SUMMARY
Bromodomain function as the acetyl-lysine binding domains to regulate gene transcription in chromatin. Bromodomains are rapidly emerging as new epigenetic drug targets for human diseases. However, owing to their transient nature and modest affinity, histone-binding selectivity of bromodomains has remained mostly elusive. Here, we report high-resolution crystal structures of the bromodomain-PHD tandem module of human transcriptional co-activator CBP bound to lysine-acetylated histone H4 peptides. The structures reveal that the PHD finger serves a structural role in the tandem module, and the bromodomain prefers lysine-acetylated motifs compromising a hydrophobic or aromatic residue at −2, and a lysine or arginine at −3 or −4 position from the acetylated-lysine. Our study further provides structural insights into distinct modes of singly and di-acetylated histone H4 recognition by the bromodomains of CBP and BRD4 that function differently as a transcriptional co-activator and chromatin organizer, respectively, explaining their distinct roles in control of gene expression in chromatin.
INTRODUCTION
The bromodomain (BrD) recognition of acetylated-lysine residues in histones is a fundamental molecular mechanism in control of gene transcription in chromatin (Dhalluin et al., 1999; Sanchez and Zhou, 2009). Bromodomain and acetyl-lysine binding serves to modulate chromatin structure opening, facilitate the recruitment of transcription factors to target gene promoter and enhancer sites, and promote the assembly and activation of the paused RNA polymerase II machinery complex for productive gene transcription (Chiang, 2009). Owing to their key functions in gene activation, the bromodomains of transcription-associated proteins such as BET (bromodomain and extra-terminal domain) proteins (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Nicodeme et al., 2010; Zhang et al., 2012; Zuber et al., 2011) and transcription co-activators CBP (Borah et al., 2011) and PCAF (Zeng et al., 2005) have been reported as new epigenetic drug targets for a wide array of human diseases including cancer and inflammation.
Acetylation at site-specific lysine residues in nucleosomal histones represents distinct biological functions to direct ordered gene transcription. For instance, single acetylation of histone H3 at lysine 14 (H3K14ac) or lysine 18 (H3K18ac) marks for chromatin remodeling, whereas di-acetylation of histone H4 at lysine 5 and lysine 8 (H4K5ac/K8ac), or lysine 12 and lysine 16 (H4K12/K16ac) signals an active state of gene transcription. In contrast to histone methyl-lysine binding protein modules such as chromodomains and PHD fingers (Patel and Wang, 2013; Yap and Zhou, 2010), bromodomain/acetyl-lysine interactions are typically transient and of modest tens-to-hundreds micromolar affinity (Filippakopoulos et al., 2012). As such, histone binding selectivity of human bromodomainsvhas remained mostly elusive. The human bromodomain family consists of 61 members resided in 46 proteins of different chromatin and transcription-associated functions (Figure 1A). Of these are two distinct subgroups representing the histone acetyltransferase transcriptional co-activators including CBP/p300 and PCAF, and the BET family proteins that are characteristic of two tandem bromodomains such as BRD4. The latter have been extensively characterized recently (Filippakopoulos et al., 2012; Moriniere et al., 2009). However, despite their equally important functions for lysine acetylation in gene activation, the histone binding selectivity of the former subgroup of bromodomains is not well understood.
