Abstract
‘Superantigens’ (SAgs) trigger the massive activation of T cells by simultaneous interactions with MHC and TCR receptors, leading to human diseases. Here we present the first crystal structure, at 2.5-Å resolution, of a complete ternary complex between a SAg and its two receptors, HLA-DR1/HA and TCR. The most striking finding is that the SAg Mycoplasma arthritidis mitogen, unlike others, has direct contacts not only with TCR Vβ but with TCR Vα.
To investigate the molecular mechanism of SAg recognition by host receptors, we determined the crystal structure of a complete ternary complex involving Mycoplasma arthritidis mitogen (MAM, the SAg in the complex), murine single-chain T-cell receptor T7 (scTCR-T7) derived from mouse CD8+ clone 2C (refs. 1,2), and a human class II major histocompatibility complex (MHC II) HLA-DR1 receptor bound to a hemagglutinin peptide (HLA-DR1/HA; peptide-bound MHC in general is abbreviated pMHC) (Fig. 1, Supplementary Methods and Supplementary Table 1 online). In the ternary complex, MAM acts as a bridge between the TCR and MHC molecules (Fig. 1a). As a consequence, the TCR-T7 interacting with MAM does not have any contacts with the MHC II and the associated HA peptide. Similar findings have also been reported for the hypothetical models of other TCR–SAg–pMHC complexes, although the overall orientations of TCRs relative to MHC II are very different from that in the TCR–MAM–MHC complex3,4. These results indicate that recognition of TCR by a SAg does not require specificity for the pMHC that presents the SAg. The structure of the T7-TCR–MAM–HLA-DR1/HA complex readily accounts for immunological evidence indicating that SAgs can activate both CD4+ and CD8+ T cells5. However, unlike the peptide antigens that are normally presented to CD8+ TCR by MHC I, SAgs stimulate CD8+ T cells via MHC II. Nevertheless, in the presence of MHC II–expressing cells, MAM can stimulate the activation of TCR-2C transfectants and other CD8+ T cells, as efficiently as MAM does for CD4+ T cells (data not shown and ref. 6).
Figure 1.
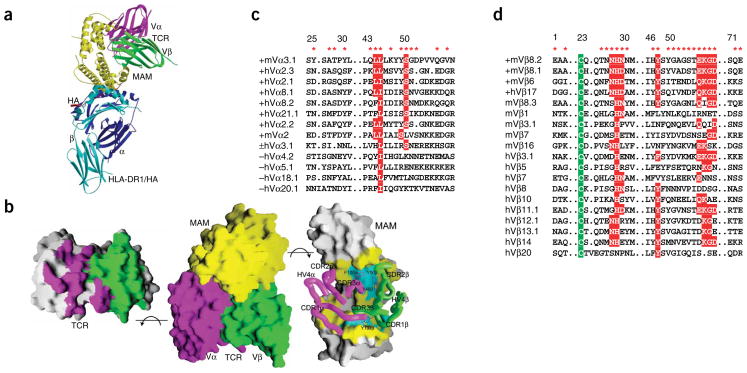
Crystal structure of scTCR–MAM–HLA-DR1/HA ternary complex. Lime, MAM; blue, DR1α; cyan, DR1β; red, HA; purple, TCR Vα; green, TCR Vβ. (a) Structure of the scTCR–MAM–HLA-DR1/HA complex. (b) Interaction surfaces of MAM and scTCR T7. Center, surface presentation of the MAM–scTCR structure; left, opened-up view of the MAM-binding surface of scTCR; right, opened-up view of the TCR-binding surface of MAM. Hydrophobic surface patches are colored in cyan. CDR and HV4 loops of TCR are shown as ‘worms’. Selected aromatic residues of TCR at the interface are shown as rods. (c) Sequence alignment of the Vα residues of MAM-reactive (indicated by +) and unreactive (−) TCRs. Conserved or conservatively substituted residues (red boxes) and MAM-contacting residues (red asterisks) are indicated. (d) Sequence alignment of MAM-contacting residues of TCR Vβs that are most frequently activated by MAM (indicated by +) and TCR Vβs that are less frequently activated by MAM. Conserved residues indicated as in c; green boxes mark strictly conserved cysteine residues.
The MAM-scTCR interaction buries a total of 3,444 Å 2 of surface area, which is substantially larger than those (1,200–2,000 Å 2) in TCR–pMHC, SAg–pMHC and other SAg–TCR complexes (Fig. 1b)3,4,7. Upon MAM binding, the Vα domain of TCR contributes a buried surface area of 622 Å 2, which accounts for more than one-third of the total contribution from TCR (Fig. 1b). The buried surface area from TCR Vα is comparable with those (600–900 Å 2) from TCR Vβ domains in other SAg–TCR-β complexes3,4. A total of 17 Vα residues, from Vα complementarity-determining region-1 (CDR1; 6% of total TCR contribution), framework region-2 (FR2; 3%), CDR2 (14%); FR3 (3%) and CDR3 (10%), are involved in both van der Waals contacts and specific hydrogen bonds at the MAM-TCR interface (Fig. 1b, Supplementary Fig. 1 and Supplementary Table 2 online).
