Summary
Sustained tumor progression has been attributed to a distinct population of tumor-propagating cells (TPCs). To identify TPCs relevant to lung cancer pathogenesis, we investigated functional heterogeneity in tumor cells isolated from Kras-driven mouse models of non-small cell lung cancer (NSCLC). CD24+ITGB4+Notchhi cells are capable of propagating tumor growth in both a clonogenic and an orthotopic serial transplantation assay. While all four Notch receptors mark TPCs, Notch3 plays a non-redundant role in tumor cell propagation in two mouse models and in human NSCLC. The TPC population is enriched after chemotherapy and the gene signature of mouse TPCs correlates with poor prognosis in human NSCLC. The unique role of Notch3 in tumor propagation may provide a therapeutic target for NSCLC.
Introduction
Lung cancer is the leading cause of cancer death worldwide (Jemal et al., 2011) and Non-small cell lung cancer (NSCLC) accounts for approximately 80% of cases. Despite aggressive frontline treatment, the 5-year survival rate remains poor for most patients. A functional hierarchy with regards to tumor propagating capacity is a well-established feature of some cancers and may account for incomplete therapeutic response. Cells with an increased capacity to sustain tumor propagation are called tumor-propagating cells (TPCs, also referred to as cancer stem cells) and can be prospectively identified using cell surface markers. TPCs have been identified in several solid tumors (Al-Hajj et al., 2003; Hermann et al., 2007; O’Brien et al., 2007; Singh et al., 2004). Additionally, TPCs have been linked to chemo- and radio-resistance as well as metastasis (Bao et al., 2006; Chen et al., 2012; Hermann et al., 2007; Phillips et al., 2006). In NSCLC, several reports have described isolation of TPCs with surface markers including CD133, CD44 or CD166 (Eramo et al., 2008; Leung et al., 2010; Zhang et al., 2012). However, other studies have yielded conflicting results (Cui et al., 2011; Meng et al., 2009; Tirino et al., 2009) and none of these markers have been shown to be functionally required for the TPC state. Furthermore, whether TPCs in NSCLC are linked to chemoresistance and if their prevalence is associated with prognosis of human NSCLC has not been determined.
The use of mouse models of cancer provides an opportunity to assess the influence of specific genotypes commonly found in NSCLC on TPC frequency. CD45−Pecam−Sca1+ have been proposed to be genotype specific surface markers of TPCs (Curtis et al., 2010; Kim et al., 2005) only in tumors with the KrasG12D; Trp53fl/fl genotype but not in tumors of the KrasG12D or EGFRT790M-L858R genotypes. However, a residual mesenchymal cell component has been reported using the CD45−Pecam−Sca1+ enrichment strategy, questioning the specificity of these markers (McQualter et al., 2009; Teisanu et al., 2009). Contamination of tumor stroma is a particularly important issue in the KrasG12D; Trp53fl/fl lung tumor model, as these tumors are characterized by a significant desmoplastic stromal component (Jackson et al., 2005).
The self-renewal pathways required for maintaining long-term tumor propagation potential in NSCLC are not well defined. The Notch pathway has been previously linked to regulation of self-renewal in TPCs of colon, breast and brain cancer (Fan et al., 2010; Harrison et al., 2010; Hoey et al., 2009). Over-expression of N1ICD in murine alveolar epithelium initiates hyperplasia and eventually lung adenomas (Allen et al., 2011). In addition, Notch1 and Notch3 signaling promote tumor cell proliferation and inhibit cell apoptosis in some NSCLC cell lines (Haruki et al., 2005; Konishi et al., 2010; Westhoff et al., 2009). In a mouse model of NSCLC with mutant Kras but wild-type for Trp53, genetic or pharmacologic blockade of signaling from all four Notch receptors leads to decreased tumorigenesis (Maraver et al., 2012). These studies support the hypothesis that Notch signaling plays an important pro-oncogenic role in NSCLC. However, whether Notch marks TPCs with relevance to chemotherapy resistance and prognosis in primary tumor models has not been established. In addition, the specific roles of each Notch receptor in lung tumorigenesis as well as the specific role of each Notch receptor in establishing heterogeneity within lung tumors has not been defined.
We sought to develop an improved approach to positively identify tumor cells and separate them from tumor stromal in order to determine whether tumors arising in either the KrasG12D or KrasG12D; Trp53fl/fl mouse models demonstrate evidence of functional heterogeneity consistent with the presence of a rare TPC population. Furthermore, we sought to determine whether loss of Trp53 alters the frequency or characteristics of the TPC population. Identification of a TPC population in mouse models of lung cancer could be important as the functional characteristics of TPCs may be common between the mouse and human disease. In particular, while some studies have suggested that TPCs are chemoresistant, there is a paucity of data confirming this phenotype in vivo. In addition, the self-renewal pathways that are critical for maintenance of TPCs in many cancers remain to be described. Therefore, a key goal of our studies was to identify critical regulators of TPC self-renewal in lung cancer. The comparative cross-species strategy employed here, combining mouse models with the analysis of primary patient-derived samples, may be applicable to other TPC studies.
RESULTS
Isolation and characterization of fluorescent labeled tumor cells from mouse models of NSCLC
Efforts to isolate TPCs are confounded by the presence of abundant stroma within the tumor (Jackson et al., 2005). To allow for positive identification of tumor cells and separation from tumor stroma, KrasG12D or KrasG12D; Trp53fl/fl mice were crossed with conditional reporter lines carrying Cre-inducible alleles of either eYFP or tdRFP (Luche et al., 2007; Srinivas et al., 2001) (Figure S1A). Analysis of the lung epithelium of these mice one week after infection with an adenovirus expressing Cre (AdCre) revealed rare, fluorescent cells distributed throughout the distal lung epithelium. These fluorescent cells showed evidence of proliferation only in mice carrying mutation of either KrasG12D or KrasG12D; Trp53fl/fl (Figure S1B). Therefore, AdCre infection of compound mutant reporter mice led to the development of lung tumors in which tumor cells were labeled with a fluorescent marker that could be positively identified and isolated by flow cytometry (FACS) (Figure S1C and S1D).
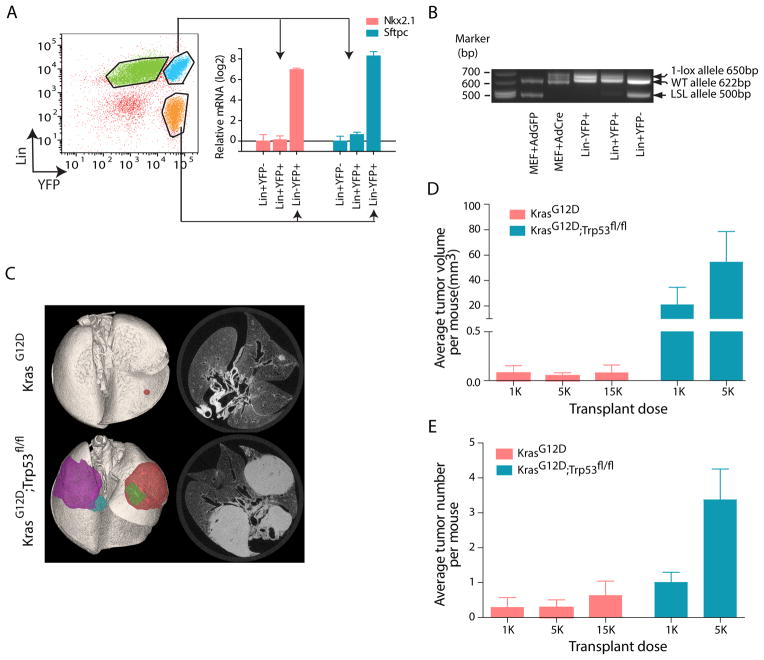
FACS analysis of lungs from these mice identified two populations of fluorescent cells, distinguished by expression of lineage markers (CD45, PECAM and Ter119) (Lin+) (Figure 1A). A majority of Lin+ YFP+ cells were positive for F4/80 (Figure S1E), suggesting that they were tumor-associated macrophages. In contrast, Lin− YFP+ cells were Epcam+, supporting an epithelial origin (Figure S1F). Lin− YFP+ cells also demonstrated higher expression of the lung epithelial markers Nkx2.1 (thyroid transcription factor 1, TTF-1) and Sftpc (surfactant protein C, SP-C), relative to Lin+YFP+ cells (Figure 1A). Finally, the tumor origin of Lin−YFP+ cells was supported by the absence of an unrecombined “two-lox”, KrasG12D-specific band (LSL allele) together with the presence of a “one-lox” recombined allele (Figure 1B). Thus, the combination of genetic labeling and cell sorting allowed for the isolation of a highly enriched population of tumor cells depleted of normal lung parenchyma, tumor stroma and immune infiltration. We use “tumor cells” to refer to this highly purified Lin−fluorescence+ population hereafter.