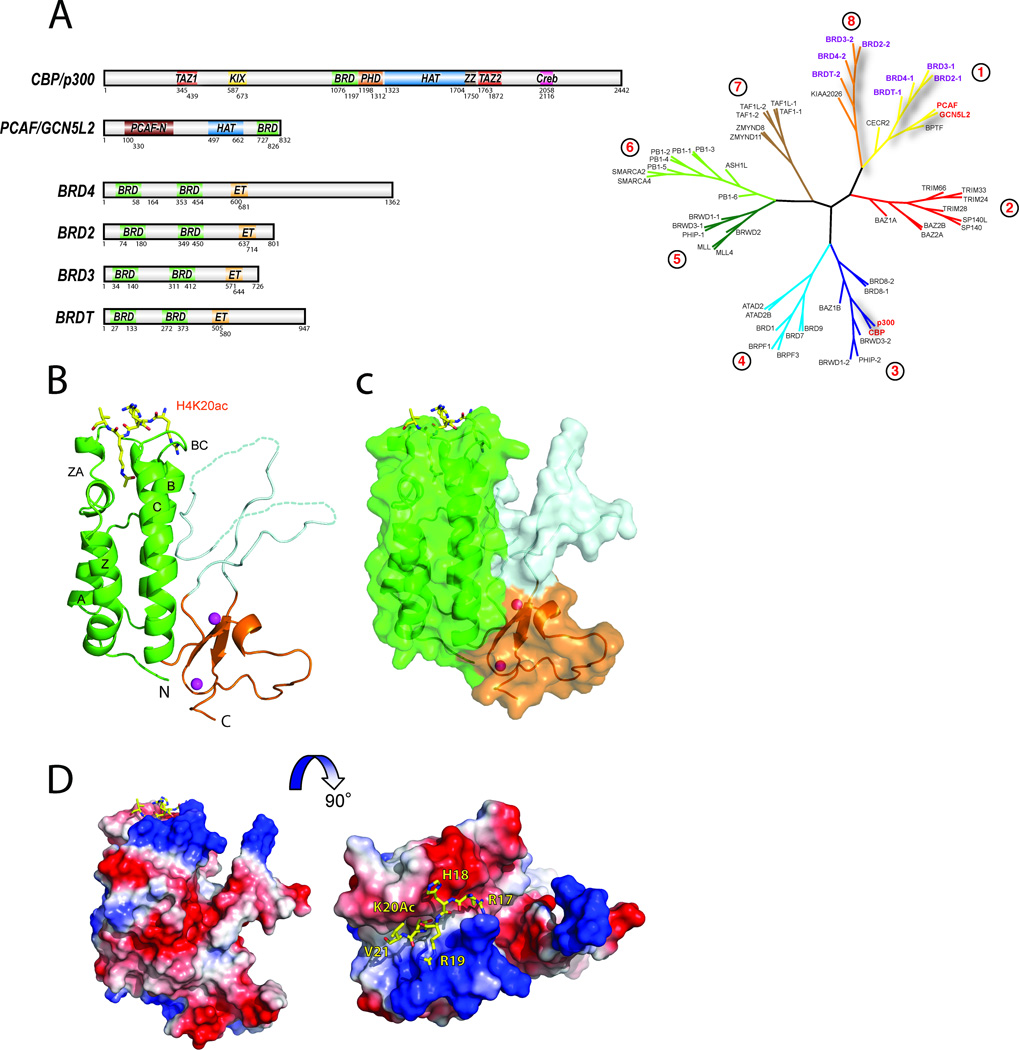
Figure 1. Crystal structure of CBP BrD-PHD in complex with H4K20ac peptide.
(A) Left, domain organization of CBP and other human bromodomain proteins. Right, phylogenetic tree of human BrD proteins.
(B) Ribbon depiction of 3D structure of the tandem BrD (green) and PHD finger (orange) of CBP bound to a histone H4K20ac peptide (shown in sticks with carbon atoms in yellow). The linker of the BrD-PHD module is colored in light cyan and regions that are lack of electron density is indicated by dots. Zn atoms are shown in magenta spheres.
(C) Surface-filled representation of the CBP BrD-PHD/H4K20ac complex structure.
(D) Electrostatic surface potential representation of the CBP BrD-PHD/H4K20ac structure.
See also Figure S1.
Notably, it has been previously reported by Ragvin and coworkers that the bromodomain-PHD finger tandem module of human co-activator p300 binds to nucleosomes with a high degree of lysine acetylation, and that this binding requires the presence of both the bromodomain and the PHD finger (Ragvin et al., 2004). Further, our recent NMR binding analysis revealed that the bromodomain of CBP, which shares 98% sequence identity to that of p300, exhibits a preference for binding to lysine 20-acetylated histone H4 (H4K20ac) (GGARKRHR-Kac-VLRDNIQ, residues 13–27) over other lysine-acetylated histone peptides, of which the molecular underpinning is not well understood (Zeng et al., 2008b). Determination of histone binding specificity of bromodomains would require detailed knowledge of the molecular basis of bromodomain recognition of functionally relevant acetylated histone peptides. In this study, we report two crystal structures of the bromodomain-PHD finger tandem module of CBP in complex with lysine-acetylated histone H4 peptides.
RESULTS AND DISCUSSION
Structures of the CBP BrD-PHD Module/Acetylated H4 Peptide Complexes
To determine histone binding selectivity of the CBP bromodomain and how it functions together with the adjacent the PHD finger, we solved the crystal structures of the bromodomain-PHD tandem module of human CBP in complex with a lysine-acetylated histone H4 peptide (residues 5–25) containing either H4K20ac (KGGKG-LGKGGAKRHR-Kac-VLRDN where Kac is acetylated-lysine) (Figure 1B–1D; Figure S1A–1B), or H4K12ac/K16ac (KGGKGLG-Kac-GGA-Kac-RHRKVLRDN) at 1.9Å and 1.83Å resolution, respectively (Table 1). The new structures revealed that the bromodomain and the PHD finger in the tandem domain establish direct interactions forming a single structural unit. Specifically, the CBP bromodomain adopts the classical left-handed four-helix bundle structure in complex with the acetylated-H4 peptide. The either H4 peptide bound structure of the bromodomain is nearly identical to that of the protein in the free state with root-mean-square-derivations of 0.46Å and 0.47Å for all Ca atoms, respectively. The PHD finger is packaged against the bromodomain between helix B and helix C at one end of the helical bundle opposite from the acetyl-lysine binding pocket. A significant portion of the long linker connecting the two modular domains including residues 1212 to 1251, and 1262 to 1270, were missing in the electron density in the X-ray crystallography data, suggesting a high degree of structural mobility in solution.
Table 1.
