Abstract
The major modules for realizing molecular biological assays in a micro total analysis system (μTAS) were developed for the detection of pathogenic organisms. The specific focus was the isolation and amplification of eukaryotic messenger RNA (mRNA) within a simple, single-channel device for very low RNA concentrations that could then be integrated with detection modules. The hsp70 mRNA from Cryptosporidium parvum was used as a model analyte. Important points of study were surface chemistries within poly(methyl methacrylate) (PMMA) microfluidic channels that enabled specific and sensitive mRNA isolation and amplification reactions for very low mRNA concentrations. Optimal conditions were achieved when the channel surface was carboxylated via UV/ozone treatment followed by the immobilization of polyamidoamine (PAMAM) dendrimers on the surface, thus increasing the immobilization efficiency of the thymidine oligonucleotide, oligo(dT)25, and providing a reliable surface for the amplification reaction, importantly, without the need for blocking agents. Additional chemical modifications of the remaining active surface groups were studied to avoid non-specific capturing of nucleic acids and hindering of the mRNA amplification at low RNA concentrations. Amplification of the mRNA was accomplished using nucleic acid sequence-based amplification (NASBA), an isothermal, primer-dependent technique. Positive controls consisting of previously generated NASBA amplicons could be diluted 1015 fold and still result in successful on-chip re-amplification. Finally, the successful isolation and amplification of mRNA from as few as 30 C. parvum oocysts was demonstrated directly on-chip and compared to bench-top devices. This is the first proof of successful mRNA isolation and NASBA-based amplification of mRNA within a simple microfluidic device in relevant analytical volumes.
INTRODUCTION
Rapid and reliable detection of microorganisms is essential for useful applications in areas such as food safety, water quality, clinical analysis, and defense against bioterrorism. Traditional microbiological methods requiring the use of culturing techniques are time-consuming, and only applicable to organisms that can be grown under laboratory conditions. For these reasons, when possible, they have been replaced by techniques that involve polymerase chain reaction (PCR) with real-time detection as these assays are highly specific, highly sensitive, and very rapid needing only hours instead of days to produce a conclusive result.1–5
Portability of these assays is also very advantageous and allows for onsite, or point-of-care, testing, which further decreases the time and cost of acquiring results. The concept of a micro total analysis system (μTAS), later indicated as a lab on a chip, was introduced by Manz et al. in the early 1990s.6,7 They proposed scaling down the size of chemical analytical devices to improve performance. An ideal μTAS requires only a small volume of sample and incorporates all necessary manipulation and analysis steps to deliver a quantitative, or in some cases qualitative, result in a simple sample-in-answer-out fashion. The μTAS concept has also been applied to biological assays, including the detection of microorganisms within microfluidic devices, and many of these systems have successfully incorporated PCR into the design.1–5 As the temperature cycling necessary in PCR greatly increases the complexity of devices that incorporate this method of amplification, isothermal amplification processes have been explored. Amplification techniques, such as helicase-dependent amplification (HDA) (Mahalanabis et al., 2010; Ramalingam et al., 2009),8,9 loop-mediated isothermal amplification (LAMP),10–12 rolling circle amplification (RCA),13,14 and nucleic acid sequence-based amplification (NASBA),15–18 have been integrated into μTAS designs, offering decreased chip complexity as there is no need for temperature cycling equipment. NASBA is a primer-dependent amplification technique that is able to amplify single-stranded RNA.19 Specifically, this process uses T7 RNA polymerase, RNaseH, avian myeloblastosis virus (AMV) reverse transcriptase, two primers specific to the target sequence, deoxynucleoside triphosphates (dNTPs), and buffers to facilitate a cyclic amplification reaction at a constant temperature that is capable of producing a 109-fold amplification in 90–120 minutes.19 NASBA was the amplification technique used in this study.
Many of these microanalytical systems that incorporate nucleic acid amplification use glass-, silicon- and quartz-based devices.20 However, the fabrication of these microchips is often expensive and time-consuming.20 Consequently, organic polymers have been used as an alternative.3,9,10,15,17,18,21–27 Poly(methyl methacrylate) (PMMA) is a commonly used substrate for microfluidic device fabrication due to its advantageous properties, including its machinability, low cost, and optical transparency,20,28 and it was explored as a new material for NASBA in this study.
Within microfluidic devices, silica structures/beads15,29–31 and paramagnetic beads32,33 are often used for nucleic acid isolation from a lysate sample. However, these methods require increased microdevice complexity, are more complicated to use, and are expensive. Furthermore, the previous microdevice that incorporated both nucleic acid isolation and NASBA had separate chambers for isolation and NASBA,15 making the device more complex to fabricate and use as well as increase the potential loss of target nucleic acids during transfer, which is especially a problem at low RNA concentrations. Here, we present a very simple single-microchannel design that uses surface chemistry to facilitate both the nucleic acid isolation and NASBA.
The protozoan parasite, Cryptosporidium parvum (C. parvum), is an organism of interest in the water quality community due to its highly infective nature in humans and other mammals.34,35 This pathogen causes the disease, cryptosporidiosis, which is a gastrointestinal disease that is potentially fatal in immunocompromised and immunosuppressed patients. 34,35 This parasite is of concern in countries relying on chlorination as a form of water treatment as it is ineffective when C. parvum is in its oocyst state. 34,35 Due to the worldwide importance of C. parvum detection, it was chosen as the model analyte for our μTAS.
In this study, we present a PMMA microfluidic device that is capable of isolating and amplifying specific messenger RNA (mRNA) from lysed C. parvum oocyst samples. Specifically, mRNA is targeted to differentiate between and only detect viable oocysts, since dead oocysts are not infectious (Bukhari and Smith, 1997). The microchannels undergo surface chemistry modifications to enable mRNA isolation directly on the channel surface, and amplification via NASBA is performed within the same channel. Surface modifications of the channels were studied to determine an optimal surface for both mRNA isolation and amplification, avoid non-specific binding of proteins and nucleic acids, and hence prevent loss of NASBA enzymes and increase availability of the mRNA to these enzymes.
