Summary
Protein synthesis is inherently a dynamic process, requiring both small- and large-scale movements of tRNA and mRNA. It has long been suspected that these movements might be coupled to conformational changes in the ribosome, and in its RNA moieties in particular. Recently, the nature of ribosome structural dynamics has begun to emerge from a combination of approaches, most notably cryo-EM, X-ray crystallography and FRET. Ribosome movement occurs both on a grand scale, as in the intersubunit rotational movements that are coupled to tRNA-mRNA translocation, and in intricate localized rearrangements such as those that accompany codon-anticodon recognition and peptide bond formation. In spite of much progress, our understanding of the mechanics of translation is now beset with countless new questions, reflecting the vast molecular architecture of the ribosome itself.
Introduction
One of the most intriguing of all biological structures is the ribosome, the ribonucleoprotein complex that is responsible for translation of the genetic code to produce proteins in all living organisms. In its simplest bacterial and archaeal forms, it is composed of 16S rRNA (~1500 nts), 23S rRNA (~2900 nts) and 5S rRNA (~120 nts), which make up about 60 per cent of its mass, and more than 50 different proteins. The small (30S) subunit, which contains 16S rRNA and about 20 proteins, binds the mRNA and is responsible for mediating codon-anticodon interaction. The large (50S) subunit, which is made up of the 23S and 5S rRNAs and more than 30 proteins, contains the peptidyl transferase catalytic site. Binding of tRNA to the A (aminoacyl), P (peptidyl) and E (exit) sites (Fig. 1a) and their translocation through the ribosome are functions to which both subunits contribute. Although each phase of protein synthesis - initiation, elongation and termination - consists of multiple steps requiring numerous extra-ribosomal translation factors, most if not all of these steps are based on molecular mechanisms of the ribosome itself. And to the extent that we presently understand ribosomal mechanisms, they all appear to involve participation of rRNA, in keeping with the presumed ancient origins of the ribosome in an RNA world [1-4].
Figure 1.
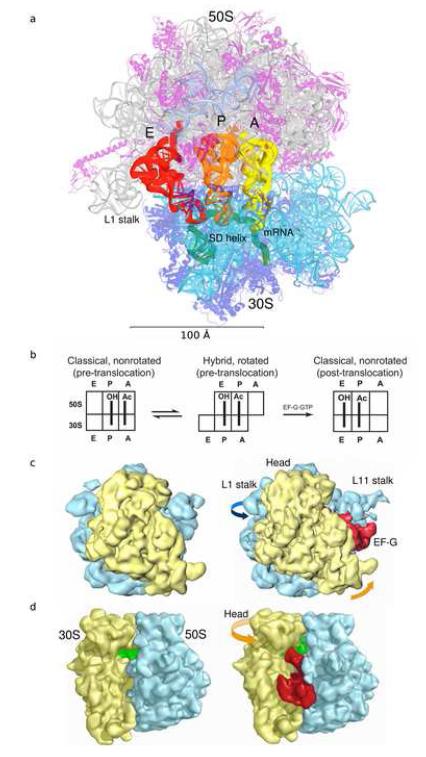
Positions and translocation of tRNA in the 70S ribosome. (a) tRNAs bound to the A (yellow), P (orange) and E (red) sites of the T. thermophilus 70S ribosome [60]. mRNA (green) and Shine-Dalgarno helix are indicated. Molecular features are indicated by color: 16S rRNA (cyan), small subunit proteins (blue), 23S rRNA (grey), 5S rRNA (light blue) and large subunit proteins (magenta). The model was constructed by superimposing the A-site tRNA and the SD helix from their corresponding 70S complexes [35,47] into the 2.8 Å structure of the T. thermophilus 70S ribosome [31]. (b) Translocation of tRNA from the A and P sites to the P and E sites, respectively. (c) 30S solvent view and (d) A-site view of cryo-EM maps for the non-ratcheted (70S•tRNA, left) and ratcheted (70S•tRNA•EF-G•GDPNP, right) states of the 70S ribosome. In panels c and d the 30S and 50S subunits are shown in yellow and light blue and the tRNA and EF-G in green and red, respectively. The figures in panels c and d were adapted from references [12,61].
