Abstract
The vertebrate gut microbiota evolved in an environment typified by periodic fluctuations in nutrient availability, yet little is known about its responses to host feeding and fasting. Because many model species (e.g., mice) are adapted to lifestyles of frequent small meals, we turned to the Burmese python, a sit-and-wait foraging snake that consumes large prey at long intervals (>1 month), to examine the effects of a dynamic nutrient milieu on the gut microbiota. We employed multiplexed 16S rRNA gene pyrosequencing to characterize bacterial communities harvested from the intestines of fasted and digesting snakes, and from their rodent meal. In this unprecedented survey of a reptilian host, we found that Bacteroidetes and Firmicutes numerically dominated the python gut. In the large intestine, fasting was associated with increased abundances of the genera Bacteroides, Rikenella, Synergistes, and Akkermansia, and reduced overall diversity. A marked postprandial shift in bacterial community configuration occurred. Between 12 hours and 3 days after feeding, Firmicutes, including the taxa Clostridium, Lactobacillus, and Peptostreptococcaceae, gradually outnumbered the fasting-dominant Bacteroidetes, and overall ‘species’-level diversity increased significantly. Most lineages appeared to be indigenous to the python rather than ingested with the meal, but a dietary source of Lactobacillus could not be ruled out. Thus, the observed large-scale alterations of the gut microbiota that accompany the Burmese python's own dramatic physiological and morphological changes during feeding and fasting emphasize the need to consider both microbial and host cellular responses to nutrient flux. The Burmese python may provide a unique model for dissecting these interrelationships.
Keywords: digestion, fasting, gastrointestinal bacteria, host-microbe interactions, nutrient deprivation, reptile
Introduction
The gut microbiotas of terrestrial mammals are among the most widely and intensively surveyed of vertebrate-hosted microbial communities. Indeed, a large and diverse microbiota inhabits the mammalian gastrointestinal tract where its composition is shaped, in part, by host diet and phylogeny (Dethlefsen et al., 2007, Ley et al., 2008a, Ley et al., 2008b and the majority of bacteria belong to one of two dominant phyla: the Bacteroidetes or Firmicutes (Ley et al., 2005, Ley et al., 2008a. Among its many important functions, the gut microbiota is involved in degrading otherwise indigestible components of the host's diet (e.g., plant polysaccharides) into compounds that the host can absorb (e.g., short-chain fatty acids) (Flint et al., 2008). In the absence of this microbial fermentation, calories present in a diverse array of complex dietary glycans would be lost to the host. Interestingly, when dietary polysaccharides are in short supply (e.g., if removed from the diet or, ostensibly, after fasting), certain gut-adapted bacteria are capable of alternatively foraging host mucosal glycans (Salyers et al., 1977; Sonnenburg et al., 2005; Bjursell et al., 2006; Martens et al., 2008; Martens et al., 2009). It has been proposed that this flexible foraging strategy may serve to enhance ecosystem stability in a dynamic nutrient environment (Yachi and Loreau, 1999; Sonnenburg et al., 2005).
In humans and mice, increased adiposity has been associated with alterations in the gastrointestinal microbiota that allow it to more efficiently liberate energy from the host's diet, an arrangement that might benefit the host if food resources were only intermittently available (Backhed et al., 2004; Ley et al., 2005; Turnbaugh et al., 2006; Samuel et al., 2008,Turnbaugh et al., 2009a). A corollary hypothesis, that the microbiota assumes a different configuration when nutrients are scarce, and that this state also confers some benefit to the host, has also been posited. While the latter, nutrient-deprived scenario is less well understood, a recent study has revealed that a 24-hr fast in mice produces a marked shift in gut microbial ecology, and that the presence of a gut microbiota while fasting enhances nutrient supply to the heart (Crawford et al., 2009). Further elucidating the interrelationships between the gut microbiota's composition and function, and the host's diet, nutritional status, and cellular physiology would benefit from the study of a wider variety of host species in which each of these parameters could be either intentionally or naturally varied over a broad range.
Many model species, including humans and mice, are adapted to lifestyles of frequent small meals. These species' gastrointestinal tracts typically contain food and are rarely empty and, as a consequence, their postprandial metabolic responses are regulated over relatively narrow ranges (Secor and Diamond, 1998). By contrast, sit-and-wait foraging snakes such as the Burmese python, Python molurus, consume enormous meals at long intervals. As an apparent adaptation to extended periods without food, pythons exhibit exceptionally large physiological and morphological responses to feeding and fasting, features that have made them an attractive model species in studies of the regulation of host processes (Secor et al., 1994; Secor and Diamond, 1995; Secor and Diamond, 1998; Starck and Beese, 2001; Secor, 2008). Here, we consider whether changes in the composition and structure of the Burmese python's gut microbiota accompany this reptile host's other, well-documented postprandial responses.
Burmese pythons are native to southeastern Asia (and invasive in Puerto Rico and southern Florida) where they feed largely on birds and mammals (Pope, 1961; Snow et al., 2007). Adult pythons are among the largest snakes, reaching 6.5 m in length and 100 kg; however, juveniles weigh only 0.1-1.0 kg and consume rats and mice, making them tractable experimental animals (Pope, 1961; Secor and Diamond, 1998). In the wild, pythons employ the sit-and-wait tactic of foraging that is characterized by long intervals (>1 month) between meals and prey that can exceed 50% of the snake's body mass (Pope, 1961; Murphy and Henderson, 1997). Accordingly, the python's gastrointestinal tract is well-adapted to long periods of quiescence punctuated by the digestion of extremely large, intact prey (Secor, 2008). Intriguingly, almost nothing is known about the composition of the Burmese python's gut microbiota or, for that matter, the gut microbiota of most reptiles (Hill et al., 2008).