EXPERIMENTAL METHODS
Microfluidic Channel Fabrication
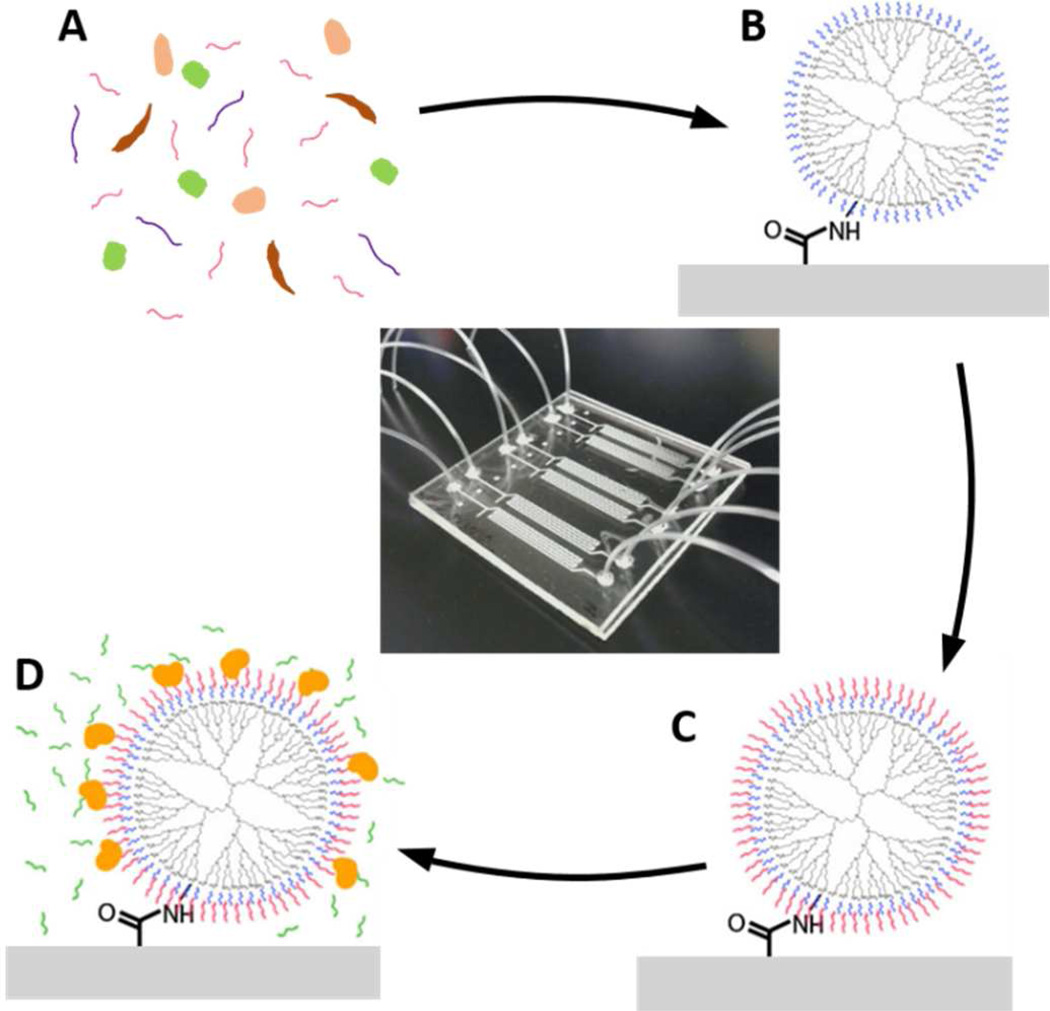
Microfluidic channels were patterned simply and rapidly in PMMA (Lucite Int., UK) via a hot embossing technique using a copper template as previously described.25 The copper template was fabricated at the Cornell Nanoscale Facility (CNF) via photolithography using KMPR 1050 (Micro-Chem Corp., MA) and copper electroplating resulting in elevated channel structures. A 5×5cm piece of PMMA was sandwiched between the copper template and a blank copper plate, and using a hot press (CarverLaminating), the PMMA was patterned at 130°C and 8000 lbs. of pressure for 10 minutes. Inlet and outlet holes were drilled in the embossed piece of PMMA. The device was bonded similarly to that previously described using UV/ozone-assisted thermal bonding.37 The embossed PMMA and a second 5×5cm piece of blank PMMA were UV/ozone-treated for 10 minutes using a UVO-Cleaner (Model 144AX, Jelight Company Inc, Irvine, USA). This treatment decreases the glass transition temperature of the surface only, which ensures bonding of the channels without channel deformation.37 They were then sandwiched between two blank copper plates, and pressed at 90°C and 5000 lbs. of pressure for 10 minutes. Tygon tubing (S-54-HL, Murdock Industrial) with an inner diameter of 0.010” and outer diameter of 0.030” was glued at the inlet and outlet holes of the channels to allow connection to 1mL syringes (Becton, Dickinson and Company) with 30-gauge, luer lock, blunted, stainless-steel needles (Small Parts, Inc.). After fabrication, the chips were stored dry, and usually used the following day. Prior to any surface chemistry, nuclease-free water was pumped into the channels by hand at a high flow rate to expel all the air and ensure no bubble trapping occurred, as well as to test for leakage. Fig. 1 shows a completely assembled microfluidic device consisting of 6 microchannels each with a volume of approximately 3.5µL, as well as a schematic representation of the mRNA isolation and NASBA within the microchannels with surface chemistry modifications described below.
Figure 1.
Image of a completed 6-channel microfluidic device and a step-by-step schematic of the isolation and NASBA process within the microchannels. A sample of C. parvum lysate with the target mRNA (A) flows into the surface-modified microchannel (B), where the mRNA is isolated (C). NASBA reagents are injected into the microchannel, and the target mRNA is amplified (D).
Surface Chemistry Modification
The UV/ozone treatment, prior to bonding, chemically modified the surface of the PMMA resulting in a carboxylated surface,38 which facilitated the surface chemistry modifications for mRNA isolation. Polyamidoamine (PAMAM) dendrimers (Dendritech Inc.) were immobilized on the channel surface to ultimately increase the immobilization efficiency of oligomer probes.39 The PAMAM dendrimers used were 5th generation dendrimers, which possess 128 amine surface groups, and they were immobilized using 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-Hydroxysuccinimide (NHS) crosslinking chemistry. This was accomplished by flowing 150µL of solution (200mM EDC (Sigma Aldrich), 200mM NHS (Thermo Scientific), and 500mM NaCl (Sigma Aldrich) in 50mM 2-(N-Morpholino) ethanesulfonic acid (MES) (FisherBiotech) pH 6.0) through the channels at 5µL/min using a syringe pump (KD Scientific, MA). EDC initially reacts with the carboxylic acid groups on the channel surface, and then it is replaced by NHS to form a more stable NHS-ester that readily reacts with amine groups. Immediately following the EDC/NHS reaction, 150µL of 2mM PAMAM dendrimers in a solution of 10mM NaCl and 50mM N-(2-Hydroxyethyl)piperazine-N'-2-ethanesulfonic acid (HEPES) (Mallinckrodt Baker) pH 8.5 were pumped through the channels at 1µL/min and allowed to incubate at room temperature overnight. The channels were then rinsed with 200µL of nuclease-free water (BDH) at 20µL/min. Phosphate-modified thymidine oligonucleotides, 5’-[Phos]-[T]25-3’ ([Phos]-oligo(dT)25) (Eurofins MWG Operon) were then immobilized on the periphery of the dendrimers using EDC as a crosslinker. Here, 150µL of solution (60µM [Phos]-oligo(dT)25, 200mM EDC, and 100mM imidazole (Sigma Aldrich) in nuclease-free water) were pumped through the channels at 1µL/min and again allowed to incubate overnight at room temperature.
