Abstract
Bacterial translation termination is mediated by release factors RF1 and RF2, which recognize stop codons and catalyze hydrolysis of the peptidyl-tRNA ester bond. The catalytic mechanism has been debated. We proposed that the backbone amide NH group, rather than the side chain, of the glutamine of the universally conserved GGQ motif participates in catalysis by H-bonding to the tetrahedral transition-state intermediate and by product stabilization. This was supported by complete loss of RF1 catalytic activity when glutamine is replaced by proline, the only residue that lacks a backbone NH group. Here, we present the 3.4 Å crystal structure of the ribosome complex containing the RF2 Q253P mutant and find that its fold, including the GGP sequence, is virtually identical to that of wild-type RF2. This rules out proline-induced misfolding, and further supports the proposal that catalytic activity requires interaction of the Gln253 backbone amide with the 3′ end of peptidyl-tRNA.
Introduction
Translation termination in bacteria is mediated by the class I release factors RF1 and RF2 (reviewed in (Dunkle and Cate, 2010; Kisselev and Buckingham, 2000; Klaholz, 2011; Korostelev, 2011; Loh and Song, 2010; Petry et al., 2008; Zhou et al., 2012)), which trigger hydrolysis of the peptidyl-tRNA ester bond in response to a stop codon presented in the ribosomal A site (Capecchi, 1967; Caskey et al., 1971; Vogel et al., 1969). RF1 recognizes UAG and UAA, and RF2 recognizes UAA and UGA (Capecchi and Klein, 1969; Scolnick et al., 1968; Scolnick and Caskey, 1969); by contrast, a single class I release factor (eRF1 or aRF1) mediates termination at all three stop codons in eukaryotic and archaebacterial cells, respectively.
Crystal structures of 70S termination complexes have provided insights into the mechanism of termination by RF1 and RF2 (Jin et al., 2010; Korostelev et al., 2008; Korostelev et al., 2010; Laurberg et al., 2008; Petry et al., 2005; Weixlbaumer et al., 2008). Both release factors recognize each nucleotide of the stop codons via specific interactions of conserved regions of domain 2, including the PVT and SPF motifs (Ito et al., 2000; Nakamura and Ito, 2002), which are involved in recognition of the second nucleotide of the respective stop codons by RF1 and RF2. Recognition of a stop codon results in rearrangements in the decoding center, including the stop codon itself and in the universally conserved nucleotides A1492, A1493 of 16S rRNA and A1913 of 23S rRNA. These rearrangements allow domain 3 of the release factor to dock into the peptidyl-transferase center (PTC) and catalyze peptidyl-tRNA hydrolysis. Catalysis is achieved by positioning the universally conserved GGQ motif located at the tip of domain 3 into the reaction center (Frolova et al., 1999; Mora et al., 2003; Seit-Nebi et al., 2001; Shaw and Green, 2007; Trobro and Aqvist, 2007). The GGQ motif is the only functional region that is conserved between RFs of bacterial, eukaryotic and archaeal origin. Based on the crystal structures of 70S termination complexes, release factors have been proposed to directly participate in peptidyl-tRNA ester-bond cleavage by providing a catalytic group (Korostelev et al., 2008; Korostelev et al., 2010; Laurberg et al., 2008; Weixlbaumer et al., 2008).
