Abstract
Sodium phenylbutyrate and glycerol phenylbutyrate mediate waste nitrogen excretion in the form of urinary phenylacetylglutamine (PAGN) in patients with urea cycle disorders (UCDs); rare genetic disorders characterized by impaired urea synthesis and hyperammonemia. Sodium phenylbutyrate is approved for UCD treatment; the development of glycerol phenylbutyrate afforded the opportunity to characterize the pharmacokinetics (PK) of both compounds. A population PK model was developed using data from four Phase II/III trials that collectively enrolled patients ages 2 months to 72 years. Dose simulations were performed with particular attention to phenylacetic acid (PAA), which has been associated with adverse events in non-UCD populations. The final model described metabolite levels in plasma and urine for both drugs and was characterized by (a) partial presystemic metabolism of phenylbutyric acid (PBA) to PAA and/or PAGN, (b) slower PBA absorption and greater presystemic conversion with glycerol phenylbutyrate, (c) similar systemic disposition with saturable conversion of PAA to PAGN for both drugs, and (d) body surface area (BSA) as a significant covariate accounting for age-related PK differences. Dose simulations demonstrated similar PAA exposure following mole-equivalent PBA dosing of both drugs and greater PAA exposure in younger patients based on BSA.
Keywords: ammonia, metabolic disorders, nitrogen, orphan disorders, phenylacetic acid
Urea cycle disorders (UCDs) constitute a collection of inherited enzyme and transporter deficiencies that impair the synthesis of urea, the body’s vehicle for waste nitrogen removal via the urine.1–4 UCD patients experience elevated levels of ammonia in blood and brain, which can cause illness ranging from subtle mental abnormalities to coma and death.1–3 Control of hyperammonemia is the main objective of treatment and most UCD patients require strict dietary protein restriction to decrease intestinal ammonia production.4,5
Sodium phenylbutyrate (trade name BUPHENYL® tablets or powder) is approved for the treatment of UCD in patients whose symptoms cannot be managed by diet alone. Glycerol phenylbutyrate, a triglyceride consisting of three molecules of 4-phenylbutyric acid (PBA) joined to glycerol by ester linkage that was approved in 2013 for treatment of UCD patients whose symptoms cannot be controlled by diet alone, has no sodium burden, and offers palatability and pharmacokinetic (PK) advantages over sodium phenylbutyrate.6,7 It is digested by pancreatic lipases, which release PBA.8 PBA delivered by either drug is converted via beta oxidation to phenylacetic acid (PAA), which is then conjugated with glutamine by enzymes in the liver and kidney to form phenylacetylglutamine (PAGN),9 which is excreted in the urine, thereby mediating excretion of waste nitrogen in UCD patients.
The present modeling was undertaken to address two issues. First, PK results in healthy adults demonstrated that plasma metabolite levels were several-fold lower following single dosing with glycerol phenylbutyrate as compared with sodium phenylbutyrate, such that the two compounds were not bioequivalent based on plasma levels even though mole-equivalent PBA doses were administered.8 However, urinary PAGN (UPAGN) excretion, which reflects nitrogen removal, was similar as would be expected based on mole-equivalent PBA dosing.8 Further, studies in UCD patients showed that the correlation between dose and plasma metabolite levels was weaker than that between dose and UPAGN, and the proportion of PBA excreted in urine as PAGN was very similar for the two compounds despite differing plasma profiles.6,7,10 These findings suggest that partial conversion of PBA to PAA and/or PAGN may occur prior to its reaching the systemic circulation.
Second, age-related differences in systemic PAA exposure were identified during dosing with glycerol phenylbutyrate compared to sodium phenylbutyrate. In adults, PAA following sodium phenylbutyrate was significantly lower,11 and in pediatric patients ages 6–17 it was higher (although not significantly).7 While statistical analyses showed no relationship between PAA levels and adverse events (AEs),12 PAA exposure is of potential clinical importance, as intravenous PAA infusion into cancer patients is reportedly associated with reversible AEs (e.g., headache, nausea, vomiting somnolence) at plasma levels ranging from 499 to 1,285 mcg/mL.14,15
Since UCDs constitute an “ultra-orphan” population with an estimated United States patient pool of 1,000 patients,3 popPK modeling and dosing simulations were performed to understand the clinical pharmacology of the two compounds and predict PAA exposure in pediatric and adult UCD patient populations.
METHODS
Software
Nonlinear mixed-effects modeling with NONMEM (version 7.2, Icon Development Solutions, Ellicott City, MD) using the differential equation solver ADVAN13 with FOCE-I or Laplacian (for BQL modeling) estimation was used to develop a popPK model. Parallelization of NONMEM runs using MPI (20 cores) was used to speed up model development reducing the ~4-day runtimes to <6 hours. Processing of NONMEM output was performed using Wings for NONMEM (WFN) software (version 7, Dr. Nick Holford, University of Auckland, NZ). Simulation work was performed using NONMEM in simulation mode. The PsN software (version 3.5.3)18 was used to perform the bootstrap analysis on the final popPK model. NONMEM dataset construction, results processing, and creation of plots were performed using S-PLUS (version 8.1, TIBCO, Palo Alto, CA).
Participants, Trial Design, and Pharmacokinetic Sampling
The data for analysis were derived from 79 UCD patients enrolled in four Phase II/III clinical trials (protocols, UP 1204-003, HPN-100-005, HPN-100-006, and HPN-100-012), the results of which have been previously reported.6,7,11,13 Patients collectively spanned ages 2 months to 72 years and were estimated to represent approximately 20% of the UCD patients in the US taking sodium phenylbutyrate (Table 1).
Table 1.