The remaining unmodified amine groups on the dendrimers were subsequently modified to eliminate the positive charges that resulted from the amine groups. Without this modification, the amine groups would attract the negatively charged DNA and RNA molecules to the surface and render them unavailable to the NASBA enzymes. The unmodified amine groups were converted to carboxylic acid groups via succinic anhydride modification. A solution of 200mM succinic anhydride (Alfa Aesar) in 700mM borate buffer adjusted to pH 9 was used, and 200µL were pumped through the channels at 20µL/min. This was followed by a 200µL wash with 1M 2-Amino-2-hydroxymethyl-propane-1,3-diol (Tris)-HCL (Sigma Aldrich) adjusted to pH 9 pumped through the channels at 20µL/min. The channels were then rinsed by pumping 200µL of nuclease-free water at 20µL/min. The microfluidic channels were then ready to be used for C. parvum isolation and amplification.
Evaluation of Surface Chemistry Modifications
To evaluate carboxylation density on the channel surface after UV/ozone treatment, an assay using Toluidine Blue O (TBO) (TCI America) was performed. TBO is a dye that adsorbs to carboxylic groups in an alkaline environment and desorbs in acidic conditions.40 After bonding the channels, 200µL of 500µM TBO in deionized water adjusted to pH 10 were pumped through the channels at 20µL/min, which caused the TBO to adsorb. The channels were washed with 200µL of deionized water adjusted to pH 10 at 20µL/min. Finally, 150µL of 50% (w/w) acetic acid were pumped through the channels at 20µL/min to cause desorption of the TBO, and the effluent was collected in a 96-well microtiter plate. The absorption at 633 nm was measured using a PowerWave XS plate reader (BioTek Instruments Inc.).
An Acid Orange 7 (AO7) assay41 was used to assess the density of amine groups, and therefore of dendrimers immobilized on the surface of the channels. The assay is analogous to the TBO assay described above. A solution of 1mM AO7 (TCI America) in deionized water adjusted to pH 3 was pumped into the channels and washed with deionized-water at pH 3. The dye was desorbed with deionized water adjusted to pH 12 and collected in a 96-well microtiter plate. Flow rates and volumes were the same as described for the TBO assay above. The absorption was measured at 460 nm using a plate reader.
The immobilization efficiency of oligo(dT)25 on a dendrimer-modified surface was compared to that on a simple carboxylated surface by measuring its capture efficiency of a fluorescent complimentary adenosine oligonucleotide, oligo(dA)25. For this comparison, amine-modified oligo(dT)25 probes were immobilized onto the carboxylated microchannel surface (no dendrimers) using EDC as a crosslinker, and this was done by pumping the EDC/NHS solution through the channels as described above, and then pumping 150µL of 60µM [NH2]-oligo(dT)25 and 10mM NaCl in 50mM HEPES at pH 8.5 through the channels at 1µl/min and allowing incubation overnight at room temperature. Fluorescein-conjugated oligo(dA)25 was prepared at 10µM in nuclease-free water, and 100µL were pumped through both types of modified channels at 5µL/min. The excess oligo(dA)25 was washed out of the channels using 30µL of washing buffer B (Invitrogen Dynabeads Kit) at 1µL/min. The channels were placed on a heating block set at 82°C, 150µL of 0.01M Tris-HCl at pH 7.5 was pumped through the channel at 20µL/min, and the effluent from each channel was collected in a 96-well microtiter plate. The fluorescence was measured using an FLx800 fluorescence plate reader (BioTek Instruments Inc.) at an absorbance wavelength of 485nm and an emission wavelength of 528nm. Carboxylated channels and dendrimer-modified channels without oligo(dT)25 were also tested as negative controls.
Heat Shock and Lysis
Live C. parvum oocysts in 1× phosphate-buffered saline (1×PBS) were obtained from Waterborne Inc., and diluted to concentrations of interest using nuclease-free water. The oocyst samples were heat-shocked in a heating block at 41°C for 5 minutes to cause a high transcription yield of the target, heat-shock protein 70 (hsp70) mRNA. The oocysts samples were subsequently diluted in Lysis Binding Buffer (Invitrogen Dynabeads Kit) and lysed via five repeated freeze-thaw cycles. The samples were stored at −80°C until needed.
mRNA Isolation within the Microchannels
A volume of 150µL of C. parvum lysate was pumped through the channels at 5µL/min. This allowed any mRNA in the sample to hybridize with the immobilized oligo(dT)25 via its poly-adenosine (poly-A) tail. Several different washing techniques were tested to clear the cellular debris, proteins, unbound nucleic acids, and lysis binding buffer out of the channels without washing out the desired target mRNA. The optimal washing parameters were to pump 100µL of washing buffer B (Invitrogen Dynabeads Kit) through the channels at 5µL/min and then 50µL of nuclease-free water at 5µL/min. Finishing the washing steps with nuclease-free water is necessary as many buffers including the lysis binding buffer and washing buffer B inhibited NASBA. Bench top controls for the isolation procedure were performed in Eppendorf tubes (VWR) using paramagnetic beads coated with oligo(dT)25 according to the manufacturer’s instruction in the Dynabeads® mRNA DIRECT™ Micro Kit (Invitrogen).
NASBA within the Microchannels
The NASBA solution consisting of enzymes, primers, and dNTPs was prepared using the NucliSENS Easy-Q Basic Kit (BioMerieux, USA) according to the manufacturer’s instructions. The sequences for both primers can be seen in Table 1. This solution was pumped into the channels by hand until the microchannels were completely filled. The microchannels were then submerged in a water bath set at 41°C for 90 minutes with gentle manual agitation every 30 minutes. Following incubation, the samples were expelled from the channels into Eppendorf tubes for off-chip detection. Positive NASBA controls were samples of NASBA amplicon diluted 1:108 in nuclease-free water. Negative NASBA controls were samples containing only nuclease-free water. These controls underwent NASBA on the bench top where the NASBA reagents and enzymes were added directly to the Eppendorf tube and incubated in a water bath at 41°C for 90 minutes. Controls accounting for cross-reactivity with other microorganisms were perform previously,42 and here, the specificity of the assay to C. parvum was confirmed.
Table 1.