Although the importance of the 2′-hydroxyl group of ribose 76 of peptidyl-tRNA for the hydrolysis reaction has been established (Brunelle et al., 2008; Shaw et al., 2012) the contribution of the release factors themselves to the catalytic mechanism has been the subject of debate. Based on its universal conservation and ubiquitous post-translational methylation (Heurgue-Hamard et al., 2002; Heurgue-Hamard et al., 2005; Heurgue-Hamard et al., 2006; Mora et al., 2003), the side chain of the glutamine of the GGQ motif was proposed to be involved in catalysis (Heurgue-Hamard et al., 2002; Heurgue-Hamard et al., 2005; Heurgue-Hamard et al., 2006; Mora et al., 2003; Song et al., 2000). Molecular dynamics studies have suggested that the glutamine side chain plays a critical role in catalysis (Song et al., 2000; Trobro and Aqvist, 2007, 2009) by positioning the attacking nucleophilic water molecule (Trobro and Aqvist, 2009). However, such proposals are inconsistent with the observation that mutations of the glutamine to shorter or bulkier side chains confer only modest effects on the rate of peptidyl-tRNA hydrolysis (Mora et al., 2003; Seit-Nebi et al., 2001; Seit Nebi et al., 2000; Shaw and Green, 2007; Shaw et al., 2012); for example, replacement of the conserved Gln with Ala confers only a 3.8-fold decrease in rate of peptidyl-tRNA hydrolysis by RF1 using an fMet release assay (Shaw and Green, 2007) and a 4.5-fold or 4.8-fold decrease for RF1 and RF2, respectively, using release of the tetrapeptide MFTI as an assay (Mora et al., 2003). An 88-fold decrease reported for the Ala mutation by (Zavialov et al., 2002), using the same MFTI release assay, might be explained by the presence of an additional A246T mutation in the RF1 construct used in that particular study. In contrast to the small effects on hydrolysis, Shaw and Green (2007) observe that the alanine mutation confers a dramatic (14,000-fold) defect in the ability of the factor to discriminate between water and other nucleophiles, which they propose is the likely role of the conserved Gln side-chain.
An alternative proposal for the catalytic contribution of the GGQ motif emerged from high-resolution crystal structures of 70S termination complexes, which revealed that the backbone amide NH group of the glutamine is positioned to donate a hydrogen bond that could stabilize both the transition-state tetrahedral intermediate and the leaving 3′-OH group of the terminal nucleotide of the deacylated tRNA (Korostelev et al., 2008; Korostelev et al., 2010; Laurberg et al., 2008). Thus, the backbone moiety of the glutamine, rather than its side chain, was proposed to participate in catalysis of peptidyl-tRNA hydrolysis. To test the involvement of the backbone amide NH group in catalysis, we previously measured the peptide release activity of a mutant version of E. coli RF1, in which the glutamine of the GGQ motif was mutated to proline, the only residue that lacks a backbone NH amide group (Korostelev et al., 2008). This mutation would thus eliminate the ability of the polypeptide backbone to donate a hydrogen bond to either the transition-state intermediate or the leaving group. Consistent with the prediction, the peptide release activity of the proline mutant was completely abolished. It remained unclear however, whether the failure of the proline mutant to catalyze peptidyl-tRNA hydrolysis was due to loss of hydrogen-bonding capability or to indirect effects of the proline mutation, such as misfolding of the GGP motif, conformational changes in the PTC or obstructing the attack by the nucleophilic water molecule.
Here, we report the 3.4 Å crystal structure of a Thermus thermophilus 70S termination complex bound with the Q253P mutant form of RF2. We show that Q253P T. thermophilus RF2 is completely inactive in catalyzing peptidyl-tRNA hydrolysis, as previously shown for the corresponding proline mutant form of E. coli RF1(Korostelev et al., 2008). The crystal structure reveals that the conformations of the factor and the PTC are virtually identical in the mutant and wild-type structures and that no obstruction to attack by the nucleophilic water is introduced by the proline substitution. Together, our findings support the proposal that the backbone amide NH moiety of the glutamine is indispensable for the catalytic mechanism of translation termination.
Results
To test the release activity of the T. thermophilus RF2 Q253P mutant, we prepared T. thermophilus 70S pre-termination complex containing mRNA, [14C]fMet-tRNA bound to the P site in response to an AUG codon, and UAA stop codon in the A site. The complex was incubated in the presence or absence of Q253P mutant or wild-type RF2. As expected, incubation with wild-type RF2 resulted in rapid hydrolysis of [14C]fMet-tRNA (Fig. S1). The Q253P mutant was, however, completely inactive, showing no detectable release of [14C]fMet, even after prolonged incubation. These results confirm and extend our previous finding that the analogous proline mutation in E. coli RF1 completely abolishes its catalytic activity (Korostelev et al., 2008).