The dynamic nature of protein synthesis can be inferred from the nature of the translation elongation cycle (Fig. 1b), which involves translocation of tRNAs through the ribosome along a path of more than 100 Ångstroms, in steps of some tens of Ångstroms (Fig. 1a). It was long ago anticipated that such large-scale molecular movements would involve corresponding structural changes in the ribosome [5,6]. Forty years later, molecular movements in the ribosome that accompany translocation are being observed directly, even in single ribosomes and in real time. Furthermore, detailed examination of its structure suggests that the ribosome is a flexible, dynamic object, very unlike the man-made machines that are often invoked in discussions of mechanism. The challenge is now to understand the many modes of ribosome dynamics, and how they enable the complex processes of translation.
Intersubunit movement
The two-subunit organization of the ribosome was first linked to its structural dynamics by Spirin [6] and Bretscher [5] who independently predicted that translocation, the coupled movement of mRNA and tRNA throught the ribosome, is based on intersubunit movement. In the late 1980s, chemical footprinting experiments showed that translocation takes place in two consecutive steps [7]. In the first step, the tRNAs move on the 50S subunit, leading to formation of a hybrid-state intermediate (Fig. 1b). In the second step, EF-G·GTP catalyzes their movement, coupled to that of mRNA, on the 30S subunit. This result, together with sedimentation and neutron scattering studies ([8,9] and references therein), was suggestive of a role for intersubunit movement in translocation.
The first direct evidence for independent movement of the ribosomal subunits during translocation came from the cryo-EM studies of Frank and Agrawal [10]. Ribosome complexes containing elongation factor EF-G bound with either a non-hydrolysable analog of GTP (GDPNP) or GDP and fusidic acid (an antibiotic that prevents release of EF-G following GTP hydrolysis) were found to have an altered conformation, in which the small subunit was rotated counter-clockwise with respect to the large subunit, and tRNA appeared to be bound in the hybrid P/E state (Fig. 1c) [10-12]. This finding prompted the proposal that translocation is driven by a ratchet-like mechanism that is coupled to intersubunit rotation. In a critical test of this model, introduction of an intersubunit disulfide bridge between protein S6 on the 30S subunit and L2 on the 50S subunit was found to specifically block translocation, showing that intersubunit movement is indeed required for translocation [13]. The axis of rotation was localized to the vicinity of intersubunit bridge 3 (residues 1483-1486 of h44 of 16S RNA and 1948-1949 of h71 of 23S rRNA). Accordingly, intersubunit bridges located near the rotation axis (B2a-c, B3, B5 and B7a), appear to be essentially maintained during intersubunit rotation, while bridges located at the extremities of the subunits (such as B1a-b, B7b and B8) are disrupted or rearranged.
Recently, intersubunit rotation has been directly observed in solution using FRET. Binding of EF-G was found to cause counterclockwise rotation of the small subunit in ribosomes containing fluorophores attached to proteins S6 (or S11) in the 30S subunit and L9 in the 50S subunit [14]. By combining chemical probing and FRET studies, it was shown that the EF-G-induced rotation corresponds to formation of the hybrid state characterized in the early chemical probing studies, and can occur in the absence of EF-G under conditions that favor spontaneous hybrid state formation [14,15]. Thus, the hybrid-state and ratchet models have converged on a common unified mechanism for translocation (Fig 1b).