C. parvum-associated Sequences42
| DNA Probe/Primer | Sequence 5’-3’ |
|---|---|
| NASBA Primer 1 | AATTCTAATACGACTCACTATAGGGAGAAGGTAGAACCACCAACCAATACA |
| NASBA Primer 2 | AGATTCGAAGAACTCTGCGCTGA |
| Reporter Probe | GTGCAACTTTAGCTCCAGTT |
| Capture Probe | AGATTCGAACTCTGCGC |
Lateral Flow Assay
A lateral flow assay (LFA) was used to detect any C. parvum target sequences in the sample solution.35 These assays used sulforhodamine B (SRB)-encapsulating liposomes tagged with C. parvum reporter probe (Table 1) that were synthesized as previously described.43 A 1µL volume of sample solution was mixed with 1µL of liposomes, 1µL of 1× HEPES sucrose saline (1×HSS), and 5µL of hybridization buffer (20% formamide, 4x saline sodium citrate (4XSSC), 0.2% Ficoll type 400, and 125mM sucrose), and this solution was incubated at 41°C for 5 minutes to allow any target sequences to hybridize with the reporter probes conjugated to the liposomes. These samples were allowed to vertically flow up nitrocellulose strips containing immobilized C. parvum capture probes (Table 1). Samples containing target sequences formed sandwich hybridizations that produced a colorimetric signal. The LFA signals were evaluated visually and scanned using a flatbed scanner. ImageJ software was then used for quantification of the obtained signal intensities. This approach was chosen over analyses via a reflectometer44 as the probing surface would not have covered the entire strip surface in the latter approach.
The entire assay for C. parvum detection takes approximately 3 hours from heat shock to colorimetric signal detection via lateral flow assay.
RESULTS AND DISCUSSION
The ultimate goal of the presented studies was to combine isolation and amplification of mRNA molecules into a single inexpensive, simple, polymer-based microfluidic device. Previously, there has only been one demonstration of nucleic acid isolation and NASBA within the same microchip15 to our knowledge, and here, solid-phase extraction via silica beads was used for nucleic acid isolation. This nucleic acid isolation technique increases chip complexity, decreases ease of use, and non-specifically binds all nucleic acids using chaotropic salts and organic solvents that inhibit NASBA. Thus, an exploration of surface modifications within the microfluidic channels to facilitate mRNA isolation was performed to realize an efficient capture method while avoiding non-specific binding and denaturing of NASBA enzymes.
Microfluidic Channel Fabrication
The use of PMMA as the substrate material has several advantages. PMMA is inexpensive, durable, very easy and inexpensive to pattern, and easily facilitates the surface chemistry for mRNA isolation. Our extremely simple single-channel design is also very beneficial. Unlike the previous design that contained separate chambers for nucleic acid isolation and amplification, our design enables simple fabrication and operation as well as the elimination of losses due to nucleic acid transfer between the isolation and amplification modules, which is especially critical for low RNA concentrations. Additionally, each chip contains six parallel microchannels that enable six samples to be processed in parallel on a single chip demonstrating the ability for multiplexing with our design.
Surface Chemistry Modification
In order to render the PMMA surface chemically active, carboxylic acid groups were generated using UV/ozone treatment, which were subsequently changed to amine groups using PAMAM dendrimers. The density of carboxylic acid groups after UV/ozone treatment and amine groups after dendrimer immobilization on the surface of the channel was measured using simple dye assays for an initial assessment of the success of the modification. TBO assays determined the carboxylic acid density to be between 5 and 33 nmol/cm2, with a relative standard deviation (RSD) up to 33%. AO7 assays were used to measure the amine-group density after channels were coated with PAMAM dendrimers and values between 70 and 105 nmol/cm2 were obtained. The RSD was between 3 and 26% for channels within the same experiment, and 20% between experiments. While being simple methods, they provided a reasonable and rapid indication of the desired surface modifications, which were then further used and more quantitatively analyzed as described below. Most importantly, it was determined that the use of PAMAM dendrimers provides a significant increase in the number of available functional group as compared to simple UV/ozone functionalization alone.
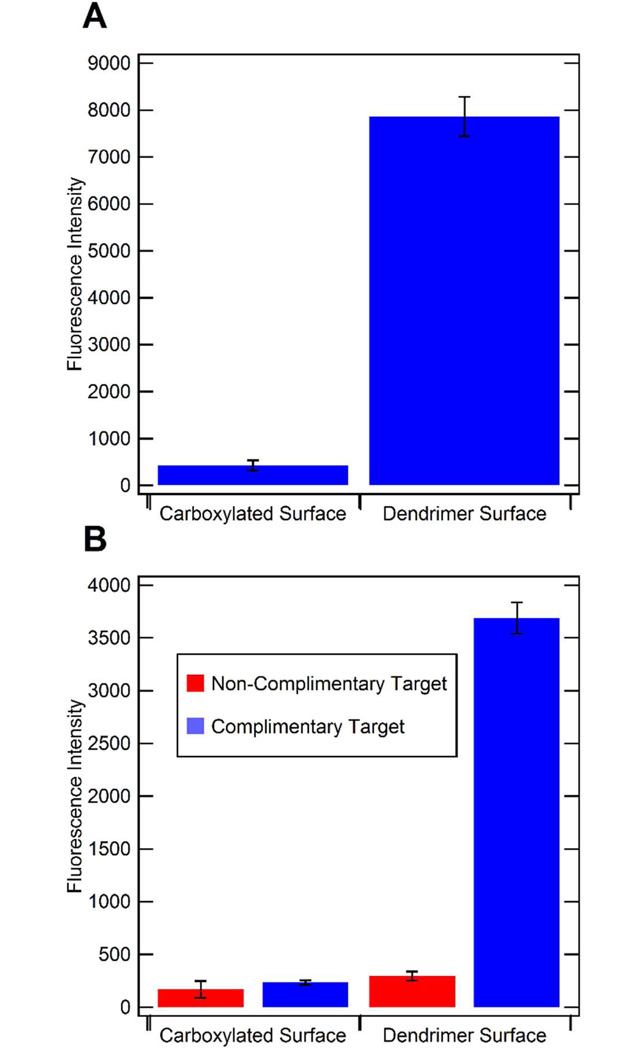
The advantage of using a dendrimer surface was then quantitatively demonstrated by testing the immobilization efficiency of oligo(dT)25 and the capture efficiency of oligo(dA)25 by immobilized oligo(dT)25 on both the carboxylated channel surface and dendrimer-modified channel surface (Fig. 2). The concentration of oligo(dT)25 on the channel surfaces was quantified using fluorescently-labelled oligonucleotides. Here, an 18-fold increase in the number of immobilized oligo(dT)25 molecules was found using a dendrimer-modified surface versus a simply carboxylated surface (Fig. 2a). This dramatic difference was still apparent when evaluating the actual hybridization functionality of the immobilized oligonucleotides. Here, fluorescently-labelled oligo(dA)25 was allowed to hybridize with the immobilized oligo(dT)25 capture probes. The dendrimer-modified surface resulted in a 16-fold increase in capture efficiency relative to the carboxylated surface (Fig. 2b). Consequently, all of the channels used for mRNA isolation had dendrimer-modified surfaces.
