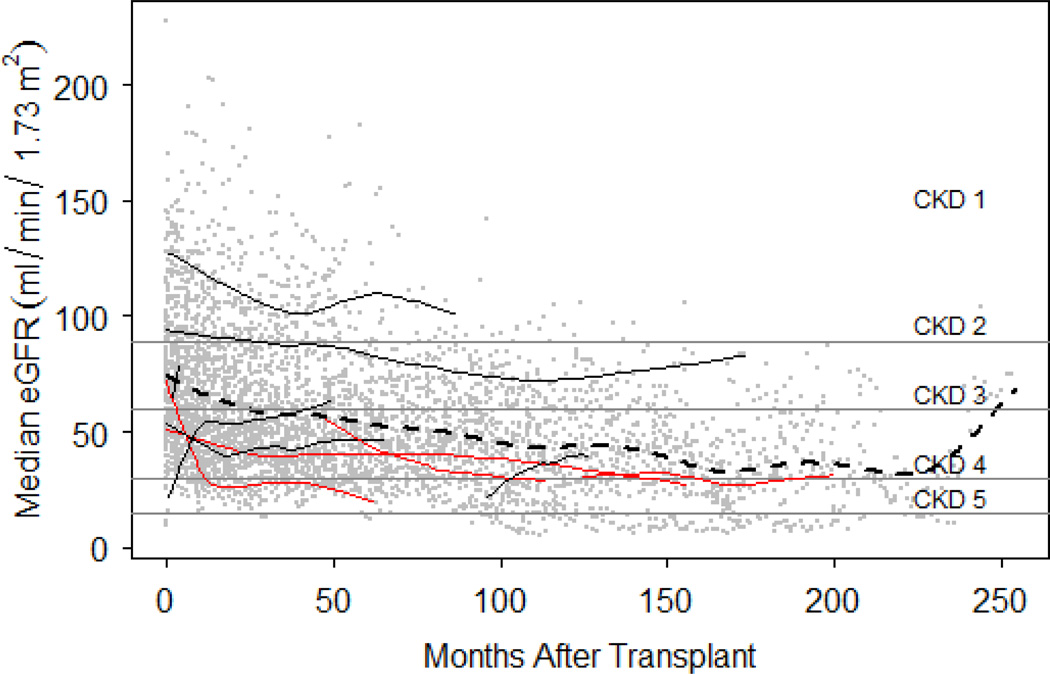
As expected for this patient population, the eGFR prior to transplant was lower than would be expected for a healthy population (median = 68.0 mL/min/1.73m2). Follow up time for these patients varied (Figure 1): median time to the final eGFR measurement in the SD was 8.8 years, and the median frequency of follow-up was 5.5 (IQR: 4.2–7.5) eGFR measurements per year. Kidney function continued to decline over time (Figure 1). In the second year (12–24 months) post-transplant 14.0, 31.4, 50.0, and 4.6 percent of individuals had median eGFR measurements that corresponded with the first four stages of CKD, respectively. By the fifth year (60–72 months), the distribution shifted towards lower median eGFR levels: 3.4, 22.4, 62.0, and 12.0 percent of individuals were observed with median eGFRs in range with the first four stages of CKD. At year ten, 11.7 and 11.7 percent of patients median eGFR measurements corresponded to CKD stages four and five, respectively.
Figure 1. Post-transplant eGFR measurements plotted on the thresholds of the five stages of chronic kidney disease.
Individual post-transplant eGFR measurements are plotted on the y-axis against time in months after transplant on x-axis as grey dots. The dashed line represents a polynomial function fit to all eGFR measurements collected in the study. Ten randomly selected patient’s eGFR profiles have fitted with loess lines and colored in red if the patient developed Chronic Kidney Disease (CKD) Stage 4 or below. Thresholds for the 5 stages of CKD are indicated: CDK1 > 90, CKD2 60–89, CKD3 30–59, CKD4 15–29, and CKD5 < 15 mL/min/1.73m2.