A shortcoming of ensemble FRET experiments is that the behavior of individual molecules is masked by averaging. This problem is overcome by single-molecule FRET (smFRET) experiments. Using smFRET, fluorescently-labeled tRNAs bound to pre-translocation ribosomes were observed to move spontaneously relative to one another, interpreted as fluctuations between the classical and hybrid states [16-18]. This was shown directly in smFRET experiments using fluorescently-labeled ribosomal subunits, in which pre-translocation ribosomes containing deacylated tRNA in the P site were seen to fluctuate spontaneously between two rotational conformations corresponding to the classical and hybrid states [19]. In contrast, post-translocation ribosomes, containing peptidyl-tRNA in the P site were fixed predominantly in the classical, non-rotated state. Analysis of equilibria between the two intersubunit conformations, based on FRET and chemical probing data, shows that stabilization of the rotated, hybrid state is influenced both by movement of the acceptor stem of deacylated tRNA into the 50S E site and by binding of EF-G [19,20].
Binding of initiation factor IF2 [21], release factor RF3 [14,22,23] or ribosome recycling factor RRF [24] also causes movement into the rotated, hybrid state, extending the involvement of intersubunit rotation to the initiation and termination phases of protein synthesis. Moreover, counterclockwise rotation of the small subunit was also seen in cryo-EM reconstructions of EF-2-containing complexes of the eukaryotic ribosome [25,26]. Thus, intersubunit movement and the hybrid-state intermediate appear to be universal features of translation that account for conservation of the two-subunit organization of ribosomes among all branches of life.
Movement of the head of the small subunit
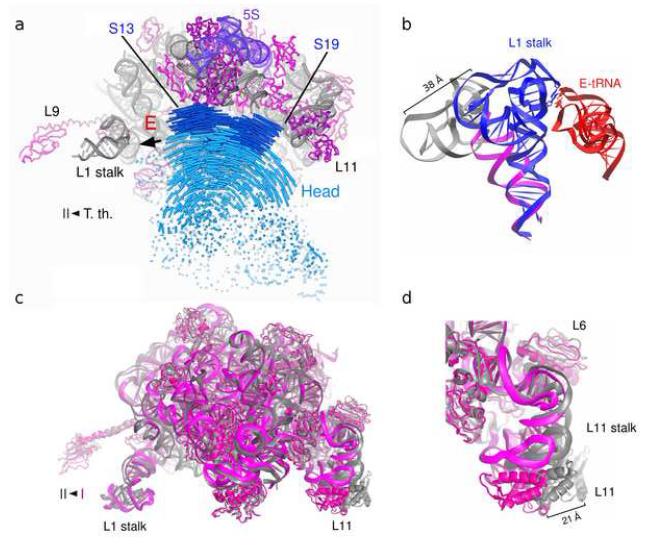
Careful inspection of the conformations of the ribosomal subunits shows that the intersubunit rotation described above is not a simple, rigid-body movement, but includes large- and small-scale structural rearrangements within both subunits. Moreover, there is a growing list of examples of localized conformational changes that are found throughout the ribosome during different steps of protein synthesis. A prime example is the movement of the head of the small subunit, an autonomous structural domain that is connected to the rest of the subunit by a neck consisting of a single helix (h28) of 16S rRNA. Comparison of X-ray structures of vacant ribosomes from E. coli and tRNA-containing ribosomes from T. thermophilus shows a 12° counterclockwise rotation (viewed from the top) (Fig. 2a) [27]. Cryo-EM reconstructions of EF-G-containing ribosomal complexes show that the aforementioned intersubunit rotation is accompanied by a similar 12-15° rotation of the head (Fig. 1c) [11,12,28]. A comparable rotational movement of the head was observed in eukaryotic ribosomes containing EF-2 [25]. This movement is most likely required to maintain proper contact between the tRNAs and the two ribosomal subunits upon transition into the hybrid state. A further movement of the head is predicted to occur during the second step of translocation to open the gap between residues G1338-U1341 in the head and A790 in the platform to create the ~20Å clearance that is needed for the ASL of the tRNA to pass from the P site to the E site of the small subunit [27]. Interestingly, binding of a mRNA and a tRNA anticodon stem-loop to the P site reverses this movement of the 30S head [29] resulting in a small subunit conformation similar to that found in classical-state ribosomes containing full-length tRNAs [3-5].