Crystallography Data and Refinement Statistics
| CBP-BP/H4K20ac | CBP-BP/H4K12ac/K16ac | |
|---|---|---|
| PDB Code | --- | --- |
| Data collection | ||
| Space group | C121 | C121 |
| Cell dimensions | ||
| a, b, c (Å) | 92.46, 59.31, 53.44 | 92.01, 59.46, 53.88 |
| α, β, γ (°) | 90, 102.96, 90 | 90, 102.68, 90 |
| Resolution (Å) (highest resolution shell) | 30.00-1.9 (1.95–1.9) | 30.00-1.83 (1.88–1.83) |
| Rmerge (%) | 5.1 (32.2) | 4.9 (48.9) |
| I/σI | 35.9 (5.6) | 26.7 (2.7) |
| Completeness(%) | 98.7 (93.8) | 98.4 (96.9) |
| Redundancy | 7.4 (7.1) | 4.9 (4.4) |
| Refinement | ||
| Resolution (Å) | 19.6-1.9 | 19.69-1.83 |
| No. reflections | 20,884 | 23,279 |
| Rwork/ Rfree (%) | 23.2/28.3 | 20.9/24.2 |
| No. atoms | 1,709 | 1,785 |
| RMSD | ||
| Bond lengths (Å) | 0.016 | 0.02 |
| Bond angles (°) | 1.8 | 1.7 |
Notably, this structural observation of the CBP bromodomain-PHD module was confirmed by a newly reported crystal structure of the p300 catalytic core by Delvecchio and colleagues (Delvecchio et al., 2013), which consists of the bromodomain-PHD finger-HAT tandem module, solved in the absence of an acetylated histone peptide. This latter structure reveals that the region of residues 1168–1242 in p300 comprises a CH2-containing RING domain, which interacts with the HAT catalytic domain and was speculated to play a role in control of the HAT activity towards a biological ligand. In our construct of the tandem bromodomain-PHD finger of CBP, we observed only partial electron density of the RING domain. The region that we could trace electron density to the backbone atoms of several amino acid residues with confidence adapts a different conformation as compared to that in the p300 catalytic core structure, suggesting that this RING domain is very flexible. Nevertheless, it is worth noting that these two crystal structures represent the first high-resolution crystal structures of the bromodomain-PHD finger tandem module of CBP, as well as of the CBP bromodomain bound to lysine-acetylated histone peptides. Therefore, our structural feature analysis as described in detail below provides new insights into the molecular functions of CBP in control of gene transcription in chromatin.
Bromodomain-PHD Finger Interactions
The PHD finger of CBP has the canonical PHD finger fold that is composed of a small two-stranded β sheet packaged on one side by a short α helix that is stabilized by two zinc atoms coordinated by Cys1199, Cys1200, His1291, and Cys1294, and Cys1283, Cys1286, Cys1308 and Cys1311, respectively (Figure 2A). The two zinc coordination centers also serve a stable base for an extended interface established between the PHD finger and the bromodomain. This inter-domain interface comprises a network of hydrophobic/aromatic and electrostatic interactions involving Asp1149 with Lys1289, Trp1151 with Phe1280, Asp1155 with Lys1203, Trp1158 with Tyr1204, and Tyr1198 with Arg1288 of the bromodomain and the PHD finger, respectively (Figure 2B).
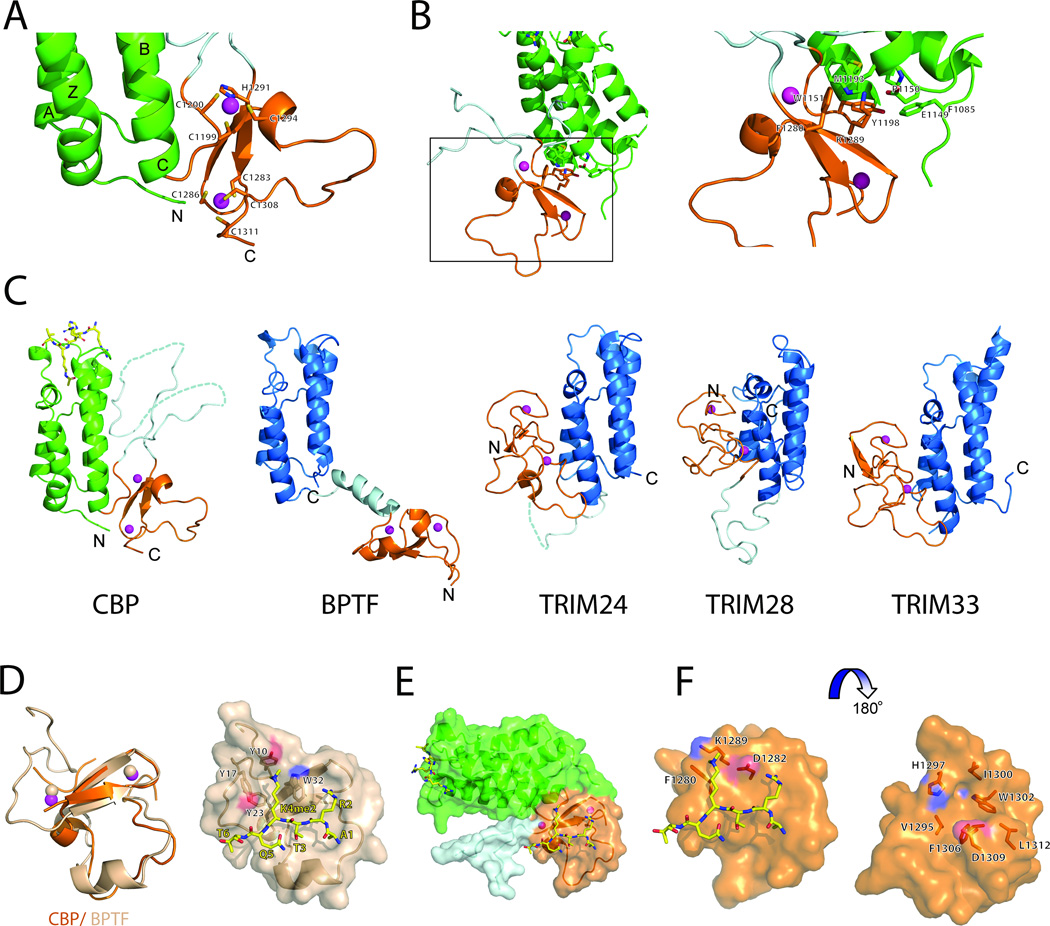
Figure 2. Molecular basis of histone recognition of the BrD-PHD module of CBP.
(A) A close-up view of the PHD finger of CBP. The residues coordinating the Zn atoms (in magenta spheres) are in sticks and labeled.
(B) Inter-domain interactions of the bromodomain and the PHD finger of CBP.
(C) Structural comparison of the tandem BrD-PHD finger modules of CBP, BPTF, TRIM24, TRIM28 and TRIM33. PHD modules are colored in orange, BrDs in blue and green for CBP, linkers between domain are colored in light cyan. Zn atoms are shown in magenta spheres.