In the fasted state, the Burmese python's gut and other organs exhibit dramatic alterations in structure and function (Secor, 2008). The ingestion of a meal signals the python's stomach to begin secreting digestive acid and enzymes (Secor, 2003) and for the atrophied small intestine to upregulate brush-border digestive enzymes and nutrient transporters (Secor and Diamond, 1995; Secor, 2008). During the digestion of a meal, the small intestine doubles in mass, reflecting both hyperplastic and hypertrophic responses in its epithelium. These changes within the gut show regional specificity: a comparable increase in mass does not occur in the large intestine. Feeding and fasting also have extra-intestinal manifestations: heart, liver, pancreas and kidney masses increase by 40-106% within 3 days after feeding, although the roles played by hyperplastic and hypertrophic mechanisms in these responses remain undefined (Secor and Diamond, 2000; Andersen et al., 2005).
Peak changes in small intestinal morphology [a 5-fold lengthening of the microvilli (as documented by transmission electron microscopy)] and function (a 3- to 13-fold increase in hydrolase activity and nutrient uptake capacity) occur after 2-3 days of digestion, at which point unabsorbed material begins to fill the python's large intestine (Secor and Diamond, 1995; Lignot et al., 2005; Cox and Secor, 2008; Secor, 2008). Six to seven days after feeding, as the last of the meal exits the stomach and traverses the small intestine, downregulation begins, and by day 10, enzyme activities, nutrient uptake rates, and organ masses have decreased significantly (Secor and Diamond, 1995; Lignot et al., 2005; Cox and Secor, 2008; Secor, 2008). Thus, downregulation with the final passage of the meal appears to occur as quickly as upregulation upon ingestion.
In the present study, we surveyed bacterial communities resident within the small and large intestines of fasted Burmese pythons and at 8 time points following the ingestion of a rodent meal weighing 25% of the snake's body mass. Prior to experimentation, the 32 juvenile pythons we examined had fasted for a minimum of 30 days. Post-feeding samples were collected at 0.25, 0.5, 1, 2, 3, 4, 6, and 10 days, and 3-4 snakes were sacrificed at each time point. Contents were harvested from the proximal and distal regions of the small and large intestine and from the cecum, an extension of the large intestine that forms a blind-end pouch at the junction with the small intestine. We also characterized the bacterial assemblages consumed with the python's rodent meal. For each sample, bacterial 16S rRNA genes were PCR-amplified using a primer set with a unique error-correcting barcode (Hamady et al., 2008). Coupling this barcoding approach with high-throughput pyrosequencing, we were able to survey the gut microbiota of a non-mammal vertebrate host at an unprecedented level of detail. We were also able to examine the effects of an extreme fluctuation in nutritional status on the gut bacterial communities of a host species uniquely adapted to this lifestyle.
Materials and Methods
Animals
Captive-born hatchling pythons were purchased commercially and housed individually in 20-liter plastic containers at 28-32°C under a 14h:10h light:dark cycle. Snakes were fed laboratory rats every 2 weeks and had continuous access to water. Rats (RodentPro.com® 7-wk old CD strain fed proprietary chow) were purchased frozen and thawed in clean warm water prior to feeding. Prior to experimentation, food was withheld from snakes for a minimum of 30 days to ensure that the snakes were postabsorptive. Burmese pythons are known to complete their digestion within 10-14 days after feeding (Secor and Diamond, 1995). Snakes used in this study were of both sexes, between 12 and 36 months old, and had body masses averaging 817 ± 38 g (s.d.). The University of Alabama Institutional Animal Care and Use Committee approved all procedures involving the animals used in this study.
Sample collection
Snakes were killed by severing the spinal cord immediately posterior to the head. Following death, a mid-ventral incision was made to expose the gastrointestinal tract, which was removed. The contents of the cecum and the proximal and distal regions of the small and large intestine were emptied into sterile vials, immediately frozen in liquid N2, and stored at -80°C. Gut content mass was determined by weighing the organ before and after emptying. The average masses of contents harvested from the large and small intestine at each time point are shown in Figure 1a and Supplementary Figure 3a, respectively. In fasted snakes, the small intestine contains a small amount of clear fluid, presumably secretions from the gall bladder, pancreas, and intestine. Here, several milliliters of sterile saline were used to flush the fasted small intestinal contents into collection vials. After feeding and before day 10, the small intestine contained enough material to be squeezed directly out into vials. The large intestine typically contains some material, including residual feces, fluid, and urate originating from the kidneys in fasted snakes. In the present study, material from the meal began to enter the large intestine ∼1 day post-feeding, with the cecum filling at 2 days. At day 3 and beyond, the large intestine and cecum were fairly filled with material.
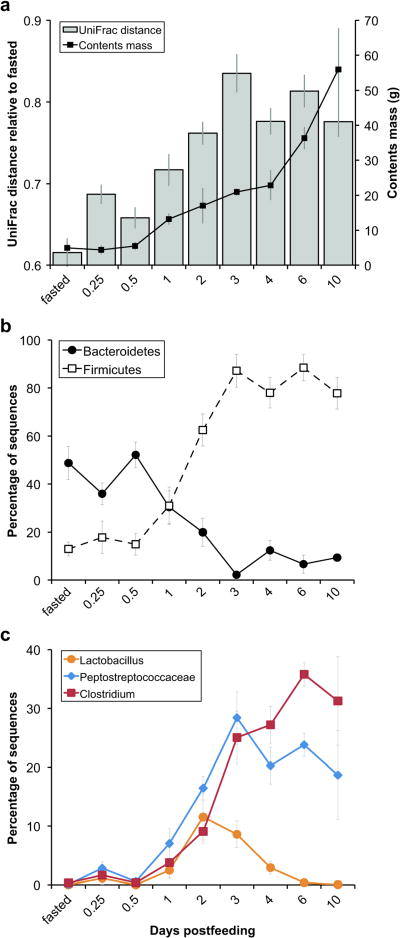
Figure 1.