Because of the restriction of backbone torsion angles flanking proline residues, it cannot be ruled out that loss of catalytic activity of the Q253P mutant might be explained by distortion of the fold of the catalytic domain of the release factor; alternatively, substitution of the glutamine by proline might result in distortion of neighboring elements of the peptidyl transferase center of the ribosome. To examine these possibilities we determined the 3.4 Å crystal structure of the mutant Q253P RF2 bound to the T. thermophilus 70S ribosome in response to a UAA stop codon.
The overall position and conformation of Q253 RF2 in the 70S ribosome are similar to those in previously reported X-ray and cryo-EM structures of RF1 (Laurberg et al., 2008; Petry et al., 2005; Rawat et al., 2006; Rawat et al., 2003) and virtually identical to those of the wild-type RF2 complex (Korostelev et al., 2008; Petry et al., 2005) (Fig. 1). Domain 1 reaches from the beak of the small subunit to the L11 stalk of the large subunit; domains 2 and 4 form a superdomain, which is bound to the decoding center; and domain 3 spans the interface cavity between the decoding and peptidyl-transferase centers, with its GGP motif inserted into the PTC. The all-atom root-mean-square difference between the Q253P mutant and WT RF2 bound to the ribosome programmed with a UAA codon (Korostelev et al., 2008) is 0.7 Å, similar to the coordinate error at 3.4 Å resolution.
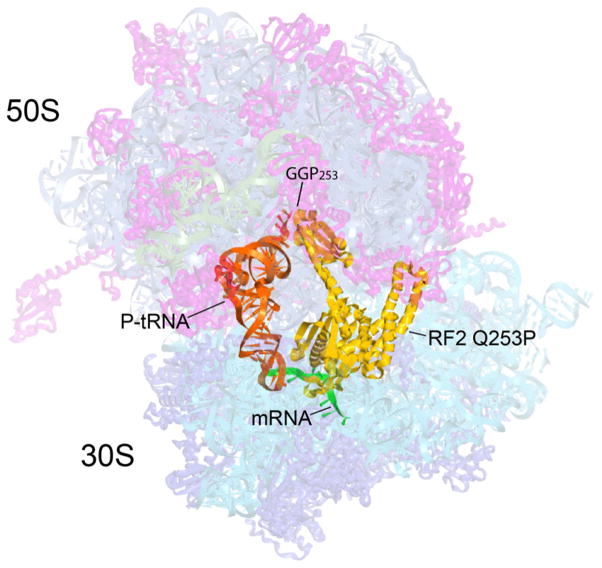
Figure 1. Overall position and orientation of RF2 Q253P in the 70S ribosome.
The positions of RF2 Q253P (yellow), P-site tRNA (orange) and M0-27 mRNA (green) are shown with the structure of the complete T. thermophilus 70S ribosome, showing 30S and 50S ribosomal proteins (blue and magenta), 16S, 5S and 23S rRNA (cyan, blue-grey and grey) in semi-transparent rendering. The position of the site of mutation in RF2, GGP253, is indicated.
Mutant RF2 interacts with the UAA stop codon in exactly the same way as wild-type RF2 (Korostelev et al., 2008) (Figs. S2, S3). The backbone of helix α5 and the conserved SPF motif form hydrogen bonds with the first (U1) and second (A2) nucleotides of the stop codon, respectively. The third codon nucleotide is separated from the second by His215, which stacks on A2. The binding pocket for the third nucleotide is formed by A530 of 16S rRNA, which stacks on A3, and the conserved Val203 and Thr216, which recognize the Hoogsteen face of A3. Thus, stop codon recognition is unaffected by the mutation.
The decoding center nucleotides A1492, A1493 and G530 of 16S rRNA are rearranged as previously observed for the interaction of the UAA stop codon with RF1 and RF2 (Korostelev et al., 2008; Laurberg et al., 2008) and the UAG codon with RF1 (Korostelev et al., 2010): A1492 is flipped out of helix 44 to stack on Trp320 of RF2, and A1913 of 23S rRNA is stacked on A1493 within helix 44 of 16S rRNA. This arrangement of the nucleotides in the decoding center allows the packing of the switch loop of RF2, which connects helix α7 of domain 3 with domain 4, as described previously (Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). Interactions of the switch loop with the decoding center and helix 69 of 23S rRNA place helix α7 in a position that allows docking of the GGP motif into the peptidyl-transferase center (Fig. 1). In summary, the conformations of the codon-recognition domain 2 of the Q253P mutant and the decoding center of the ribosome are undisturbed by the mutation.