Summary of Patient Characteristics and PK Sampling by Study
| Protocol Number | UP 1204-003 (N = 9) |
HPN-100-005 (N = 11) |
HPN-100-006 (N = 44) |
HPN-100-012 (N = 15) |
|
|---|---|---|---|---|---|
| Design | Open-label, fixed-sequence, switch-over |
Open-label, fixed-sequence, switch-over |
Pivotal, randomized, double-blind, active-controlled crossover, active-controlled |
Open-label, fixed-sequence, switch-over |
|
| Sex, n (%) | Male | 5 (35.7) | 1 (9.1) | 14 (31.1) | 8 (53) |
| Female | 9 (64.3) | 10 (90.9) | 31 (68.9) | 7 (47) | |
| Weight (kg) | Mean (SD) | 83.1 (33.5) | 41.8 (20.1) | 75.1 (26.3) | 15.3 (4.7) |
| BSA (m2) | Mean (SD) | 1.92 (0.42) | 1.22 (0.34) | 1.83 (0.34) | 0.63 (0.14) |
| Age (years) | Mean (SD) | 35.71 (16.3) | 10.18 (3.9) | 32.73 (13.5) | 2.87 (19) |
| UCD subtype, n (%) | OTC | 12 (85.7) | 9 (81.8) | 40 (88.9) | 3 (20) |
| CPS 1 | 0 | 0 | 2 (4.4) | 0 | |
| ARG | 0 | 0 | – | 1 (7) | |
| ASS | 1 (7.1) | 1 (9.1) | 3 (6.7) | 3 (20) | |
| ASL | 0 | 1 (9.1) | – | 8 (53) | |
| HHH | 1 (7.1) | 0 | – | 0 | |
| Daily dose of NaPBA prior to study (g)a | Mean (SD) | 13.49 (6.08) | 12.41 (4.39) | 14.54 (6.81) | 5.28 (2.45) |
| Dose during study (grams of PBA/day) | NaPBA | 12.22 (4.05) | 10.90 (3.86) | 12.33 (5.58) | 4.63 (2.16) |
| Glycerol phenylbutyrate | 12.36 (3.92) | 11.10 (3.81) | 12.50 (5.53) | 4.78 (2.15) | |
| Measurable data points (percent of values BLQ) for both treatments | Plasma PBA | 159 (31) | 117 (48) | 468 (67) | 65 (92) |
| Plasma PAA | 150 (41) | 128 (34) | 494 (60) | 64 (93) | |
| Plasma PAGN | 189 (0) | 153 (1) | 682 (7) | 104 (27) | |
| Urinary PAGN | 54 (NA) | 42 (NA) | 345 (NA) | NC | |
ARG, arginase deficiency; ASS, argininosuccinate synthetase deficiency; ASL, argininosuccinate lyase deficiency; CPS, carbamoyl phosphate synthetase deficiency; HHH, ornithine translocase deficiency; OTC, ornithine transcarbamylase deficiency; SD, standard deviation; UCD, urea cycle disorder; BLQ, below limits of quantitation; NA, not applicable; NC, not collected.
Each gram of NaPBA contains approximately 0.88 g of PBA.
Prior to study start, patients were taking sodium phenylbutyrate at breakfast, lunch, and dinner with infants and very young children often receiving sodium phenylbutyrate more frequently depending on feedings. Each clinical trial involved a switchover design, whereby patients received a mole-equivalent PBA dose of glycerol phenylbutyrate taken with meals. PK sampling was performed during steady state dosing with either sodium phenylbutyrate or glycerol phenylbutyrate, and prior to switching to the alternate treatment.
Blood sampling was done as frequently as 0, 0.5, 1, 2, 4, 5, 6, 8, 10, 12, 16, 20, and 24 hours following the first dose of the day, although only some of the sampling times were common to all the trials. Each plasma sample was analyzed for PBA, PAA, and PAGN concentration. Twenty-four hour output of urinary PAGN (UPAGN) was used as a surrogate measure of effectiveness, assessed as waste nitrogen removal, that is, 1 mol of PAGN excreted in urine removes 2 mol of nitrogen from the body. Because timed urine collections were not possible for young pediatric UCD patients under the age of 6 (Table 1, protocol HPN-100-012), UPAGN output was not available for this group.
Model Development
The purpose of the present work was to develop an integrated compartmental PK model wherein plasma PBA, PAA, PAGN concentration-time, and urinary PAGN amount-time data following treatment of pediatric and adult UCD patients with either sodium phenylbutyrate or glycerol phenylbutyrate were analyzed simultaneously using nonlinear mixed effects modeling. This approach of modeling different compounds and their data simultaneously was needed to estimate the rates of conversion from prodrug to PBA, PBA to PAA, PAA to PAGN, and PAGN to UPAGN, that is, parent-metabolite relationships. Urinary PAGN data was used to close the mass-balance relationship between treatment dose and final metabolite (PAGN) to improve parameter identifiability. Sodium phenylbutyrate and glycerol phenylbutyrate doses are predominantly (~70%) eliminated as PAGN in the urine.10,11
Compartmental structures evaluated during model development included 1-and 2-compartment models for PBA, PAA, and PAGN considering both systemic disposition and combined systemic and presystemic (conversion to downstream analytes occurs prior to entry into the systemic circulation) disposition. Also tested were linear and saturable formation and elimination processes for PAA and PAGN.
Because the number of parameters being estimated could become large (between 22 and 36) among tested models, every attempt was made during model development to provide sufficient parameter identifiability. In addition, pharmacokinetic measurements below the lower limit of quantitation (BLQ) were included in the NON-MEM dataset and either modeled19–21 or set to one-half the lower limit of quantitation for each analyte (1 mcg/mL). Inclusion of BQL data afforded the model to account for structure based on support that after certain time points for sodium phenylbutyrate and glycerol phenylbutyrate data was not available.
Parameter values were assumed to have a log-normal distribution and between-subject variability (BSV) was modeled as an exponential random-effect model to constrain the individual parameter values to positive numbers. Residual error was tested using a statistical model with additive and/or proportional components.
Some covariate analysis was performed using demographic covariates including body surface area (BSA), body weight, and age; while, disease-specific covariates investigated included plasma glutamine levels, dietary protein intake, and patient age when UCD was diagnosed. These covariates were tested in developing the final model if relationships between the covariate and variability for the PK parameters with BSV (for the base model) showed a clear trend suggesting that inclusion of the covariate might reduce the amount of BSV.
Because of the age range among the patients from the four trials, body size covariates (BSA and body weight) were tested as part of the base model development. A maturation parameter was not included in the modeling due to the sparseness of the data in young patients. Additionally, inclusion of such a parameter to account for maturing metabolic systems was felt unnecessary since initial modeling indicated that age-related changes in conversion were adequately explained by changes in BSA and, presumably, BSA-related changes in organ functional capacity. The effects of weight and BSA were evaluated on CL/F, V/F, and presystemic parameters for PBA, PAA, and PAGN. The influence of body size was tested using linear and power models with the individual’s covariate centered using the typical population median for the covariate, that is, 70 kg for body weight and 1.67 m2 for BSA. For body weight, the exponent is typically fixed to 0.75 for clearance parameters and 1.0 for apparent volume of distribution parameters. Similarly, for BSA, the exponent may be fixed to 0.67. Models were also tested that allowed for the exponent to be estimated.