Figure 2.
Comparison of oligo(dT)25 immobilization efficiency and oligo(dA)25 capture efficiency by oligo(dT)25 on carboxylated channel surfaces and dendrimer-modified channel surfaces. Fluroescently-labelled oligo(dT)25 was immobilized on carboxylated surfaces containing no dendrimers and dendrimer-modified surfaces, and the fluorescence intensities were compared for the different surfaces (A). Fluorescently-labelled oligo(dA)25 (complimentary target) was captured on both carboxylated and dendrimer-modified surfaces with immobilized oligo(dT)25 capture probes, and the fluorescence was quantified (B: blue bars). Fluorescent oligo(dT)25 (non-complimentary target) was also pumped through both of these channels to determine the non-specific binding to each type of surface (B: red bars).
In previous reports of microfluidic NASBA, blocking of the device surface using BSA or yeast tRNA was necessary to prevent non-specific adsorption of the NASBA enzymes.15–18 This requires additional steps, which can increase the overall assay time, or if added to the NASBA mixture can present potential interferents, which can affect NASBA. However, blocking agents are not necessary in the device presented here; the surface modifications necessary for efficient and simple mRNA isolation provide a suitable surface for NASBA, making it therefore a significantly more robust device.
mRNA Isolation within the Microchannels
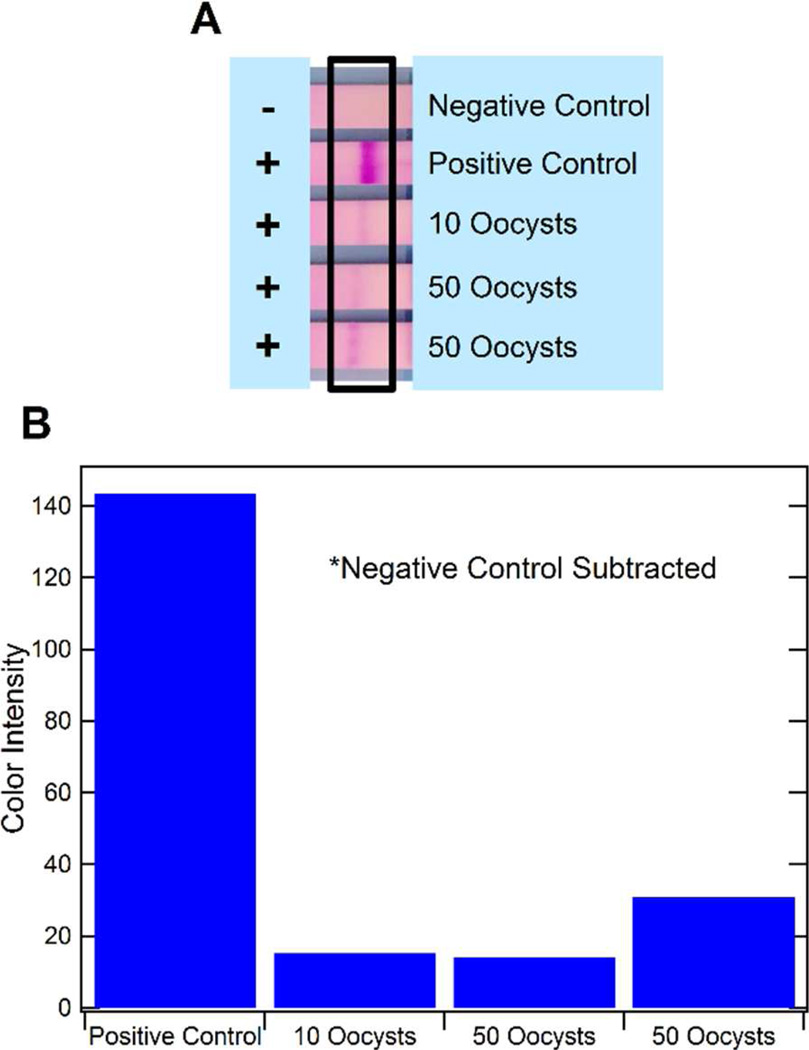
Initially, isolation of the mRNA from lysed C. parvum oocyst samples within the functionalized channels was demonstrated independently from an on-chip NASBA reaction. Here, following the on-chip mRNA isolation procedure, the captured mRNA was dehybridized and expelled from the channels into Eppendorf tubes by pumping 20µL of nuclease-free water at 20µL/min while the channels were submerged in a water bath at 65°C. The NASBA reaction solution was then added to each tube and they were incubated in a water bath at 41°C for 90 minutes following conditions previously published for C. parvum hsp70 mRNA.45 It was quickly determined that only the surfaces containing the dendrimers enabled a truly reliable and successful mRNA isolation. While some success was also obtained with carboxylated surfaces, the dendrimer-modified surface proved to be more reliable for both mRNA isolation and amplification within the same channel. Here, when challenging the procedure to low analyte concentrations, successful mRNA isolation could be demonstrated for as few as 10 and 50 oocysts (Fig. 3). Signals obtained are lower than that of the positive control (a 1:108 dilution of positive NASBA amplicon) as a low mRNA concentration is to be expected from 10 and 50 oocysts, since the RNA was highly diluted when expelled from the microfluidic channel (into a 20µL, instead of 5µL, volume) and because we assume that isolation and dehybridization reactions did not provide a 100% yield.
Figure 3.
Results from mRNA isolation within the microchannels and bench top NASBA. The results of on-chip isolation and off-chip NASBA for 10 and 50 C. parvum oocysts are shown by the LFA strips (A). An ImageJ quantification of the color intensity of the strips is also shown (B). The positive control is NASBA amplicon diluted by 108.
NASBA within the Microchannels
Secondly, NASBA was realized within the microchannels separately from mRNA isolation from lysed oocysts. Initially, a set-up was determined that would result in a temperature-controlled environment to ensure appropriate NASBA conditions. While ultimately an internal heating element will be desirable, a water bath was chosen to avoid temperature fluctuations and facilitate easy access to the microfluidic channel for desired manipulations. The seal achieved through the device fabrication ensured a no-leakage set up. Samples chosen initially were dilutions of NASBA amplicons. In the bench top positive control, the amplicon is diluted by a factor of 108. For assessing NASBA within the channels, dilutions of 1010, 1011, 1013, and 1015 were tested. These samples were mixed with the NASBA reaction solution and pumped into the channels. The channels were then incubated at 41°C for 90 minutes with gentle agitation every 30 minutes. Initial experiments were carried out in microfluidic channels with varying surface chemistries including carboxylic acid groups, dendrimers (amine groups), as well as dendrimer surfaces blocked with 0.01–1% (w/v) polyvinyl pyrrolidone, 0.1–1% (w/v) bovine serum albumin (BSA), 0.01–1% (w/v) gelatin, and 1.11–111ng of extracted Escherichia coli (E. coli) RNA. However, in all of these cases only insufficient or unreliable amplification was observed. It was assumed that enzyme adsorption to the channels and/or loss of mRNA through non-specific binding led to unsuccessful amplification reactions.