Figure 2.
Large-scale intrasubunit movements. (a) Movement of the 30S subunit head. Trajectories of phosphorus (blue) and Cα (dark blue) atoms demonstrating movement of the head between two crystallographically determined positions for tRNA-containing T. thermophilus ribosomes [35] and vacant E. coli [27] ribosomes. The figure is adapted from reference [27]. (b) Movement of the L1 stalk of the 50S subunit. Interaction of the elbow of E-site tRNA [60] with 23S rRNA in the L1 stalk [34] causes a large-scale displacement of the stalk (blue) relative to its position in the vacant ribosome (magenta; grey shows modeled portion of L1 stalk rRNA). (c) Movement of the L1 and L11 stalks between conformations I (magenta) and II (grey) of vacant E. coli ribosomes [27]. (d) Detailed view of (c) showing movement of the L11 stalk [27].
The head of small subunit also moves during initiation and termination of protein synthesis. In the X-ray structure of a 70S ribosome termination complex containing release factor RF1 [30] the beak of the head moves 4.5Å closer to the A site than that in the A-site tRNA bound complex [31]. Movement of the head of the eukaryotic small ribosomal subunit is also induced by binding of initiation factors eIF1 and eIF1A [32].
Movement of the L1 stalk of the large subunit
The largest movement within the 50S subunit is that of the L1 stalk, a feature that comprises helices 76, 77 and 78 of 23S rRNA and protein L1. Cryo-EM and X-ray studies show the L1 stalk in at least three different orientations. In X-ray structures of ribosomes with a vacant E site, the L1 stalk is observed in an “open” conformation leaning away from the body of the subunit (Figs. 2b, 2c) [27,29,33]. When deacylated tRNA is bound in the classical E/E state, formation of the contact between the L1 stalk and the elbow of the tRNA requires the stalk to move inward by 30-40 Å relative to its open conformation (Fig. 2b) [31,34,35]. In cryo-EM reconstructions of EF-G-containing complexes, preserving contact with the elbow of the hybrid-state P/E tRNA requires the stalk to move by an additional 15-20 Å closer to the small subunit, relative to its position in the E/E state complex (Fig. 1c) [12,28]. Normal mode analysis and molecular dynamics simulations suggest that movement of the L1 stalk is inherently coupled to movement of the head of the small subunit during intersubunit rotation [36-38]. Recently, spontaneous fluctuations in smFRET between fluorescently labeled tRNA and protein L1 have provided evidence for inward movement of the L1 stalk upon movement of deacylated tRNA into the 50S E site during translocation or spontaneous hybrid state formation [39]. No significant differences in tRNA-L1 FRET were observed between the classical E/E and hybrid P/E states. It appears that the “closed” conformations of the L1 stalk are stabilized by its interactions with E-site tRNA [34,39]. It remains unclear what triggers opening of the L1 stalk, which is thought to facilitate release of deacylated tRNA from the E site following translocation.
Movement of the L11 stalk
Another highly dynamic feature of the 50S subunit is the L11 stalk, the so-called “GTPase-associated center” (GAC), located on the opposite side of the 50S subunit from the L1 stalk (Figs 2a, 2c). The L11 stalk is formed from helices 42, 43 and 44 of 23S rRNA and protein L11. At the base of the stalk is the binding site for protein L10, to which are bound two (E. coli) or three (T. maritima) dimers of L7/L12 [40], a protein which has been extensively implicated in the functions of the GTP-dependent translation factors [41].