Model Evaluation
Model selection was based on mechanistic considerations, model parameter estimates, assessment of goodness of fit plots, and statistical estimators. The minimum value of the objective function is typically used to compare models and determine which model explains the data better, but because both nested and non-nested models were tested, the Akaike Information Criterion (AIC) was also used as a statistical estimator among the models being tested. The AIC penalizes a more complex model by taking into account the difference in number of estimated parameters between the compared models.
The final model was evaluated using visual predictive check (VPC) simulations to compare model predicted PAA plasma levels with those observed in the trials. This was achieved by plotting the calculated median and 90% prediction interval of the PAA concentration-time simulated data and superimposing the observed data. In addition, a bootstrap analysis was performed to confirm the standard errors of the estimated model parameters. This was done by creating 250 datasets from the original dataset using a bootstrap algorithm, and using NONMEM to analyze each dataset. PsN was used to calculate the empirical standard error and other summary statistics for each estimated parameter.
Dosing Simulations
In order to predict a range of PAA exposures expected during patient dosing, simulations were performed using the final model and parameter estimates over a range of sodium phenylbutyrate and glycerol phenylbutyrate doses and body sizes. Systemic PAA exposure was assessed both as plasma 24-hour area under the curve (AUC) and as maximal PAA concentration (Cmax). The exposure metrics were compared for different ages, that is, body sizes, and glycerol phenylbutyrate doses as mole-equivalent doses of sodium phenylbutyrate.
The final parameter estimates were used to generate distributions of PK parameters based on population variability and residual measurement error. From these distributions, random draws for simulated PK parameters and simulation scenario specific BSA and doses were used to simulate the concentration-time profile for 1,000 virtual patients. From these simulated data, PAA AUC and Cmax were calculated. Each simulation scenario was comprised of treatment schedules, sampling times, a specified dose for sodium phenylbutyrate and glycerol phenylbutyrate, and a BSA that reflected the age category being explored.
The BSA covariate was simulated using the values from the four clinical trials and additional data on body size in UCD patients from a longitudinal study sponsored by the NIH-funded UCD Consortium.3,17 Assignment of BSA to age categories was done by matching the simulated BSA with BSA ranges based on standard height and weight growth charts (Height & Weight—website).16 Two different doses, including the PBA equivalent of the highest labeled dose for sodium phenylbutyrate (13 g/m2), and a dose equivalent to half the lower end of the labeled sodium phenylbutyrate range (4.9 g/m2) were used as the dose range for simulating exposure.
RESULTS
Patient Demographics and NONMEM Dataset
A summary of the demographic characteristics of the data is presented in Table 1. A higher percentage of female patients took part in three of the four studies. In the ≤6-year-old study (HPN-100-012) the percentage of female patients was similar to male patients (47% vs. 53%). This gender distribution is consistent with the predominance among the older patients of the ornithine transcarbamylase subtype, which is X-linked. The adult studies had mean body weights above the 70 kg typical weight for male adults and the mean age for these studies was >30 years of age. Because PK differences based on sex were not anticipated given the metabolism path for glycerol phenylbutyrate and sodium phenylbutyrate, body size covariates were used to reflect any changes due to females being smaller than males. The predominant UCD subtype was OTC (>81%) for all but the ≤6-year-old study where the ASL subtype (53%) was most common among patients.
The NONMEM dataset was created using dose, concentration, and covariate information from 79 UCD patients ages 2 months to 72 years. The NONMEM dataset contained a total of 3,942 plasma PBA, PAA, PAGN and urine PAGN data points with 3,214 measurable and 728 BQL levels from the sodium phenylbutyrate and glycerol phenylbutyrate treatments. A detailed breakdown of sampling is presented in Table 1.
Population PK Model
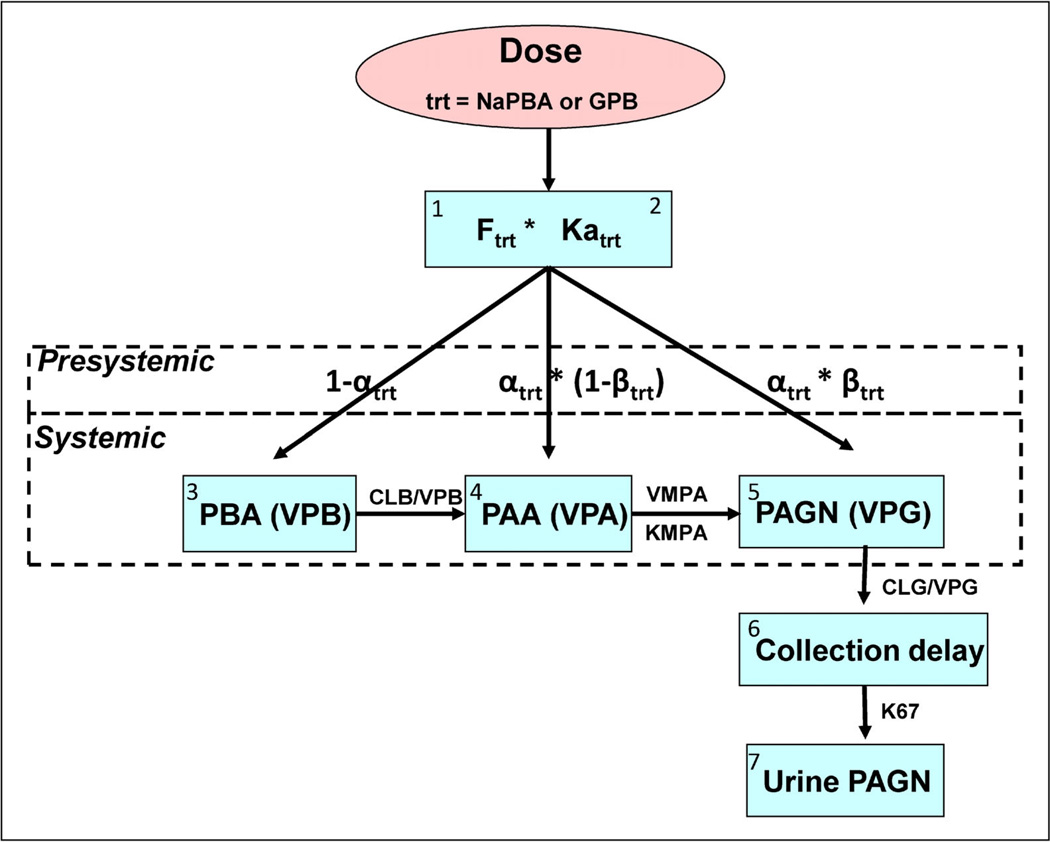
The model best describing the PK for plasma PBA, PAA, PAGN, and UPAGN levels for glycerol phenylbutyrate and sodium phenylbutyrate is shown schematically in Figure 1. This semi-mechanistic model represented each analyte as a one-compartment model parameterized in terms of clearances and volumes. The systemic metabolism of PAA to PAGN was described using a concentration-limited relationship while PBA to PAA metabolism and elimination of PAGN were described using linear processes. Separate first-order absorption parameters were used for each treatment. Because the extent of total dose absorbed (recovery percentage), as reflected by UPAGN output, only varied slightly by treatment and adult versus pediatric populations, the F for each treatment was fixed to mean recovery values using results from studies HPN-100-005 (pediatric) and HPN-100-006 (adult). In addition, gastrointestinal absorption also used a presystemic submodel for metabolism processes that allowed for intestinal/hepatic metabolism of PBA to PAA and PAGN prior to PBA reaching the bloodstream. This submodel used two parameters (α and β) to estimate presystemic conversion, where α estimates the distribution of PBA to plasma and presystemic compartments, and β estimates the proportion of PBA that is converted presystemically to PAA and then to PAGN. Separate α and β parameters were estimated for each treatment.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where PBAin, PAAin, and PAGNin are defined as
| (8) |
| (9) |
| (10) |
Figure 1.