3.2 Time to CKD Stage 4 and 5 Survival Analysis
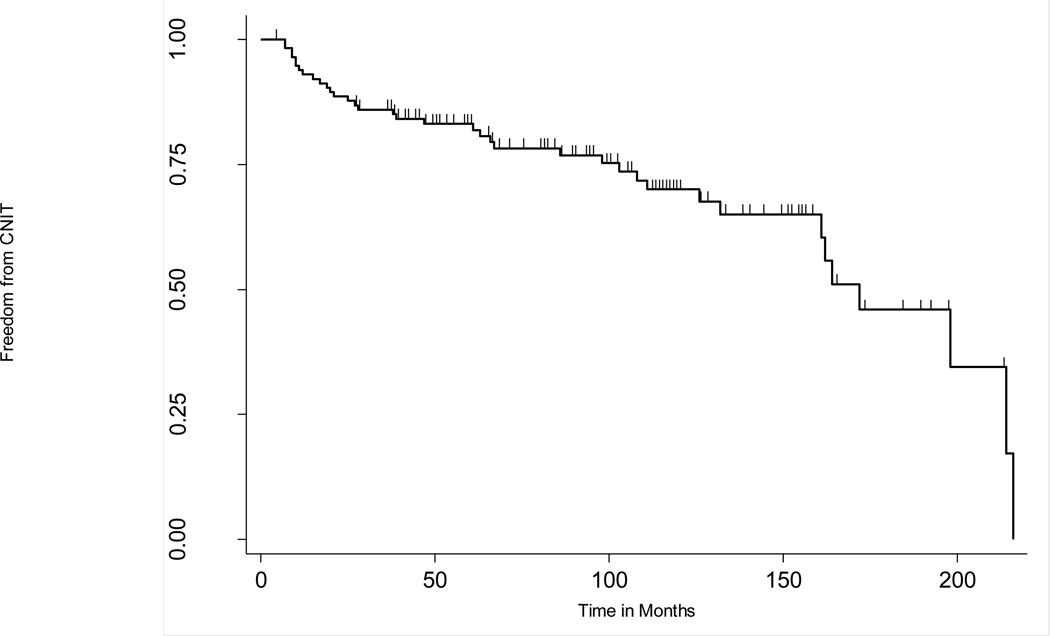
Figure 2 displays the development of CNIT in this study population in months post-heart transplant. Thirty-seven out of 115 patients (25.2) in this heart transplant cohort met the CNIT case definition. By twelve months, eight individuals (7.0%) met the criteria for CNIT, 19 (16.5%) by 60 months, and 28 (24.3%) by 120 months. From among the various clinical variables tested for an association with CNIT (the three most significant PCAs, gender, systolic and diastolic blood pressure, pre-transplant diabetes, pre-transplant hypertension, pre-transplant chronic kidney disease, age at transplant, pre-transplant eGFR, BMI, and prescribed calcineurin inhibitor), only pre-transplant eGFR, pre-transplant CKD status, pre-transplant diabetes mellitus status, and age at transplant met a significance threshold of p < 0.05 (Table 2).
Figure 2. Kaplan-Meier plot describing the proportion of non-nephrotoxic heart transplant recipients over time.
The y-axis indicates the proportion of event-free subjects and tick marks on the plot indicate where individuals are censored from the analysis.
Table 2.
Results of CNIT Analysis in European Americans
| Predictor | Hazard Ratio (95% CI) | P-value |
|---|---|---|
| Univariate Clinical Variable Model | ||
| Recipient Age per year | 1.05 (1.01–1.08) | 9.85 × 10−3 |
| Pre-transplant CKD | 3.69 (1.36–10.01) | 0.01 |
| Pre-Transplant eGFR per ml/min/1.73m2 | 0.96 (0.94–0.98) | 1.03 × 10−3 |
| Prior Diabetes Mellitus | 6.92(2.64–18.54) | 8.33 × 10−5 |
| Multivariable Genetic Model | ||
| DPYD rs1801265 | 0.45 (0.22–0.93) | 0.03 |
| UGT2B17 rs1902023 | 2.23 (1.21–4.11) | 0.01 |
| SLCO1B1 rs4149056 | 0.38(0.14–0.96) | 0.03 |
| SLC22A1 rs34305973 | 2.14(1.18–3.90) | 0.01 |
First, in the European American subset (n=99 heart transplant recipients with 35 cases of CKD stages 4 and 5) we tested the 49 Illumina ADME Core Panel markers that passed quality control for association with CNIT outcome. In unadjusted analysis, no markers were associated with CNIT after adjustment for multiple testing (p < 1.02 × 10−3). Variants in SLC22A1 rs34305973 and UGT2B17 rs1902023 trended toward significance in the unadjusted model (p = 0.02 and p=0.02, respectively). In models adjusted for pre-transplant CKD, pre-transplant diabetes mellitus, age at transplant, and the three most significant PCAs, UGT2B17 rs1902023 was the most significant (p = 0.01) among all the tested ADME Core Panel markers (Table 2). Secondly, we expanded our analysis to the full dataset regardless of race/ethnicity (n=115 heart transplant recipients with 37 cases of CKD stage 4 and 5) and the results were largely unchanged (data not shown). In the adjusted models for the full dataset, DPYD rs1801265 was the most significant (p = 9.24×10−3HR: 0.39, CI: 0.19–0.79) among all the tested ADME Core Panel markers. No marker was associated with CNIT in unadjusted or adjusted models after correction for multiple testing when the data were limited to cyclosporine treated only patients (n=95 heart transplant recipients with 27 cases of CKD stage 4 or 5) or tacrolimus treated only patients (n=79 heart transplant recipients with 18 cases of CKD stage 4 or 5; data not shown).