Figure 1. Analysis of tumor-propagating cell frequency using fluorescent reporters and genetic models of NSCLC.
(A) Representative FACS plot of tumor cells isolated from KrasG12D; eYFP mice (left). Expression of lung markers Nkx2.1 (red bars) and Sftpc (blue bars) in tumor cells (Lin−YFP+) and lineage + cells (Lin+YFP− and Lin+YFP+) were assessed by RT-PCR (right) (relative to Hprt). Expression of each transcript in Lin+YFP− cells is set to 1 (n=3, mean±SEM). (B) Representative allele-specific PCR of indicated sorted populations from tumor bearing KrasG12D; eYFP. MEFs (mouse embryonic fibroblast) derived from KrasG12D embryos were infected with high titer cre-carrying adenovirus in vitro as positive controls. Lower band: knock-in Lox-stop-Lox (LSL) KrasG12D allele without recombination (500 bp); middle band: wild-type Kras allele (622 bp), top band: recombined LSL allele with one loxP site (650 bp). (C) Representative ex-vivo CT scan of tumors arising from intatracheal transfer of 5,000 FACS-sorted KrasG12D; tdRFP and KrasG12D; Trp53fl/fl; eYFP tumor cells into recipient nu/nu mice. Left, pseudocolored 3-dimensional reconstruction. (D) Quantification of average tumor volume per recipient mouse (mean±SEM) in secondary tumors from KrasG12D; tdRFP or KrasG12D; Trp53fl/fl; eYFP tumor cells. (E) Quantification of average tumor number per recipient mouse (mean±SEM) in secondary tumors derived from KrasG12D; tdRFP or KrasG12D; Trp53fl/fl; eYFP tumor cells. See also Figure S1.
To establish the tumorigenic potential of tumor cells from the KrasG12D or KrasG12D; Trp53fl/fl models, Lin− fluorescence+ cells were transplanted into recipient immunocompromised mice via intratracheal intubation. As KrasG12D mice develop tumors more slowly than KrasG12D; Trp53fl/fl, different time points were used to isolate cells from each model (21 and 12 weeks respectively). At these time points, the vast majority of the tumor cells in both models are from well-established, large adenomas and adenocarcinomas rather than hyperplasias (Figure S1G). Dispersion of Lin− fluorescence+ cells in the lung parenchyma as single cells was confirmed by immunofluorescence (Figure S1H). Secondary tumors formed from KrasG12D or KrasG12D; Trp53fl/fl primary tumor cells were readily detectable 16–21 weeks after transplantation. The histology of the secondary tumors from both models resembled the primary tumors from which they were derived (data not shown). The vast majority of transplanted tumor cells failed to initiate tumors, suggesting that tumor cells from either model contain a rare fraction of TPCs. Both tumor number and tumor volume per recipient mouse were significantly greater in KrasG12D; Trp53fl/fl tumors (Figure 1C–E). Approximately 1 out of 2,012 KrasG12D; Trp53fl/fl tumor cells gave rise to secondary tumors, while only ~1 in 40,860 KrasG12D tumor cells formed tumors (p=0.0001) (Figure S1I). Therefore, only a small fraction of tumor cells from either model are capable of tumor propagation and loss of Trp53 is a significant contributor to TPC frequency. Next, we sought to identify cell surface markers that could enrich for the TPC population.
Spheroid culture to propagate TPCs in NSCLC
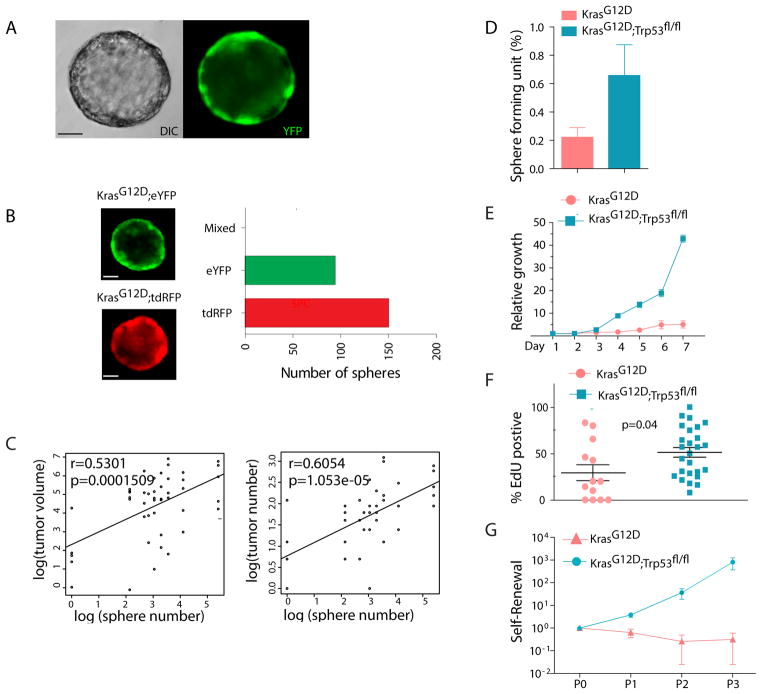
Spheroid culture has been successfully applied to propagate TPCs from solid tumors (Bao et al., 2006; Vermeulen et al., 2008). We developed a spheroid culture system to propagate tumor cells from both the KrasG12D and the KrasG12D; Trp53fl/fl models. Lin−YFP+ single cells from freshly dissociated tumor-bearing lungs were plated in a semi-solid matrix (see methods). As expected, spheroids were composed only of fluorescent cells, demonstrating that they originated from genetically labeled tumor cells (Figure 2A). In contrast, Lin−YFP− cells formed very few spheres (Figure S2A). Spheroids were not detected from single cell suspensions of normal lung, indicating that non-transformed normal lung progenitor or epithelial cells do not contribute to sphere formation (data not shown). Spheres were hollow (Figure S2B) and stained positive for E-cadherin, ZO-1 and several other epithelial markers (Figure S2C–E). Sphere clonality was confirmed by plating a 1:1 mixture of YFP+ and tdRFP+ tumor cells (Figure 2B). When FACS-sorted tumor cells were divided and used for either pulmosphere assay in vitro or orthotopic transplantation in vivo, a significant correlation between frequency of sphere and tumor formation was observed (Figure 2C), suggesting that the pulmosphere assay could serve as a screening tool to identify markers likely to confer increased tumor-propagating ability in vivo.
Figure 2. Sphere culture enriches for lung NSCLC tumor-propagating cells.
(A) Representative live cell image of a pulmosphere arising from a single cell suspension derived from a KrasG12D, Trp53fl/fl; eYFP tumor bearing mouse (6 days). Nomarski DIC (differential interference contrast) image of sphere is shown on the left. Fluorescence image of eYFP+ sphere is shown on the right. (B) EYFP and tdRFP mixing experiments to assess clonal origin of pulmospheres. Single cell suspensions were derived from the lungs of KrasG12D; tdRFP and KrasG12D; eYFP tumor bearing mice. (C) Correlation between in vitro sphere forming ability and in vivo tumor formation capacity by linear regression analysis. (D) Frequency of sphere-forming cells in FACS-sorted Lin−Fluorescence+ tumor cells in KrasG12D and KrasG12D; Trp53fl/fl genotypes. (E) Relative quantification of cell growth in pulmospheres measured by cell titer blue (Promega). Reading at day one (24 hours after plating) is set to 1 (n=3, mean±SEM) (F) Percentage of proliferating spheroid cells identified by EdU staining in pulmospheres with KrasG12D and KrasG12D; Trp53fl/fl genotype. Pulmospheres were labeled with EdU for 12 hours. (G) Self-renewal of KrasG12D and KrasG12D; Trp53fl/fl pulmospheres were determined by passaging of single cells from enzyme dissociated spheres (see methods). Total numbers of pulmospheres after secondary and tertiary culture are normalized against the primary culture (n=3, mean±SEM). Scale bars represent 50 μm in all panels. See also Figure S2.