Simultaneous modeling of parent and metabolite data in plasma and urine following administration of two different treatments. The model approximated the biotransformation of GPB and NaPBA; including the absorption of PBA, conversion of PBA to PAA via β-oxidation, enzymatic conjugation of PAA with glutamine to form PAGN and the urinary excretion of PAGN. Conversion of PAA to PAGN is concentration-limited, saturable and described using a Michaelis–Menten relationship; whereas, the rate of β-oxidation of PBA to PAA was hypothesized to be linear due to the widespread prevalence of the enzymes. One hundred percent PBA to PAA conversion via β-oxidation was assumed, that is, no alternative metabolic pathways were included. The elimination rate for PAGN (CLG/VPG) was determined to be a linear process, consistent with renal elimination of PAGN. A delay rate constant (K67) was used to adjust for the delay between appearance of PAGN in the urine compartment and bladder emptying. This model incorporating partial presystemic conversion of PBA to PAA and subsequently to PAGN best described the data.
Mass balance for plasma PBA, PAA, PAGN, and UPAGN was maintained by using three expressions, one for each analyte in the plasma—PBA: (1 − αtrt), PAA: αtrt × (1 − βtrt), and PAGN: αtrt × βtrt—and constraining 0 < α < 1 and 0 < β < 1. Figure 1 is represented mathematically by a system of differential equations (Eqs. 1–10) that simultaneously track changes in amount [A(n), where n is the compartment] of plasma PBA, PAA, PAGN, and urinary PAGN for presystemic and systemic disposition processes.
Body size (expressed as BSA/1.73) was significant on parameters of clearance, volume, and presystemic conversion (α and β) resulting in small BSA individuals having smaller PK values for these parameters compared to individuals with larger BSA values. Thus, younger patients would have lower CL and accumulate PAA more compared to adults. Covariate models for body weight did not produce significant changes in objective function value or AIC compared to covariate models using BSA. Exploratory plots of covariate versus parameter BSV did not indicate significant trends for covariate modeling for the other covariates.
A combined residual error model was used to describe measurement error and model misspecification for measurable levels of each analyte. The additive component was fixed to the BQL (1 mcg/mL) to reduce the number of estimated parameters. Because the percentage of BQL values was high (Table 1), accounting for BQL data were also explored. PBA and PAA following sodium phenylbutyrate treatment had a higher percentage of BQL data, a finding consistent with the faster elimination of these analytes following sodium phenylbutyrate treatment compared to glycerol phenylbutyrate treatment resulting in a higher proportion of BQL measurements at later time points. Using the Beal M3 method resulted in improved objective function values and AIC compared to treating BQL as fixed to ½ BQL levels.
Model Evaluation
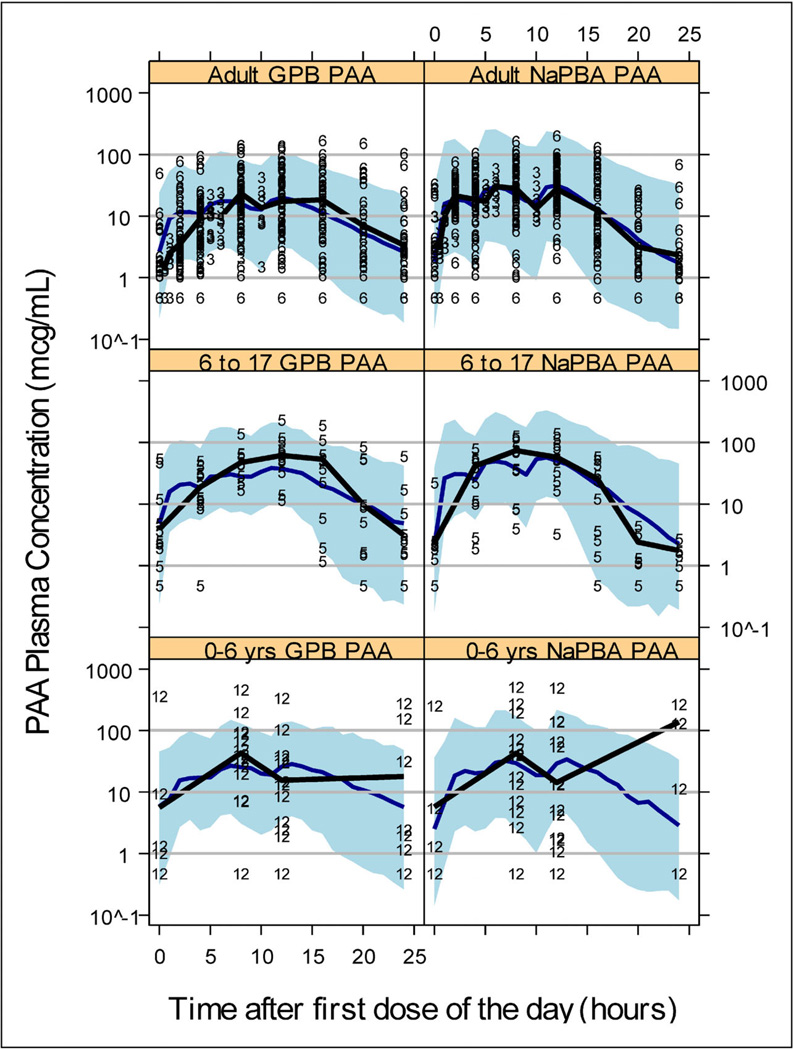
The VPC used an enriched dataset with the same number of individuals and doses as in the original dataset, but plasma sampling was simulated every hour for 24 hours instead of the sampling times used in the actual studies. This dataset was simulated 100 times to calculate a 90% prediction interval based on the estimated noise from BSV and residual error. Figure 2 is the VPC for a 24-hour PAA concentration-time profile stratified by treatment and age group; from UP 1204-003 and HPN-100-006 (Adult), HPN-100-005 (6–17 years), and HPN-100-012 (0–6 years). The adult and 6-to 17-year-old panels capture the spread and central tendency of the data well as seen by comparing the black and blue lines representing median observed and simulated PK profiles; respectively. The VPC of the 0-to 6-year olds also looks good given the data sparseness. The VPC in Figure 2 suggests the model describes PAA levels across treatments and ages from 2 months to 72 years by accounting for differences in absorption processes, amount of presystemic conversion that occurs, and accounting for body size impact on disposition of these nitrogen-scavenging compounds.