3.2 Modeling Post-Transplant eGFR
As a secondary analysis of post-transplant kidney function, the repeated eGFR measurements were analyzed directly using mixed effects models to account for the within subject correlation. In univarate analyses of covariates among European Americans, only cyclosporine use (coef(S.E) = -17.05(7.13), p = 0.02), median BMI (coef(S.E) = -1.27(0.62), p < 0.05), and age at transplant (coef(S.E) = -1.01(0.15), p = 1.55x 10−8) were associated with eGFR over time. No SNP met the significance threshold for multiple testing in unadjusted or adjusted analyses. However, in unadjusted analyses, two of the three SNPs that met a threshold of 0.05 have previously been associated with post-transplant renal function: CYP2C19 rs4244285 (coef(S.E) = 13.28(6.17),p = 0.03) and CYP3A5 rs776746 (coef(S.E) = 21.94(8.37),p = 0.01). SNP CYP2A6 rs28399433 also met the 0.05 threshold (coef(S.E) = 20.91(3.46), p = 0.02) in unadjusted analyses. Two of these associations maintained significance at the 0.05 threshold in the multivariate models CYP3A5 rs776746 (coef(S.E) = 14.60(6.41), p = 0.03) and CYP2A6 rs28399433 (coef(S.E) = 17.14(8.24), p = 0.04)[19]. In analyses extended to the full dataset regardless of race/ethnicity, only CYP2A6 rs28399433 (coef(S.E) = 17.46(6.70), p = 0.01) approached significance in the adjusted analysis (data not shown).
4. Discussion
4.1. Summary and Relevance
We used a biorepository linked to de-identified electronic medical records to identify heart transplant patients for pharmacogenomic studies. The two outcomes of interest in the present pharmacogenomics study was the development of advanced nephropathy (CKD Stage 4 or 5) in the setting of calcineurin-inhibitor therapy post-transplant and post-transplant eGFR over time. In this study, we have demonstrated that EMR-based cohorts linked to DNA samples provide ample opportunity to identify adverse drug reactions (ADR). This specific study focused on a common ADR to calcineurin-inhibitor therapy among heart transplant recipients. While there are several studies that have explored the relationship between a patient’s genetic profile and calcineurin-inhibitor dosing [5; 20; 21], this is the first study of our knowledge utilizing an EMR-based cohort of heart transplant patients to examine the pharmacogenetics of calcineurin-induced nephrotoxicity.
Our most significant result in the time to CNIT survival analysis was DPYD rs1801265, which approached our corrected p-value (p = 9.24 × 10−3) in the full dataset regardless of race/ethnicity. DPYD rs1801265 defines the DPYD *9A haplotype and encodes a cysteine to arginine missense mutation in the 29th position of the protein that some studies have suggested to be without significant enzymatic activity [22]. The gene is located in the centromeric region of chromosome one between 1p22 and 1q21 [23]. While the variant did not meet our multiple-testing threshold, larger studies may confirm its role in CNIT. It is interesting to note that CYP3A5 variants, which have been strongly associated with tacrolimus dosing in multiple studies [5], were not associated with CNIT but one marker in this gene trended towards significance in modeling eGFR directly. This marker rs776746 defines the CYP3A5*3 allele, a nonexpressing variant of the gene found a high frequency in populations of European descent[24]. In this study we found the functional CYP3A5*1 allele, which we found at comparable frequency to other studies (MAF = 0.06), to be positively associated with eGFR post-transplant [25].