Digestion alters bacterial community membership and taxon relative abundances within the Burmese python large intestine. (a) Average mass of large intestinal contents harvested at each time point (right axis) and unweighted UniFrac distance between fasted and fed communities and, for the fasted time point, between fasted samples only (left axis). (b and c) Average proportional abundance of sequences classified as (b) Bacteroidetes or Firmicutes or as (c) selected Firmicutes taxa at each time point. Because other lineages can also be present, the proportions need not add up to 100%. Error bars indicate one standard error of the mean. Fasted snakes were fed a rodent meal equal to 25% of their body mass. Contents mass shown in (a) does not include the cecum, which contained ∼3-4 g of material on days 2 through 10.
Frozen feeder rats were thawed overnight to 4°C. Each thawed rat was homogenized for 3-5 min in 750 ml of autoclaved water using a 1-gal commercial blender on the highest speed setting (Waring model CB10, Torrington, CT, USA). Immediately after blending, sterile cotton swabs were dipped into the whole rat homogenates (5 replicates per animal), packaged into sterile tubes, and frozen at -80°C.
DNA extraction
Snake gut contents (100 mg) were suspended in 500 μl of buffer A (200 mM Tris, pH 8.0, 200 mM NaCl, 20 nM EDTA), followed by addition of 210 μl of 20% SDS and 500 μl of a 1:1 mixture of phenol/chloroform. Five hundred microliters of zirconium beads were added to each tube, which were then placed in a bead beater on the highest speed setting for 2 min (Biospec, Bartlesville, OK, USA). The mixture was centrifuged at 6,000 × g at 4°C for 3 min and DNA was purified from the resulting supernatant via phenol/chloroform extraction, followed by ethanol precipitation.
For rat homogenate samples, genomic DNA was extracted from cotton swabs using the MO BIO PowerSoil DNA Isolation kit (Carlsbad, CA, USA) with modifications. Like the extraction protocol described above, this method involves the disruption of cells by bead beating in the presence of a buffered SDS solution. The cotton tips of frozen swabs were broken off directly into bead tubes to which 60 μl of Solution C1 had been added. Tubes were incubated at 65°C for 10 minutes and then shaken horizontally at maximum speed for 2 min using the MO BIO vortex adapter. The remaining steps were performed as directed by the manufacturer.
PCR amplification of the V2 region of bacterial 16S rRNA genes
We amplified variable region 2 (V2) of the 16S rRNA gene using a previously published primer set (Fierer et al., 2008). The forward primer (5′-GCC TTG CCA GCC CGC TCA GTC AGA GTT TGA TCC TGG CTC AG-3′) contained the 454 Life Sciences primer B sequence, the broadly conserved bacterial primer 27F, and a two-base linker sequence (‘TC’). The reverse primer (5′-GCC TCC CTC GCG CCA TCA GNN NNN NNN NNN NCA TGC TGC CTC CCG TAG GAG T-3′) contained the 454 Life Sciences primer A sequence, a unique 12-nt error-correcting Golay barcode used to tag each PCR product (designated by NNNNNNNNNNNN), the broad-range bacterial primer 338R, and a ‘CA’ linker sequence inserted between the barcode and the rRNA primer.
For python samples, PCR reactions were carried out in quadruplicate 20 μl reactions with 0.3 μM forward and reverse primers, 100 ng gel purified template DNA (Qiaquick kit, Qiagen, Valencia, CA, USA), and 1× HotMasterMix (5 PRIME, Gaithersburg, MD, USA). Thermal cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 20 sec, 52°C for 20 sec, and 65°C for 1 min. Replicate amplicons were pooled and cleaned using Ampure magnetic purification beads (Agencourt, Danvers, MA, USA).
For rat samples, PCR reactions were carried out in triplicate 25 μl reactions with 0.6 μM forward and reverse primers, 3 μl template DNA, and 1× HotMasterMix. Thermal cycling consisted of 94°C for 3 min, followed by 35 cycles of 94°C for 45 sec, 50°C for 30 sec, and 72°C for 90 sec, with a final extension of 10 minutes at 72°C. Replicate amplicons were pooled and cleaned using the UltraClean-htp 96-well PCR Clean-up kit (MO BIO).
Amplicon quantitation, pooling, and pyrosequencing
Amplicon DNA concentrations were determined using either the bisbenzimide H assay (Sigma, St. Louis, MO, USA; used for snake samples) or the Quant-iT PicoGreen dsDNA reagent and kit (Invitrogen, Carlsbad, CA, USA; used for rat samples). Following quantitation, amplicons were pooled in equimolar ratios and cleaned. Amplicon pyrosequencing was carried out using primer A on a 454 Life Sciences Genome Sequencer FLX instrument (Roche, Branford, CT, USA).
Sequence analysis
Sequences were processed using the QIIME software package (Caporaso et al., 2010). Sequences were removed from the analysis if they had a mean quality score <25, were <200 or >300 nt in length, contained ambiguous characters, did not contain the primer sequence, contained a homopolymer run exceeding 8 nt, or contained an uncorrectable barcode. The remaining sequences were assigned to samples by examining the 12-nt barcode. Similar sequences were clustered into operational taxonomic units (OTUs) using cd-hit (Li and Godzik, 2006) with a minimum coverage of 99% and a minimum identity of 97%. A representative sequence was chosen from each OTU by selecting the most abundant sequence. Representative sequences were aligned against the Greengenes (DeSantis et al., 2006) core set using PyNAST (Caporaso et al., 2009) with a minimum alignment length of 150 and a minimum identity of 75%. The PH Lane mask was used to screen out hypervariable regions after alignment. A phylogenetic tree was inferred using FastTree (Price et al., 2009) with Kimura's 2-parameter model. Taxonomy was assigned to each unique sequence (i.e., sequences representative of OTUs picked at 100% sequence identity) using the RDP classifier with a minimum support threshold of 80% and the RDP taxonomic nomenclature (Wang et al., 2007). Statistical analyses were performed in R. The mean (± s.e.m.) is reported in the text unless otherwise noted.