Figure 2.
Model qualification plots using visual predictive check for PAA and stratified by treatment and age group (0–6 years, 6–17 years, and adults). The symbols in the plots represent the study number from which the data were derived (3 = UP-1204-003, 5 = HPN-100-005, 6 = HPN-100-006, and 12 = HPN-100-012; see Table 1). The blue solid line is the VPC median profile and the thicker solid black line is the median profile from the study data contained in the respective plot. The shaded region represents the 90% prediction interval. LLOQ = 1 mcg/mL and symbols below LLOQ are presented as ½ LLOQ for display purposes.
Failure to produce a variance-covariance matrix made NONMEMcalculation of parameter uncertainty impossible. Instead, the relative standard errorwas calculated empirically from a bootstrap analysis using 250 samples of the original dataset. Table 2 compares the NONMEM final parameter estimates with those calculated from the bootstrap analysis. The low %CV for the bootstrap results suggests parameter uncertainty was small. The bootstrap calculated %BSV was lower compared to what NONMEM estimated for the patient-to-patient variability; and the NONMEM estimated and bootstrap calculated mean for the PK parameters were similar. The VPC and bootstrap parameter values both showed that the model was a good description of the data.
Table 2.
Final Parameter Estimates and Results from Bootstrap Analysis
| NONMEM | Bootstrap | |||||||
|---|---|---|---|---|---|---|---|---|
| Parameters | Units | Status | Mean | %BSV | Mean | %CV | %BSV | %CV |
| Presystemic | ||||||||
| F1 NaPBA pediatric | % | Fixed | 69 | 16 | ||||
| F2 GPB pediatric | % | Fixed | 66 | 16 | ||||
| F1 NaPBA adult | % | Fixed | 71 | 18 | ||||
| F2 GPB adult | % | Fixed | 69 | 17 | ||||
| KA1 NaPBA | 1/h | 1.4 | 71 | 1.407 | 5 | 50 | 12 | |
| KA2 GPB | 1/h | 0.331 | 55 | 0.333 | 5 | 30 | 14 | |
| Alpha NaPBA × (BSA/1.73) | 0.109 | – | 0.11 | 5 | – | – | ||
| Alpha GPB × (BSA/1.73) | 0.352 | – | 0.35 | 5 | – | – | ||
| Beta NaPBA × (BSA/1.73) | 7890 | – | 7850 | 6 | – | – | ||
| Beta GPB × (BSA/1.73) | 4.39 | – | 4.39 | 6 | – | – | ||
| Systemic | ||||||||
| PBA | ||||||||
| VPB × (BSA/1.73) | L | 12.4 | 71 | 12.3 | 6 | 51 | 9 | |
| CLB × (BSA/1.73) | L/h | 12.7 | 34 | 12.6 | 5 | 12 | 12 | |
| Residual proportional | % | 66 | 44 | 9 | ||||
| Residual additive | µM | Fixed | 6.1 | |||||
| PAA | ||||||||
| VPA × (BSA/1.73) | L | 30.8 | 62 | 31.0 | 8 | 38 | 13 | |
| VMPA × (BSA/1.73) | µmol/h | 5260 | – | 5274 | 3 | |||
| KMPA | µM | 193 | – | 194.1 | 4 | |||
| Residual proportional | % | 53 | 27 | 8 | ||||
| Residual additive | µM | Fixed | 7.3 | |||||
| PAGN | ||||||||
| VPG × (BSA/1.73) | L | 23.4 | 31 | 23.5 | 4 | 10 | 9 | |
| CLG × (BSA/1.73) | L/h | 10.6 | 21 | 10.9 | 5 | 5 | 9 | |
| Residual proportional | % | 34 | 12 | 10 | ||||
| Residual additive | µM | Fixed | 3.8 | |||||
| UPAGN | ||||||||
| K67 | 1/h | 1.02 | – | 1.02 | 5 | |||
| Residual proportional | % | 50 | 24 | 9 | ||||
| Residual additive | µM | Fixed | 3.8 | |||||
PK Parameters
Table 3 compares PK differences between age groups (calculated from the empirical Bayes estimates of each patient’s parameter values). The absorption rate for each treatment across age groups showed that glycerol phenylbutyrate absorption rate (KA2) was always slower compared to the sodium phenylbutyrate absorption rate (KA1). As expected with body-sized adjusted PK parameters, the parameter estimate increased as the age increased. The presystemic conversion across all age groups was greater following glycerol phenylbutyrate compared to sodium phenylbutyrate treatment. The conversion from drug to analytes presystemically and the slower absorption of glycerol phenylbutyrate is a likely cause for the analyte plasma profiles following dosing with glycerol phenylbutyrate being lower compared to the analyte plasma profiles following dosing with sodium phenylbutyrate. That is, the slower absorption of PBA when delivered as glycerol phenylbutyrate “trickles” the analytes into the systemic circulation compared to a “bolus-like” input of analytes into the systemic circulation following sodium phenylbutyrate treatment.
Table 3.
