Abstract
Background. Low-frequency nevirapine (NVP)–resistant variants have been associated with virologic failure (VF) of initial NVP-based combination antiretroviral therapy (cART) in women with prior exposure to single-dose NVP (sdNVP). We investigated whether a similar association exists in women without prior sdNVP exposure.
Methods. Pre-cART plasma was analyzed by allele-specific polymerase chain reaction to quantify NVP-resistant mutants in human immunodeficiency virus–infected African women without prior sdNVP who were starting first-line NVP-based cART in the OCTANE/A5208 trial 2. Associations between NVP-resistant mutants and VF or death were determined and compared with published results from women participating in the OCTANE/A5208 trial 1 who had taken sdNVP and initiated NVP-based cART.
Results. Pre-cART NVP-resistant variants were detected in 18% (39/219) of women without prior sdNVP exposure, compared to 45% (51/114) with prior sdNVP exposure (P < .001). Among women without prior sdNVP exposure, 8 of 39 (21%) with NVP-resistant variants experienced VF or death vs 31 of 180 (17%) without such variants (P = .65); this compares with 21 of 51 (41%) vs 9 of 63 (14%) among women with prior exposure (P = .001).
Conclusions. The risk of VF on NVP-based cART from NVP-resistant variants differs between sdNVP-exposed and -unexposed women. This difference may be driven by drug-resistance mutations emerging after sdNVP exposure that are linked on the same viral genome.
Clinical Trials Registration NCT00089505.
Keywords: minor drug-resistant variants, single-dose nevirapine, antiretroviral therapy failure
Mutations in the human immunodeficiency virus type 1 (HIV-1) genome have been hypothesized to occur at all nucleotide positions, including those that confer drug resistance, as the result of high rates of HIV-1 replication, mutation, and recombination [1–3]. Consequently, after exposure to one antiretroviral drug such as nevirapine (NVP), given as a single dose to prevent mother-to-child HIV-1 transmission, drug-resistant variants can rapidly emerge [4–7]. It is well established that such drug-resistant variants, when detected by standard genotype, can compromise virologic responses to combination antiretroviral therapy (cART) [8–10]. For example, in the OCTANE/A5208 trial 1, conducted in African women with prior exposure to single-dose NVP (sdNVP) and who subsequently initiated NVP-based cART, NVP resistance detected by standard genotype at study entry was associated with virologic failure (VF) or death, and lopinavir/ritonavir (LPV/r) was superior to NVP-based cART in sdNVP-exposed women [11]. The impact of minor populations of drug-resistant variants on the response to initial cART has been more controversial, with some studies reporting their association with VF and others finding no association [12–19]. We reported that the risk of VF with NVP-based cART was significantly associated with low frequency (>1%), NVP-resistant variants in African women with prior exposure to sdNVP (OCTANE/A5208 trial 1) [20]. In women who had never been exposed to sdNVP (OCTANE/A5208 trial 2), however, NVP- or LPV/r-based cART showed equivalent virologic efficacy [21]. To investigate the different outcomes of the NVP-based cART arms of trial 1 and trial 2, we performed allele-specific polymerase chain reaction (PCR) on pre-cART samples from women without prior sdNVP exposure (trial 2), to quantify frequencies of the 3 most common NVP resistance mutations (K103N, Y181C, and G190A) and their association with VF or death.
METHODS
Study Design and Participants
The A5208/OCTANE study consisted of 2 parallel, randomized, open-label cART trials. A5208/OCTANE was approved by all relevant institutional review boards and ethics committees, and all participants provided written informed consent. Trial 1 enrolled women who had previously received sdNVP ≥6 months before enrollment. Trial 2 enrolled women who had no prior sdNVP exposure. Details of primary analyses and results from trial 1 and trial 2 have been previously published [11, 21]. In brief, participants were HIV-1–infected women from 10 sites in 7 countries of sub-Saharan Africa. Women had a screening CD4 count <200 cells/µL, were cART-naive, and were followed until 48 weeks. Women were randomized to cART with lopinavir/ritonavir (LPV/r) or NVP, each with tenofovir/emtricitabine (TDF/FTC).
Definition of Primary Endpoint
The primary study endpoint was time from randomization to the earlier of death or VF. VF was defined as <1 log10 copies/mL drop in plasma HIV-1 RNA copies/mL from baseline by week 12 or HIV-1 RNA >400 copies/mL at or after 24 weeks.