Comparison of the X-ray structures of vacant E. coli ribosomes crystallized in two different conformations [27] reveals an inward movement of the L11 stalk by more than 15 Å toward the A site (Fig. 2d). Likewise, the L11 stalk is observed in both its “inward” [35,42] and “outward” [34] positions in X-ray structures of similar tRNA-containing complexes of ribosomes from T. thermophilus. In X-ray structures of RF1-and RF2-containing termination complexes, the L11 stalk is seen in the inward position [30,43]. Various degrees of movement of the L11 stalk were seen in cryo-EM reconstructions of ribosomal complexes containing EF-G [11,28], EF-Tu [44], and RF3 [23]. However, the low resolution of cryo-EM data, possible structural heterogeneity and differences in interpretation of the maps for EF-G complexes by different groups [11,28] indicate that further study will be required to gain a full understanding of how the dynamics of the L11 stalk are linked to ribosome function.
Dynamics of the Shine-Dalgarno helix on the 30S subunit
During initiation of protein synthesis in bacteria and archaea, selection of the mRNA start site is facilitated by base pairing between the Shine-Dalgarno (SD) sequence upstream from the mRNA start codon and the anti-Shine-Dalgarno (anti-SD) sequence at the 3′-end of 16S RNA [45]. Two crystallographic studies have provided insights into the dynamics of the SD helix. In one study, the SD helix was observed in two distinct positions relative to proteins S2 and S18: in one conformation, the 3′ end of 16S RNA is displaced by 10 Å toward protein S18 and away from protein S2 relative to the other conformation [42]. It was proposed that the implied movement occurs upon unwinding the SD helix during early steps of the elongation phase. Another study used translation-libration-screw (TLS) refinement of X-ray data, which yields parameters describing potential anisotropic displacements of mobile regions of the structure. TLS analysis of a 70S ribosome complex showed that the SD helix is predisposed to a screw-like motion along its helical axis [46,47].
The structural basis for hinge-like movements of rRNA
Remarkably, the putative hinge regions for movement of the 30S head and the L1 and L11 stalks contain three specific structural motifs known to facilitate the flexibility of RNA: G-U wobble base pairs[48], kink turns [49-52] and G-ribo motifs [53]. By comparison of the structures of two conformers of the 70S ribosome, Schuwirth et al. localized the hinge point for one type of head movement within the G-C pairs 929:1388 through 932:1385 in helix 28 of 16S rRNA [27]. We note that two or more consecutive wobble base pairs (base pairs 924:1391 and 926:1390) are always found flanking the bulged G926 in helix 28 of 16S rRNA, suggesting that this conserved feature may be the point for other modes of movement of the 30S subunit head. Clusters of G-U pairs are also found in helix 76, which forms the base of the L1 stalk (base pairs 2099:2190 and 2100:2099) and in helix 42 in the base of the L11 stalk (base pairs 1035:1120 and 1036:1119). K-turns are found in the hinge regions of the L1 stalk at the junction of helices 75, 76 and 79 of 23S rRNA, and in the L11 stalk at the internal loop of helix 42. Additionally, the recently characterized G-ribo motif [53] found at the very base of the L11 stalk at nucleotide 1024 in the helix 41 – helix 42 junction may also contribute to the conformational flexibility of the L11 stalk. It should be noted that the importance of these features for enabling hinge-like movements of rRNA has yet to be tested by mutational analysis.
Localized rearrangements in the decoding center
Besides these dramatic large-scale inter- and intrasubunit movements, protein synthesis involves numerous localized conformational changes in rRNA. One of the most vivid examples is found in the decoding center of the small ribosomal subunit, where the universally conserved nucleotides G530, A1492, A1493 of 16S rRNA and A1913 of 23S rRNA undergo conformational rearrangements during initiation, aminoacyl-tRNA selection and termination, as shown by biochemical, genetic and crystallographic studies [2,30,54,55]. Depending on the functional state of the ribosome, these nucleotides are found in at least four different conformations (Fig. 3).
Figure 3.