Pulmosphere-forming cells were rare among both KrasG12D; Trp53fl/fl and KrasG12D tumor cells, although loss of Trp53 increased sphere-forming capacity approximately three-fold (Figure 2D). As expected, KrasG12D spheres proliferated more slowly than KrasG12D; Trp53fl/fl spheres (Figure 2E–F). When primary spheres were passaged, only a small percentage of cells formed secondary spheres, suggesting that self-renewal is a feature of only a subset of tumor cells. Notably, the total number of pulmospheres from KrasG12D mice did not increase with passage, whereas the number of pulmospheres derived from KrasG12D; Trp53fl/fl primary spheres increased over several passages (Figure 2G). Therefore, Trp53 appears to play an inhibitory role in the regulation of self-renewal.
Identification of CD24+ITGB4+Notchhi cells as TPCs in murine NSCLC
Next we sought to identify surface markers with the ability to enrich for sphere-forming cells using in vitro assay. Several previously described cell surface markers, including CD133, CD44 and CD166, failed to show significant enrichment of sphere forming capacity (Figure S3A). In contrast, CD24, which has been previously been shown to mark TPCs in other tissues (Lee et al., 2011), marked a population of cells with increased ability to form spheres (p=0.0002, Figure S3A). CD24 enriched for sphere-forming capacity independent of genotype, as CD24+ tumor cells were highly enriched for sphere forming capacity in both KrasG12D and KrasG12D; Trp53fl/fl models (Figure S3B). However, CD24+ cells are a high percentage of the total tumor number (~50%) in tumors from both models, suggesting that further enrichment of TPCs could be achieved with additional markers.
To identify additional markers to enrich for TPCs, microarray profiling was used to identify differentially expressed genes between CD24+ and CD24− tumor cells (Table S1, Figure S3C). Among the genes coding for surface markers with increased expression in CD24+ cells were those for insulin growth factor receptor 1(igf1r), CD133 (Prominin1), integrin β4 (CD104, Itgb4) and Notch3 (Notch3) (Table S2). Differential expression of these genes in CD24+ and CD24− tumor subpopulations was validated by qRT-PCR using FACS-sorted tumor cells from both genotypes (Figure S3D). As Igf1r showed modest change in expression level, and CD133 had previously failed to enrich for spheres (Figure S3A), only ITGB4 and Notch3 were further tested individually for their ability to enrich for pulmosphere-forming tumor cells in the KrasG12D; Trp53fl/fl; eYFP model. Both ITGB4+ and Notch3hi tumor cells had significantly higher sphere-forming capacity than their marker negative counterparts (Figure S3E–F). Given the structural similarity and functional overlap between Notch receptors, we extended our marker analysis to the other three Notch paralogs. Notch1, Notch2 and Notch4 also enriched for sphere forming capacity in the KrasG12D; Trp53fl/fl; eYFP model (Figure S3G–I). In addition, tumor cells sorted for each of the Notch receptors had higher expression of the canonical Notch target genes Hey1 and DTX1, and lower expression of the notch antagonists Numb and Numb-Like (Figure S3J).
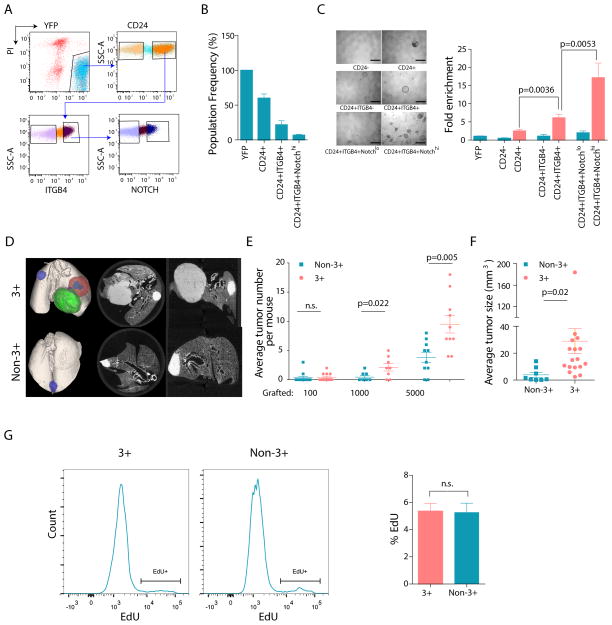
Next, a sorting strategy combining CD24, ITGB4 and Notch 1–4 (CD24+ITGB4+Notchhi) was used to determine if the combination could further enrich for pulmosphere-forming capacity (Figure 3A & 3B). Plating of CD24+ITGB4+ tumor cells led to a ~2.5 fold increase in pulmosphere-forming capacity compared to CD24+ tumor cells (p=0.0036) while plating of the CD24+ITGB4+Notchhi tumor population led to an additional 2.8-fold increase (p=0.0053) in sphere formation relative to CD24+ITGB4+ tumor cells. Overall, CD24+ITGB4+Notchhi tumor cells showed a ~17.2-fold enrichment over the bulk Lin−YFP+ tumor cells (p<0.0001) (Figure 3C). A similar step-wise increase in sphere-forming ability for CD24+ITGB4+Notchhi cells was also observed in the KrasG12D; eYFP model (Figure S3K–L). Thus, the combination of these three markers identified a rare subpopulation of cells with increased ability to form spheres in vitro, independent of Trp53 loss. Enrichment for TPCs using these markers was reproduced on three different FACS sorters, indicating robustness to technical variability (Figure S3M–N).
Figure 3. Combined sorting for CD24+ITGB4+Notchhi identifies TPCs in NSCLC.
(A, B) Representative FACS gating scheme (left) and the frequency of each population as a percentage of the total YFP labeled tumor cells (right) from KrasG12D; Trp53fl/fl; eYFP mice (n=5). (C) Representative images for pulmosphere formation by each indicated population are shown on the left. Relative enrichment of pulmosphere forming ability after step-wise addition of markers is shown on right. Y-axis is the fold enrichment of pulmospheres normalized by bulk tumor cells (Lin−YFP+) in KrasG12D; Trp53fl/fl genotype. Data is the average of at least four independent experiments using tumor cells from either a single mouse or pooled donor mice. Scale bars represent 500 μm. (D) Representative ex vivo CT scan images for mouse lungs grafted with FACS-sorted Lin−YFP+CD24+ITGB4+Notchhi primary tumor cells (3+, top) and remainder of bulk tumor cells (non-3+, bottom) with KrasG12D; Trp53fl/fl; eYFP genotype at a 1,000 cell transplantation dose. (E) Average number of secondary tumors in primary grafting with 3+ and non-3+ tumor cells at different dose. (F) Average size of secondary tumors. (G) Frequency of EdU+ cycling cells in 3+ and non-3+ tumor subpopulations in KrasG12D; Trp53fl/fl; eYFP donor mice. Representative FACS plots from 3+ and non-3+ cells are shown as histogram on left and the bar chart on right is the average of four mice. Error bars indicate mean±SEM. p-values are from 2-tailed t-test. See also Figure S3.
To assess the tumor-propagating potential of CD24+ITGB4+Notchhi tumor cells (3+ cells) relative to the remainder of tumor cells (non-3+ cells) in vivo, ex-vivo CT scanning was used to quantify tumor number and size in recipient mice after transplantation (Figure 3D). Given the consistency of the pulmosphere assay results between KrasG12D and KrasG12D; Trp53fl/fl models (Figure 3C and Figure S3L) and the more robust tumor grafting in the KrasG12D; Trp53fl/fl model (Figure 1C–E), only the KrasG12D; Trp53fl/fl; eYFP model was used for in vivo studies. Mice grafted with 1000 and 5000 3+ tumor cells began to show signs of morbidity earlier than those grafted with non-3+ cells at the same dose due to a heavier tumor burden, leading to the euthanasia of some animals ~4–6 weeks before the planned end point (data not shown). Despite the shorter time frame in the 3+ grafts, the average tumor number per recipient mouse was significantly higher in this group (Figure 3E) and the average tumor size was 7.4 times larger (p=0.02) than that of the non-3+ group (Figure 3F). Thus, sorting for 3+ cells enriches for a population of cells with a more robust tumorigenic capacity. Furthermore, regression analysis revealed a highly significant enrichment in tumor-propagating cells in the 3+ population relative to the non-3+ cells (p=0.0007). (Figure S3O). This difference in tumor propagating capacity was not due to differential proliferation rates, as there was no difference in the frequency of EdU+ proliferating cells was observed between TPC (3+) and non-TPC (Non-3+) tumor cells (p=0.8365)(Figure 3G). Thus, 3+ cells are enriched for TPCs in vivo.