Allele-Specific PCR
All samples were tested without knowledge of primary study endpoint or standard genotype results. Testing of samples was approved by the National Institutes of Health Office of Human Subjects Protection. HIV-1 RNA was extracted and PCR amplified using reported methods [20, 22, 23]. PCR products were then analyzed by real-time subtype-specific allele-specific PCR (ASP) to quantify mutant frequencies down to 0.1% for codon 103 and 190 and 0.3% at codon 181 in reverse transcriptase. Equivalent reactions were performed using 2 primer sets to quantify mutant and wild-type frequencies in a background of total HIV-1 DNA. A common forward primer was used in combination with different reverse primers that are allele specific at the 3′ end but mismatch all templates at the penultimate base. The penultimate mismatch minimizes the opportunities for base pairing and reduces amplification (up to 1000-fold) when the 3′ end of the primer is also mismatched [24–26]. In parallel, total HIV-1 DNA was quantified to accurately determine the percent variant at each allele as well as detect sequence polymorphism that may have affected PCR efficiency. Each ASP assay is performed at annealing temperatures of ≥5 degrees above the melting temperature of each selective primer, resulting in an average negative control background of 0.01% for assays measuring 103N and 190A and 0.05% for 181C assays.
To minimize the possibility of obtaining false-positive assay results, the estimated mutant frequency for reporting a sample as positive for any mutation had to be at least 3.10 standard deviations above the negative control background for 0% mutant (100% wild-type) standards. Each positive sample was retested, and the mean value was used for confirmed positives (unconfirmed positives were counted as negative). To account for polymorphisms that might affect amplification efficiencies of wild-type and mutant sequence equally, positive values were normalized to the sum of the species detected. If the sum of the species detected was <25%, the assay was considered indeterminate at that codon. Negative values were never normalized. We have previously reported that this method measures mutant frequencies accurately compared to other methods that detect minority variants, such as single-genome sequencing and 454 deep sequencing [27–30].
Data Analyses
Trial 2 data from sdNVP-unexposed women were compared to published data from sdNVP-exposed women in trial 1, which were generated by the same laboratory and methods described above. All analyses were intent-to-treat for women who initiated treatment. A composite measure across the 3 codons was created as the sum of the percentages at each codon. The composite was used because it is not possible to determine whether the mutations were on the same or different viral genomes. The categories of mutation frequencies were the same as those in the published analyses for sdNVP-exposed women in trial 1 [20]. The Fisher exact test and the Jonckheere-Terpstra test were performed to compare the proportions of women in each mutation frequency category between the 2 trials. The Wilcoxon rank-sum test was employed to compare the frequency of mutations between the 2 trials. The mutant population size was obtained by multiplying the percentage of mutant frequency by the HIV-1 RNA copy number. Samples from both trials were analyzed for mutant frequency and mutant population size, then analyzed for each mutation separately. The Fisher exact test and exact logistic regression analyses were used to evaluate whether there was a significant trend in odds of VF or death with increasing category of mutation frequency within each trial, and whether the trends in association differed between the 2 trials (results for the latter give interaction P values). Parallel analyses using proportional hazards models gave similar conclusions. All P values are 2-sided and all analyses were done using SAS software, version 9.1 (SAS Institute, Cary, North Carolina).
RESULTS
Of the 249 women without prior sdNVP exposure randomized to the NVP/TDF/FTC treatment arm of trial 2, baseline samples were not available for ASP analysis from 17. Samples from the remaining 232 women were tested by ASP with results obtained from 219. These results were compared to the published ASP results from 114 women with prior sdNVP exposure randomized to the NVP/TDF/FTC arm of trial 1 [20]. Women in the 2 trials had similar pretreatment median HIV-1 RNA in plasma (5.17 vs 5.21 log10 copies/mL, respectively; P = .62), but had a significant but small difference in median CD4 count (119 vs 138 cells/µL; P = .025) and median age (35 vs 30 years; P < .001).
