Abstract
Objective
Mutations in profilin-1 (PFN1) have recently been identified in patients with amyotrophic lateral sclerosis (ALS). Because of the considerable overlap between ALS and the common subtype of frontotemporal dementia, which is characterized by transactive response DNA-binding protein 43 pathology (FTLD-TDP), we tested cohorts of ALS and FTLD-TDP patients for PFN1 mutations.
Methods
DNA was obtained from 342 ALS patients and 141 FTLD-TDP patients at our outpatient clinic and brain bank for neurodegenerative diseases at the Mayo Clinic Florida, Jacksonville, USA. We screened these patients for mutations in coding regions of PFN1 by Sanger sequencing. Subsequently, we used TaqMan genotyping assays to investigate the identified variant in 1167 control subjects.
Results
One variant, p.E117G, was detected in 1 ALS patient, 1 FTLD-TDP patient, and 2 control subjects. The mutation frequency of patients versus control subjects was not significantly different (p-value = 0.36). Moreover, PFN1 and TDP-43 staining of autopsy material did not differ between patients with or without this variant.
Conclusion
The p.E117G variant appears to represent a benign polymorphism. PFN1 mutations, in general, are rare in ALS and FTLD-TDP patients.
Keywords: Amyotrophic lateral sclerosis, frontotemporal dementia, profilin-1, TDP-43, genetics
Introduction
Recently, exome sequencing revealed mutations in profilin-1 (PFN1) in patients with amyotrophic lateral sclerosis (ALS) (1). ALS is the most common motor neuron disease, and is caused by progressive loss of both upper- and lower motor neurons (2). In the last twenty years, mutations in many genes have been reported in ALS patients, including mutations in superoxide dismutase-1 (SOD1), angiogenin (ANG), TAR DNA-binding protein (TARDBP), fused in sarcoma / translated in liposarcoma (FUS), vesicle-associated membrane protein B (VAPB), optineurin (OPTN), and valosin-containing protein (VCP) (3, 4).
Frontotemporal dementia (FTD) is the third most common cause of early-onset dementia, and it results from degeneration of the frontal and temporal cortex (5–8). A pathological link between ALS and FTD has been established in 2006, when transactive response DNA-binding protein 43 (TDP-43) was identified as the ubiquitinated pathological protein in ALS and in the majority of FTD patients, now referred to as frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) (9, 10). In these FTLD-TDP patients, mutations in VCP and progranulin (GRN) have already been described (11, 12).
The recent discovery of hexanucleotide repeat expansions in chromosome 9 open reading frame 72 (C9ORF72) (13, 14), which are present in ~34% of patients with familial ALS and also in ~26% of patients with familial FTD (15), provided further evidence for a shared underlying pathogenesis. In this study, we assessed the frequency of PFN1 mutations in both ALS patients (n = 342) and FTLD-TDP patients (n = 141).
Material and Methods
Study population
Our ALS study population consisted of 342 patients. Material was obtained through the Mayo Clinic Florida (Clinic and Brain Bank). Patients were clinically diagnosed according to the El Escorial criteria (16). Their diagnosis was pathologically confirmed in 52 patients (15%) and 43 of those were known to have TDP-43 pathology (83%). TDP-43 subtypes were determined for 30 patients, all of whom showed TDP-43 subtype 3 pathology (100%). We included 166 females (49%) and 176 males (51%); 314 were Caucasian (91%), 22 were African/American (6%), 3 were Hispanic (1%), 1 was Asian (<1%), 1 was Arabic (<1%), and 1 was Native Hawaiian/Pacific Islander (<1%). Thirty-seven patients were known to have a positive family history for motor neuron diseases (12%), of whom 12 had C9ORF72 repeat expansions (32%), and 3 had SOD1 mutations (8%). Our entire ALS cohort included 36 patients with known mutations (11%): 28 had C9ORF72 repeat expansions (8%), 4 had SOD1 mutations (1%), 2 had FUS mutations (<1%), and 2 had TARDBP mutations (<1%). We refer to Table 1 for more information regarding our patient characteristics.
Table 1.