Serial transplantation reveals a stable TPC phenotype in the CD24+ITGB4+Notchhi population
Prior work has suggested significant plasticity in the TPC phenotype such that non-tumorigenic cells transit into TPCs under certain conditions (Marjanovic et al., 2013). To determine whether 3+ cells could give rise to non-3+ cells and vice versa, secondary 3+ and non-3+-derived tumors were analyzed by FACS. Secondary tumors derived from 3+ cells regenerated both 3+ and non-3+ populations recapitulating the heterogeneity of the original tumors. Non-3+ derived tumors also contained some 3+ cells, although in a smaller fraction, suggesting that some plasticity exists in TPC marker expression (Figure S4).
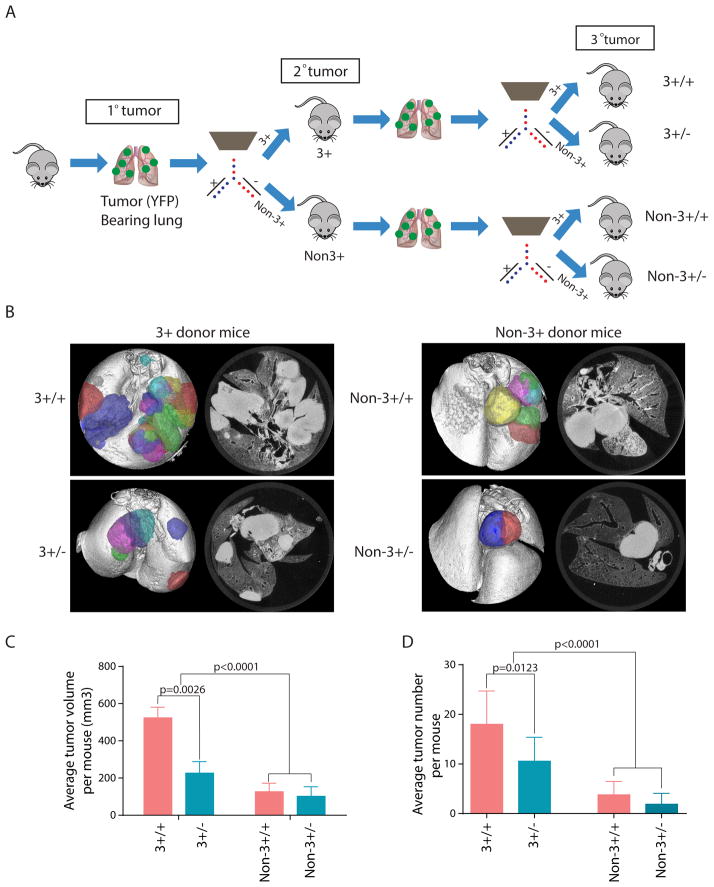
To assess the functional stability of TPCs, we conducted serial transplantation of secondary tumors. Secondary tumors derived from either 3+ cells (3+ donor) or non-3+ cells (non-3+ donor) were FACS sorted and transplanted into recipient mice (Figure 4A). After 16 weeks, ex--vivo CT imaging revealed that 3+ cells from 3+ donors (3+/+) were most potent in generating tertiary tumors (Figure 4B) when compared with non-3+ cells from the same donor group (3+/−) (Figure 4C and 4D). Statistical analysis indicated that both 3+ and non-3+ cells from a 3+ donor (3+/+ and 3+/−) had increased tertiary tumor formation when compared with tumor cells originating from a non-3+ primary donor (non-3+/+ and non-3+/−) (p<0.0001 for both tumor volume and tumor number). Taken together, these results strongly support the existence of a hierarchy in which 3+ cells give rise over time to non-3+ cells with progressively diminished capacity to propagate tumors. While non-3+ cells can give rise to 3+ cells, these “reverted” 3+ cells do not have the same tumor initiating capacity as the original TPCs.
Figure 4. CD24+ITGB4+Notchhi tumor cells display a stable TPC phenotype after serial transplantation.
(A) Schematic illustration of serial transplantation experiment. (B) Representative ex-vivo CT scan of tertiary tumors arising from 3+/+, 3+/−, Non-3+/+ and Non-3+/− groups (see A for definitions). (C–D) Quantification of average tumor volume (C) and tumor number (D) in secondary transplantation study. Error bars indicate mean±SEM. See also Figure S4.
The CD24+ITGB4+Notchhi TPC population contributes to chemoresistance and disease prognosis
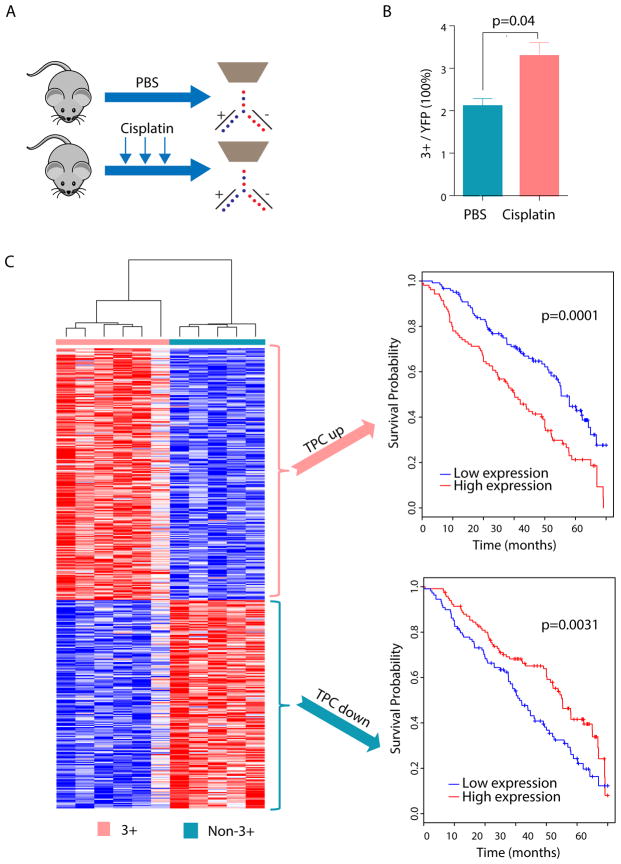
TPCs have been proposed to be associated with chemoresistance and tumor relapse (Chen et al., 2012; Shafee et al., 2008). However, few studies have confirmed this phenotype in vivo in autochthonous mouse models or in human cancers. To determine whether the TPC population was enriched after treatment of tumor-bearing mice with chemotherapy, tumor-bearing KrasG12D; Trp53fl/fl mice were repeatedly dosed with cisplatin or vehicle. In this tumor model, this regimen leads to a ~50% tumor growth inhibition (Hegde et al., 2013). Cisplatin or vehicle treated tumors were analyzed by FACS to assess the relative frequency of the TPC population within the bulk tumor. Repeated doses of cisplatin led to a significant increase in TPC frequency (p=0.038, n=8 mice) (Figure 5A–B). These results suggest that the TPC population is intrinsically more chemoresistant compared to the non-TPC population.
Figure 5. Mouse NSCLC TPCs are enriched after chemotherapy and their gene signature predicts prognosis in human NSCLC.
(A) Schematic illustration of repeated dose of chemotherapy drug treatment regimen in KrasG12D; Trp53fl/fl mice. Tumor bearing mice were treated with 3 dose of cisplatin (7mg/kg bodyweight) or control phosphate-buffered saline on a weekly basis. Mice were sacrificed and FACS analysis was performed 72 hours after the last treatment. (B) Average frequency of CD24+ITGB4+Notchhi population (3+ cells) within the bulk tumor (Lin−YFP+) in cisplatin (n=5) or PBS (n=3) treated mice. Error bars indicate mean±SEM. p-value is from 2-tailed t-test. (C) Genes differentially expressed in TPC and non-TPC cells are correlated to human patient survival. Left: Heatmap of genes differentially expressed in TPC cells when compared with non-TPC cells. Right upper: Kaplan-Meier curve indicating that early stage NSCLC patients with higher expression of genes upregulated in the TPC population have a worse overall survival (p = 0.0001). Right lower: Kaplan-Meier curve indicating that NSCLC patients with lower expression of genes down-regulated in the TPC population have a worse overall survival (p = 0.0031). P-value for survival analysis was derived using a cox proportional hazards model.