PK Parameters
| Age Groups (N) | |||||||
|---|---|---|---|---|---|---|---|
| 0–2 (3) | 3–5 (10) | 6–11 (7) | 12–17 (4) | Adult (53) | |||
| Description | Parameter | Units | Mean (CV%) | ||||
| Formulation-dependent parameters | |||||||
| Absorption rate constant (NaPBA) | KA1 | 1/h | 1.34 (56) | 1.45 (40) | 1.88 (39) | 1.8 (33) | 1.43 (51) |
| Absorption rate constant (GPB) | KA2 | 1/h | 0.31 (48) | 0.42 (24) | 0.44 (39) | 0.3 (53) | 0.35 (54) |
| PBA % metabolized presystemically (NaPBA) | Alpha/(1 + alpha) × 100 | % | 3 (25) | 4 (15) | 6 (17) | 9 (19) | 10 (17) |
| PBA % metabolized presystemically (GPB) | Alpha/(1 + alpha) × 100 | % | 8 (24) | 12 (14) | 17 (15) | 24 (16) | 27 (14) |
| PAA % metabolized to PAGN presystemically (NaPBA) | Beta/(1 + beta) × 100 | % | 100 | 100 | 100 | 100 | 100 |
| PAA % metabolized to PAGN presystemically (GPB) | Beta/(1 + beta) × 100 | % | 53 (13) | 63 (6) | 72 (5) | 79 (4) | 82 (3) |
| PBA | |||||||
| Clearance for PBA | CLB | L/h | 3.5 (23) | 5.9 (31) | 6.6 (27) | 11.8 (23) | 13.6 (38) |
| Volume of distribution for PBA | VPB | L | 3.6 (39) | 6.2 (100) | 9.2 (99) | 9.2 (40) | 14.7 (52) |
| PAA | |||||||
| Vmax for PAA to PAGN conversion | VMPA | µmol/h | 1368 (26) | 2092 (16) | 3166 (18) | 4697 (21) | 5559 (19) |
| Km for PAA to PAGN conversion | KMPA | µM | 193 | 193 | 193 | 193 | 193 |
| Clearance for PAA calculated as Vmax/Km (linear portion) | L/h | 7.1 | 10.9 | 16.4 | 24.4 | 28.9 | |
| Volume of Distribution for PAA | VPA | L | 6.6 (44) | 12.8 (35) | 15.7 (38) | 57.6 (85) | 47.2 (71) |
| PAGN | |||||||
| Clearance for PAGN | CLG | L/h | 3 (23) | 5 (22) | 7.7 (12) | 12.7 (18) | 11.9 (25) |
| Volume of distribution for PAGN | VPG | L | 6.1 (26) | 8.7 (20) | 18.7 (46) | 20.9 (18) | 24.6 (24) |
Note: Km is the same for all age groups because no variability parameter (BSV) was estimated for KMPA.
The clearances and volumes for PBA and PAGN combined with the saturable formation of PAGN (i.e., PAA clearance) impact the ability of a UCD patient to clear PAA from the circulation. These PK processes are also scaled by body size and account to some degree for the higher PAA levels seen in smaller patients, that is, smaller body size ~ less capacity to convert PAA to PAGN. Even with these PK explanations of the differences seen in exposures among the age groups, it is interesting to note that accumulation of PAA is not appreciable among any of the age groups.9,11,13
Dosing Simulations
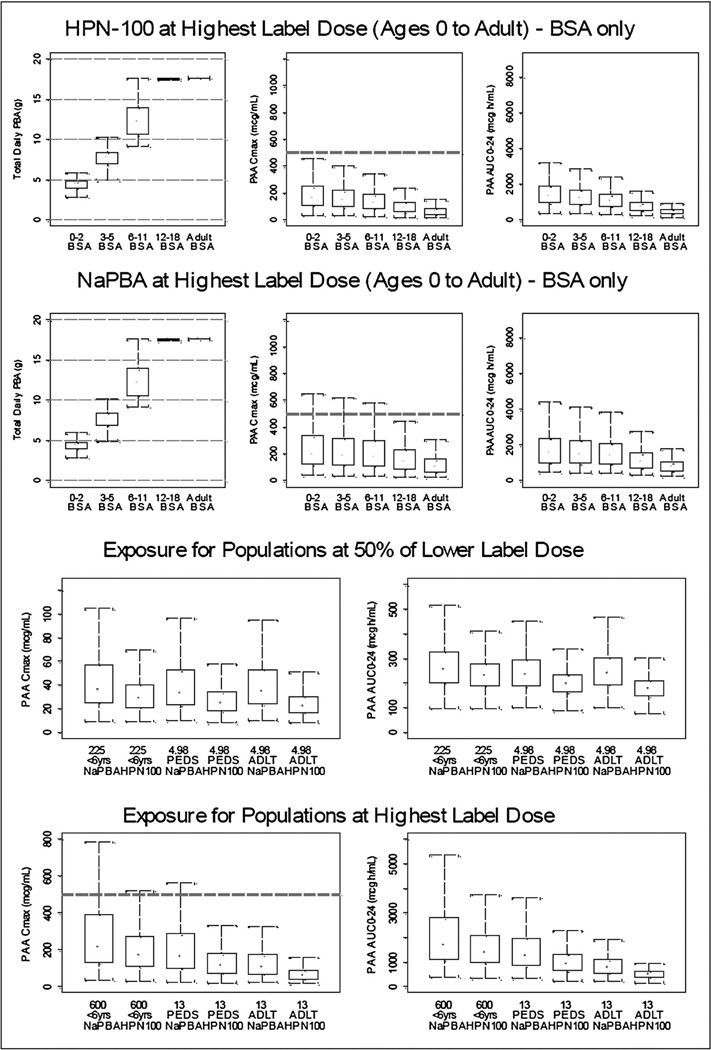
Figure 3 depicts the PAA exposure across 5 age categories, newborns through adults, dosed at the PBA equivalent amount at the top of the labeled range (top panel) as well as at half the lower end of the labeled range. Dosing at the PBA equivalent of one half of the lower end of the sodium phenylbutyrate labeled range yields predicted median PAA exposure, assessed as Cmax, which is similar for the two drugs and many-fold below the range (499–1,285 mcg/mL) at which reversible AEs have been reported in cancer patients.14,15 Even the upper 95% confidence interval for PAA Cmax for both drugs is below 200 mcg/mL. Dosing at the PBA equivalent of the upper end of the range also yields median PAA exposure, which is similar for the two drugs (albeit tending to be lower with glycerol phenylbutyrate) and generally below 200 mcg/mL for all age groups. However, the upper 95% CI for Cmax frequently extends above 500 mcg/mL in the pediatric age groups, particularly among the younger patients and during dosing with sodium phenylbutyrate. The predicted urinary output of PAGN for UCD patients ≥6 years of age agreed generally well with that measured by non-compartmental analysis.6,7,12
Figure 3.