(D) Superimposition of CBP and BPTF PHD modules. The domain represented in cartoons and colored in orange and salmon for CBP and BPTF, respectively. Zn atoms are shown in magenta and salmon spheres for CBP and BPTF, respectively. On the right, surface representation of BPTF PHD finger bound to H3K4me2 peptide (shown in sticks with carbon atom labeled in yellow). The residues comprising aromatic cage are shown in sticks.
(E) A surface representation of a model of CBP bound to H3K4me2 peptide. The orientation is as in A.
(F) Close-up view of a model of the CBP PHD finger bound to H3K4me2 peptide. The residues close to the methylated Lys and line-up the groove of the domain are shown in stick and labeled.
See also Figures S2 and S3.
Notably, while many of these residues involved in inter-domain interactions are conserved in CBP or p300 from different species from humans to mouse, they are not all present in the bromodomains from many different types of transcription-associated proteins, particularly BET proteins and those that do not contain bromodomain and PHD finger tandem modules (Figure S2). Domain-domain interactions or orientation even in the bromodomain-PHD finger or PHD finger-bromodomain tandem modules varies widely, as shown in comparison of such tandem module structures from BPTF (Li et al., 2006), TRIM24 (Tsai et al., 2010), TRIM28 (Zeng et al., 2008a) and TRIM33 (Xi et al., 2011) (Figure 2C). This suggests that domain-domain orientation in the tandem modules is likely related to specific functions of the individual domains or of the overall tandem modules, which have diverse functionality in molecular interactions with histones in control of gene transcription. Indeed, unlike the other tandem modules that interact with histones, the tandem PHD finger-bromodomain of human co-repressor TRIM28 (also known as KRAB-associated protein, KAP1) functions as a unique small ubiquitin-like modifier (SUMO) E3 ligase for gene transcription silencing (Zeng et al., 2008a).
The Structural Role of the PHD Finger in the Tandem Module
We performed extensive NMR 15N-HSQC titration of the 15N-labeled tandem bromodomain-PHD finger of CBP as well as the individual PHD finger with a set of histone H3 and H4 peptides in the free form or with site-specific lysine methylation such as H3K4me3, H3K9me3, H3K27me3, and H4K20me3. We did not observe any specific histone peptide binding by the PHD finger, either in the tandem module or in its individual construct (Figure S3). Our results are consistent with a recent study of the CBP PHD finger that does not interact with histones (Park et al., 2013). To investigate the structure-function relationship of the CBP PHD finger, we compared its structural features to that of the BPTF PHD finger that is known to bind to H3K4me3 (Li et al., 2006). Notably, while the overall structures of the two PHD fingers are very similar with r.m.s.d. of 2.8Å for all Ca atoms (Figure 2D), and a possible H3 peptide binding site in the CBP bromodomain-PHD finger tandem module would also be accessible, the key aromatic cage residues such as Tyr10, Tyr17 and Trp32 in the BPTF for the H3K4me3 recognition are absent in the CBP PHD finger (Figure 2E). This explains its lack of interactions with the histone peptides. Moreover, the overall structure of the CBP bromodomain-PHD module bound to the acetylated H4K20ac peptide is very similar to that in the free form structure of the p300 bromodomain-PHD finger-HAT module (Delvecchio et al., 2013). Taken together, these results support our notion that this PHD finger plays a structural role in the bromodomain-PHD finger tandem module, which serves as one functional unit in assisting substrate recognition for the histone acetyltransferase activity of CBP.