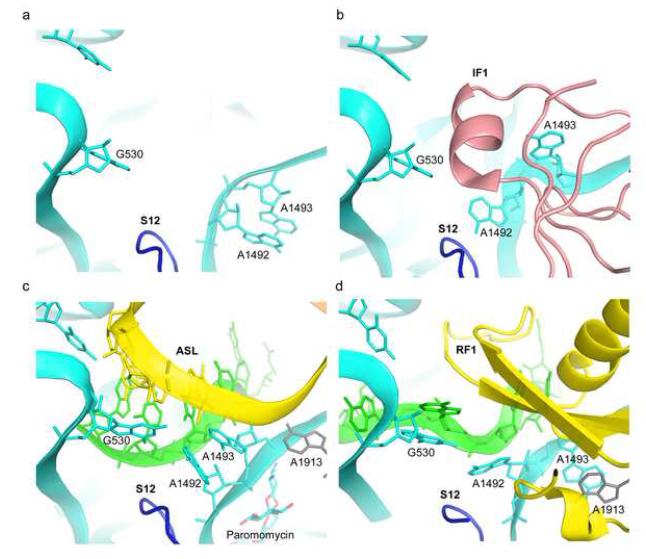
Rearrangements of nucleotides in the decoding center of the 30S subunit. (a) Vacant A site of the 30S subunit [62] (PDB 1J5E). (b) IF1 (pink) bound to the 30S subunit [56] (PDB 1HR0). (c) Cognate tRNA (yellow) bound to its mRNA codon (green) in the A site of the 70S ribosome in the presence of paromomycin [31] (PDB 2J00, 2J01). (d) Release factor RF1 (yellow) bound to a UAA stop codon (green) in a 70S termination complex [30]. 16S rRNA nucleotides G530, A1492 and A1493 are shown in cyan and 23S rRNA nucleotide A1913 is shown in grey.
In free 30S subunits, A1492 and A1493 are stacked within h44 of 16S rRNA (Fig. 3a) [2]. When initiation factor IF1 is bound to the 30S subunit, A1492 and A1493 become unstacked, flipping out of h44 and into pockets in IF1 (Fig. 3b) [56]. During tRNA selection, A1492 and A1493 again flip out from h44, but remain stacked on each other while contacting the minor groove of the codon-anticodon helix (Fig. 3c) [31,57]. G530 flips from the syn to the anti conformation, forming an A-G base pair with A1492, while A1913 of 23S rRNA interacts with the codon-anticodon helix. G530, A1492 and A1493 appear to discriminate Watson-Crick pairing by a precise steric fit involving A-minor interactions with the codon-anticodon helix. This induced-fit RNA mechanism plays a major role in selection of cognate aminoacyl-tRNA (reviewed in [2]). In the RF1 termination complex [30], A1492 and G530 flip out in a way similar to that seen in the decoding complex, but A1493 and A1913 are found in dramatically different conformations. A1913 occupies the site within h44 vacated by A1492, stacking on A1493, which remains in its ground-state position within h44 (Fig. 3d). These rearrangements appear to be critical for triggering a conformational change in RF1 that occurs upon recognition of the stop codon, leading to peptidyl-tRNA hydrolysis [30]. Thus, interactions with IF1, tRNA or class I release factors cause characteristically different induced-fit conformations of these conserved nucleotides in the decoding site.