Frequency of NVP-Resistant Mutations by Prior sdNVP Exposure
Results from ASP testing on pretreatment samples are shown in Table 1. ASP detected NVP-resistant variants in 18% (39/219) of women without sdNVP exposure, significantly less than the 45% (51/114) detected among women with prior exposure (P < .001). Women without prior sdNVP exposure who had NVP resistance detected had a similar range of mutant frequencies as women with prior exposure who had NVP resistance detected (0.1%–96.3% vs 0.1%–100%), but had a significantly lower median mutant frequency (0.8% vs 2.9%; P = .008). Fewer women without sdNVP exposure had the 103N mutation detected (8% [17/219] vs 39% [45/114]; P < .001), and the median frequency of 103N among women with 103N detected was significantly lower than among unexposed women (0.6% vs 2.1%; P = .017). By contrast, there were no significant differences in the proportions of women without or with prior exposure to sdNVP who had 181C or 190A mutations (6% vs 12%; P = .13 and 6% vs 8%; P = .49, respectively). A smaller fraction of women without sdNVP exposure had multiple NVP resistance mutations (10% [4/39]) compared to women with sdNVP exposure (25% [13/51]), but was not statistically significant (P = .10). The multiple NVP mutations were usually 103N combined with 181C or 190A.
Table 1.
Nonnucleoside Reverse Transcriptase Inhibitor Resistance Mutations Detected at Entry by Allele-Specific Polymerase Chain Reaction for Women Without and With Prior Single-Dose Nevirapine Exposure
| Category | ASP Result | K103N | Y181C | G190A | All Mutations |
|---|---|---|---|---|---|
| (>0.1%)a | (>0.3%)a | (>0.1%)a | |||
| No prior sdNVP exposure | No. with results | 219 | 217 | 218 | 219 |
| No. with mutation | 17 | 14 | 13 | 39 | |
| % | 8% | 6% | 6% | 18% | |
| Median | 0.60% | 0.70% | 4.40% | 0.80% | |
| Frequencyb (range %) | (0.1–96) | (0.3–5) | (0.1–27) | (0.1–96) | |
| Prior sdNVP exposure | No. with results | 114 | 109 | 109 | 114 |
| No. with mutation | 45 | 13 | 9 | 51 | |
| % | 39% | 12% | 8% | 45% | |
| Median | 2.10% | 0.60% | 0.80% | 2.90% | |
| Frequencyb(range %) | (0.1–100) | (0.3–18) | (0.1–7) | (0.1–100) |
Abbreviations: ASP, allele-specific polymerase chain reaction; sdNVP, single-dose nevirapine.
a Signifies the limit of sensitivity for each assay.
b Frequency among women with a mutation detected.
Size of Mutant Populations by Prior sdNVP Exposure
We also calculated the size of the virus population containing each mutation. As expected, mutant population sizes were strongly correlated with mutant frequencies (Pearson correlation, 0.74; P < .001). Among women with any NVP-resistant mutation detected, the size of the mutant viral populations differed according to prior sdNVP exposure. The median mutant population size across the 3 codons assayed was significantly lower among women without vs with prior sdNVP exposure (1044 vs 5550 copies/mL; P = .002). This difference reflected, in part, a lower median K103N mutant population among women with the K103N mutation (1350 vs 5550 copies/mL; P = .008). By contrast, when the analysis was limited to only women with the Y181C mutation detected, the median population sizes were not significantly different (1007 vs 1297; P = .25). The median mutant population sizes were also not significantly different among women with the G190A mutation detected (2606 copies/mL vs 3429; P = .50). The differences in mutant population were not due to differences in median viremia (149 187 vs 162 582 copies/mL; P = .62) in women with and without prior sdNVP exposure.