Baseline characteristics of ALS cohort
| Cohort (N) | Male/Female (N)(%) (N=342) | Age at Onset (y)(CI) (N=312) | Bulbar Site of Onset (N)(%) (N=313) | Duration (y)(CI) (N=312) | FH Motor Neuron Disease (N)(%)(N=313) | FH Neuro-degeneration (N)(%)(N=313) | Neuropathology (N)(%) (N=342) | TDP-43 Type (1/2/3)(%) (N=30) | Known Mutations (N)(%) |
|---|---|---|---|---|---|---|---|---|---|
| 342 | 176/166 (51/49) | 58.8 (57.6–60.0) | 74/239 (24/76) | 3.4 (3.1–3.8) | 37/276 (12/88) | 157/156 (50/50) | 52/290 (15/85) | 0/0/30 (0/0/100) | 36 (11%) |
Abbreviations: ALS = amyotrophic lateral sclerosis, N = number, y = years, CI = 95% confidence interval, and FH is family history. To estimate disease duration, intervals between age at onset and age at death or between age at onset and current age were calculated.
In addition, we included 141 FTLD-TDP patients from the Mayo Clinic Jacksonville Brain Bank (Table 2), primarily ascertained through The State of Florida Alzheimer’s Disease Initiative funded by the Department of Elder Affairs, The Einstein Aging Study, The Udall Center for Excellence in Parkinson’s Disease Research, CurePSP/The Society for Progressive Supranuclear Palsy, the Mayo Alzheimer’s Disease Patient Registry (ADPR) and the Florida Alzheimer’s Disease Research Center (ADRC). TDP-43 proteinopathy subtypes were determined for 128 patients (91%): type 1 was observed in 77 patients (60%), type 2 in 23 patients (18%), and type 3 in 28 patients (22%). Sixty-five were female (46%), and 76 were male (54%). Among patients with known ethnicities, 139 were Caucasian (99%), and 1 was Hispanic (<1%). Of the 141 FTLD-TDP patients in our cohort, 54 patients had known mutations (38%): 28 had C9ORF72 repeat expansions (20%), 24 had GRN mutations (17%), 1 had a VCP mutation (<1%), and 1 had a leucine-rich repeat kinase 2 (LRRK2) mutation (<1%). Thirty-four patients showed signs of motor neuron disease on autopsy (24%).
Table 2.
Baseline characteristics of FTLD-TDP and control cohorts
| Method | Cohort | Number (N) | Male/Female (N)(%) | Age * (y)(CI) | Neuropathology (N)(%) | TDP-43 Type (1/2/3)(%) | Known Mutations (N)(%) |
|---|---|---|---|---|---|---|---|
| Sequencing | FTLD-TDP | 141 | 76/65 (54/46) | 73.1 (71.3–74.9) | 141/0 (100/0) | 77/23/28 (60/18/22) | 54 (38%) |
| TaqMan | CON | 1167 | 536/631 (46/54) | 70.8 (70.2–71.5) | N/A | N/A | N/A |
Abbreviations: FTLD-TDP = frontotemporal lobar degeneration with TDP-43 pathology, CON = control subjects, and N/A = not applicable.
Age at death is shown for FTLD-TDP patients, and age at sampling for control subjects.
The PFN1 p.E117G (c.350-351AA>GT) mutation identified in this study was tested in an additional cohort of 1167 control subjects (Table 2). These subjects were Caucasian and ascertained through the Mayo Clinic in Jacksonville, Florida, and Scottsdale, Arizona.
Sequencing analysis
Coding regions of PFN1 were screened for mutations with M13-tailed primers (eTable 1). For exon 2 and 3, amplicons were generated with a 60°C – 50°C touchdown PCR using Apex Taq and Standard Buffer (Genesee Scientific, San Diego, California). For exon 1, the temperature was raised to 67°C and reactions contained Q-solution (Qiagen, Hilden, Germany). All PCR products were purified with the Agencourt AMPure system (Beckman Coulter, Brea, California), and then sequenced in both directions using M13 sequencing primers and BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, California). After purification with Agencourt CleanSEQ (Beckman Coulter), products were run on an ABI 3730xl Genetic Analyzer (Applied Biosystems). Sequence analysis was performed using Sequencher 4.8 software (Gene Codes, Ann Arbor, Michigan). The impact of mutations on the structure and function of PFN1 was predicted with PolyPhen-2 (PolyPhen-2 version 2.2.2; http://genetics.bwh.harvard.edu/pph2/), PMut (http://mmb.pcb.ub.es/PMut/PMut.jsp), and SIFT (http://sift.jcvi.org/www/SIFT_seq_submit2.html).