Site-Specific Acetylated H4 Recognition by the BRD Bromodomain
The crystal structures of the bromodomain-PHD module bound to the two acetylated-H4 peptides reveal the atomic level details of the molecular basis of histone binding selectivity by the CBP bromodomain. Both H4 peptides contain the same H4 residues 5–25, but carry different lysine acetylation sites, i.e. H4K20ac of KGGKGLGKGGAKRHR-Kac-VLRDN and H4K12ac/K16ac of KGGKGLG-Kac-GGA-Kac-RHRKVLRDN. Notably, only a small portion of each H4 peptide (underlined in the H4 peptides) was observed in the electron density, which is centered on one acetylated lysine bound to the CBP bromodomain (Figure 3A). Despite the different amino acid residues in their sequence, the two H4 peptides bound to the bromodomain adopt almost an identical conformation across the surface-exposed ligand-binding pocket formed between the ZA and BC loops. Similarly, little structural derivations of the bromodomain when bound to the two H4 peptides were observed for backbone or side-chain conformation of several key residues in the acetyl-lysine binding pocket. Of the most notable is the conserved Asn1168 that forms an essential hydrogen bond to the acetyl group of a single bound acetylated-lysine, H4K20ac or H4K12ac. Notably, the side chains of His18 in the H4K20ac peptide, and Leu10 in the H4K12ac/K16ac peptide are orientated in a nearly identical position and interact with Pro1123 of the CBP bromodomain. Additionally, the side-chain amino group of Lys8 in the H4K12ac/K16ac peptide is engulfed in electrostatic interactions with a cluster of backbone oxygen atoms of Trp1165, Leu1166, Tyr1167 and Asn1168 that form the last C-terminal turn of helix B (Figure 3B). This set of interactions appears partially fulfilled by Arg19 in the H4K20ac peptide (Figure 3C). Collectively, these new structural insights suggest that the CBP bromodomain prefers to interact with singly lysine-acetylated motifs compromising a hydrophobic or aromatic amino acid at −2, and a lysine or arginine at −3 or −4 position from the acetylated-lysine. This consensus recognition sequence agrees with our previous NMR structural analysis of the CBP bromodomain recognition of H4K20ac (Zeng et al., 2008b), as well as activated, acetylated tumor repressor p53 at K382ac (QSTSRHK-Kac-LMFKTEG, residues 375–389) (Mujtaba et al., 2004). The latter has been shown to play an important role for p53 recruitment of the HAT co-activator CBP to its target gene transcription activation in response to DNA damage repair (Mujtaba et al., 2004; Mujtaba et al., 2006).
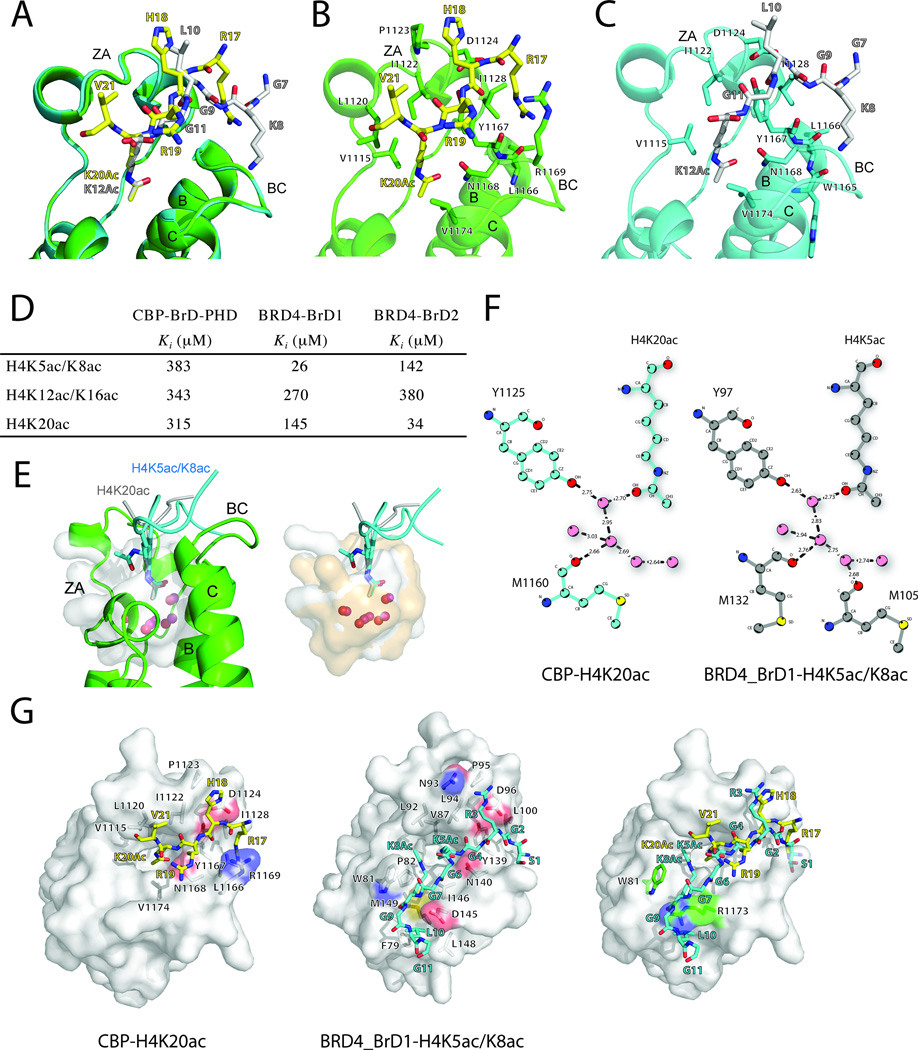
Figure 3. Molecular insights into H4 recognition by the BrDs of CBP versus BRD4.