Discussion
The crystal structure of the 70S termination complex containing the Q253P mutant form of RF2 is closely similar to that of the previously determined wild-type complex (Korostelev et al., 2008) (Figs. 2, 3, S4), supporting our proposal for the involvement of the backbone amide NH group of Q253 in catalysis (Korostelev et al., 2008; Laurberg et al., 2008). The root-mean-square difference between all atoms in RF2 and the ribosome in the core of the PTC (residues within 5 Å from the GGP motif, including the GGP motif itself) between the mutant and wild-type structures is 0.7 Å, with calculated maximum-likelihood-based coordinate errors of 0.5Å and 0.6Å–0.8Å for the structures of the mutant and wild-type complexes, respectively.
Figure 2. Superimposition of the wild-type and Q253P mutant versions of RF2.
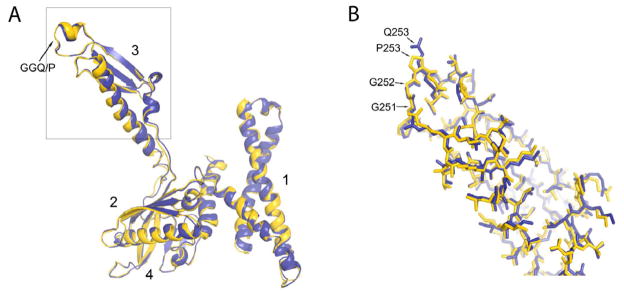
(A) Structures of wild-type (blue) and Q253P mutant (yellow) versions of RF2 from their respective termination complexes are superimposed to indicate their close conformational similarities. The all-atom rmsd value for the factors is 0.7Å. Domains 1, 2, 3, and 4 are indicated. (B) All-atom view of the boxed region of domain 3 of RF2. The structure of wild-type RF2 is from ref. (Korostelev et al., 2008). See also Figures S2 and S3.
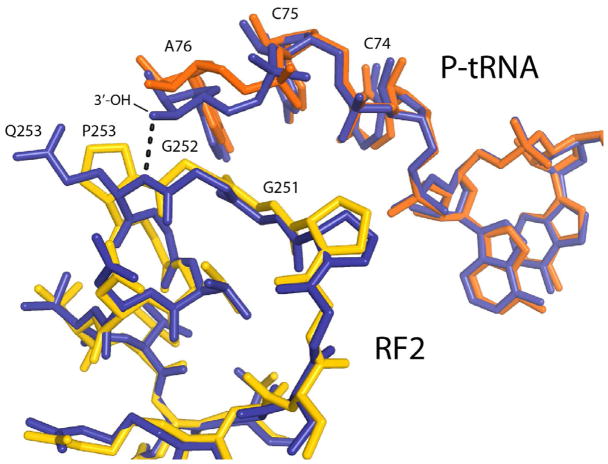
Figure 3. Detailed view of the structures of the catalytic sites of wild-type and Q253P mutant RF2.
Interactions between the GGQ and GGP motifs of wild-type (blue) and mutant (yellow) RF2 with P-site tRNA in the two corresponding termination complex structures (blue and red, respectively). The close conformational similarity between the wild-type and mutant structures is evident. A dotted line indicates H-bonding between the α-NH group of Q253 in wild-type RF2 and the 3′-OH group of the terminal A76 ribose of P-site tRNA (Korostelev et al., 2008). The bulk of proline 253 results in a ~1Å movement of A76 away from RF2. See also Figure S4B for stereo view of the 2Fo - Fc electron density map around the GGP motif and Figure S4C for stereo view of all-atom representations of the structures of wild-type (Korostelev et al., 2008) and Q253P mutant (this work) versions of release factor RF2 termination complexes. See also Figures S4A, S4B and S4C.