We hypothesized that human NSCLCs with a molecular signature more similar to TPCs may have a worse prognosis given the relative chemoresistance of this population in mice. To test this hypothesis, microarrays were used to identify differentially expressed genes between 3+ and non-3+ tumor cells (Table S3). A gene expression signature consisting of 245 up- (“TPC up”) and 205 down-regulated (“TPC down”) genes in the 3+ TPC cells was generated (Table S4) (see methods) (Figure 5C, left). Using the human orthologues of these genes we determined the ability of the “TPC up” or “TPC down” gene signature to predict early stage lung adenocarcinoma patient prognosis by employing a previously described approach for separation of samples based on gene signatures (Vicent et al., 2012). Patients with increased expression of the “TPC up” or decreased expression of the “TPC down” gene signatures had a worse prognosis (p<0.0001 for “TPC up” and p=0.0031 for “TPC down”) (Figure 5C, right panels). Together, the increase in TPCs after chemotherapy treatment and the worse prognosis of human samples with a “TPC like” signature strongly suggest the clinical relevance of the 3+ TPC population to human NSCLC.
Notch signaling regulates the self-renewal of non-small cell lung cancer TPCs
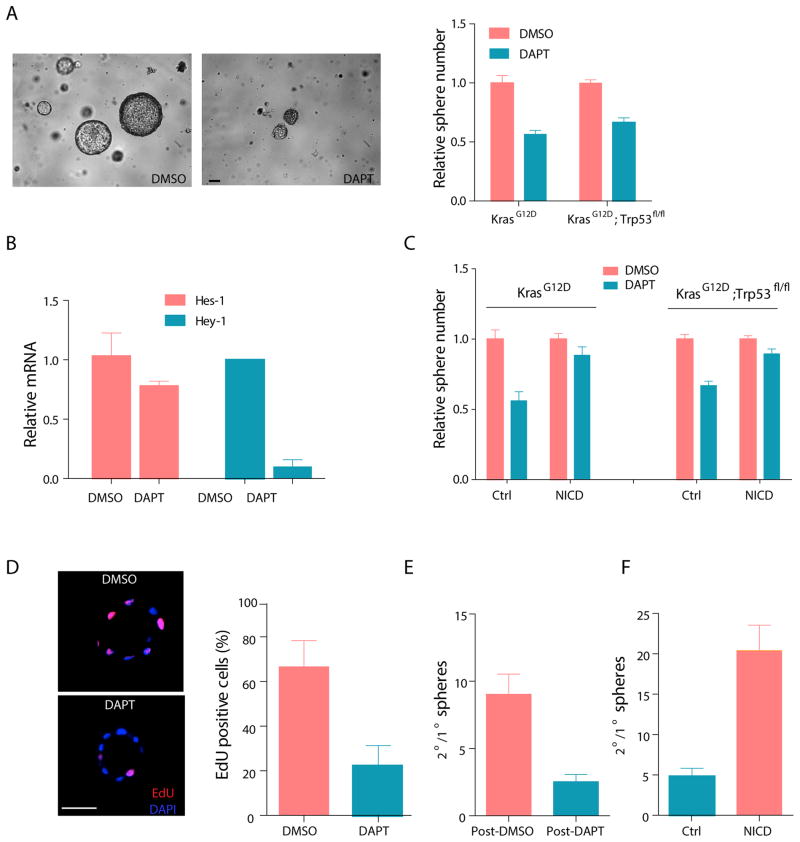
Notchhi cells have increased expression of Notch target genes (Figure S3J), suggesting that the Notch pathway may play a role in maintaining NSCLC TPCs. To determine whether sphere formation is Notch-dependent, we plated freshly isolated tumor cells in the presence or absence of DAPT, a gamma-secretase inhibitor (GSI) that blocks proteolytic processing of Notch. DAPT-treatment significantly decreased the number of primary pulmospheres (Figure 6A). Consistent with the inhibition of Notch signaling, DAPT treatment led to decreased expression of Notch target genes Hes1 and Hey1 (Figure 6B). Furthermore, inhibition of primary pulmosphere growth was rescued by retroviral transduction of a Notch intracellular domain (NICD), confirming that the effect of DAPT on primary pulmosphere number was mediated by inhibition of canonical Notch signaling (Figure 6C). In addition, EdU labeling demonstrated a decrease in global proliferation of pulmosphere cells in the presence of DAPT (Figure 6D).
Figure 6. Notch signaling is functionally important for TPC self-renewal.
(A) Effect of DAPT on primary pulmosphere growth from KrasG12D; eYFP and KrasG12D; Trp53fl/fl; eYFP models. Left panel is representative image for primary pulmosphere growth after 7-day DAPT or DMSO treatment. The scale bar represents 100 μm. Quantification of total number of primary pulmospheres from both genotypes is shown on the right. (B) Expression of canonical Notch target genes Hes-1 and Hey-1 in DMSO or DAPT treated pulmospheres determined by qRT-PCR. The y-axis is the relative quantification of each transcript normalized against Hprt and the expression of each transcript in DMSO treated pulmospheres is set to 1. (C) Quantification of relative number of primary pulmospheres in DAPT and DMSO treated culture. Freshly isolated tumor cells from KrasG12D (n=3) or KrasG12D; Trp53fl/fl (n=3) mice were infected with control or NICD expressing retroviral vector before plating for pulmospheres. (D) Cell proliferation in KrasG12D; Trp53fl/fl pulmospheres was assessed by a six-hour EdU pulse labeling followed by immunofluorescence (left). Quantification of the frequency of EdU positive KrasG12D; Trp53fl/fl cells in each pulmosphere is shown on right. The scale bar represents 50 μm. (E) Effect of DAPT on pulmosphere self-renewal in spheres isolated from KrasG12D; Trp53fl/fl model (n=3 mice). Y-axis is the ratio of total number of secondary spheres over number of primary spheres. (F) Effect of constitutively active NICD on KrasG12D; Trp53fl/fl pulmosphere self-renewal (n=3 mice). Y-axis is the ratio of total number of secondary spheres over number of primary spheres. Error bars indicate mean±SEM. All p-values are from 2-tailed t-test.
Only a small percentage of tumor cells within pulmospheres have the capacity to self-renew and give rise to secondary pulmospheres (Figure 2G). As Notch enriches for TPCs (Figure 3C), Notch may also be functionally required for their self-renewal. To address this, primary KrasG12D; Trp53fl/fl pulmospheres grown in the presence of DAPT or vehicle control were dissociated and replated for secondary pulmosphere growth in the absence of DAPT. DAPT pre-treatment led to a 3.5 fold reduction in the self-renewal ability of primary spheres (p=0.0007) (Figure 6E). Conversely, primary tumor cells overexpressing the NICD demonstrated a 4-fold increase in self-renewal upon secondary passage (p=0.0009) (Figure 6F). Taken together, these data suggest that Notch activation is functionally important for the self-renewal of TPCs in NSCLC.
Notch3 plays a critical role in non-small cell lung cancer TPC self-renewal
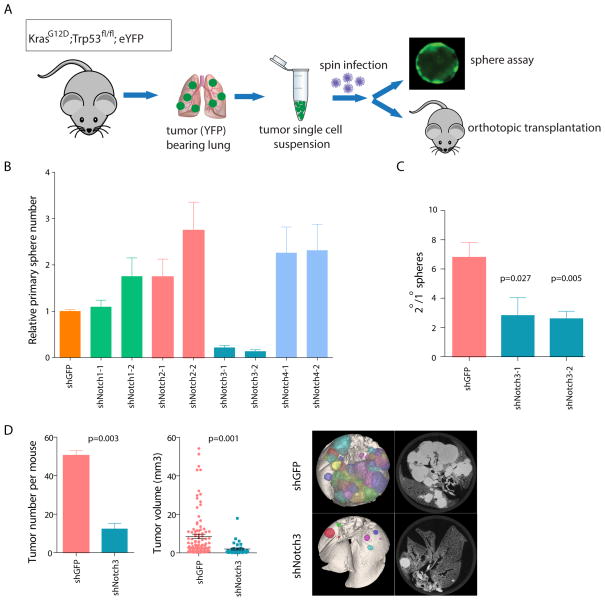
GSIs inhibit activation of all Notch receptors and lead to gastrointestinal toxicity, limiting their clinical effectiveness. Therefore, identification of a non-redundant role for specific Notch receptors could provide a potential therapeutic opportunity in NSCLC. To determine whether specific Notch receptors play non-redundant roles in TPCs, tumor cells isolated from KrasG12D; Trp53fl/fl; eYFP mice were transduced with two independent shRNAs against each of the four Notch genes (Figure S5A–D) or a control hairpin against GFP and plated to assess sphere growth (Figure 7A). None of the hairpins against Notch1, Notch2 or Notch4 significantly decreased primary pulmosphere number (Figure 7B). In contrast, knockdown of Notch3 led to a 5–6 fold reduction in primary sphere number (p<0.0001 for both hairpins) (Figure 7B). In addition, knockdown of Notch3 with the two independent shRNAs led to a 60 and 70% reduction in sphere self-renewal efficiency (p=0.027 and p=0.0049) upon passaging to secondary spheres, suggesting that Notch3 plays a critical role in self-renewal of spheres (Figure 7C).