A custom TaqMan SNP genotyping assay (Applied Biosystems) was designed to test the p.E117G (c.350-351AA>GT) mutation, and data was analyzed on ABI Prism 7900 Detection System (Applied Biosystems) using SDS version 2.2.2 software (Applied Biosystems). For statistical analysis, Chi-square tests were calculated (p-value < 0.05).
Immunohistochemistry
Immunohistochemistry for PFN1 (1:750, rabbit polyclonal, Novus Biologicals, Littleton, Colorado) and TDP-43 (1:2,500, rabbit polyclonal, Mayo Clinic, Jacksonville, Florida) (17) was performed on sections from the spinal cord and temporal cortex. Five μm thick sections were used, cut from formalin-fixed paraffin-embedded blocks, deparaffinized in xylene, rehydrated with washes in a graded series of ethanol, and thoroughly washed in distilled water. All stains were processed on DAKO Autostainer Plus (DAKO, Carpinteria, California) with DAKO EnVision™ + System-horseradish peroxidase (diaminobenzidine). Normal goat serum (1:20 in Tris-Buffered Saline and Tween 20 [TBST]; Sigma, St. Louis, Missouri) was added to slides prior to the primary antibody to block nonspecific antibody binding.
Results
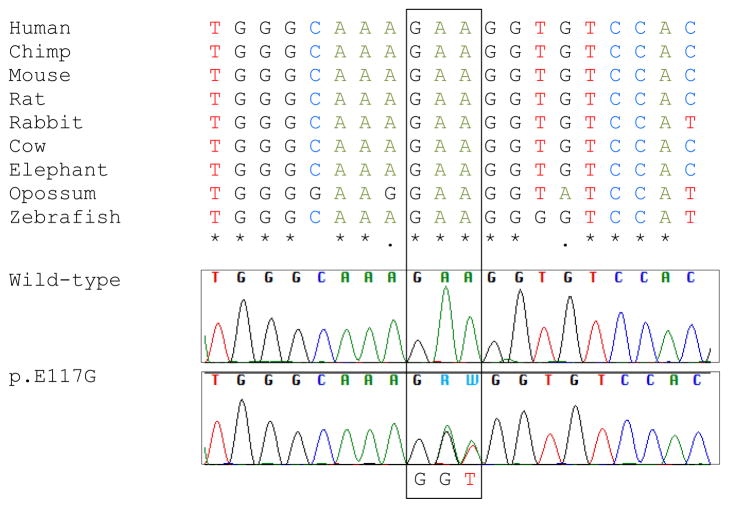
In our cohort of 342 ALS patients and 141 FTLD-TDP patients, we only detected the previously reported consecutive base-pair change p.E117G (c.350-351AA>GT). This variant is located in a relatively well conserved region (Figure 1); although SIFT predicts p.E117G to affect protein function, both PolyPhen-2 and PMut predict benign effects. We identified p.E117G in one ALS patient and one FTLD-TDP patient (Table 3). The apparently sporadic ALS patient was of Caucasian origin; she did not show any signs of FTD, and died at 81 years of age. The FTD patient was of Caucasian origin as well, and she developed the behavioral variant of FTD at 69 years of age. She died without signs of motor neuron disease at 75 years of age; no family members with FTD were reported. Material of both these patients was obtained through the Mayo Clinic Brain Bank, and more detailed clinical information was unavailable.
Figure 1. Conservation and chromatogram of p.E117G.
Conservation of amino-acid residues across species was generated using Clustal Omega online tool, http://www.ebi.ac.uk/Tools/msa/clustalo/.
Table 3.
Clinical characteristics of subjects with p.E117G variants
| Cohort | Diagnosis | Gender | Age at Onset (y) | Age at Death (y) | Age at Sampling (y) | Family History |
|---|---|---|---|---|---|---|
| ALS | ALS | Female | N/A | 81 | 81 | No |
| FTLD-TDP | bvFTD | Female | 69 | 75 | 75 | No |
| CON | Normal | Female | N/A | N/A | 65 | N/A |
| CON | Normal | Female | N/A | N/A | 87 | N/A |
Abbreviations: bvFTD = behavioral variant of frontotemporal dementia. More detailed clinical information was unavailable for these patients.