In wild-type release factors, the two glycines of the GGQ motif adopt backbone conformations that are disallowed for other amino acids, which helps to explain the drastic (up to 104-fold) effects of mutations at either glycine (Frolova et al., 1999; Shaw and Green, 2007). The glycines of the Q253P mutant are found in the same conformations as those of the wild-type GGQ motif (Fig. 3). A further explanation for the severe effect of substitution of these glycines is that introduction of a side chain would block access of the main-chain amide of the glutamine to the reaction center (Korostelev et al., 2008). The backbone torsion angles for the proline residue (ϕ = −64°, ψ = −25°) are similar to those found for glutamine 253 in the wild-type RF2 complex (ϕ = −48° to −67° and ψ = −7° to −37°) (Korostelev et al., 2008; Weixlbaumer et al., 2008) (Jin et al., 2010) consistent with the absence of conformational distortion by the proline mutation. The sole difference observed is a slight shift in the position of the polypeptide backbone of the GGP motif and the ribose of A76, caused by the added bulk of the proline ring (Fig. 3). Whereas the main-chain nitrogen of the wild-type RF2 glutamine is positioned within hydrogen bonding distance (3.0 Å) from the 3′-OH group of A76, the distance between the backbone N of proline 253 and the 3′-OH of A76 is increased to ~4 Å (Figs. 3, S4). The ~1Å distance difference exceeds the coordinate error of the structure (0.5 Å) and thus reflects a rearrangement between structures of the ribosome bound with Q253P RF2 and wild-type RF2(Korostelev et al., 2008).
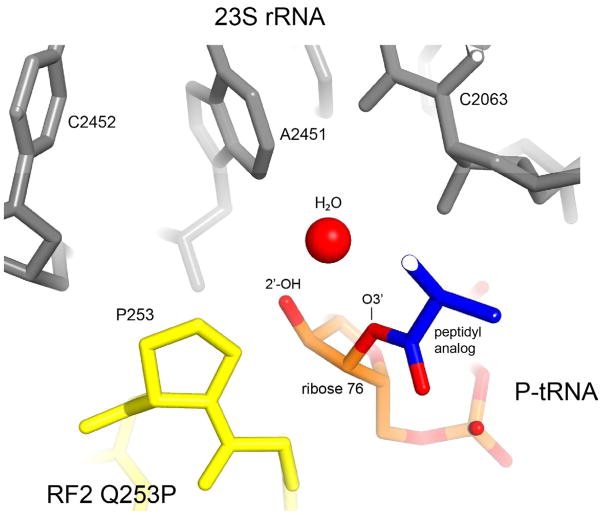
No major rearrangements are found in the conformation of Q253P RF2 or in the peptidyl transferase center of the ribosome, ruling out the possibilities that loss of peptide release activity of the Q253P mutant is due to conformational distortion or binding defects. Another potential effect of the mutation could be steric interference of the proline residue with the attacking nucleophilic water molecule. To address this possibility, we superimposed the Q253P structure with that of the 70S·RF2 complex formed in the presence of a peptidyl-tRNA substrate analog (Jin et al., 2010) (Fig. 4). In the latter study, a putative nucleophilic water molecule was modeled in the vicinity of the 2′-OH and 3′-ester groups of A76. The superposition shows that the water molecule is positioned more than 4 Å away from the proline, making it unlikely that the Q253P mutation would perturb the position of the nucleophile. The apparent lack of steric clash between the bulky proline residue and the water molecule is consistent with the high peptide release activity of a mutant version of RF1 in which the glutamine of the GGQ motif is replaced with the even more bulky tryptophan (Shaw and Green, 2007). Since none of the tested replacements of glutamine, except for the proline mutant, completely abolished peptide release activity (Korostelev et al., 2008; Mora et al., 2003; Seit-Nebi et al., 2001; Seit Nebi et al., 2000; Shaw and Green, 2007; Shaw et al., 2012), our findings strongly suggest that the complete loss of peptide release activity by the Q253P mutant is due to the loss of a catalytic group. We conclude that the defect of the proline mutant results from the inability of the backbone nitrogen of proline to donate a hydrogen bond to the leaving 3′-OH group and/or the developing oxyanion of the transition-state intermediate. Analogous mechanisms have been proposed for catalysis by other hydrolases, including proteases, esterases and GTPases, in which the α-NH group stabilizes formation of the product and the developing negative charge of the transition state (Jaeger et al., 1999; Wilmouth et al., 2001).