(A) A close-up view of superimposed crystal structures of CBP BrD-PHD bound to H4K20ac (yellow) and H4K12ac/K16ac (white). The H4 peptides are color-coded by atom types.
(B), (C) A close-up views of acetylated H4-binding sites in the individual structure of CBP BrD-PHD bound to H4K20ac (yellow) and H4K12ac/K16ac (white). Key protein residues interacting with the peptides are shown in sticks.
(D) Binding affinity of lysine-acetylated H4 peptides by the BrDs of CBP and BRD4, determined in a fluorescence anisotropy assay. The H4 peptides H4K5ac/K8ac, H4K12ac/K16ac, and H4K20ac consist of H4 residues 1–13, 5–25 and 17–23, respectively.
(E) Superimposition of ligand-binding pockets of CBP BrD and BRD4-BrD1. The ligand binding sites are shown in transparent surfaces and colored in grey and light orange for CBP BrD and BRD4-BrD1, respectively. H4K5ac/K8ac peptide of BRD4-BrD1-H4K5ac/K8ac complex and H4K20ac peptide of CBP-BrD-PHD/H4K20ac complex are colored in cyan and gray, respectively. Acetyl-lysine residues are shown in sticks. Water molecules tightly bound in the ligand binding sites are shown in spheres. On the left panel, CBP protein chain is shown in cartoon and colored in green.
(F) The role of water molecules in acetyl-lysine recognition by the BrDs. Schematic diagram of the interaction between water molecules in acetyl-lysine binding sites of CBP-H4K20ac and BRD4-BrD1-H4K5ac8Kac complexes. Water molecules are shown in spheres, hydrogen bonds are drawn in dashed line, and donor-acceptor distances are given. The figure was generated by using the program LIGPLOT.
(G) Structural comparison of singly and dually lysine-acetylated H4 peptides by CBP (left) and BRD4 BrDs (middle), respectively. CBP and BRD4 are shown in light grey surfaces, carbon atoms of H4K20Ac and H4K5Ac8Ac peptides are colored in yellow and cyan respectively, residues interacting with peptides are shown in sticks and carbon atoms are in light grey. Right, structure superimposition of CBP-BrD-PHD/H4K20ac and BRD4-BrD1-H4K5ac/K8ac complexes. Surface representation of CBP is shown only. Residues involved in discriminating of binding singly and di -acetylated H4 are shown in stick and carbon atoms colored in green.
See also Figure S4.
Differences of Histone Binding Specificity by CBP and BRD4 Bromodomains
We next performed structural comparison of lysine-acetylated-histone H4 recognition by the bromodomains of CBP and BRD4 that have distinct functions in gene transcriptional regulation in chromatin. Unlike the CBP bromodomain (Figure 3D; Figure S4), the two bromodomains of BRD4, as well as other members of the BET protein family, have been shown to prefer characteristic di-acetylated histone H4 sequence, particularly H4K5ac/K8ac, of which H4K5ac serves as the anchoring acetyl-lysine hydrogen-bonded to the conserved Asn140 in BRD4-BrD1 (Filippakopoulos et al., 2012; Moriniere et al., 2009). Strikingly, five stably bound water molecules in the acetyl-lysine binding pocket in the bromodomains of CBP and BRD4-BrD1 are located at almost identical positions (Figure 3E), of which one water forms two hydrogen bonds, one to oxygen of the acetyl-lysine, and the other to the phenoxy group of the conserved Tyr1125 in CBP and Tyr97 in BRD4 (Figure 3F). The second acetylated-lysine H4K8ac in the H4K5ac/K8ac peptide adds hydrophobic and aromatic interactions with Trp81, Pro82, Leu92, and Ile146 that form a shallow cavity located in the ZA channel and the WPF shelf (Figure 3G). Notably, the long side-chain of Arg1173 in the CBP bromodomain would block off H4K8ac binding due to a steric collision with the H4K5acK8ac peptide, thus explaining that the CBP bromodomain binds likely only singly acetylated-lysine in histones, whereas the BRD4 bromodomains could accommodate interactions with di-acetylated histone H4.