The application of a heart transplant cohort for the pharmacogenetics of calcineurin-inhibitor nephrotoxicity has advantages over kidney and liver transplantations, as it eliminates the potential for donor-recipient gene interactions. The donor genetic information of kidney and liver transplant may play crucial roles in the susceptibility of nephrotoxicity. The liver is the primary site of drug metabolism, and in the case of liver transplants, the donor’s genome becomes the driver of metabolism. Its own unique genetic variation may lead to a different pharmacokinetic profile of calcinineurin-inhibitor metabolism compared with the recipient. The donor genome in the case of kidney transplant may also be a factor in developing nephrotoxicity [26]. Therefore studies designed at identifying these interactions are presented with experimental design challenges unlikely to be overcome in blood sample focused biorepository [27].
4.2. Limitations
Small sample size is a pervading challenge to pharmacogenetic study design. Even in an immense resource such as BioVU with over 160,000 samples as of July 2013, we were only able to identify 167 patients who met the study criteria, and of those, only 35 of those samples developed CKD stage 4 over the course of calcineurin drug therapy. This finding highlights the need for very large repositories when studying uncommon outcomes of medical interest. While survival analysis did afford us more power opposed to treating the data as strict case-control and performing logistic regression, we were still underpowered to detect an association. For example, assuming a dominant genetic model with an allele with a frequency of 0.5, a sample size of 191 cases of CKD stage 4 would be required to detect an association with a moderately sized hazard ratio of 1.5 at an alpha of 0.05 [28].
Heterogeneity marked another challenge when defining this study population and modeling the association. Clinically, heart transplant recipients are a very diverse population in regards to co-morbidities and medications. Further complicating the issue is that CNIT is not the only cause of CKD in this population: other factors include the decline of kidney function with age, diabetes, hypertension, heart disease, other medication exposures, and latent infection of the BK virus [29]. In this study, we ignored phenotypic heterogeneity to increase the sample size and overall power of the study. Also, to avoid increasing the type II error rate, we were parsimonious in our covariate selection for our statistical model to maximize statistical power [30]. Indeed, large multi-center studies may be required to fully model the relationship between heart disease and kidney function. Large studies will also be required to fully address the phenotype heterogeneity problem or to explore more susceptible subpopulations such as high dose patients, a strategy successfully used to identify genetic variants associated with statin-induced ADRs [31].
4.3. Conclusions
Despite the relatively small sample size for a genetic association study, the current study represents a fairly large sample size for pharmacogenomics studies of ADRs. We have demonstrated here that the EMR, rich in clinical data, is an excellent and logical resource to establish pharmacogenomics studies for less common ADRs such as CNIT. While the genetic association results presented here require replication and downstream functional and biological interpretation, the existence of other biobanks linked to DNA samples in the United States [32] and across the world [33] makes this future direction possible for CNIT as well as other ADRs with a suspected genetic risk factor.
Contributor Information
Matthew Oetjens, Email: matthew.t.oetjens@vanderbilt.edu, Center for Human Genetics Research.
William S. Bush, Email: william.s.bush@vanderbilt.edu, Department of Biomedical Informatics, Center for Human Genetics Research.
Kelly A. Birdwell, Email: kelly.birdwell@vanderbilt.edu, Department of Medicine.
Holli H. Dilks, Email: holli.dilks@chgr.mc.vanderbilt.edu, Vanderbilt Technologies for Advanced Genomics Core Facility.
Erica A. Bowton, Email: erica.a.bowton@vanderbilt.edu, Office of Personalized Medicine.
Joshua C. Denny, Email: josh.denny@vanderbilt.edu, Department of Biomedical Informatics.
Russell A. Wilke, Email: russell.a.wilke@vanderbilt.edu, Department of Medicine, Department of Pharmacology.
Dan M. Roden, Email: dan.roden@vanderbilt.edu, Department of Medicine, Department of Pharmacology, Office of Personalized Medicine.
Dana C. Crawford, Email: crawford@chgr.mc.vanderbilt.edu, Department of Molecular Physiology and Biophysics, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Ave, Nashville, TN 37212, United States of America.