Figure 4. Position of a putative attacking nucleophilic water molecule in the context of the Q253P mutant RF2 complex.
The structure of a termination complex containing a peptidyl-tRNA analog, in which a nucleophilic water molecule was modeled in the vicinity of the 2′- and 3′-OH groups of A76 of the peptidyl-tRNA (Jin et al., 2010), was superimposed on the structure of the Q253P RF2 termination complex. It is evident that the proline would not present a steric block to the approach of the water molecule to its target.
Experimental Procedures
Chemicals and buffers
Nucleoside triphosphates and tRNAfMet were purchased from Sigma-Aldrich, radioisotopes from Perkin-Elmer, PfuTurbo DNA polymerase from Agilent Technologies and other chemicals from Sigma-Aldrich or Fisher Scientific. Ribosome and RF2 reaction and storage buffer, used in all experiments, contained 25 mM TrisOAc (pH 7.0), 10 mM Mg(OAc)2 (if not otherwise stated), 50 mM KOAc (pH 7.0) and 10 mM NH4OAc. The mRNA M027 [GGC AAG GAG GUA AAA AUG UAA AAA AAA] was chemically synthesized (IDT) (Korostelev et al., 2008; Laurberg et al., 2008).
Mutagenesis and purification of Thermus thermophilus RF2
In vitro site-directed mutagenesis was performed to replace the glutamine at position 253 of Thermus thermophilus RF2 with proline using Stratagene QuickChange. RF2 Q253P mutant protein was purified following the procedure described for RF1 (Laurberg et al., 2008).
Purification of ribosomes
Ribosomes were isolated from wild-type Thermus thermophilus HB27 cells, grown in a 120 L fermentor and harvested at mid-log phase, essentially as described in (Cate et al., 1999). Cell disruption and ribosome purification, dissociation and reassociation were conducted as described (Laurberg et al., 2008).
Aminoacylation of tRNAfMet
Aminoacylation and formylation of tRNAfMet was conducted using [14C]fMet for 30 min at 37°C as described (Lancaster and Noller, 2005). The extent of aminoacylation was assessed by TCA precipitation of small aliquots and the 14C content was quantified using scintillation counting as described (Ivanova et al., 2004).
Termination assay
Ribosomal pre-termination complexes were generated by incubating Thermus thermophilus 70S ribosomes diluted to 5 μM final concentration with a 1.5-fold molar excess of mRNA (M0-27) and a 0.2 molar equivalent of [14C]-fMet-tRNA for 30 min at 37° in 50 mM K-HEPES (pH 7.5), 20 mM MgCl2, 75 mM NH4Cl, and 1 mM DTT buffer. The complexes were then diluted five-fold in the same buffer and concentrated by ultrafiltration through 100 kD cutoff Microcon spin-concentrators to remove unbound [14C]-fMet-tRNA. The dilution and concentration steps were repeated three times. Purified pre-termination complexes were divided into three reactions and a 5-fold molar excess of wild-type or Q253P RF2 or no release factor was added to each reaction. The reactions were incubated for 30 minutes at 37°. Half of each reaction was quenched with 5% (v/v) formic acid (final concentration) after 90 seconds and the remaining amount after 30 minutes. The resulting precipitate, containing ribosomes with still bound [14C]-fMet-tRNA (as well as other components of the translation mix) was recovered by centrifugation and dissolved in 20 μl 1 M NaOH. The amount of released [14C]-fMet in the supernatant and the amount of remaining [14C]-fMet-tRNA bound to the ribosome in the pellet were determined by scintillation counting. We define 100% as the sum of [14C]-fMet released plus the [14C]-fMet remaining bound to the ribosome.