Alpha and beta diversity
To evaluate the amount of diversity contained within samples (alpha diversity), we constructed rarefaction plots from both branch length-based phylogenetic diversity (PD) measurements (Faith, 1992) and OTU-based measures using QIIME. For each sample, the mean of 10 iterations per sub-sampling interval was calculated. Sample means were then averaged within each days-post-feeding category (0-0.5, 1-2, and 3-10). The 95% confidence interval was calculated using the critical value for the two-tailed t distribution.
To determine the amount of diversity shared between two samples (beta diversity) we employed the UniFrac metric (Lozupone and Knight, 2005; Lozupone et al., 2007; Hamady et al., 2010) in QIIME. UniFrac distances are based on the fraction of branch length shared between two communities within a phylogenetic tree inferred from the 16S rRNA gene sequences from all communities being compared. We used unweighted UniFrac, in which only the presence or absence of lineages is considered (community membership) and weighted UniFrac, which also accounts for relative abundance (community structure). Principal coordinates plots were visualized using the KiNG graphics program (http://kinemage.biochem.duke.edu/software/king.php). Hierarchical clustering based on UniFrac distances between composite communities was performed using the unweighted pair group method with arithmetic mean (UPGMA).
Data deposition
The bacterial 16S rRNA gene sequences reported in this paper were deposited in the GenBank Short Read Archive (accession number SRA012490).
Results and Discussion
Dominance of Firmicutes and Bacteroidetes in the python gut
From 109 gut content samples collected from 32 juvenile Burmese pythons (Supplementary Table S1), we obtained ∼280,000 high quality, classifiable 16S rRNA gene sequences with an average read length of 230 nt and an average count per sample of 2,560 ± 480 (s.d.) (∼9000 sequences per animal). To our knowledge, this constitutes the most comprehensive survey of a reptilian microbiota performed to date. We detected members of 12 bacterial phyla in the python gastrointestinal tract. Surprisingly, as observed in humans, mice, and a wide variety of other mammals (Eckburg et al., 2005; Ley et al., 2005,Ley et al., 2008a), most of the sequences detected in the python gut were classified as either Firmicutes (61.8%) or Bacteroidetes (20.6%). Proteobacteria (10.1%) and Deferribacteres (3.9%) were also abundant, and less abundant phyla included the Actinobacteria (0.6%), Verrucomicrobia (0.6%), Fusobacteria (0.5%), and Lentisphaerae (0.01%). When sequences were classified to the highest taxonomic level to which they could be confidently assigned, the following taxa were abundant in the overall pool of python gut sequences: the class Clostridiales (Firmicutes, 4.2%); the families Clostridiaceae and Peptostreptococcaceae (Firmicutes, 19.0% and 18.5%, respectively); and the genera Bacteroides and Rikenella (Bacteroidetes, 7.0% and 5.6%, respectively), Lactobacillus (Firmicutes, 5.6%), and Synergistes (Deferribacteres, 3.9%). Thus, the Burmese python appears to have in common with mammals several gross taxonomic features of its gut microbiota. These results raise the question of whether the last common ancestor of the amniotes (reptiles, birds and mammals) also harbored a gut microbiota dominated by Firmicutes and Bacteroidetes, or whether this trait has evolved more than once. Notably, 16S rRNA-based profiling of the gastrointestinal bacteria of several wild-captured pit vipers (3 individuals from 2 species) also detected the genera Lactobacillus, Bacteroides, and Fusobacterium (Hill et al., 2008). Later in this paper we examine whether the mammal-like membership of the python's gut microbiota stems from an input of bacteria from the snake's rodent meal.
Fasting is associated with expanded Bacteroidetes and reduced diversity in the large intestine
After fasting for >30 days, Burmese pythons harbored gut bacterial communities dominated by members of the phylum Bacteroidetes [48.8% ± 7.0% (N = 7 samples from 3 individuals; different regions, including the cecum, were similar; hence, we averaged their values and refer to them jointly as large intestine)]. Due, in part, to the difficulty in obtaining adequate quantities of microbial biomass from the fasted small intestine, only a single sample was available for analysis (distal region; 34.4% Bacteroidetes). Thus, our analysis of the fasted microbiota is focused on the large intestine. Abundant Bacteroidetes-related taxa included the genera Bacteroides (16.0% ± 1.1%) and Rikenella (12.7% ± 2.5%), as well as lineages that could not be confidently assigned beyond the phylum level (15.7% ± 3.7%). In addition to the Bacteroidetes, the fasted communities also featured abundant Proteobacteria (14.3% ± 6.7%), a relatively small proportion of Firmicutes (13.2% ± 2.6%), and Deferribacteres and Verrucomicrobia of exclusively two genera, Synergistes (6.5% ± 1.5%) and Akkermansia (4.6% ± 1.9%), respectively.
In addition to our findings, an expanded representation of Bacteroidetes has been observed in mouse ceca following a 24-hr fast (Crawford et al., 2009) and, as noted in the Introduction, host-derived glycans can present a consistent nutritional foundation for certain members of this phylum (e.g., Bacteroides thetaiotaomicron) in the absence of dietary glycans (Sonnenburg et al., 2005; Bjursell et al., 2006; Martens et al., 2008). Our results raise the interesting possibility that Bacteroides spp. may function in a similar manner in the Burmese python, a host that is distantly related to mammals and a strict carnivore. Notably, Akkermansia muciniphila is also capable of subsisting on host mucus as a sole source of carbon and nitrogen (Derrien et al., 2004), and this genus has recently been found to proliferate in the ceca of active (i.e., non-hibernating) hamsters subjected to a 96-hr fast (Sonoyama et al., 2009).