The specific MAM-Vα contacts could imply that MAM-TCR recognition has sequence specificity for the TCR Vα, a property that could be manifested in skewed Vα expression by MAM-reactive T cells. Although the issue has not been systematically investigated, several functional studies have indicated that recognition of MAM is associated with selective use of particular Vα regions8,9. Sequence alignment indicates that two residues, Leu/Ile46 and Ser51, are conserved or nearly conserved among MAM-reactive α chains (Fig. 1c). In addition, Leu45 is a dominant residue for the MAM-reactive α chains, but it is not present in the MAM-unreactive ones. Upon MAM binding, these residues also make large contributions to the buried surface area and/or form hydrogen bonds with MAM residues (Supplementary Fig. 1 and Supplementary Table 2). Therefore, these residues may represent a MAM-binding motif on TCR Vα and could be crucial for MAM recognition.
The crystal structure of the ternary complex clearly indicates that MAM directly interacts with the TCR α chain. This finding is quite distinct from previous studies of SAgs, which have defined their superantigen properties as a result of interactions with only the TCR Vβ region3, and it significantly advances understanding of SAg structures and functions. Thus, certain SAgs may have specificity for not only TCR Vβ but also Vα. It is even possible that certain SAgs, such as staphylococcal enterotoxin H (SEH)10, may have only TCR Vα specificity. Direct interactions similar to those we observed between MAM and TCR-T7 could exist between TCR Vα10 and SEH, leading to skewed Vα expression.
In our complex, MAM also binds the TCR β chain, with a buried surface area of 1,131 Å 2 (Fig. 1b). A total of 27 Vβ residues, from FR1 (2% of total TCR contribution), CDR1 (19%), CDR2 (20%), FR3 (9%), HV4 (4%) and CDR3 (10%), contact MAM (Supplementary Fig. 1 and Supplementary Table 2). The crystal structure readily accounts for mutational and immunological evidence implicating Vβ CDR3 residues, Tyr107 in particular, in MAM recognition (Fig. 1b)8.
Other than the involvement of Vβ CDR3 (CDR3β), the MAM-binding site on TCR Vβ nearly overlaps with that for staphylococcal enterotoxins B and C3 (SEB and SEC3) and streptococcal pyrogenic exotoxin A (SpeA)3,4. However, the mechanisms for SAg recognition by TCRs are quite different for MAM and for SEB and SEC3. It has been hypothesized that recognition of SEB and SEC3 by TCR is conformation dependent but may be highly independent of the TCR Vβ sequence3.
In contrast, MAM forms numerous specific hydrogen bonds and van der Waals interactions with both main chain and side chain atoms of Vβ8.2 (Supplementary Fig. 1 and Supplementary Table 2), implying that MAM recognition by TCR is sequence dependent. Indeed, MAM recognizes fewer TCR β chains than do SEB and SEC3. T cells bearing murine Vβ8.1, Vβ8.2 or Vβ6, or human Vβ17, are most frequently activated by MAM. Sequence alignment indicates that several MAM-contacting residues are conserved among these Vβ regions (Fig. 1d), including Asn28, His29 and Asn/Asp30 of CDR1, Tyr48 and Gln/Glu56 of CDR2, and Lys57, Gly58 and Asp59 of FR3. Five of these residues are completely or nearly completely buried upon MAM binding, and ten hydrogen bonds are formed via the side chain atoms of these residues (Supplementary Fig. 1 and Supplementary Table 2). Therefore, the conserved residues may represent a MAM-binding motif and account, at least in part, for the TCR Vβ specificity in MAM recognition. Sequence alignment of TCR Vβ regions less frequently activated by MAM supports the notion that these residues, collectively, account for the binding specificity of the Vβ regions in the interaction with MAM, as none of the less frequently activated Vβs have all of the conserved motif residues (Fig. 1d).
In agreement with our crystal structure, marked structural differences between MAM-reactive and unreactive TCR Vβ domains have been observed at the CDR1, CDR2 and FR3 regions11. It should be noted that the latter two regions also contribute most strongly to the binding of pyrogenic SAgs, suggesting a common recognition mechanism that permits MAM and other SAgs to interact with various Vβ families. Accordingly, CDR2 and FR3 regions of TCR β chains are required for binding and may determine the SAg specificity.
The TCR binds in a deep, curved groove between the two domains of MAM (Fig. 1b). A total of 45 residues from both the MAM N- and C-terminal domains (NTD and CTD) are involved in the contacts. Involvement of the CTD is consistent with previous observations that two C-terminal deletion mutants of MAM, terminated at positions 132 and 176, respectively, do not trigger T cell activation12. The structure can also reconcile the finding that peptides composed of MAM residues 14–31 or 11–38 inhibit MAM-induced T-cell activation, as residues 18–29 contact TCR and residues 11–18 interact with pMHC13.
Superimposition of the preformed pMHC–MAM (NTD only) binary complex13 onto the ternary complex resulted in only a small r.m.s. deviation of 0.65 Å (Fig. 2a). No large movement of the domains relative to each other occurs in either the MAM NTD or pMHC. This indicates that, upon TCR binding, the preformed MAM–pMHC binary complex preserves its binding geometry.