Formation of ribosomal termination complexes for crystallization
To form ribosomal complexes containing Q253P RF2, Thermus thermophilus 70S ribosomes diluted to 3 μM final concentration were incubated with a 2-fold molar excess of mRNA (M0-27) and a 2-fold molar excess of deacylated tRNAfMet for 30 min at 37° in 25 mM Tris acetate (pH 7.0), 10 mM magnesium acetate, 10 mM ammonium acetate and 50 mM potassium acetate (pH 7.0) buffer. A 4-fold molar excess of the Q253P RF2 mutant release factor was then added to the ribosomal complex in the same buffer, and the reaction was incubated for an additional 20 min. Prior to crystallization, Deoxy Big Chaps (Hampton Research) diluted in the reaction buffer was added slowly to the ribosomal complexes to a final concentration of 1.7 μM.
Crystallization and cryoprotection
Crystallization was performed essentially as described (Zhu et al.) by optimizing around previously reported conditions (Korostelev et al., 2008; Laurberg et al., 2008). Following optimization, crystals were grown by the sitting-drop vapor-diffusion method using drops containing 1 μl ribosome complex mixed with 1 μl of reservoir solution [100 mM Tris-OAc, pH 7.0, 200 mM KSCN, 3.6–5 % PEG 20,000, 3 – 11.9 % v/v PEG 200] for three to four weeks at 22.5°C. The reservoir solution giving the best-diffracting crystals contained 100 mM Tris-OAc, pH 7.0, 200 mM KSCN, 3. 95% PEG 20,000, 7.5 % (w/v) PEG 20K. Cryoprotection and crystal freezing were performed as described (Laurberg et al., 2008).
Data collection and structure determination
Crystals were screened at beamlines 7.1, 9.1, 9.2, 11.1 and 12.2 at the Stanford Synchrotron Radiation Laboratory, and at the SIBYLS beam line 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. X-ray diffraction data were collected at beamline 12.2 at the Stanford Synchrotron Radiation Laboratory using the PILATUS 6M PAD detector, at an X-ray wavelength of 1.0332 Å and an oscillation angle of 0.2°. For determining the structure of the 70S·M027·tRNAfMet•RF2(Q253P) complex, one dataset obtained from a single crystal was integrated and scaled using XDS (Kabsch, 2010). 2% of the unique reflections were marked as test-set (Rfree set) reflections and used for cross-validation throughout refinement. The previously determined X-ray structure of the 70S·RF2 termination complex obtained from the same crystal form (Korostelev et al., 2008) was used as a starting model for molecular replacement. Initial Fobs-Fcalc and 3Fobs-2Fcalc difference maps were calculated using the structure of the 70S ribosome, excluding RF2, mRNA and the CCA end of the P-site tRNA to avoid model bias. The density for the omitted parts of the structure was clearly visible in unbiased Fobs-Fcalc maps. Structure determination was carried out using PHENIX (Adams et al., 2002) and CNS (Brunger et al., 1998) as described (Korostelev et al., 2008). Coot (Emsley and Cowtan, 2004) was used for structure visualization and calculation of maps averaged using two-fold non-crystallographic symmetry (Figs. S3 and S4). Figures were rendered using PyMOL (DeLano, 2002). Information on data collection and refinement statistics can be found in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |||||
|
| |||||
| Space group | P212121 | ||||
| Cell dimensions | |||||
| a, b, c (Å) | 212.07, 454.4, 618.45 | ||||
| α, β, γ (°) | 90, 90, 90 | ||||
| Resolution (Å) | 3.4 (3.4 – 3.5) * | ||||
| Rp.i.m# | 0.152 (1.1) | ||||
| CC(1/2)## | 99.3 (43) | ||||
| I/σI | 7.84 (1.12**) | ||||
| Completeness (%) | 100 (100) | ||||
| Redundancy | 7.0 (7.2) | ||||
| Refinement | |||||
| Resolution (Å) | 50 – 3.4 | ||||
| No. reflections | 807,927 | ||||
| Rwork/Rfree | 0.2343/0.2681 | ||||
| No. atoms | 294,088 | ||||
| Ions modeled as Mg2+ | 2410 | ||||
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.004 | ||||
| Bond angles (°) | 0.621 | ||||
Values in parentheses are for highest-resolution shell.