References
- 1.Radovancevic B, Konuralp C, Vrtovec B, et al. J. Heart Lung Transplant. 2005;24:156. doi: 10.1016/j.healun.2003.11.399. [DOI] [PubMed] [Google Scholar]
- 2.Naesens M, Kuypers DR, Sarwal M. Clin. J. Am. Soc. Nephrol. 2009;4:481. doi: 10.2215/CJN.04800908. [DOI] [PubMed] [Google Scholar]
- 3.Hamour IM, Omar F, Lyster HS, Palmer A, Banner NR. Nephrol. Dial. Transplant. 2009;24:1655. doi: 10.1093/ndt/gfn759. [DOI] [PubMed] [Google Scholar]
- 4.Murray B, Hawes E, Lee RA, Watson R, Roederer MW. Pharmacogenomics. 2013;14:783. doi: 10.2217/pgs.13.68. [DOI] [PubMed] [Google Scholar]
- 5.Birdwell KA, Grady B, Choi L, et al. Pharmacogenet. Genomics. 2012;22:32. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haufroid V, Mourad M, Van KV, et al. Pharmacogenetics. 2004;14:147. doi: 10.1097/00008571-200403000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Hesselink DA, van GT, van Schaik RH. Pharmacogenomics. 2005;6:323. doi: 10.1517/14622416.6.4.323. [DOI] [PubMed] [Google Scholar]
- 8.Kuypers DR, Naesens M, de JH, Lerut E, Verbeke K, Vanrenterghem Y. Ther. Drug Monit. 2010;32:394. doi: 10.1097/FTD.0b013e3181e06818. [DOI] [PubMed] [Google Scholar]
- 9.Oetjens MT, Denny JC, Ritchie MD, et al. Pharmacogenomics. 2013;14:735. doi: 10.2217/pgs.13.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roden DM, Pulley JM, Basford MA, et al. Clin. Pharmacol. Ther. 2008;84:362. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Clin. Transl. Sci. 2010;3:42. doi: 10.1111/j.1752-8062.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Jiang M, Oetjens M, et al. J. Am. Med. Inform. Assoc. 2011;18:387. doi: 10.1136/amiajnl-2011-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poggio ED, Wang X, Greene T, Van LF, Hall PM. J. Am. Soc. Nephrol. 2005;16:459. doi: 10.1681/ASN.2004060447. [DOI] [PubMed] [Google Scholar]
- 14.Abboud H, Henrich WL. N. Engl. J Med. 2010;362:56. doi: 10.1056/NEJMcp0906797. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, et al. Am. J Hum. Genet. 2007;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Pac. Symp. Biocomput. 2010;315 [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Nat. Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. Genet. Med. 2010;12:648. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boso V, Herrero MJ, Bea S, et al. Drug Metab Dispos. 2013;41:480. doi: 10.1124/dmd.112.047977. [DOI] [PubMed] [Google Scholar]
- 20.Ware N, MacPhee IA. Curr. Opin. Mol. Ther. 2010;12:270. [PubMed] [Google Scholar]
- 21.Ferraresso M, Tirelli A, Ghio L, et al. Pediatr. Transplant. 2007;11:296. doi: 10.1111/j.1399-3046.2006.00662.x. [DOI] [PubMed] [Google Scholar]
- 22.Vreken P, Van Kuilenburg AB, Meinsma R, Van Gennip AH. Hum. Genet. 1997;101:333. doi: 10.1007/s004390050637. [DOI] [PubMed] [Google Scholar]
- 23.Takai S, Fernandez-Salguero P, Kimura S, Gonzalez FJ, Yamada K. Genomics. 1994;24:613. doi: 10.1006/geno.1994.1680. [DOI] [PubMed] [Google Scholar]
- 24.Lamba J, Hebert JM, Schuetz EG, Klein TE, Altman RB. Pharmacogenet. Genomics. 2012;22:555. doi: 10.1097/FPC.0b013e328351d47f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de DS, Zakrzewski M, Barhdadi A, et al. J. Heart Lung Transplant. 2011;30:326. doi: 10.1016/j.healun.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Hauser IA, Schaeffeler E, Gauer S, et al. J. Am. Soc. Nephrol. 2005;16:1501. doi: 10.1681/ASN.2004100882. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie MD, Denny JC, Crawford DC, et al. Am. J Hum. Genet. 2010;86:560. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoenfeld DA. Biometrics. 1983;39:499. [PubMed] [Google Scholar]
- 29.Bloom RD, Reese PP. J. Am. Soc. Nephrol. 2007;18:3031. doi: 10.1681/ASN.2007040394. [DOI] [PubMed] [Google Scholar]
- 30.Pirinen M, Donnelly P, Spencer CC. Nat. Genet. 2012;44:848. doi: 10.1038/ng.2346. [DOI] [PubMed] [Google Scholar]
- 31.Link E, Parish S, Armitage J, et al. N. Engl. J Med. 2008;359:789. [Google Scholar]
- 32.McCarty CA, Chisholm RL, Chute CG, et al. BMC. Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris JR, Burton P, Knoppers BM, et al. Eur. J. Hum. Genet. 2012;20:1105. doi: 10.1038/ejhg.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]