Finally, we found that fasted snakes harbored large intestinal bacterial assemblages with significantly lower overall phylogenetic diversity (PD; 17.0 ± 0.6 in fasted vs. 20.5 ± 0.6 in fed at 1400 sequences per sample; P = 0.001; two-tailed t-test with unequal variance) and a significantly lower number of ‘species’-level OTUs defined at ≥ 97% sequence similarity (253 ± 18 in fasted vs. 375 ± 12 in fed at 1400 sequence per sample; P < 0.001) when compared to recently fed snakes. Taken together, these results support the notion that in the absence of dietary nutrients, such as after extended periods of fasting, the Burmese python's gut microbiota, like that of mammals, alters its structure.
Temporal analysis of the postprandial reconfiguration of the large intestinal microbiota
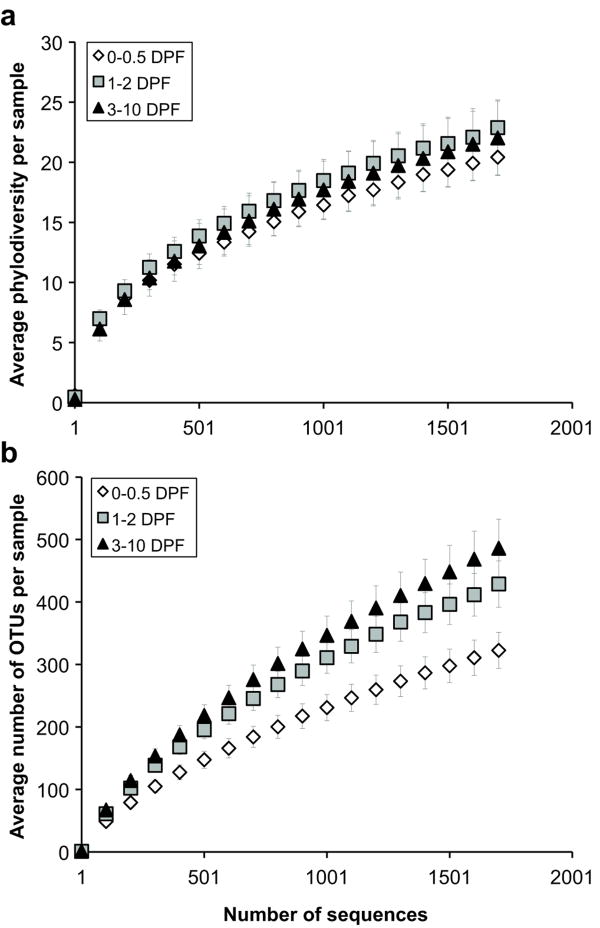
Next, we examined whether the Burmese python gut microbiota remodels itself when the host digests a meal. Fasted snakes were fed rats weighing 25% of the snake's body mass (see Introduction and Material and Methods for details). We assessed changes in overall bacterial community composition as a function of time post-feeding using a phylogeny-based metric, UniFrac (Lozupone and Knight, 2005; Lozupone et al., 2007). A relatively small UniFrac distance implies that two communities are similar, consisting of closely related lineages. UniFrac analysis confirmed that relative to the fasted state, a substantial postprandial shift in large intestinal bacterial community membership (Figure 1a and Supplementary Figure S1b) and structure (Supplementary Figure S2) occurred. Alteration in bacterial community composition appeared to begin at ∼1 day post-feeding with the entry of material from the meal, and to peak at ∼3 days post-feeding, corresponding with peaks in the python's digestive form and function (Secor, 2008) and the accumulation of unabsorbed material (e.g., hair) in the large intestine. Changes in overall community membership and structure were underpinned by a large postprandial shift in the proportional abundances of Bacteroidetes and Firmicutes in the python large intestine (Figure 1b). In the fasted state and up to 12 hours post-feeding, Bacteroidetes were dominant (45.6% ± 5.6% of the community; N = 24 samples from 10 individuals) while Firmicutes were less abundant (15.3% ± 4.7%). Beginning at 12 hours post-feeding, the proportional representation of the Firmicutes increased significantly over time [R2 = 0.65, 35 d.f. (degrees of freedom), P < 0.001] at the expense of the Bacteroidetes, which decreased significantly (R2 = 0.48, 35 d.f., P < 0.001), until 3 days post-feeding. From 3 days onward, Firmicutes were dominant (82.8% ± 6.3% of the community; N = 20 samples from 14 individuals) and Bacteroidetes were less abundant (7.7% ± 2.5%). Firmicutes taxa that increased in relative abundance during digestion included Lactobacillus spp. (max. of 11.5% on day 2), Peptostreptococcaceae (max. of 28.4% on day 3), and Clostridium spp. (max. of 35.8% on day 6) (Figure 1c). We also observed a modest postprandial increase in the average overall phylogenetic diversity (PD) of the large intestinal microbiota (Figure 2a), coupled with a significant increase in the average number of OTUs defined at ≥ 97% sequence similarity (Figure 2b). This suggests that the dominant lineages (primarily Firmicutes) that appeared in the digesting python's large intestine were more diverse at the ‘species’-level than those present during fasting (primarily Bacteroidetes).
Figure 2.
Digestion increases bacterial community diversity and ‘species’ richness within the Burmese python large intestine. (a) Rarefaction curves for phylogenetic diversity measured by the average total branch length in a phylogenetic tree per sample after a specified number of sequences was observed. (b) Rarefaction curves for ‘species’ richness measured by determining the average number of OTUs defined at ≥ 97% sequence similarity per sample. Samples were partitioned into three post-feeding time intervals based on results shown in Figure 1b. Bars indicate 95% confidence intervals. DPF, days post-feeding.