I/σI=2.0 at 3.65 Å
Rp.i.m. is the precision-indicating merging R factor (Weiss, 2001)
CC(1/2) is the percentage of correlation between intensities from random half-datasets as defined by Karplus and Diederichs (Karplus and Diederichs, 2012)
Supplementary Material
Highlights.
New evidence for the catalytic role of the backbone amide group of the release factors
Th. thermophilus RF2 Q253P mutant lacks catalytic activity
Unaltered conformation and ribosome binding of Th. thermophilus RF2 Q253P
Acknowledgments
This work was supported by grants from the Worcester Foundation for Biomedical Research and the UMMS Center for AIDS Research (to A.A.K) and from the NIH and the Agouron Foundation (to H.F.N.). We would like to thank M. Soltis, T. Doukov and the beamline staffs at Stanford Synchrotron Light Source and at beamline 23-ID at the Advanced Photon Source as well as S. Classen at beamline 12.3.1 at the Advanced Light Source for expert advice and support during screening and data collection, and L. Lancaster for comments on the manuscript. Atomic coordinates and structure factors have been deposited at the PDB with accession numbers 4KFH, 4KFI, 4KFL and 4KFK.
Footnotes
Supplemental information includes two figures and one table and is available online at http//…
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Shaw JJ, Youngman EM, Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. Rna. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci U S A. 1967;58:1144–1151. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR, Klein HA. Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harb Symp Quant Biol. 1969;34:469–477. doi: 10.1101/sqb.1969.034.01.053. [DOI] [PubMed] [Google Scholar]
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci U S A. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. Rna. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. Embo J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Mora L, Merkulova-Rainon T, Kisselev LL, Buckingham RH. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J Biol Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem. 2006;281:36140–36148. doi: 10.1074/jbc.M608571200. [DOI] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annual review of microbiology. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci U S A. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev LL, Buckingham RH. Translational termination comes of age. Trends Biochem Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- Klaholz BP. Molecular recognition and catalysis in translation termination complexes. Trends Biochem Sci. 2011;36:282–292. doi: 10.1016/j.tibs.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Loh PG, Song H. Structural and mechanistic insights into translation termination. Curr Opin Struct Biol. 2010;20:98–103. doi: 10.1016/j.sbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K. A tripeptide discriminator for stop codon recognition. FEBS Lett. 2002;514:30–33. doi: 10.1016/s0014-5793(02)02330-x. [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick EM, Caskey CT. Peptide chain termination. V. The role of release factors in mRNA terminator codon recognition. Proc Natl Acad Sci U S A. 1969;64:1235–1241. doi: 10.1073/pnas.64.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit Nebi A, Frolova L, Ivanova N, Poltaraus A, Kiselev L. Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol Biol (Mosk) 2000;34:899–900. [PubMed] [Google Scholar]
- Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JJ, Trobro S, He SL, Aqvist J, Green R. A Role for the 2′ OH of peptidyl-tRNA substrate in peptide release on the ribosome revealed through RF-mediated rescue. Chem Biol. 2012;19:983–993. doi: 10.1016/j.chembiol.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. Mechanism of the translation termination reaction on the ribosome. Biochemistry. 2009;48:11296–11303. doi: 10.1021/bi9017297. [DOI] [PubMed] [Google Scholar]
- Vogel Z, Zamir A, Elson D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–5168. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130– 135. [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmouth RC, Edman K, Neutze R, Wright PA, Clifton IJ, Schneider TR, Schofield CJ, Hajdu J. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat Struct Biol. 2001;8:689–694. doi: 10.1038/90401. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- Zhou J, Korostelev A, Lancaster L, Noller HF. Crystal structures of 70S ribosomes bound to release factors RF1, RF2 and RF3. Curr Opin Struct Biol. 2012 doi: 10.1016/j.sbi.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Korostelev A, Costantino DA, Donohue JP, Noller HF, Kieft JS. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci U S A. 108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.