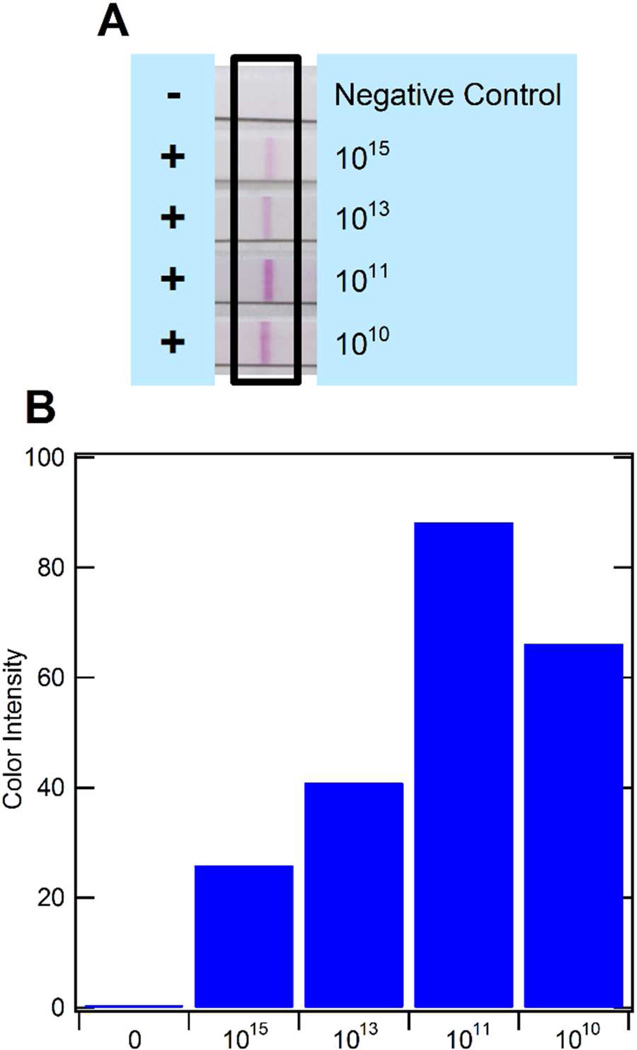
General protein adsorption to the channel surface (and possibly Tygon tubing) was therefore determined through a model protein, horseradish peroxidase (HRP). Here, a 0.1µg/mL concentration of HRP was pumped through the microfluidic channel and the activity was determined in the outflow. However, the possible loss that may have occurred in the channels could not be differentiated from the original solution. It was therefore assumed that inaccessibility of mRNA molecules for NASBA amplification was likely the main challenge. Thus, the dendrimers’ peripheral amine groups were modified using succinic anhydride to convert the amine groups to carboxylic acid groups. This provided repelling surfaces for mRNA molecules and assisted in achieving an appropriate pH of approximately 8.5 for the NASBA reaction. When utilizing this optimized dendrimer surface coupled to oligo(dT)25 capture probes, highly reliable and sensitive amplification could be realized (Fig. 4). The dilution of the positive NASBA amplicon to 1015 is pushing the limit of RNA molecules present in a sample as NASBA typically provides a 109 – 1011-fold amplification of RNA molecules.46 Thus, on-chip NASBA was demonstrated to be highly successful and could now be combined with on-chip mRNA isolation.
Figure 4.
Results from NASBA within microchannels using diluted amplicon. The results of on-chip NASBA for amplicon dilutions of 1010–1015 are shown by the LFA strips (A). An ImageJ quantification of the color intensity of the strips is also shown (B). The negative control is a sample of nuclease-free water (no amplicon) that underwent NASBA.
mRNA Isolation and NASBA within the Microchannels
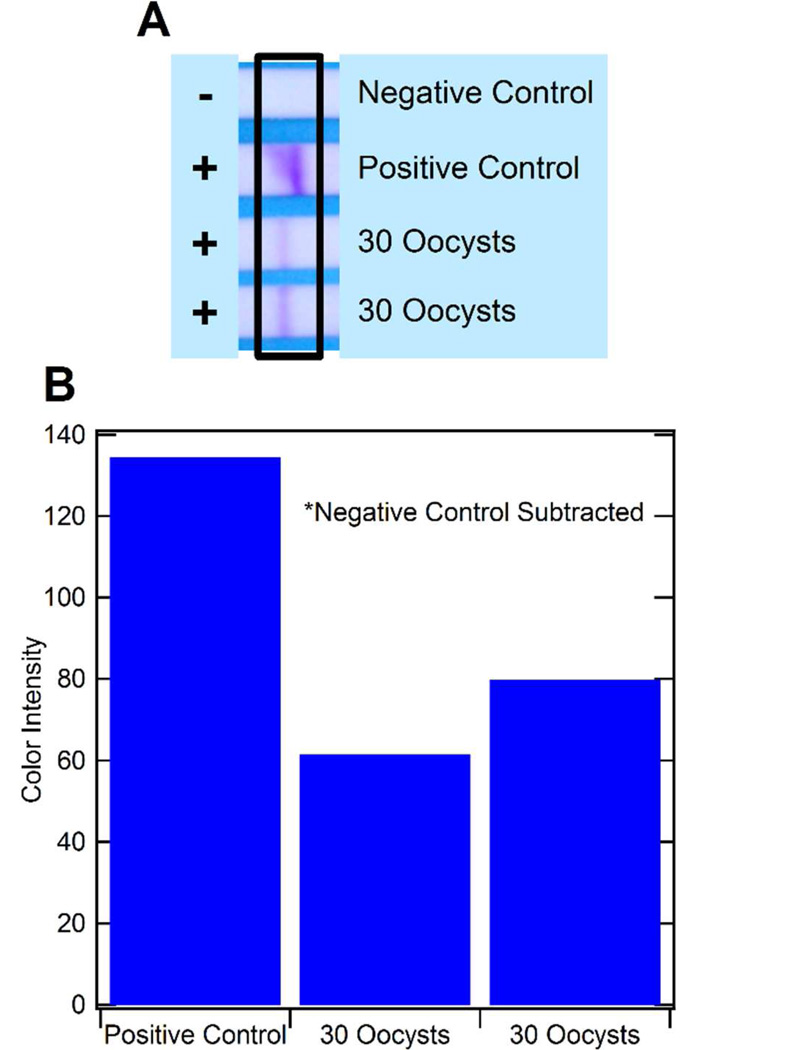
Finally, on-chip isolation of hsp70 mRNA from lysed C. parvum oocyst samples with subsequent on-chip NASBA was successfully demonstrated in the functionalized microfluidic channels within the same device. Various assay conditions were investigated including different washing volumes, flow rates, and buffers. It was found that these factors highly influenced the reliability of the on-chip isolation and NASBA reaction once the dendrimer-oligo(dT)25 surface was optimized. Chaotropic salts and organic compounds in the lysis buffer and washing buffer needed to be removed from the microfluidic channels prior to realizing a successful NASBA reaction. At the same time, captured mRNA needed to remain hybridized on the dendrimer-oligo(dT)25 surface and accessible for NASBA enzymes and primer binding. In the end, the most reproducible results were obtained by washing with 100µL of washing buffer B at 5µL/min, and then 50µL of nuclease-free water at 5µL/min. On-chip isolation and on-chip amplification of mRNA from just 30 oocysts was successfully demonstrated (Fig. 5). These results are a significant improvement over the limit of detection reported in the previous microfluidic device incorporating nucleic acid isolation and NASBA, where detection of 100 Escherichia coli cells was reported.15 This suggests an increased performance using surface modifications for nucleic acid purifications over solid-phase isolation as well as a single isolation and amplification chamber to eliminate nucleic acid losses during transport to the amplification chamber. In fact, we have determined that isolation and amplification of higher concentrations of cells (data not shown) demands less defined surface conditions, and leads to easier success, although working at very low RNA (and cell) concentrations requires additional attention. In addition, the dendrimer surface provided a highly reliable surface for avoiding false negative results.