Localized rearrangements in the peptidyl-transferase center
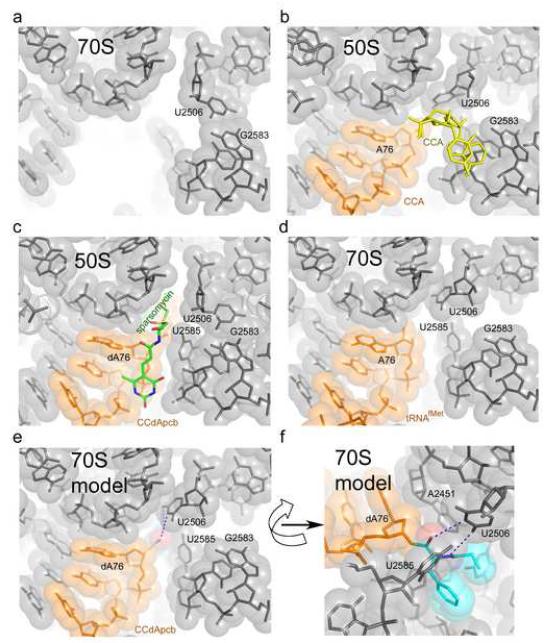
Comparison of crystal structures of different 50S subunit and 70S ribosome complexes shows that the largest rearrangements in the PTC occur in the pocket that binds the 3′ end of the aminoacyl-tRNA in the 50S A site. Based on structures of 50S complexes containing aminoacyl-tRNA analogs, it was inferred that the conformation of this pocket, formed by U2506, G2583, U2584 and U2585, depends on the occupancy of the A site [58]. In 50S subunits in which the A site is vacant or occupied by a CA dinucleotide, the universally conserved U2506 forms a base pair with G2583 (Fig. 4a). When a CCA trinucleotide is bound, uracil 2506 rotates by 90° towards U2585, leading to disruption of the U-G pair (Fig. 4b). During this transition, the Watson-Crick edge of U2506 can travel by up to 9 Å. This observation was interpreted to mean that the presence of C74 of the A-site tRNA is required to induce conformational changes in the PTC needed for optimal positioning of the substrates [58]. Surprisingly, however, in a 2.8 Å 70S ribosome structure, where deacylated tRNA is bound to the P site but the 50S A site is unoccupied [31], the positions of U2506 and U2585 were close to those seen in 50S subunit complexes containing the CCA analog (Fig. 4d). Superposition of the 70S-tRNA structure [31] on the 50S subunit with an aminoacyl-tRNA analog bound to the P site [59] (Fig 4c) suggests that U2506 is positioned to interact with the backbone of the aminoacyl moiety (Figs. 4e and 4f). This suggests that the rearrangements in the PTC may be the result of binding of tRNA to the P site rather than of aminoacyl-tRNA to the A site, as proposed [58]. U2506 may serve to position the aminoacyl moiety in the peptidyl-transferase center and indirectly protect the P-site peptidyl-tRNA ester bond from hydrolysis, thus preventing spontaneous termination of peptide synthesis.
Figure 4.
Changes in the conformation of the binding pocket for the peptidyl moiety of the P-site tRNA in the peptidyl transferase center. (a) 70S ribosome with vacant P and A sites [27] (PDB 2AW4). (b) 50S subunit with CCA trinucleotide analogs of P-site (orange) and A-site (yellow) tRNAs [59] (PDB 1QVG). (c) 50S subunit with peptidyl-CCdApcb (orange) mimicking peptidyl-tRNA bound to the P site in the presence of sparsomycin (green) [27] (PDB 1VQ8). (d) 70S ribosome with deacylated tRNA bound to the P site (orange) (The 50S region of A-site tRNA was disordered) [31] (PDB 2J00 and 2J01). (e) and (f) Two views of a model in which the peptidyl-tRNA analog CCdApcb in (c) was superimposed on the 70S structure shown in (d). The model suggests a possible interaction of U2506 with the peptidyl moiety of the P-site tRNA (orange). The O4 and N3 positions of U2506 and the backbone amide and carbonyl groups of the aminoacyl moiety could move to within hydrogen bonding distance of each other upon minor repositioning of U2506 and the aminoacyl moiety. 23S rRNA is shown in grey.
Conclusions
Recent advances in the study of ribosome structure have begun to reveal the remarkable conformational flexibility of the ribosome. The most dramatic structural rearrangement so far observed occurs in formation of the hybrid-state intermediate of the translocation cycle, in which large-scale intersubunit rotational movement is coupled to a 40 Å movement of the L1 stalk and a 12° rotation of the 30S head. In view of the high structural and sequence conservation of the ribosome throughout all branches of life, it seems likely that most of the conformational changes observed in bacterial and archaeal ribosomes will be found in all ribosomes. Most of the conformational changes of the ribosome characterized to date involve structural rearrangements of rRNA, consistent with the idea that protein synthesis originated with an all-RNA ribosome.