CONCLUSION
Bromodomain/acetyl-lysine recognition is a fundamental molecular mechanism that modulates a variety of protein-protein interactions between histones and non-histone proteins required for controlled gene transcriptional activation in chromatin. Because of the large size of human bromodomain family, it is likely that different sub-groups of bromodomains possess different functions in gene transcription. Due to the transient nature and modest affinity of individual bromodomain/acetyl-lysine interactions, in vitro biochemical binding analysis alone is not adequate to determine binding specificity of bromodomains to histones and non-histone transcriptional proteins. In this study, we present the high-resolution X-ray crystal structures of the CBP bromodomain-PHD finger tandem module in complex with two different acetylated histone H4 peptides, which contain H4K20ac and H4K12ac/K16ac, respectively. Our detailed structural analysis reveals that the PHD finger plays a structural role in the bromodomain-PHD tandem module, and that the CBP bromodomain clearly exhibits a preference for a singly-acetylated sequence motifs compromising a hydrophobic or aromatic amino acid at −2, and a lysine or arginine at −3 or −4 position from the acetylated-lysine.
Our study further provides structural insights into distinct modes of singly and di-acetylated histone H4 recognition by the bromodomains of CBP and BRD4 that function differently as a transcriptional co-activator and a chromatin organizer, respectively. Given that bromodomains are rapidly emerging as new epigenetic drug targets for treatment of a wide array of human diseases, and that the functional differences of CBP as a major transcription co-activator and BRD4 as a key chromatin organizer, this new structural knowledge is expected to facilitate future mechanistic investigation of the role of site-specific lysine acetylation in histones and transcription proteins in control of gene transcription in chromatin.
EXPERIMENTAL PROCEDURES
Preparation of Proteins and Peptides
DNA fragment encoding the bromodomain-PHD finger module of human CBP (residues 1081–1316) was amplified by PCR and sub-cloned into the pET28a-LIC vector, downstream of the poly-histidine coding region. The CBP BrD-PHD protein was over expressed in E. coli BL21 (DE3) codon plus RIL strain (Stratagene) by addition of 1 mM isopropyl-1-thio-D-galactopyranoside and incubated overnight at 18°C. Harvested cells were re-suspended in 50 mM sodium phosphate buffer, pH 7.4, supplemented with 500 mM sodium chloride, 5 mM imidazol, 2 mM β-mercaptoethanol, and 5% glycerol, and lysed using a microfluidizer (Microfluidics Corp.) at 20,000 psi. After clarification of the crude extract by high-speed centrifugation, the lysate was loaded onto a 5 mL HiTrap chelating column (GE Healthcare), charged with Ni2+. The column was washed, and the protein was eluted with 20 mM Tris-HCl buffer, pH 8.0, 250 mM sodium chloride, 250 mM imidazole, and 5% glycerol. The protein was next purified on a Superdex200 column (26×60) (GE Healthcare), equilibrated with 20 mM Tris-HCl buffer, pH 8.0, and 150 mM sodium chloride. The protein was treated with thrombin (Sigma) to cleave off its His-tag, and further purified to homogeneity by ion-exchange chromatography. The BrDs of BRD4 were purified by affinity chromatography on a nickel-IDA column (Invitrogen), followed by the removal of poly-His tag by thrombin cleavage using a procedure described previously(Zhang et al., 2012). Isotope-labeled proteins of the individual BrDs and the PHD, and the CBP bromodomain-PHD finger were prepared from cells grown on a minimal medium containing 15NH4Cl. The synthetic histone peptides were synthesized by GenScript, and confirmed by LC-MS analysis.
NMR Study
The protein (0.2 mM) and histone peptides (1.2 mM) were prepared in a 100 mM sodium phosphate buffer of pH 7.4, containing 5 mM perdeuterated DTT and 0.5 mM EDTA in H2O/2H2O (9/1). All two-dimensional 1H-15N HSQC (heteronuclear single quantum correlation) NMR spectra were collected with a 500 MHz or 600 MHz NMR spectrometer at 25°C.