Figure 5.
Results from mRNA isolation and NASBA within the microfluidic channels. The results of on-chip mRNA isolation and NASBA for C. parvum lysate are shown by the LFA strips (A). An ImageJ quantification of the color intensity of the strips is also shown (B). The negative control for NASBA is a sample of nuclease-free water.
CONCLUSION
In biological and chemical analysis, μTASs are increasingly becoming the end goal of developing assays for on-site and point-of-care use. The portability and simplicity of these devices is crucial if they are to be used in resource-limited areas and in situations where experts are not readily available. In this study, we have presented a simple PMMA-based microfluidic device that can isolate and amplify C. parvum mRNA from lysed oocysts through surface chemistry modifications and NASBA within the same channel. By incorporating PAMAM dendrimers, a significant increase in the immobilization efficiency of the oligo(dT)25 capture probe was achieved, which provided a 16-fold increase in its capture efficiency. The resulting surface also provided a suitable surface for amplification without necessitating the use of blocking reagents that have always been needed with past on-chip NASBA reactions. Through the exploration of different washing regimes, 30 C. parvum oocysts were successfully detected, and this represents a significant improvement in detection compared to the previous device incorporating nucleic acid isolation and NASBA. The isolation and amplification steps of the assay are very important and will be central to a complete μTAS for C. parvum oocyst detection, which will also incorporate previously developed oocyst lysis (Rheonix Inc.)47 and electrochemical detection48 steps. The presented device is envisioned to be a functional module in a μTAS integrating immunomagnetic separation42 upstream and electrochemical detection48 downstream of the module. It is therefore designed to be a highly portable device that is inexpensive, due to the materials ($0.22 in material cost) and fabrication methods used, as well as easy to use, due to the designed simplicity of the assay. An example integration of this device is the automated liquid-handling system, such as the fluidic CARD™ platform developed by Rheonix Inc.47 The proposed system can be used to detect any eukaryotic microorganism through the mRNA isolation mechanism. This is the first demonstration of C. parvum mRNA isolation and amplification using NASBA within a very simple microfluidic device. While being a relevant analyte in itself, C. parvum is a prime example of a difficult biological analyte that can be detected with high sensitivity in chip-based devices. The combination of isolation and amplification into the same microfluidic channel avoids loss and contamination otherwise obtained in multi-step procedures, and therefore has the potential as an alternative in the bioanalysis of low-concentration analytes.
ACKNOWLEDGMENTS
The authors would like to acknowledge the support of the Cornell University, College of Engineering Lester B. Knight Fellowship. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECCS-0335765). Also, this publication was developed in part under the auspices of the Cornell University Center for Life Science Enterprise, a New York State Center for Advanced Technology supported by New York State and industrial partners. The authors also acknowledge partial support by a subcontract with Rheonix, Inc. and 1U01 A1082448-01 from the National Institutes of Health. Any opinions, findings and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of Rheonix Inc. nor those of the National Institutes of Health.
Funding Sources
Footnotes
Author Contributions
The research described in this manuscript was carried out by all authors and they have given approval to the final version of the manuscript.