Acknowledgements
We apologize to our many colleagues whose studies have not been mentioned because of space limitations. We thank Jamie Cate and Joachim Frank for providing figures of their work, and Martin Laurberg for helpful discussions. Work in the Noller laboratory was supported by grants from the NIH and NSF. D.N.E. was supported by a NATO-NSF postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green R, Noller HF. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 3.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 4.Yarus M. Boundaries for an RNA world. Curr Opin Chem Biol. 1999;3:260–267. doi: 10.1016/S1367-5931(99)80041-6. [DOI] [PubMed] [Google Scholar]
- 5.Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 6.Spirin AS. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harb Symp Quant Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 8.Spirin AS, Baranov VI, Polubesov GS, Serdyuk IN, May RP. Translocation makes the ribosome less compact. J Mol Biol. 1987;194:119–126. doi: 10.1016/0022-2836(87)90720-0. [DOI] [PubMed] [Google Scholar]
- 9.Spirin AS. Ribosomal translocation: facts and models. Prog Nucleic Acid Res Mol Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- 10.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 11.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 12.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 13.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci U S A. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HD, Puglisi J, Chu S. Fluctuations of tRNAs between classical and hybrid states. Biophys J. 2007 doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybridstate conformation of the 70S ribosome. Rna. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- 23.Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 24.Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. Embo J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. Embo J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 28.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 29.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci U S A. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008 doi: 10.1038/nature07115. In press. [DOI] [PubMed] [Google Scholar]
- 31.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 32.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 33.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 34.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 35.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 36.Tama F, Valle M, Frank J, Brooks CL., 3rd Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryoelectron microscopy. Proc Natl Acad Sci U S A. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trylska J, Tozzini V, McCammon JA. Exploring global motions and correlations in the ribosome. Biophys J. 2005;89:1455–1463. doi: 10.1529/biophysj.104.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. J Struct Biol. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Fei J, Kosuri P, MacDougall DD, Gonzalez RL. Coupling of Ribosomal L1 Stalk and tRNA Dynamics during Translation Elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Rodnina MV, Stark H, Savelsbergh A, Wieden HJ, Mohr D, Matassova NB, Peske F, Daviter T, Gualerzi CO, Wintermeyer W. GTPases mechanisms and functions of translation factors on the ribosome. Biol Chem. 2000;381:377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 42.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 43.Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. Embo J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shine J, Dalgarno L. Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3′-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur J Biochem. 1975;57:221–230. doi: 10.1111/j.1432-1033.1975.tb02294.x. [DOI] [PubMed] [Google Scholar]
- 46.Korostelev A, Noller HF. Analysis of structural dynamics in the ribosome by TLS crystallographic refinement. J Mol Biol. 2007;373:1058–1070. doi: 10.1016/j.jmb.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korostelev A, Trakhanov S, Asahara H, Laurberg M, Lancaster L, Noller HF. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci U S A. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos A, Varani G. Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. Embo J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 51.Razga F, Zacharias M, Reblova K, Koca J, Sponer J. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 2006;14:825–835. doi: 10.1016/j.str.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Goody TA, Melcher SE, Norman DG, Lilley DM. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. Rna. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinberg SV, Boutorine YI. G-ribo motif favors the formation of pseudoknots in ribosomal RNA. Rna. 2007;13:1036–1042. doi: 10.1261/rna.495207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 56.Carter AP, Clemons WM, Jr., Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 57.Ogle JM, Brodersen DE, Clemons WM, Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 58.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 59.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. Rna. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christodoulou J, Larsson G, Fucini P, Connell SR, Pertinhez TA, Hanson CL, Redfield C, Nierhaus KH, Robinson CV, Schleucher J, et al. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 2004;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci U S A. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]