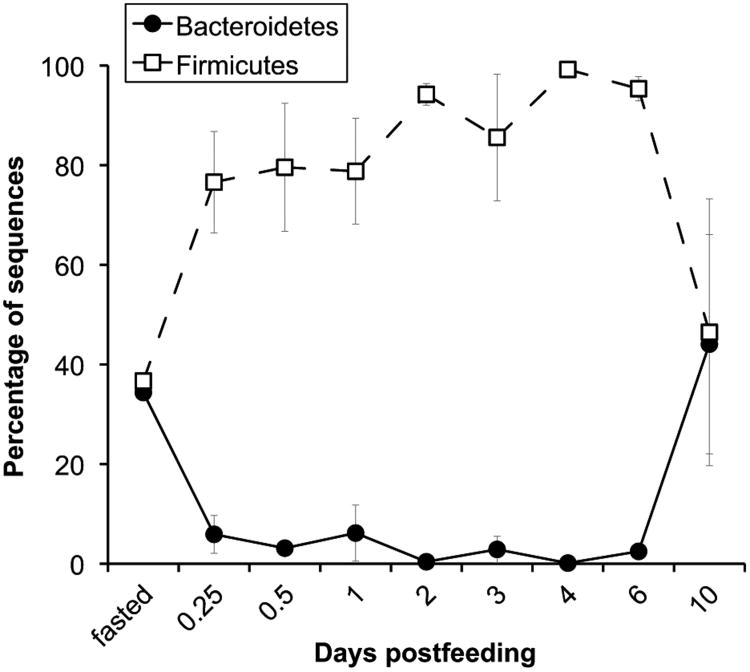
By contrast, over time in the small intestine (between 6 hours and 6 days post-feeding) the initially dominant Firmicutes increased only modestly in proportional abundance (Figure 3). Yet, like in the large intestine, dominant taxa in the small intestine during digestion included the Firmicutes family Peptostreptococcaceae (30.8%) and the genera Clostridium (19.6%) and Lactobacillus (8.3%), and these taxa fluctuated in their proportional abundances over time (Supplementary Figure S3b). A small number of available samples from fasted snakes and from 10 days post-feeding had similar membership (Supplementary Figure S1a) and suggest that Bacteroidetes may increase in proportional abundance in the nutrient-deprived small intestine as well (Figure 3). If this is indeed the case, postprandial shifts in small intestinal microbial community structure may occur in less than 6 hours. However, this may not be surprising given that the python's small intestine also doubles its microvillus length, amino acid uptake rates, and aminopeptidase-N activity within 6 hours of feeding, even before it receives a significant amount of chyme from the stomach, which contains the amino acids and peptides known to trigger upregulation (Secor et al., 2002; Secor, 2008). Within 24 hours of feeding ∼25% of the meal has entered the small intestine and within 2 to 3 days, only ∼25% of the meal remains in the stomach, including portions of the trunk vertebrae, hind limbs, tail and hair (Secor and Diamond, 1995). The latter time point corresponds with maximal changes in small intestinal anatomy (a 5-fold lengthening of the microvilli) and physiology (a 3- to 13-fold increase in hydrolase activity and nutrient uptake capacity) and the accumulation of unabsorbed material in the python's cecum and large intestine where, unlike in the small intestine, large-scale changes in morphology and function are not apparent during digestion (Secor and Diamond, 1995; Ott and Secor, 2007; Cox and Secor, 2008). Unabsorbed material continues to fill the cecum and large intestine until the last remains of the meal (mainly hair) pass from the stomach and through the small intestine, usually by 6-7 days post-feeding. Pythons typically defecate within 2-3 weeks of consuming a meal, and by day 30 the stomach, small intestine and large intestine are fairly empty (Secor and Diamond, 1995). It is interesting to note that the observed changes in large intestinal bacterial community composition at 6 hours post-feeding (Figure 1a) appear to precede the entry of unabsorbed material into the large intestine and, subsequently, occur at several transition points including the onset of the Firmicutes bloom at 1 day post-feeding and the later peaks in Lactobacillus (2 days post-feeding), Peptostreptococcaceae (3 days post-feeding) and Clostridium (6 days post-feeding) relative abundance (Figure 1). The underlying causes of these shifts are unknown, but may relate to changes in the quantity and quality of unabsorbed nutrients passing into the large intestine, as well as to the timing and rate of their transit relative to microbes, or to other physiological changes in the host.
Figure 3.
Average proportional abundance of sequences classified as Bacteroidetes or Firmicutes from the Burmese python small intestine at each time point. Because other lineages can also be present, the proportions need not add up to 100%. Error bars are one standard error of the mean. For fasted, N = 1.
Overall, our results indicate that large-scale alterations in the Burmese python's gut microbiota accompany its extensive physiological and morphological responses to feeding and fasting. While many aspects of the host environment change during digestion, bacterial community modification may be driven, in part, by a shift from host glycan foraging to the degradation of unabsorbed material derived from the prey. The snake's carnivorous diet dictates that much of this material will be soluble or insoluble animal protein, but it may also include small amounts of carbohydrate. In the case of the Burmese python, post-feeding increases in the relative abundance of the Firmicutes may depend on the bacteria's ability to rapidly exploit unabsorbed, labile amino acids and peptides from the python's diet. Indeed, many of the lineages with increased relative abundance during digestion are closely related to isolates with known proteolytic activity, including Clostridium, Lactobacillus, and Peptostreptococcus. Firmicutes may also contribute to the degradation of recalcitrant animal protein such as hair, which has a relatively long transit time in the python gut. Keratin is a main component of hair and several keratinolytic Firmicutes have been isolated (although most are from the genus Bacillus). The metabolic impact of the observed postprandial increase in the ratio of Firmicutes to Bacteroidetes on python biology remains unclear. It has been suggested that the python's large intestine plays a relatively limited role in taking up nutrients that have passed unabsorbed through the small intestine, and a larger role in absorbing water, salts, and trace nutrients synthesized by intestinal bacteria (Secor and Diamond, 1995); however, Firmicutes-dominated gut microbiotas have been found to enhance energy harvest from the diets of other animals (Ley et al., 2006; Turnbaugh et al., 2006; Turnbaugh et al., 2009a.