Simulated PAA exposure for different age groups. The top panel depicts dose (left panels) as well as PAA exposure assessed as Cmax (middle panels), and PAA AUC (right panels) for various age groups based on dosing at the PBA equivalent of the maximum labeled range for NaPBA (13 g/m2/day). The bottom panel depicts side by side comparisons of PAA exposure assessed as Cmax (left panels), and PAA AUC (right panels) at the PBA equivalents of half the lower end of the labeled NaPBA range (4.98 g/day; top) and at the top of the NaPBA labeled range (13 g/m2/day). The bottom and top of each box represents the 25th and 75th percentiles for the distribution. The ends of the whiskers approximate the 5th and 95th percentiles. The black dot is the median PAA Cmax and AUC for the conditions of the simulations. The dashed horizontal line represents a Cmax of 500 mcg/Ml HPN-100 = Glycerol Phenylbutyrate.
DISCUSSION
The final model described the behavior of the two drugs using a presystemic model; that is, a model allowing for first pass or presystemic conversion of PBA to PAA and/or PAGN. The presystemic, first pass metabolismmost likely occurs in the liver, which is enzymatically equipped both to convert PBA to PAA and/or PAGN.9
Presystemic conversion explains the unusual PK behavior of both drugs; that is, the variability in plasma metabolite profiles and differences in systemic exposure despite similar urinary excretion of PAGN.6,8,10 The model further demonstrates that PBA is absorbed (appears in the systemic circulation) more slowly when administered as glycerol phenylbutyrate as compared with sodium phenylbutyrate. This difference in absorption rate likely reflects the fact that unlike sodium phenylbutyrate, which is a salt that requires no digestion, glycerol phenylbutyrate is a triglyceride, the digestion of which requires pancreatic lipases and likely occurs largely after the drug has exited the stomach.8 This difference in PBA absorption also explains the lesser fluctuation in plasma metabolite levels, more even day versus night urinary excretion of PAGN as well as the better overnight ammonia control during dosing with glycerol phenylbutyrate as compared with sodium phenylbutyrate.6,7,11,13
This difference in PBA absorption rate also helps explain the difference in plasma metabolite levels observed with single dose administration of the two drugs.8 That is, the slower absorption of PBA when delivered as glycerol phenylbutyrate may better “meter” PBA release into the splanchnic circulation such that the liver can more effectively metabolize to PAA and/or PAGN as compared with “bolus-like” release of PBA which occurs with sodium phenylbutyrate dosing.
The metabolite clearance rates and estimated apparent volumes of distribution for PBA and PAGN combined with the saturable conversion of PAA to PAGN (i.e., PAA clearance) affect the ability of a UCD patient to clear PAA from the circulation. The PAA to PAGN conversion rate also scales with body size and helps account for the higher PAA levels seen in smaller patients, likely because of smaller organs, in particular liver, and, therefore, lesser metabolic capacity. Even with the slower PAA metabolism in children, it is interesting to note that PAA reaches steady state in ~3 days with no further accumulation.
The dosing simulations are useful in predicting PAA exposure for various age groups, and, in particular, assessing the potential for plasma PAA concentrations in the range reportedly associated with transient AEs and overall PAA exposure, assessed as AUC. The saturable, BSA-dependent conversion of PAA to PAGN is manifested as the generally higher PAA exposure observed in smaller (lower BSA) patients during maximal dosing. While median PAA levels are well below 500 mcg/mL, even at the maximal dose and in the youngest patients, the upper 95% confidence intervals suggest the theoretical possibility that some pediatric patients would be exposed to PAA values exceeding 500 mcg/mL. Although interrogation of the clinical data from patients participating in the studies suggests no relationship between PAA levels and these transient AEs,12 the nonspecific and common nature of the AEs (e.g., headache, nausea) is such that it is difficult to exclude PAA as a contributing factor to such AEs in an individual patient, and monitoring of PAA levels may be appropriate during dosing of either drug in selected circumstances, for example, compatible signs and symptoms of PAA toxicity in a pediatric patient receiving a high drug dose which are not explained by elevated blood ammonia or other intercurrent illnesses.
Finally, the present work illustrates the utility of popPK modeling and dosing simulations to understand drug behavior in “ultra orphan” populations, where large pharmacokinetic studies are not feasible because of limited patient numbers.
Acknowledgments
The authors gratefully acknowledge and thank the efforts of the Investigators who made these trials possible, including J. Bartley (Long Beach Memorial Hospital), A. Feigenbaum (Hospital for Sick Children, Toronto, Ontario, Canada), N. Longo (The University of Utah), W. Berquist (Stanford University), R. Gallagher (Children’s Hospital Colorado), D. Bartholomew (Nationwide Children’s Hospital), C.O. Harding (Oregon Health Sciences University), M.S. Korson (Tufts Medical Center), S.E. McCandless (Case Western Reserve University), W. Smith (Maine Medical Center), J. Vockley (Children’s Hospital of Pittsburgh), S. Bart (SNBL), D. Kronn (Westchester Medical Center), R. Zori (University of Florida), Sandesh Sreenath-Nagamani (Baylor College of Medicine), M. Summar (Children’s National Medical Center) and the study staffs, including N. Schrager (Mount Sinai School of Medicine), A. Donovan, J. Crawford, Pediatric TRU Staff, K. Defouw, J. Balliet (The Medical College of Wisconsin), M. Keuth, N. O’Donnell (Long Beach Memorial Hospital), M. Hussain, E. Bailey, A. Orton, M. Ambreen (The Hospital for Sick Children, University of Toronto, ON, Canada), C. Bailey, A. Lang (The University of Utah), J. Perry, V. de Leon, A. Niemi, K. Cusmano (Stanford University), T. Carlson, J. Parker (University of Minnesota), S. Burr (Children’s Hospital Colorado), K. Simpson (Children’s National Medical Center), K. Regis (Nationwide Children’s Hospital), A. Behrend, T. Marrone (Oregon Health Sciences University), N. Dorrani (University of California, Los Angeles), C. Heggie (Case Western Reserve University), S. Mortenson (Maine Medical Center), S. Deward (Children’s Hospital of Pittsburgh), K. Bart, C. Duggan (SNBL), K. Murray, C. Dedomenico (Tufts Medical Center), C. Gross (University of Florida), L. Brody (Seattle Children’s Hospital), M. Mullins, S. Carter, A. Tran, J. Stuff, TCH General Clinical Research Center nursing staff (Baylor), Kathy Lisam (Hyperion), as well as the Clinical and Translational Science Awards/General Clinical Research Center Grants (Baylor College of Medicine, M01RR00188; Case Western Reserve University, UL1RR024989; Clinical and Translational Science Institute at Children’s National Medical Center NIH/NCRR, UL1RR31988; Medical College of Wisconsin, UL1RR31973; Mount Sinai School of Medicine, UL1RR29887; Oregon Health & Science University, UL1RR24140; Stanford University, UL1RR25744; Tufts University, UL1RR25752; University of California, Los Angeles, UL1RR33176; University of Colorado, UL1RR25780; University of Florida, UL1RR29890; University of Minnesota, UL1RR33183; University of Pittsburgh, UL1RR24153, UL1TR000005; University of Utah, UL1RR25764; University of Washington, UL1RR25014), the Urea Cycle Disorders Consortium (NIH Grant U54RR019453) and grants from the O’Malley Foundation, Kettering Fund, and Rotenberg Fund which provided support. S.C.S. Nagamani is an awardee of the National Urea Cycle Disorders Foundation Research Fellowship.