REFERENCES
- 1.Belgrader P, Young S, Yuan B, Primeau M, Christel LA, Pourahmadi F, Northrup MA. Anal. Chem. 2001;73:286–289. doi: 10.1021/ac000905v. [DOI] [PubMed] [Google Scholar]
- 2.Northrup MA, Benett B, Hadley D, Landre P, Lehew S, Richards J, Stratton P. Anal. Chem. 1998;70:918–922. doi: 10.1021/ac970486a. [DOI] [PubMed] [Google Scholar]
- 3.Sauer-Budge AF, Mirer P, Chatterjee A, Klapperich CM, Chargin D, Sharon A. Lab Chip. 2009;9:2803–2810. doi: 10.1039/b904854e. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C, Xu J, Ma W, Zheng W. Biotechnol. Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Xing D. Nucleic Acids Res. 2007;35:4223–4237. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manz A, Graber N, Widmer HM. Sensors Actuators B Chem. 1990;1:244–248. [Google Scholar]
- 7.Reyes DR, Iossifidis D, Auroux P-A, Manz A. Anal. Chem. 2002;74:2623–2636. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- 8.Mahalanabis M, Do J, ALMuayad H, Zhang JY, Klapperich CM. Biomed. Microdevices. 2010;12:353–359. doi: 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramalingam N, San TC, Kai TJ, Mak MYM, Gong H-Q. Microfluid. Nanofluidics. 2009;7:325–336. doi: 10.1007/s10404-008-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang X, Liu Y, Kong J, Jiang X. Anal. Chem. 2010;82:3002–3006. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
- 11.Wang C-H, Lien K-Y, Wang T-Y, Chen T-Y, Lee G-B. Biosens. Bioelectron. 2011;26:2045–2052. doi: 10.1016/j.bios.2010.08.083. [DOI] [PubMed] [Google Scholar]
- 12.Wang C-H, Lien K-Y, Wu J-J, Lee G-B. Lab Chip. 2011;11:1521–1531. doi: 10.1039/c0lc00430h. [DOI] [PubMed] [Google Scholar]
- 13.Konry T, Smolina I, Yarmush JM, Irimia D, Yarmush ML. Small. 2011;7:395–400. doi: 10.1002/smll.201001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato K, Tachihara A, Renberg B, Mawatari K, Sato K, Tanaka Y, Jarvius J, Nilsson M, Kitamori T. Lab Chip. 2010;10:1262–1266. doi: 10.1039/b927460j. [DOI] [PubMed] [Google Scholar]
- 15.Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O’Kennedy R, Smith TJ, Ricco AJ, Lee LP. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 16.Gulliksen A, Solli L, Karlsen F, Rogne H, Hovig E, Nordstrøm T, Sirevåg R. Anal. Chem. 2004;76:9–14. doi: 10.1021/ac034779h. [DOI] [PubMed] [Google Scholar]
- 17.Gulliksen A, Solli LA, Drese KS, Sörensen O, Karlsen F, Rogne H, Hovig E, Sirevåg R. Lab Chip. 2005;5:416–420. doi: 10.1039/b415525d. [DOI] [PubMed] [Google Scholar]
- 18.Tsaloglou M-N, Bahi MM, Waugh EM, Morgan H, Mowlem M. Anal. Methods. 2011;3:2127. [Google Scholar]
- 19.Compton J. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 20.Graβ B, Neyer A, Jöhnck M, Siepe D, Eisenbeiβ F, Weber G, Hergenröder R. Sensors Actuators B. 2001;72:249–258. [Google Scholar]
- 21.Castaño-Alvarez M, Fernández-Abedul MT, Costa-Garcia A. Anal. Bioanal. Chem. 2005;382:303–310. doi: 10.1007/s00216-005-3151-2. [DOI] [PubMed] [Google Scholar]
- 22.Effenhauser CS, Bruin GJ, Paulus A, Ehrat M. Anal. Chem. 1997;69:3451–3457. doi: 10.1021/ac9703919. [DOI] [PubMed] [Google Scholar]
- 23.Martynova L, Locascio LE, Gaitan M, Kramer GW, Christensen RG, MacCrehan WA. Anal. Chem. 1997;69:4783–4789. doi: 10.1021/ac970558y. [DOI] [PubMed] [Google Scholar]
- 24.McCormick RM, Nelson RJ, Alonso-Amigo MG, Benvegnu DJ, Hooper HH. Anal. Chem. 1997;69:2626–2630. doi: 10.1021/ac9701997. [DOI] [PubMed] [Google Scholar]
- 25.Nugen SR, Asiello PJ, Baeumner AJ. Microsyst. Technol. 2008;15:477–483. [Google Scholar]
- 26.Qi S, Liu X, Ford S, Barrows J, Thomas G, Kelly K, McCandless A, Lian K, Goettert J, Soper SA. Lab Chip. 2002;2:88–95. doi: 10.1039/b200370h. [DOI] [PubMed] [Google Scholar]
- 27.Roberts MA, Rossier JS, Bercier P, Girault H. Anal. Chem. 1997;69:2035–2042. doi: 10.1021/ac961038q. [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Brittain WJ. Macromolecules. 2001;34:3255–3260. [Google Scholar]
- 29.Bhattacharyya A, Klapperich CM. Anal. Chem. 2006;78:788–792. doi: 10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- 30.Cady NC, Stelick S, Batt CA. Biosens. Bioelectron. 2003;19:59–66. doi: 10.1016/s0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 31.Hagan KA, Bienvenue JM, Moskaluk CA, Landers JP. Anal. Chem. 2008;80:8453–8460. doi: 10.1021/ac8011945. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Lien K, Weng C. Biomed. Microdevices. 2009;11:339–350. doi: 10.1007/s10544-008-9240-1. [DOI] [PubMed] [Google Scholar]
- 33.Berry SM, Alarid ET, Beebe DJ. Lab Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 35.Connelly JT, Nugen SR, Borejsza-Wysocki W, Durst RA, Montagna RA, Baeumner AJ. Anal. Bioanal. Chem. 2008;391:487–495. doi: 10.1007/s00216-008-1967-2. [DOI] [PubMed] [Google Scholar]
- 36.Bukhari Z, Smith HV. Epidemiol. Infect. 1997;119:105–108. doi: 10.1017/s0950268897007590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nugen SR, Asiello PJ, Connelly JT, Baeumner AJ. Biosens. Bioelectron. 2009;24:2428–2433. doi: 10.1016/j.bios.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 38.Tsao CW, Hromada L, Liu J, Kumar P, DeVoe DL. Lab Chip. 2007;7:499–505. doi: 10.1039/b618901f. [DOI] [PubMed] [Google Scholar]
- 39.Benters R, Niemeyer CM, Drutschmann D, Blohm D, Wöhrle D. Nucleic Acids Res. 2002;30:E10. doi: 10.1093/nar/30.2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang ET, Tan KL, Kato K, Uyama Y, Ikada Y. Macromolecules. 1996;29:6872–6879. [Google Scholar]
- 41.Uchida E, Uyama Y, Ikada Y. Langmuir. 1993;9:1121–1124. [Google Scholar]
- 42.Baeumner AJ, Humiston MC, Montagna RA, Durst RA. Anal. Chem. 2001;73:1176–1180. doi: 10.1021/ac001293h. [DOI] [PubMed] [Google Scholar]
- 43.Edwards KA, Baeumner AJ. Anal. Bioanal. Chem. 2006;386:1335–1343. doi: 10.1007/s00216-006-0705-x. [DOI] [PubMed] [Google Scholar]
- 44.Min J, Baeumner AJ. Anal. Biochem. 2002;303:186–193. doi: 10.1006/abio.2002.5593. [DOI] [PubMed] [Google Scholar]
- 45.Edwards KA, Baeumner AJ. In: Biosensors and Biodetection. Rasooly A, Herold KE, editors. Vol. 504. Humana Press/Springer; New York: 2009. pp. 185–215. [Google Scholar]
- 46.Baeumner AJ, Schlesinger NA, Slutzki NS, Romano J, Lee EM, Montagna RA. Anal. Chem. 2002;74:1442–1448. doi: 10.1021/ac015675e. [DOI] [PubMed] [Google Scholar]
- 47.Ramadan Q, Gijs MAM. Microfluid. Nanofluidics. 2012;13:529–542. [Google Scholar]
- 48.Goral VN, Zaytseva NV, Baeumner AJ. Lab Chip. 2006;6:414–421. doi: 10.1039/b513239h. [DOI] [PubMed] [Google Scholar]