To further assess this variant we performed TaqMan analysis for 1167 control subjects. In this cohort, we detected the p.E117G variant in two control subjects (Table 3). In total, we observed p.E117G in 2 out of 483 patients (0.4%) and in 2 out of 1167 control subjects (0.2%), and this difference was not significant (p-value = 0.36 [for ALS cohort (0.3%) versus controls (0.2%): p-value = 0.66, and for FTLD-TDP cohort (0.7%) versus controls (0.2%): p-value = 0.21]).
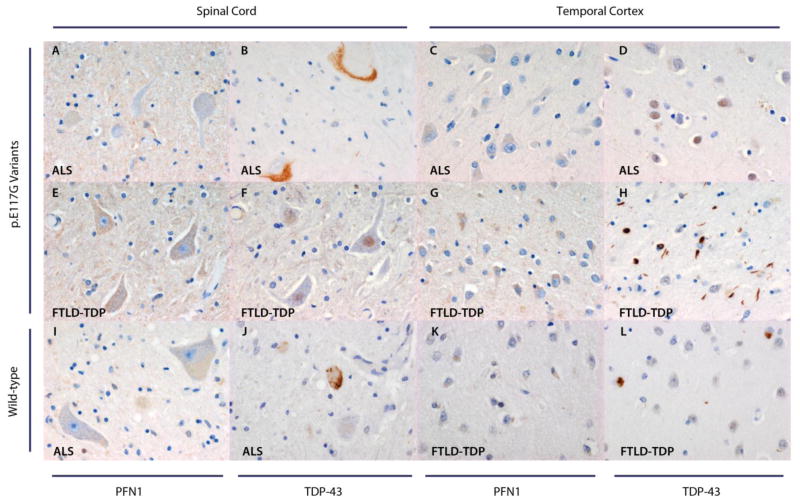
To determine whether PFN1 and TDP-43 staining varied between patients with and without the p.E117G variant, we performed immunohistochemistry on brain tissue of the two patients with this variant and two patients without this variant. Neurons did not contain PFN1-positive inclusions in patients with the p.E117G variant or in patients with wild-type PFN1, even though these patients did harbor TDP-43 positive neuronal inclusions (Figure 2).
Figure 2. PFN1 expression in patients with and without p.E117G variants.
Spinal cord motor neurons of the two patients with p.E117G variants lack PFN1 inclusions (A, E). Temporal cortex for these two patients does not demonstrate neuronal PFN1 inclusions either (C, G). Two patients without the p.E117G variant, one of whom was diagnosed with ALS (I) and one with FTLD-TDP (K), who were photographed as controls for this variant, display the same pattern as patients with p.E117G variants in the spinal cord and temporal cortex, respectively. For the two patients with p.E117G variants, TDP-43 staining is shown both in the spinal cord (B, F) and temporal cortex (D, H): the ALS patient demonstrates TDP-43 positive inclusions in the spinal cord (TDP-43 type 3, B), whereas the FTLD-TDP patient shows TDP-43 positive inclusions in the temporal cortex (TDP-43 type 2, H). TDP-43 positive inclusions are also present in the spinal cord of the wild-type ALS patient (TDP-43 type 3, J), and in the temporal cortex of the wild-type FTLD-TPD patient (TDP-43 type 1, L). Magnification: 40×.
Discussion
In this study, we screened 342 ALS patients and 141 FTLD-TDP patients for mutations in PFN1, and identified p.E117G in two patients (0.4%). The p.E117G variant, however, was also present in 2 out of 1167 control subjects (0.2%), and consequently, there was no significant difference in mutation frequency between patients and control subjects (p-value = 0.36). Although our study is the first to specifically include a large cohort of FTD patients with a pathologically confirmed diagnosis of TDP-43 proteinopathy, the apparent lack of pathogenic PFN1 mutations in familial and sporadic ALS and FTD patients is in agreement with several recent reports (18–23).