Association of Baseline NVP Resistance With Virologic Failure or Death
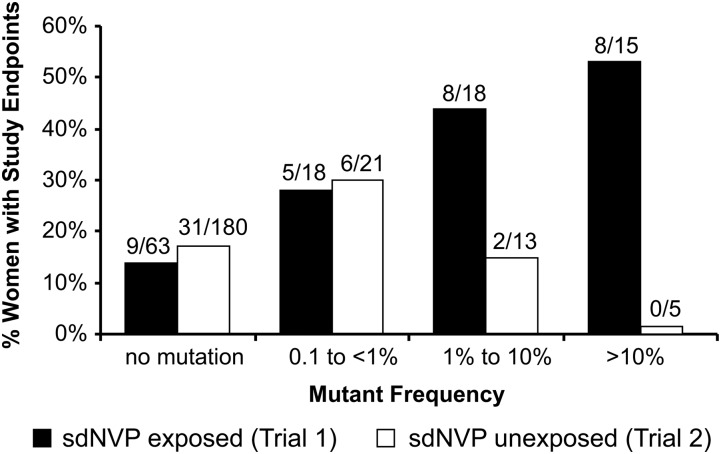
Among women without prior sdNVP exposure, there was no significant difference in the proportion of women reaching VF or death between those with any baseline NVP resistance detected (21% [8/39]) and those without baseline NVP resistance (17% [31/180]) (P = .65; Table 2). By contrast, as previously reported and summarized in Table 2, among women with prior sdNVP exposure, low-frequency NVP-resistant variants at baseline were significantly associated with increased risk of a primary endpoint (P = .001) [20]. The difference in association between trials approached statistical significance (interaction P = .056). To better understand the difference between the trials, we divided women from both trials into 4 groups (no mutation, <1%, 1% to <10%, and ≥10%) according to their combined mutant frequencies (103N + 181C + 190A) and assessed the proportion experiencing a study endpoint (Figure 1). Among women with prior sdNVP exposure, we found that there was a highly significant trend relating mutant frequency to the proportion of women who experienced a primary endpoint (P< .001) [20]. No such relationship was observed among women with no prior sdNVP exposure (P = .93; Figure 1). Furthermore, there was significant evidence that the association between mutation frequency and outcome in nonexposed women was different from that in sdNVP-exposed women (interaction P = .025 comparing trends between exposure groups). The difference in trends between the 2 trials was driven by more frequent study endpoints among sdNVP-exposed women with mutations detected, particularly in the 1% to <10% and ≥10% categories. In addition, nearly all events were virologic failures: only 5 events were deaths among women with no prior sdNVP exposure, and only 4 events were deaths among women with prior exposure. After censoring deaths, there was still no significant association between mutation frequency and time to VF among sdNVP-unexposed women (P = .95), but the association remained significant among sdNVP-exposed women (P = .005). There was no significant difference between trials in the proportion of women with no mutations detected who experienced study endpoints (Figure 1; P = .69).
Table 2.
Study Endpoints of Virologic Failure or Death by Prior Single-Dose Nevirapine Exposure
| Prior sdNVP Exposure | No. (%) | No. Failed | P Valuea |
|---|---|---|---|
| No (n = 219) | |||
| bASP+ | 39 | 8 (21%) | .65 |
| cASP− | 180 | 31 (17%) | |
| Yes (n = 114) | |||
| bASP+ | 51 | 21 (41%) | .001 |
| cASP− | 63 | 9 (14%) | |
Abbreviations: ASP, allele-specific polymerase chain reaction; sdNVP, single-dose nevirapine.
a Fisher exact test.
b ASP positive for 103N (aac,aat),181C, and/or190A.
c ASP negative for 103N (aac,aat),181C, and/or 190A.
Figure 1.
Proportion of women in trial 1, women with single-dose nevirapine (sdNVP) exposure; and trial 2, women without sdNVP exposure reaching a study endpoint of virologic failure or death in each of 3 mutant frequency categories detected at entry compared to women with no mutation in each trial. Trial 1: trend P < .001; trial 2: trend P = .93; comparison of trends between trials: interaction P = .025; comparison of the rates of “no mutations” between trial 1 and trial 2: P = .69, P values from exact logistic regression.
Mutant Population Size
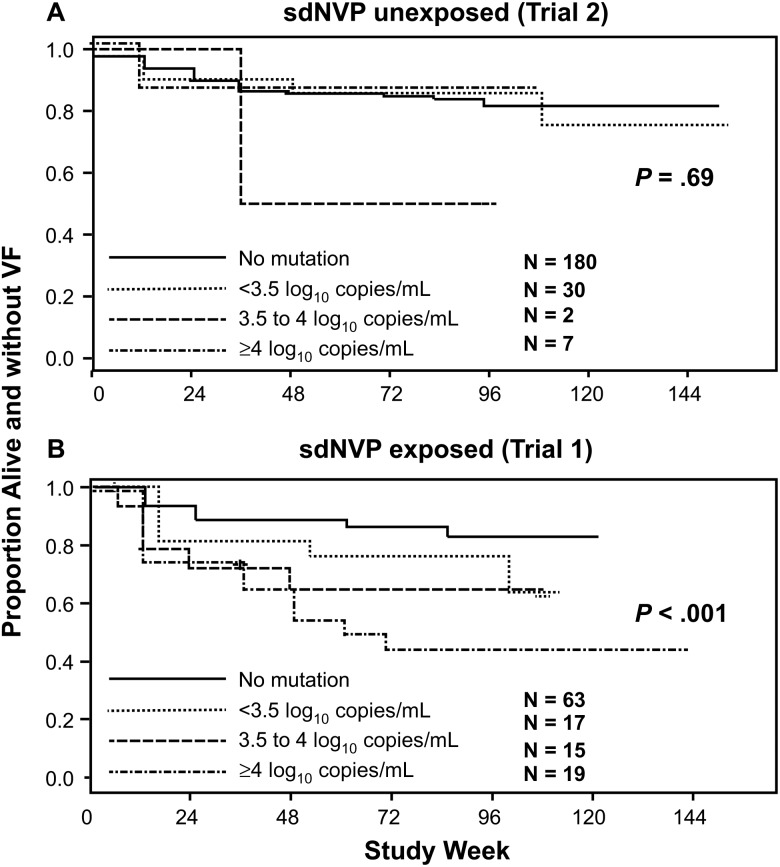
We also analyzed the association between the size of the mutant populations and time to VF or death among women without and with prior sdNVP exposure (Figure 2). Women were divided into each of 3 categories of mutant population size (<3.5 log10 copies/mL, 3.5 to <4 log10 copies/mL, or ≥4 log10 copies/mL) and compared to the no mutation detected group (Figure 2). We again found a significant trend between the size of the mutant population and the proportion of women who experienced a primary endpoint among women with prior sdNVP exposure (P < .001; Figure 2B) but not among women without sdNVP exposure (P = .69; Figure 2A).