Figure 7. Notch3 regulates TPC self-renewal in vitro and in vivo.
(A) Schematic illustration of knock-down studies using mouse primary tumor cells. Freshly isolated mouse primary tumor cells were infected with shRNA-carrying lentivirus and tumor cells were then plated in vitro or orthotopically transplanted into recipient mice via intratrachael intubation. (B) Effect of Notch1–4 shRNA on primary KrasG12D; Trp53fl/fl; eYFP pulmosphere growth (n=3, mean±SEM). (C) Effect of Notch3 knock-down on KrasG12D; Trp53fl/fl; eYFP pulmosphere self-renewal (n=3, mean±SEM). (D) Effect of Notch3 knock-down on the tumor-propagating capacity of TPCs in KrasG12D; Trp53fl/fl; eYFP mice by orthotopic transplantation study. Average number of secondary tumors formed per recipient mouse is shown on the left, individual tumor sizes are plotted in the middle and representative ex vivo CT images are on the right. All p-values are from 2-tailed t-test. See also Figure S5.
The importance of Notch3 for TPC function was further investigated in vivo. Freshly isolated primary tumor cells from KrasG12D; Trp53fl/fl;eYFP mice were immediately transduced with lentivirus carrying hairpins against either Notch3 or GFP and grafted into nude mice by orthotopic transplantation as described before (without any culture in vitro). Notch3 knockdown led to a 4-fold reduction in both tumor number and tumor size of secondary tumors in recipient mice (p=0.003 and p=0.001, respectively) (Figure 7D). Thus, Notch3-dependent signaling is essential for tumor maintenance.
Integrity of Notch3 signaling is important for human NSCLC
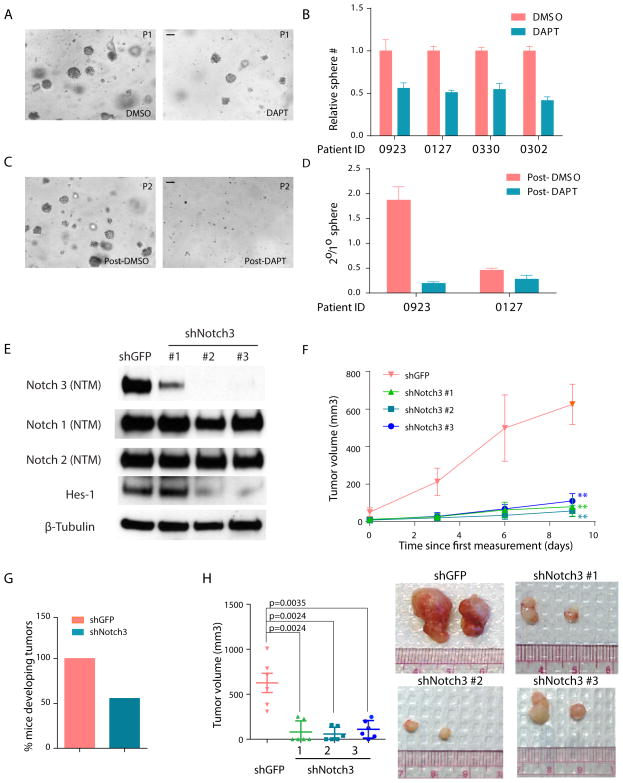
To determine whether Notch signaling plays a role in human NSCLC, the pulmosphere assay was applied to grow primary human NSCLC cells isolated directly from patient samples. Approximately 30% of primary patient samples were successfully cultured as pulmospheres and of these ~50% could be passaged efficiently. Successfully established human pulmospheres were morphologically similar to those from mouse tumors (Figure 8A). DAPT treatment resulted in an average of 2-fold reduction in the number of human primary spheres (p<0.0001) (Figure 8A & 8B). To determine whether DAPT treatment blocked self-renewal, the human pulmospheres were dissociated and passaged for secondary growth. DAPT pretreated spheres showed an average of 70% reduction in self-renewal (p=0.038) (Figure 8C & 8D). Two of the samples tested were from patients with mutations in oncogenic Kras, the third had an EGFR mutation and the fourth was wild-type for both Kras and EGFR (Figure S6A). Thus, the effect of DAPT is not limited only to tumors with Kras mutation. These data support a functional role for Notch signaling in human adenocarcinoma, independent of genotype.
Figure 8. Notch signaling is important for the self-renewal of primary tumors cells isolated from human NSCLC.
(A) Representative images of DAPT and DMSO treated primary pulmospheres derived from human NSCLC patients at day 14 of ex vivo culture. The scale bar represents 100 μm. (B) Effect of DAPT on total number of human primary pulmospheres in four primary patient samples. Y-axis is the total sphere number normalized to DMSO treated culture. (C) Representative image of secondary spheres passaged from DAPT and DMSO pre-treated samples at day 14. The scale bar represents 100 μm. (D) Effects of DAPT on human TPC self-renewal in primary patient samples. (E) Western blot of lysates from primary PDX tumor cells infected with hairpins against either GFP (as control) or Notch3. (F) Primary tumor cells derived from two PDX tumors were infected with hairpins against human Notch3 or GFP and injected into NSG mice subcutaneously. Graphs show average tumor volume at indicated time points. (G) Percentage of injections that formed palpable tumors at the end of xenograft experiment with PDX samples. (H) Average tumor size at the end of xenograft experiment using PDX samples (left). Representative images for tumors in indicated group at the end of xenograft experiment are shown on right. Error bars indicate mean ± SEM. All p-values are from a 2-tailed t-test. **, p<0.01. See also Figure S6.
As with the mouse TPCs, we sought to determine whether Notch3 plays a specific role in human NSCLC propagation. First, Notch3 knockdown was assessed in three established NSCLC cell lines. NCI-H358 harbors a G12C mutation in Kras, NCI-H1650 and NCI-H1975 are both Kras wild type. Interestingly, knockdown of Notch3 with three independent hairpins resulted in significant impairment of cell growth in all three cell lines (Figure S6B). The functional role of Notch3 in human NSCLC propagation was further tested with patient-derived xenograft (PDX) models in vivo. Two independent tumors established from a single PDX sample were infected with one of three Notch3 shRNAs or a GFP shRNA control (Figure 8E). Notch3 knock-down had no effect on protein expression of Notch 1 or Notch 2, yet was sufficient to ablate expression of Hes-1, a canonical downstream target for Notch signaling, suggesting that Notch3 signaling is non-redundant in these cells. Moreover, diminished Notch3 expression dramatically impaired tumor development in vivo (Figure 8F). At the end of the study, there was a 100% tumor incidence in the control group whereas only 55% of the injections from tumor cells infected with Notch3 hairpins formed tumors (Figure 8G). Furthermore, a ~6–10 fold reduction in tumor size was observed in the Notch3 knockdown group as compared with control (Figure 8H). Taken together, these data strongly support a non-redundant role for Notch3 –mediated signaling in both mouse models of NSCLC and in human NSCLC.