Fluorescence Anisotropy Binding Assay
Binding affinity of lysine-acetylated histone peptides for the BrDs of CBP and BRD4 was assessed in a fluorescence anisotropy competition assay using a fluorescein isothiocyanate (FITC)-labeled bromodomain inhibitor as an assay probe as described previously (Zhang et al., 2012). Competition experiments were performed with a protein (0.25–1 µM) and the fluorescent probe (80 nM), and increasing concentration of unlabeled competing ligand in a PBS buffer (pH 7.4) in total volume of 80 µL. Measurements were obtained after a 1 hour incubation of the fluorescent ligand and the protein at 25°C with Safire 2 microplate reader (Tecan). In a competition-binding assay, fluorescent ligand concentration was ≤ 2Kd, and protein concentration was set at which 50–80% of fluorescent ligand is bound. Dissociation constant of a competing ligand was calculated with the correction to Cheng-Prussoff equation introduced by Nicolovska-Coleska and colleagues(Nikolovska-Coleska et al., 2004). Assuming one-site competitive binding model, the equation used to calculate Ki’s from IC50 values recovered from fitting data using Prism:
where [I50] is the concentration of free inhibitor at 50% inhibition, [L50], the concentration of free labeled ligand at 50% inhibition, and [P0], concentration of free protein at 0% inhibition. Note that Kd for each protein-probe pair is the limit of resolvable Ki in a competition assay.
Crystallization of Human CBP BrD-PHD Protein
Purified CBP BrD-PHD protein (15 mg/mL) was crystallized in complex with either histone H4K20ac peptide (residues 13–26 of H4) or H4K12ac/K16ac (residues 5–25 of H4) at 1:10 molar ratio of protein:H4 using the sitting drop vapor diffusion method at 20° C by mixing 1 µL of protein solution with 1 µL of the reservoir solution. The CBP BrD-PHD/H4K20ac complex was crystallized in 25% PEG 3,350, 0.2 M magnesium chloride, and 0.1 M HEPES pH 7.5. The CBP BrD-PHD/ H4K12ac/K16ac complex was crystallized in 20% PEG MME 2,000, 0.2 M trimethylamine N-oxide, and 0.1 M Tris pH 8.5. Crystals were soaked in the corresponding mother liquor supplemented with 20% glycerol as cryoprotectant before freezing in liquid nitrogen.
Data Collection and Structure Determination
X-ray diffraction data were collected at 100K at beamline X6A of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. Data were processed using the HKL-2000 suite(Otwinowski, 1997). The structure of the CBP BrD-PHD was solved by molecular replacement using the program MOLREP(Vagin, 1997). The crystal structure of CBP BrD (PDB code 3DWY) was used as the search model. ARP/wARP(Perrakis et al., 2001) was used for automatic model building. REFMAC (Murshudov et al., 1997) was used for structure refinement. Graphics program COOT(Emsley, 2004) was used for model building and visualization. Crystal diffraction data and refinement statistics for the structure are displayed in Table 1.
Supplementary Material
The CBP bromodomain-PHD finger tandem module functions as a single structural unit.
The CBP PHD finger has a structural role, and does not interact with histones.
The CBP bromodomain recognizes only singly acetylated-histone H4.
The CBP and BRD4 bromodomains have distinct modes of histone H4 recognition.
ACKNOWLEDGMENTS
We wish to thank Dr. Lei Zeng at the NMR facility at Icahn School of Medicine at Mount Sinai, and Dr. Jean Jakoncic and the staff at the X6A beamline of the National Synchrotron Light Sources at the Brookhaven National Laboratory for assisting X-ray data collection. This work was supported in part by the research grants from the National Institutes of Health (R01HG004508 and R01CA87658 to MMZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. Structure factors and coordinates for the CBP bromodomain-PHD finger in complex with histone H4K20ac or H4K12ac/K16ac peptide have been deposited at the Protein Data Bank under PDB ID codes 4N3W and 4N4F, respectively.
AUTHOR CONTRIBUTIONS
A.N.P. and M.-M. Z. conceived the idea and designed the experiments for this project. S.Y. and T.J.Z. prepared the protein samples and carried out crystallization. A.N.P. solved the crystal structures. S.Y. performed the NMR study. A.F. made cDNA constructs, and E.R. performed fluorescence anisotropy binding study. A.N.P. and M.-M.Z. wrote the manuscript.
References
- Borah JC, Mujtaba S, Karakikes I, Zeng L, Muller M, Patel J, Moshkina N, Morohashi K, Zhang W, Gerona-Navarro G, et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem Biol. 2011;18:531–541. doi: 10.1016/j.chembiol.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallographica. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Modulating molecular functions of p53 with small molecules. Cell Cycle. 2006;5:2575–2578. doi: 10.4161/cc.5.22.3464. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park S, Martinez-Yamout MA, Dyson HJ, Wright PE. The CH2 domain of CBP/p300 is a novel zinc finger. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland K, Breen K, ØYan A, Eberharter A, Gibson T, Becker P, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. Journal of molecular biology. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Li J, Muller M, Yan S, Mujtaba S, Pan C, Wang Z, Zhou MM. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J Am Chem Soc. 2005;127:2376–2377. doi: 10.1021/ja044885g. [DOI] [PubMed] [Google Scholar]
- Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ, 3rd, Zhou MM. Structural insights into human KAP1 PHD finger -bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008a;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008b;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua J, et al. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.