Footnotes
Declaration of Conflicting Interest
D.F. Coakley, K. Dickinson, M. Mokhtarani, and B.F. Scharschmidt were employees of Hyperion at the time of the study. None of the other authors have a financial interest in Hyperion, although payments were made by Hyperion to Mt. Sinai School of Medicine (G. Diaz, PI), University of Minnesota (S.A. Berry, PI), Children’s National Medical Center (U. Lichter-Konecki, PI) and Baylor College of Medicine (B. Lee, PI) for services provided in the conduct of the study. The analyses performed by Pharsight Corporation (J. Monteleone) were supported by Hyperion Therapeutics.
References
- 1.Brusilow SW, Maestri NE. Urea cycle disorders: diagnosis, pathophysiology, and therapy. Adv Pediatr. 1996;43:127–170. [PubMed] [Google Scholar]
- 2.Summar ML, Dobbelaere D, Brusilow S, Lee B. Diagnosis, symptoms, frequency and mortality of 260 patients with urea cycle disorders from a 21-year, multicentre study of acute hyper-ammonaemic episodes. Acta Paediatr. 2008;97:1420–1425. doi: 10.1111/j.1651-2227.2008.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuchman M, Lee B, Lichter-Konecki U, et al. Cross-sectional multicenter study of patients with urea cycle disorders in the United States. Mol Genet Metab. 2008;94:397–402. doi: 10.1016/j.ymgme.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batshaw ML, Brusilow S, Waber L, et al. Treatment of inborn errors of urea synthesis: activation of alternative pathways of waste nitrogen synthesis and excretion. N Engl J Med. 1982;306:1387–1392. doi: 10.1056/NEJM198206103062303. [DOI] [PubMed] [Google Scholar]
- 5.Enns GM, Berry SA, Berry GT, Rhead WJ, Brusilow SW, Hamosh A. Survival after treatment with phenylacetate and benzoate for urea-cycle disorders. N Engl J Med. 2007;356:2282–2292. doi: 10.1056/NEJMoa066596. [DOI] [PubMed] [Google Scholar]
- 6.Lee B, Rhead W, Diaz GA, et al. Phase 2 comparison of a novel ammonia scavenging agent with sodium phenylbutyrate in patients with urea cycle disorders: safety, pharmacokinetics and ammonia control. Mol Genet Metab. 2010;100:221–228. doi: 10.1016/j.ymgme.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichter-Konecki U, Diaz GA, Merritt JL, II, et al. Ammonia (NH3) control in children with urea cycle disorders (UCDs); Comparison of sodium phenylbutyrate and glycerol phenylbutyrate. Mol Genet Metab. 2011;103:323–329. doi: 10.1016/j.ymgme.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGuire BM, Zupanets I, Lowe ME, et al. Pharmacology and safety of glycerol phenylbutyrate in healthy adults and adults with cirrhosis. Hepatology. 2010;51:2077–2085. doi: 10.1002/hep.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moldave K, Meister A. Synthesis of phenylacetylglutamine by human tissue. J Biol Chem. 1957;229:463–476. [PubMed] [Google Scholar]
- 10.Mokhtarani M, Diaz GA, Rhead W, et al. Urinary phenylacetylglutamine as dosing biomarker for patients with urea cycle disorders. Mol Genet Metab. 2012;107:308–314. doi: 10.1016/j.ymgme.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz GA, Krivitzsky LS, Mokhtarani M, et al. Ammonia control and neurocognitive outcome among urea cycle disorder patients treated with glycerol phenylbutyrate. Hepatology. 2013 doi: 10.1002/hep.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mokhtarani M, Diaz GA, Rhead W, et al. Elevated phenylacetic acid (PAA) levels appears linked to neurological adverse events in healthy adults but not in urea cycle disorder (UCD) patients. Mol Genet Metab. 2012;105:342. (Abstract presented at the 2012 meeting of the Society for Inherited Metabolic Diseases in Charlotte, March 31–April 3, 2012). [Google Scholar]
- 13.Smith W, Diaz GA, Lichter-Konecki U, et al. Ammonia (NH3) control in children ages 2 months through 5 years with urea cycle disorders (UCDs); Comparison of sodium phenylbutyrate and glycerol phenylbutyrate. J Pediatr. 2013;162(6):1228–1234. doi: 10.1016/j.jpeds.2012.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thibault A, Cooper M, Figg W, et al. A phase I and pharmacokinetic study of intravenous phenylacetate in patients with cancer. Cancer Res. 1994;54:1690–1694. [PubMed] [Google Scholar]
- 15.Thibault A, Samid D, Cooper MR, et al. Phase I study of phenylacetate administered twice daily to patients with cancer. Cancer. 1995;75:2932–2938. doi: 10.1002/1097-0142(19950615)75:12<2932::aid-cncr2820751221>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Height and Weight Growth Charts. ( http://www.halls.md/chart/child-growth/pediatric.htm). [Google Scholar]
- 17.Summar M, Tuchman M. Proceedings of a consensus conference for the management of patients with urea cycle disorders. J Pediatr. 2001;138:S6–S10. doi: 10.1067/mpd.2001.111831. [DOI] [PubMed] [Google Scholar]
- 18.Lindbom L, Pihlgren P, Jonsson EN. PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed. 2005;79(3):241–257. doi: 10.1016/j.cmpb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Duval V, Karlsson M. Impact of omission or replacement of data below the limit of quantification on parameter estimates in a two-compartment model. Pharm Res. 2002;19(12):1835–1840. doi: 10.1023/a:1021441407898. [DOI] [PubMed] [Google Scholar]
- 20.Ahn JE, Karlsson M, Dunne A, Ludden T. Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokinet Pharmacodyn. 2008;35:401–421. doi: 10.1007/s10928-008-9094-4. [DOI] [PubMed] [Google Scholar]
- 21.Bergstrand M, Karlsson M. Handling data below the limit of quantification in mixed effect models. AAPS J. 2009;11(2):371–380. doi: 10.1208/s12248-009-9112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]