Figure 2.
Conformational changes in MAM and TCR upon complex formation. (a) Superposition of the MAM–HLA-DR1/HA complex (green) determined in our earlier study13 onto its counterpart (red) in the ternary complex. The MAM CTD is not included in superposition. (b) Superposition of the MAM CTD in the previous MAM–HLA-DR1/HA binary complex13 onto its counterpart (red) in the ternary complex. (c) Superposition of the ligand-free TCR-2C (ref. 2), and TCR-2C in complexes with H-2Kb MHC I with an agonist14 and a superagonist peptide15, onto TCR T7 in our ternary complex. Light gray, non-CDR regions; purple, CDRs of Vα; green, T7 Vβ; lime, free 2C; pink, 2C with agonist peptide; cyan, 2C with superagonist peptide.
In contrast with the MAM-pMHC interaction, substantial domain movement in MAM is associated with TCR binding (Fig. 2a), although no global change was observed for the VαVβ geometry of TCR. The MAM CTD rotates about 11.8° toward the MAM NTD upon TCR binding. This differs from conventional TCR–pMHC and other TCR–SAg–pMHC complexes, in which no major domain movements of the interacting species have been observed3,7. Thus, such a domain movement in the ligand seems to be required for optimization of domain orientation to favor TCR binding, but not for signaling.
In addition to global domain movement, both MAM and TCR undergo substantial main chain conformational changes at the binding interface (Fig. 2b,c). In contrast, no major rearrangements in the polypeptide backbones of the interacting species have been observed in the pyrogenic SAg-mediated complexes3,4. In TCR, three of six CDRs (CDR1α, CDR3α and CDR3β), as well as Vα HV4, undergo marked conformational changes upon MAM binding (Fig. 2c). The conformational changes imply an induced-fit mechanism for MAM-TCR recognition.
Notably, CDR1α, CDR3α and HV4α of TCR-2C (ref. 2) also undergo substantial conformational changes of similar magnitude upon pMHC I binding7, but these conformational changes are quite different from those found in the TCR-MAM interaction (Fig. 2c). Thus, although the MAM–HLA-DR1/HA complex and the MHC I peptide/H-2Kb complex14 are very different ligands for TCR-2C, they both induce conformational changes in TCR, and these unique conformations provide sufficient binding energy to yield productive signaling.
Supplementary Material
Acknowledgments
This research was supported by grants AI50628 (to H.L.) and AI064611 (to D.M.K.) from the US National Institutes of Health. W.M. is supported by grants from the Arthritis Society of Canada, the Canadian Arthritis Network and the Canadian Institutes of Health Research. We thank A. Verschoor for critical reading of the manuscript, L.J. Stern (University of Massachusetts Medical School) for the gift of HLA-DR1 expression plasmids, and core facilities at the Wadsworth Center, including the Peptide Synthesis, Molecular Genetics and Macromolecular Crystallography cores, for synthesis of the HA peptide, DNA sequencing and crystal evaluation. Data for this study were collected at beamline X4A of the National Synchrotron Light Source, which is supported by the US Department of Energy, by grants from the US National Institutes of Health and by the New York Structural Biology Center.
Footnotes
Accession codes. Protein Data Bank: Coordinates have been deposited with accession code 2ICW.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 2.Garcia KC, et al. Science. 1996;274:209–219. [Google Scholar]
- 3.Li H, Llera A, Malchiodi EL, Mariuzza RA. Annu Rev Immunol. 1999;17:435–466. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Sundberg EJ, Li Y, Mariuzza RA. Curr Opin Immunol. 2002;14:36–44. doi: 10.1016/s0952-7915(01)00296-5. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann T, MacDonald HR. Semin Immunol. 1993;5:33–39. doi: 10.1006/smim.1993.1005. [DOI] [PubMed] [Google Scholar]
- 6.Matthes M, et al. Eur J Immunol. 1988;18:1733–1737. doi: 10.1002/eji.1830181112. [DOI] [PubMed] [Google Scholar]
- 7.Rudolph MG, Standfield RL, Wilson IA. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 8.Hodtsev AS, Choi Y, Spanopoulou E, Posnett DN. J Exp Med. 1998;187:319–327. doi: 10.1084/jem.187.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Sun GR, Tumang JR, Crow MK, Friedman SM. J Clin Invest. 1994;94:2525–2531. doi: 10.1172/JCI117624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersson K, Pettersson H, Skartved NJ, Walse B, Forsberg G. J Immunol. 2003;170:4148–4154. doi: 10.4049/jimmunol.170.8.4148. [DOI] [PubMed] [Google Scholar]
- 11.Li H, et al. Protein Sci. 2005;14:3025–3038. doi: 10.1110/ps.051748305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langlois MA, Etongue-Mayer P, Ouellette M, Mourad W. Eur J Immunol. 2000;30:1748–1756. doi: 10.1002/1521-4141(200006)30:6<1748::AID-IMMU1748>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, et al. Structure. 2004;12:277–288. doi: 10.1016/j.str.2004.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia KC, et al. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 15.Degano M, et al. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.