Discussion
Using genetically engineered murine models of non-small cell lung cancer, we identified a subpopulation of tumor cells with increased tumorigenicity based on their expression of a newly identified panel of cell-surface markers; CD24, ITGB4 and Notch (3+ cells). We demonstrate that 3+ cells have increased self-renewal properties in vitro and in vivo, which requires Notch3. Furthermore, this requirement is conserved in human NSCLC TPCs, as GSI treatment and Notch3 knockdown severely attenuate self-renewal and tumor propagation in NSCLC cell lines and primary patient tumors. 3+ cells are selected for after repeated doses of chemotherapy, suggesting an important role for these cells as a reservoir for chemoresistant tumor re-initiation. Importantly, we show that the gene signature that distinguishes mouse TPCs has prognostic significance in NSCLC patients
CD24, ITGB4 and Notch are surface markers that enrich for TPCs
We developed and employed an in vitro sphere assay to identify markers that would allow us to enrich for TPCs. Starting with a previously reported TPC marker, CD24, we observed an enrichment in sphere forming capacity over bulk tumor cells. The stepwise addition of ITGB4 and Notch1–4 resulted in a progressive increase in sphere formation (Figure 3A–C). We then went on to show that sphere forming capacity of 3+ and non-3+ cells closely correlated with their in vivo tumor forming capacity (Figure 3D–F).
Intriguingly, two of the markers we identified (Notch and ITGB4) have been suggested to be involved in lung development and normal lung stem cells. ITGB4 has been used to isolate a multipotent epithelial stem/progenitor population from the adult mouse lung (Chapman et al., 2011; McQualter et al., 2010). However, it was demonstrated that ITGB4 was dispensable for their stem cell function in vitro. Given the known role of integrins in adhesions and cell-cell interactions in the normal stem cell niche, it would be interesting to assess the role of ITGB4 in TPCs in vivo.
Notch signaling is also known to play an important role in lung development. Notch1 and Notch ligands Jag1/2, as well as Notch target genes Rbpjk, Hes1, Hes5 and Hey2, are expressed in early murine lung epithelium and are important for the specification of proximal lung structures (Tsao et al., 2008). Furthermore, transgenic mice over-expressing the Notch intracellular domain in the lung epithelium show arrested differentiation of distal lung progenitors, with impaired development of Type I and II pneumocytes (Dang et al., 2003; Guseh et al., 2009). Thus, Notch activity blocks differentiation of the normal alveolar epithelium. This is consistent with our finding that Notch3 is essential for the self-renewal activity of the TPCs and suggests that it may act by maintaining them in a less differentiated state. Consistent with this notion, we find decreased expression of the alveolar type II cell marker, Sftpc, in the 3+ cells (data not shown) suggesting they may be in a less differentiated state than the non-3+ cells.
The identification of CD24, ITGB4 and Notch as robust surface markers provides an opportunity to isolate murine TPCs prospectively. In future studies it will be interesting to determine whether these markers also play a role in identifying a population of human tumor cells with increased self-renewal capacity. In addition, although highly enriched for TPCs, the 3+ population is still heterogeneous and likely only a subset of these cells are true TPCs. It is therefore possible that other markers could further enrich for TPCs. Efforts to identify these additional markers are ongoing.
Critical and non-redundant role of Notch3 signaling in TPC self-renewal
Notch has been implicated in multiple human cancers but its role in oncogenesis is highly context-dependent, acting as either an oncogene or tumor suppressor depending on the disease setting (Ranganathan et al., 2011). Previous studies have shown an association between NSCLC disease progression and Notch pathway activation (Dang et al., 2000; Mimae et al., 2012; Westhoff et al., 2009) Consistent with previous studies, our findings using chemical blockade with DAPT demonstrate a global decrease in tumor cell proliferation. Furthermore, DAPT-treatment decreases TPC self-renewal, while conversely, increased Notch pathway activity results in enhanced self-renewal (Figure 6). Thus, our study underscores the role of Notch signaling in sustaining tumor propagation in NSCLC. Such a role of Notch signaling in NSCLC seems to be independent of Kras or Trp53 genotype, in contrast to the Trp53-dependent role of Notch in leukemia (Beverly et al., 2005; Guo et al., 2008).
It is interesting that all four Notch receptors enrich for TPCs as surface markers but only Notch3 seems to be functionally non-redundant (Figure 7B–D and 8E–H). This result may reflect discordance between surface expression of Notch receptors and activity of Notch signaling pathways. Indeed, recent studies demonstrate low intracellular Notch signaling in AML cells despite high expression of Notch2 at the cell surface (Kannan et al., 2013; Lobry et al., 2013). Interestingly, Notch2 seems to suppress AML upon reactivation, highlighting the necessity of characterizing the output of Notch signaling, instead of merely Notch expression. Such a discordance may be rooted in availability of Notch ligands in the TPC niche. Alternatively, other post-translational regulations, such as glycosylation and endocytosis inside TPC cells, may fine-tune specific Notch activation. Further elucidation of paralog-specific activity will lead to a better understanding of Notch signaling in NSCLC.
Relevance of Murine TPCs to Human NSCLC
Unsupervised hierarchical clustering based on gene expression profiles of 3+ and non-3+ cells clearly demonstrates that they are distinct tumor cell populations (Figure 5C). Importantly, the “TPC up” gene expression signature identified in our study correlates to poor prognosis in human NSCLC, while the “TPC down” signature correlates to good prognosis (Figure 5C), strongly suggesting that genes involved in determining the TPC state also drive aggressive human disease. This hypothesis is supported by our observation that the TPCs in mouse lung cancer are enriched after repeated doses with chemotherapy, suggesting that the TPC molecular machinery may contribute to chemoresistance (Figure 5B). These studies underscore the need to combine studies of chemotherapy response with an understanding of tumor cell heterogeneity if we hope to increase our ability to modulate chemoresistance.
In addition, we show that the Notch-mediated self-renewal observed in mouse TPCs is conserved in human NSCLC, as evidenced by inhibition of sphere formation and self-renewal in multiple DAPT-treated patient samples (Figure 8A–D). In the mouse, Notch3 knockdown, but not knockdown of Notch1,2 or 4, impaired the self-renewal of TPCs (Figure 7C) and dramatically reduced both the incidence and size of secondary tumors in vivo (Figure 7D). Importantly, such a non-redundant role of Notch3 signaling is also conserved in at least some human NSCLC, as knocking-down Notch3 expression in a PDX sample also dramatically decreases tumor formation in vivo (Figure 8).
Simultaneous inhibition of Notch1 and Notch2 has been shown to underlie the GI toxicity associated with GSIs in clinical trials (van Es et al., 2005) Thus, finding a non-redundant Notch pathway in NSCLC has important therapeutic implications, as it may help narrow the target and decrease toxicity. Taken together, our results strongly support a critical role for Notch3 in NSCLC and specifically in TPCs and suggest that specific inhibition of Notch3 may be a promising target for treatment of this devastating disease.
EXPERIMENTAL PROCEDURES
Mice
Lox-stop-Lox-KrasG12D (Jackson et al., 2001) (129 Sv/Jae), Trp53fl/fl (Jonkers et al., 2001) (FVB), Rosa26-LSL-eYFP (Srinivas et al., 2001) and Rosa26-LSL-tdRFP (Luche et al., 2007) (C57BL/6J) mice were maintained in viral-free environment. Mice were intranasally infected with adenoviral-Cre (University of Iowa) between ages of four- to ten-weeks (Jackson et al., 2001). Tumor bearing mice were sacrificed ~12 weeks after infection for KrasG12D; Trp53fl/fl; eYFP and 21 weeks after infection for KrasG12D; eYFP. All animal experiments were approved by the Stanford University School of Medicine Committee on Animal Care (APLAC).
FACS sorting
Cell sorting was performed on either a BD FACSVantage SE, or a BD FACSAria or a BD Influx, and data were analyzed with FlowJo software. Gates were established by comparing FACS plot against fluorescence minus one (FMO) sample with a cut-off of 0.5% positive cells in the FMO sample as previously described (Herzenberg et al., 2006). Negative gates were set at least one half-log apart from positive gates to avoid ambiguous distinctions between weak positive and true negative cells (see Supplemental Experimental Procedures).
Pulmosphere culture
Live tumor cells with desired density in single cell suspension were re-suspended in 100ul of medium containing 50% matrigel (see Supplemental Experimental Procedures). For self-renewal assays, single cell suspension from primary pulmospheres was replated with the conditions described above. Self-renewal was assessed as the ratio the number of secondary pulmospheres divided by that of primary pulmospheres DAPT (EMD chemicals) containing stem cell medium was changed every other day.
Orthotopic transplantation
Intratracheal intubation was performed using a mouse intubation pack (Hallowell EMC). Tumor cells were introduced into the trachea at indicated doses. Tumor burden was assessed 16 weeks after tracheal intubation by Micro-CT analysis of lungs (Caunt et al., 2008) (see Supplemental Experimental Procedures).