The rodent meal is an unlikely source of most python gut bacteria
The gut is not a closed system, and animals rarely consume microbe-free meals. In principle, the python provides a model for determining the extent to which an established host microbiota can be invaded by microbial consortia associated with consumed prey. Therefore, we investigated the degree to which bacteria observed in the python's gut after feeding were derived from the mammal it ingested. Because the composition of the vertebrate gut microbiota is influenced by host phylogeny (Ley et al., 2008b), we predicted that the python and rat would contain relatively distinct bacterial assemblages; however, it is also feasible that particular indigenous lineages could be conserved (or convergently acquired) in the two hosts.
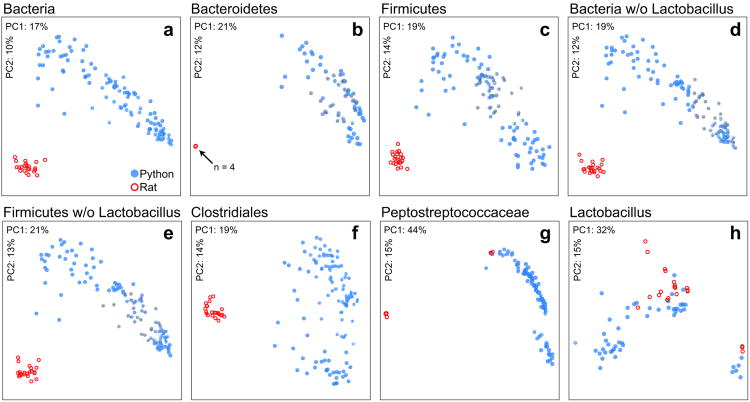
To determine whether the potentially large bacterial loads associated with intact prey could be detected in the python's intestine, we first obtained >30,000 16S rRNA gene sequences from 23 aliquots of whole rat homogenate prepared from 5 individuals. Aliquots yielded an average of 1,320 ± 210 (s.d.) sequences (∼6,000 sequences per animal), with an average read length of 250 nt. The direct transfer of bacteria from the rat meal to the python gut should result in shared identical 16S rRNA gene sequences between the two hosts. To find these exact matches, all sequences from the rat and python samples were pooled and de-replicated (i.e., nearly identical sequences were clustered by picking OTUs at 100% sequence similarity after prefix-based pre-clustering at 200 nt). This procedure resulted in 90,461 OTUs of which, 85,354 were exclusive to the python gut, 4,370 were exclusive to the rat, and 737 were found in both hosts. To be deemed present in both hosts, the OTU had to contain at least one sequence from a python sample and one sequence from a rat sample. Over 50% of the shared OTUs were classified as Lactobacillus (Supplementary Table S2). Next, we used lineage-specific unweighted UniFrac and principal coordinates analysis (PCoA) to compare the memberships of the whole rat and python gut microbiotas. We found that rats and pythons harbored compositionally distinct assemblages at each of several key taxonomic levels (Figure 4). Notably, Lactobacillus, which was abundant in whole rat assemblages (51.4% ± 2.8%), did not appear to be host specific (Figure 4h) and also exhibited a modest postprandial spike in the python's large intestine (Figure 1c). Although we did not detect Lactobacillus in the fasted large intestine, it did appear in the fasted small intestine (0.9% of the community; N = 1), leaving open the possibility that this common gut inhabitant is indigenous to both the python and rat. Similarly, members of the Peptostreptococcaceae, which were also shared between a subset of the rat and python samples (Figure 4g), were also present in the fasted python's small and large intestine, averaging 4.9% and 0.06% of the communities, respectively. In summary, while difficult to ascertain due to the potential for conserved or convergent community membership and carry over from past meals, our results suggest that most of the python gut bacteria observed in this study were indigenous to the snake rather than input with the meal. However, we cannot rule out that certain lineages such as Lactobacillus were delivered to the python's intestine via the diet.
Figure 4.
Python intestine and rodent meal (i.e., whole rat) bacterial communities appear to have largely divergent membership. Communities clustered using PCoA of unweighted UniFrac distance matrices. Each circle corresponds to a sample colored according to host species. The percentage of the variation explained by the plotted principal coordinates is indicated on the axes. Each panel shows a different taxon-specific analysis. These taxa are: (a) all Bacteria, (b) all Bacteroidetes (note that the four rat samples appear superimposed in this panel), (c) all Firmicutes, (d) all Bacteria but excluding Lactobacillus, (e) all Firmicutes but excluding Lactobacillus, (f) Clostridiales, (g) Peptostreptococcaceae, (h) Lactobacillus. Comparisons of panels a and d, and c and e, show that the Lactobacillus component, which was not host specific, did not greatly influence the clarity of separation in the snake and rat communities. Samples were excluded if they contained fewer than 30 sequences from the specified taxon.
Prospectus: The Burmese python as a model for host-microbe interactions during feeding and fasting
The Burmese python presents an attractive model organism for investigating the interplay between gut microbial communities, diet, nutritional status, and host postprandial responses. While the latter are well characterized in the python [e.g., (Secor, 2008)], the present study provides an initial foundation for understanding the gut microbiota's response to feeding and fasting in a host specifically adapted to consuming large intact prey after long periods without food. The results presented here indicate that python gut assemblages are dominated by bacteria belonging to the phyla Bacteroidetes and Firmicutes and are thus broadly comparable to mammalian assemblages. We also find that the python gut microbiota is highly responsive to feeding and fasting, exhibiting wide-ranging alterations in composition and diversity in concert with the host's own dramatic postprandial physiological and morphological changes.