Profilins were discovered in the seventies (24), and are one of the most important actin-binding proteins (25–27). They are also engaged in transcriptional activity of RNA polymerases, chromatin remodeling, and nuclear stability (28–30). Furthermore, interactions with survival motor neuron (SMN), fragile X mental retardation protein (FMRP), VCP, and huntingtin (htt) have been reported (31–39). It seems likely, therefore, that profilins are also involved in various aspects of RNA processing pathways, which have already been implicated in the pathogenesis of both ALS and FTD (40, 41).
Recently, exome sequencing identified PFN1 mutations (p.C71G and p.M114T) in two large ALS families with dominant inheritance patterns (1). Sequence analysis of all available family members, suggested a high degree of penetrance. Subsequent screening of 272 familial ALS cases revealed five other cases with PFN1 variants (2%). All PFN1 mutants displayed an onset in the limbs, and among these mutants was one ALS patient with a p.E117G variant who developed symptoms at 40 years of age. Screening of 816 sporadic ALS cases identified two additional patients with this p.E117G variant who developed limb onset ALS at 63 and 33 years of age, and thus, this variant was present in 3 out of 1090 ALS patients (0.3%). One subject with a p.E117G variant was also found in a cohort of 1089 control subjects (0.1%), indicating that p.E117G may represent a less pathogenic mutation. Western blot analysis of transfected N2A cells expressing this variant, which were subjected to NP-40-soluble and insoluble fractionation, did indeed display a pattern similar to wild-type PFN1 (1). Moreover, primary motor neurons (PMNs) expressing this mutant did not demonstrate aggregates, and immunoprecipitation and western blot analysis of cells transfected with p.E117G did not show a reduction of bound actin relative to wild-type PFN1, as opposed to other PFN1 mutants (1). It was also shown, however, that p.E117G was able to form aggregates in N2A cells, and that exposure to proteasome inhibitor MG132, both in N2A cells and in PMNs, caused moderate aggregate levels and raised insoluble protein levels, whereas wild-type PFN1 only displayed minimal aggregate- and insoluble protein levels (1). To further investigate this p.E117G variant, we performed immunohistochemistry on autopsy material of two patients with this variant and two patients without it. We showed that PFN1 and TDP-43 staining did not differ between patients with and without p.E117G.
To date, six studies have also investigated the frequency of PFN1 mutations in ALS and/or FTD patients (18–23). This resulted in the identification of one novel PFN1 mutation, p.T109M (19), but no other PFN1 mutations were detected, apart from the p.E117G variant. The p.E117G variant was present in two additional ALS patients and three additional FTD patients (18, 19, 22), one of whom also carried a pathogenic frameshift mutation in GRN (p.A303GfsX14) (22). When all PFN1 studies are combined, the p.E117G variant has been reported in 10 out of 4737 patients with ALS and/or FTD (0.2%, including 6 out of 3867 patients with a primary diagnosis of ALS [0.2%] and 4 out of 870 patients with a primary diagnosis of FTD with or without ALS [0.5%]), and in 6 out of 4607 control subjects (0.1%).
In conclusion, PFN1 mutations appear to be uncommon in ALS patients and FTLD-TDP patients. We did detect one PFN1 variant, p.E117G, but there was no significant difference in mutation frequency between our patients and control subjects. Furthermore, PFN1 and TDP-43 staining of autopsy material demonstrated a comparable pattern in patients with and without this variant. Although in silico prediction programs and previously reported functional studies generated conflicting results, our present study suggests that p.E117G represents a rare benign polymorphism.
Acknowledgments
We acknowledge financial support from NIH grants P50 AG016574 (RP, BB, DSK, NRG-R, DWD, RR), P50 NS072187 (ZKW, DWD, RR), R01 NS065782 (RR), R01 NS080882 (RR), R01 AG026251 (RR), R01 AG037491 (KAJ), and R01 AG031581 (RC), the ALS Therapy Alliance (RR), the Consortium for Frontotemporal dementia (CFR [RR]), and the Mayo Clinic Center for Regenerative Medicine (ZKW).
Footnotes
Financial Disclosure: none reported.