Virus preparation and transduction of primary tumor cells
Retrovirus was made by transfecting 293FT cells with MigR1-IRES-GFP or MigR1-ICN-IRES-GFP (gift of Dr. Warren Pear, University of Pennsylvania) together with packaging plasmids. PLKO.1 based Notch shRNAs (Open Biosystems) were transfected together with packaging plasmids in 293FT cells using FuGeneHD (Promega). High titer virus was generated by pooling virus-containing medium and spinning at 25,000rpm for 2 hours to ensure a MOI > 20. Freshly isolated tumor cells were transduced using a 30-minute spin infection followed by incubation at 37°C for 4–6 hours prior to plating or orthotopic transplantation.
Gene expression analysis
Microarray analysis was performed with Agilent Whole Mouse Genome 4×44K arrays. Agilent Feature Extraction software 9.5 was used to analyze acquired array images and obtain individual log2 ratios of background-subtracted signal intensities. Significance analysis of microarrays (SAM) and Linear Models for Microarray Data (LIMMA) were used to identify differentially expressed genes between experimental groups. Genesets were used for survival analysis using publicly available data (Shedden et al., 2008) collected from four different centers as described (Vicent et al., 2012). Cox hazard ratios were calculated to estimate the relative risk along with confidence interval (CI) and log-rank tests (see Supplemental Experimental Procedures).
EdU labeling
Mice were injected intraperitoneally with EdU (Invitrogen) at a dose of 100 mg/kilogram body weight for 16 hours before euthanasia. For EdU labeling of spheres, 1mM EdU was used to label spheres for designed time before fixation and embedding.
Human Primary NSCLC Samples
This study complied with federal, state, and local regulations of Human Research Protection Program (HRPP) and was approved by Stanford Institutional Research Board. Informed consent was obtained from all patients included in the study.
Establishment of Patient-Derived Xenograft tumors from Primary Human NSCLC
Surgically removed human NSCLC tumor tissues were kept in ice-cold HBSS (Life technologies) until use. Tumors were cut into 1mm pieces and implanted in the subrenal capsules in NOD-SCID-IL2Rγ (NSG) mice (Jackson Laboratory, Bar Harbor, Maine).
Tumor study with Patient-Derived NSCLC xenograft model
0.5 × 106 NSCLC tumor cells from PDX tumors were resuspended in 100uL stem cell medium containing 25% matrigel and injected into the lower flank of NSG mice. Tumor size was measured with a digital caliper, and tumor volume was calculated using volume = 0.5 × (length) × (width)2.
Supplementary Material
Significance.
A distinct population of tumor cells capable of tumor propagation has been identified in some cancers. However, the molecular mechanisms that drive self-renewal and propagation in these cells remain incompletely understood. Here we identified a set of surface markers that greatly enrich for cells with tumor-propagating ability in two mouse models of NSCLC. This tumor propagating cell (TPC) subpopulation is enriched after chemotherapy and the gene expression signature of TPCs is predictive of poor prognosis in human NSCLC. Furthermore, we demonstrate a specific and non-redundant role for Notch3 in mediating propagation and self-renewal of TPCs. Thus Notch receptors play specific roles in NSCLC and blockade of Notch3 may be a useful therapeutic strategy for some cases of NSCLC.
Acknowledgments
We thank Julien Sage, Steve Artandi, Laura Attardi, Tom Rando, Michael Clarke and all members of the Sweet-Cordero lab for helpful discussions. We thank Patty Lovelace for expert FACS advice. We thank the staff of Stanford shared FACS facility and the FACS core facility of Institute of Stem Cell Biology and Regenerative Medicine for technical support. We thank Ms. Anuradha Tathireddy for expert help with mouse husbandry. EASC was supported by a Research Scholar Grant from the American Cancer Society (RSG-08-132-01-CDD), and a grant from National Cancer Institute (5R01CA157510). YZ was supported by a Senior Research Fellowship from the American Lung Association (RT-82480-N). CAC was a recipient of a California Institute for Regenerative Medicine (CIRM) Bridges Award (TB1-01194).
Footnotes
Accession Number
Microarray results (GSE46438 and GSE46439) are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 2011;71:6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–7168. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- Caunt M, Mak J, Liang WC, Stawicki S, Pan Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J, et al. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer cell. 2008;13:331–342. doi: 10.1016/j.ccr.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, Vu TH. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012 doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Wang J, Chen D, Chen YJ. CD133 is a temporary marker of cancer stem cells in small cell lung cancer, but not in non-small cell lung cancer. Oncol Rep. 2011;25:701–708. doi: 10.3892/or.2010.1115. [DOI] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK, Kim CF. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Lasky JL, Chang CJ, Mosessian S, Lewis X, Xiao Y, Yeh JE, Chen JY, Iruela-Arispe ML, Varella-Garcia M, Wu H. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453:529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136:1751–1759. doi: 10.1242/dev.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, Carbone DP, Dang TP. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- Hegde GV, de la Cruz CC, Chiu C, Alag N, Schaefer G, Crocker L, Ross S, Goldenberg D, Merchant M, Tien J, et al. Blocking NRG1 and Other Ligand-Mediated Her4 Signaling Enhances the Magnitude and Duration of the Chemotherapeutic Response of Non-Small Cell Lung Cancer. Sci Transl Med. 2013;5:171ra118. doi: 10.1126/scitranslmed.3004438. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA, Tung J, Moore WA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- Hoey T, Yen WC, Axelrod F, Basi J, Donigian L, Dylla S, Fitch-Bruhns M, Lazetic S, Park IK, Sato A, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature Genetics. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, Akers LJ, Hammitt RA, McMurray JS, Kornblau SM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. The Journal of experimental medicine. 2013;210:321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Konishi J, Yi F, Chen X, Vo H, Carbone DP, Dang TP. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene. 2010;29:589–596. doi: 10.1038/onc.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Terence Kin W, Castilho A, Cheung Vincent Chi H, Tang Kwan H, Ma S, Ng Irene Oi L. CD24+ Liver Tumor-Initiating Cells Drive Self-Renewal and Tumor Initiation through STAT3-Mediated NANOG Regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD, Fink LM, Ma Y, Wong MP. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5:e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. The Journal of experimental medicine. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, Mulero F, Megias D, Sanchez-Carbayo M, Shen J, et al. Therapeutic Effect of gamma-Secretase Inhibition in Kras(G12V)-Driven Non-Small Cell Lung Carcinoma by Derepression of DUSP1 and Inhibition of ERK. Cancer Cell. 2012;22:222–234. doi: 10.1016/j.ccr.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clinical chemistry. 2013;59:168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells. 2009;27:623–633. doi: 10.1634/stemcells.2008-0866. [DOI] [PubMed] [Google Scholar]
- McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci U S A. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Li M, Wang X, Wang Y, Ma D. Both CD133+ and CD133− subpopulations of A549 and H446 cells contain cancer-initiating cells. Cancer science. 2009;100:1040–1046. doi: 10.1111/j.1349-7006.2009.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimae T, Okada M, Hagiyama M, Miyata Y, Tsutani Y, Inoue T, Murakami Y, Ito A. Upregulation of notch2 and six1 is associated with progression of early-stage lung adenocarcinoma and a more aggressive phenotype at advanced stages. Clin Cancer Res. 2012;18:945–955. doi: 10.1158/1078-0432.CCR-11-1946. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. Journal of the National Cancer Institute. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, Stanbridge EJ, Lee EY. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teisanu RM, Lagasse E, Whitesides JF, Stripp BR. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells. 2009;27:612–622. doi: 10.1634/stemcells.2008-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirino V, Camerlingo R, Franco R, Malanga D, La Rocca A, Viglietto G, Rocco G, Pirozzi G. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009;36:446–453. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- Tsao PN, Chen F, Izvolsky KI, Walker J, Kukuruzinska MA, Lu J, Cardoso WV. Gamma-secretase activation of notch signaling regulates the balance of proximal and distal fates in progenitor cells of the developing lung. J Biol Chem. 2008;283:29532–29544. doi: 10.1074/jbc.M801565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent S, Sayles LC, Vaka D, Khatri P, Gevaert O, Chen R, Zheng Y, Gillespie AK, Clarke N, Xu Y, et al. Cross-Species Functional Analysis of Cancer-Associated Fibroblasts Identifies a Critical Role for CLCF1 and IL-6 in Non-Small Cell Lung Cancer In Vivo. Cancer Res. 2012;72:5744–5756. doi: 10.1158/0008-5472.CAN-12-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.