Previous animal models, notably gnotobiotic mice, have been used to show that gut microbial communities have a marked effect on organ size: for example, transplantation of a gut microbiota from conventionally raised mice to germ-free mice produces an increase in the mass of the heart and adipose tissue (Backhed et al., 2004; Crawford et al., 2009), stimulates cell proliferation in the gut, and alters the small intestine morphology, including the density of the submucosal capillary network (Stappenbeck et al., 2002; Backhed et al., 2007; Turnbaugh et al., 2009b). Similarly, gnotobiotic mouse models have shown that the gut microbiota is rapidly responsive to diet shifts and to fasting (Crawford et al., 2009; Turnbaugh et al., 2009b). However, because colonization of a germ-free animal with a microbiota is irreversible, the snake model could, provided non-lethal procedures are developed, allow us to track the same individual across multiple feeding cycles. As such, this model organism provides a unique opportunity for dissecting the interrelationships between nutrient load and availability, microbiota configuration, and host cellular proliferative and hypertrophic responses both within and outside the gastrointestinal tract. Moreover, the python responses are consistent even between genetically heterogeneous individuals, both at the level of the microbiota and of the host. These observations prompt the question of whether the microbiota plays a key role in integrating the impact of feeding and fasting on host cellular functions, including cell hyperplasia and hypertrophy, across a broad range of vertebrates.
Supplementary Material
Acknowledgments
We thank Donna Berg-Lyons, Scott Boback, Christian Cox, Jill Manchester, Brian Ott and Sabrina Wagoner for superb technical assistance, and Greg Caporaso and Daniel McDonald for valuable feedback. This work was supported by grants from the Crohn's and Colitis Foundation, the Colorado Center for Biofuels and Biorefining, the National Science Foundation (IOS 0466139 to S.M.S.), and the NIH (DK70977, DK30292, HG004872), and by HHMI.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website
References
- Andersen JB, Rourke BC, Caiozzo VJ, Bennett AF, Hicks JW. Postprandial cardiac hypertrophy in pythons. Nature. 2005;434:37–38. doi: 10.1038/434037a. [DOI] [PubMed] [Google Scholar]
- Backhed F, Crawford PA, O'Donnell D, Gordon JI. Postnatal lymphatic partitioning from the blood vasculature in the small intestine requires fasting-induced adipose factor. Proc Natl Acad Sci. 2007;104:606–611. doi: 10.1073/pnas.0605957104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Caporaso G, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics. 2009;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso G, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows integration and analysis of high-throughput community sequencing data. Nat Methods. 2010 doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CL, Secor SM. Matched regulation of gastrointestinal performance in the Burmese python, Python molurus. J Exp Biol. 2008;211:1131–1140. doi: 10.1242/jeb.015313. [DOI] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with Arb. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and phylochip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JG, III, Hanning I, Beaupre SJ, Ricke SC, Slavik MM. Denaturing gradient gel electrophoresis for the determination of bacterial species diversity in the gastrointestinal tracts of two crotaline snakes. Herpetological Review. 2008;39:433–438. [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology - human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li WZ, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lignot JH, Helmstetter C, Secor SM. Postprandial morphological response of the intestinal epithelium of the Burmese python (Python molurus) Comp Biochem Phys A. 2005;141:280–291. doi: 10.1016/j.cbpb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JC, Henderson RW. Tales of giant snakes: A historical natural history of anacondas and pythons. Krieger Publishing; Malabar FL: 1997. [Google Scholar]
- Ott BD, Secor SM. Adaptive regulation of digestive performance in the genus. Python J Exp Biol. 2007;210:340–356. doi: 10.1242/jeb.02626. [DOI] [PubMed] [Google Scholar]
- Pope CH. The giant snakes. Alfred A. Knopf; New York: 1961. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, West SEH, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from human colon. Appl Environ Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding g protein-coupled receptor, gpr41. Proc Natl Acad Sci. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor SM. Gastric function and its contribution to the postprandial metabolic response of the Burmese python. Python molurus J Exp Biol. 2003;206:1621–1630. doi: 10.1242/jeb.00300. [DOI] [PubMed] [Google Scholar]
- Secor SM. Digestive physiology of the Burmese python: Broad regulation of integrated performance. J Exp Biol. 2008;211:3767–3774. doi: 10.1242/jeb.023754. [DOI] [PubMed] [Google Scholar]
- Secor SM, Diamond J. Adaptive responses to feeding in Burmese pythons - pay before pumping. J Exp Biol. 1995;198:1313–1325. doi: 10.1242/jeb.198.6.1313. [DOI] [PubMed] [Google Scholar]
- Secor SM, Diamond J. A vertebrate model of extreme physiological regulation. Nature. 1998;395:659–662. doi: 10.1038/27131. [DOI] [PubMed] [Google Scholar]
- Secor SM, Diamond JM. Evolution of regulatory responses to feeding in snakes. Physiol Biochem Zool. 2000;73:123–141. doi: 10.1086/316734. [DOI] [PubMed] [Google Scholar]
- Secor SM, Lane JS, Whang EE, Ashley SW, Diamond J. Luminal nutrient signals for intestinal adaptation in pythons. Am J Physiol -Gastr L. 2002;283:G1298–G1309. doi: 10.1152/ajpgi.00194.2002. [DOI] [PubMed] [Google Scholar]
- Secor SM, Stein ED, Diamond J. Rapid up-regulation of snake intestine in response to feeding - a new model of intestinal adaptation. Am J Physiol. 1994;266:G695–G705. doi: 10.1152/ajpgi.1994.266.4.G695. [DOI] [PubMed] [Google Scholar]
- Snow RW, Brien ML, Cherkiss MS, L W, Mazzotti FJ. Dietary habits of Burmese pythons, Python molurus bivatticus, from Everglades National Park, Florida. Herpetol Bull. 2007;101:5–7. [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sonoyama K, Fujiwara R, Takemura N, Ogasawara T, Watanabe J, Ito H, et al. Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl Environ Microbiol. 2009;75:6451–6456. doi: 10.1128/AEM.00692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck JM, Beese K. Structural flexibility of the intestine of Burmese python in response to feeding. J Exp Biol. 2001;204:325–335. doi: 10.1242/jeb.204.2.325. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–U7. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci TM. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proc Natl Acad Sci. 1999;96:1463–1468. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.