References
- 1.Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–55. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 3.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 4.van Blitterswijk M, van Es MA, Hennekam EA, Dooijes D, van Rheenen W, Medic J, et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:3776–84. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- 5.Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and neuropathological criteria for frontotemporal dementia. The Lund and Manchester Groups. J Neurol Neurosurg Psychiatry. 1994;57:416–8. doi: 10.1136/jnnp.57.4.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 8.McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol. 2001;58:1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 9.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 10.Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–8. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohrer JD, Warren JD. Phenotypic signatures of genetic frontotemporal dementia. Curr Opin Neurol. 2011;24:542–9. doi: 10.1097/WCO.0b013e32834cd442. [DOI] [PubMed] [Google Scholar]
- 12.Seelaar H, Rohrer JD, Pijnenburg YA, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2011;82:476–86. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 13.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Blitterswijk M, Dejesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25:689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–9. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 17.Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106:7607–12. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiloca C, Ticozzi N, Pensato V, Corrado L, Del Bo R, Bertolin C, et al. Screening of the PFN1 gene in sporadic amyotrophic lateral sclerosis and in frontotemporal dementia. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingre C, Landers JE, Rizik N, Volk AE, Akimoto C, Birve A, et al. A novel phosphorylation site mutation in profilin 1 revealed in a large screen of US, Nordic, and German amyotrophic lateral sclerosis/frontotemporal dementia cohorts. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lattante S, Le Ber I, Camuzat A, Brice A, Kabashi E. Mutations in the PFN1 gene are not a common cause in patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration in France. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Zou ZY, Sun Q, Liu MS, Li XG, Cui LY. Mutations in the profilin 1 gene are not common in amyotrophic lateral sclerosis of Chinese origin. Neurobiol Aging. 2013 doi: 10.1016/j.neurobiolaging.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 22.Dillen L, Van Langenhove T, Engelborghs S, Vandenbulcke M, Sarafov S, Tournev I, et al. Explorative genetic study of UBQLN2 and PFN1 in an extended Flanders-Belgian cohort of frontotemporal lobar degeneration patients. Neurobiol Aging. 2013 doi: 10.1016/j.neurobiolaging.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Daoud H, Dobrzeniecka S, Camu W, Meininger V, Dupre N, Dion PA, et al. Mutation analysis of PFN1 in familial amyotrophic lateral sclerosis patients. Neurobiol Aging. 2013;34:1311, e1–2. doi: 10.1016/j.neurobiolaging.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Carlsson L, Nystrom LE, Lindberg U, Kannan KK, Cid-Dresdner H, Lovgren S. Crystallization of a non-muscle actin. J Mol Biol. 1976;105:353–66. doi: 10.1016/0022-2836(76)90098-x. [DOI] [PubMed] [Google Scholar]
- 25.Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–83. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 26.Yarmola EG, Bubb MR. How depolymerization can promote polymerization: the case of actin and profilin. Bioessays. 2009;31:1150–60. doi: 10.1002/bies.200900049. [DOI] [PubMed] [Google Scholar]
- 27.Schluter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 28.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–49. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 29.Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–42. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16:191–6. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Caraballo-Miralles V, Cardona-Rossinyol A, Garcera A, Villalonga P, Soler RM, Olmos G, et al. SMN deficiency attenuates migration of U87MG astroglioma cells through the activation of RhoA. Mol Cell Neurosci. 2012;49:282–9. doi: 10.1016/j.mcn.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Nolle A, Zeug A, van Bergeijk J, Tonges L, Gerhard R, Brinkmann H, et al. The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet. 2011;20:4865–78. doi: 10.1093/hmg/ddr425. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A, Lambrechts A, Hao le T, Le TT, Sewry CA, Ampe C, et al. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp Cell Res. 2005;309:185–97. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem. 1999;274:37908–14. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 35.Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31:1427–39. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, et al. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–63. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 37.Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–76. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol. 2008;28:5196–208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skare P, Kreivi JP, Bergstrom A, Karlsson R. Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res. 2003;286:12–21. doi: 10.1016/s0014-4827(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 40.Baloh RH. How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other? Curr Opin Neurol. 2012;25:701–7. doi: 10.1097/WCO.0b013e32835a269b. [DOI] [PubMed] [Google Scholar]
- 41.van Blitterswijk M, Landers JE. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics. 2010;11:275–90. doi: 10.1007/s10048-010-0239-4. [DOI] [PubMed] [Google Scholar]