Figure 2.
Kaplan-Meier plots showing the proportion of women in trial 2 (A, women without single-dose nevirapine [sdNVP] exposure) and trial 1 (B, women with sdNVP exposure) alive and without virologic failure (VF) in each of 3 categories of mutant copy number detected at entry compared to women with no mutations detected. P values from proportional hazards models.
Analysis of Specific Resistance Mutations
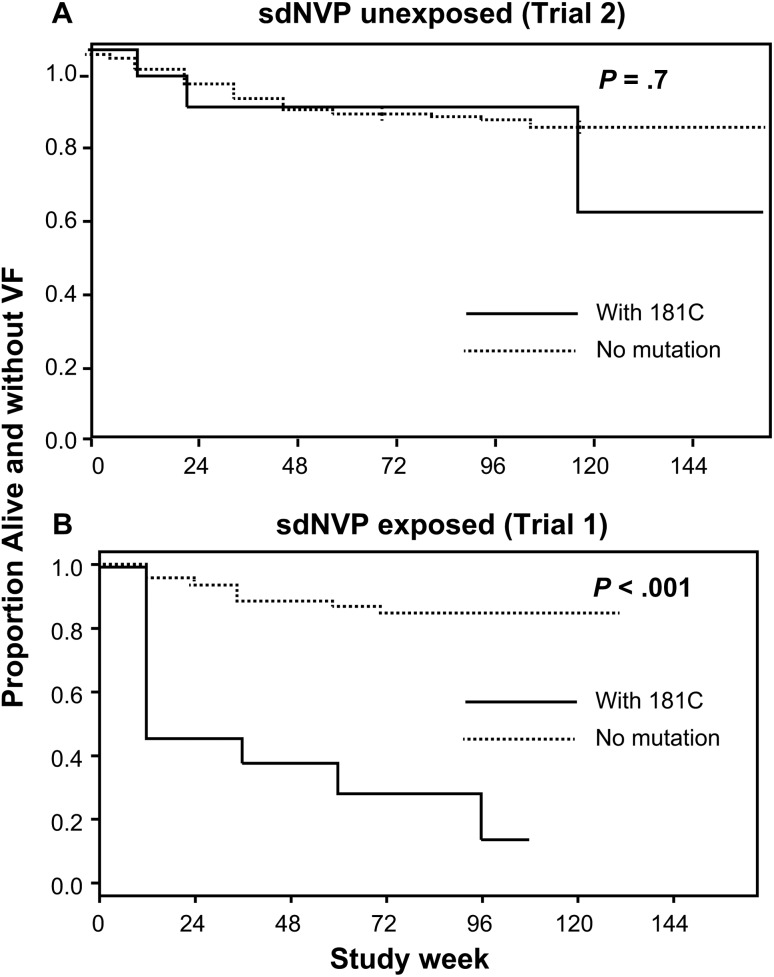
We also analyzed the association between mutations at specific codons and primary study endpoints. Among women with no prior sdNVP exposure, there was no association between the detection of the 103N mutation at baseline and study endpoints (18% [3/17] of women with 103N at entry failed vs 18% [36/202] of women without 103N; P = 1.00; Table 3). By contrast, in trial 1 [20] there was a significant association between the detection of the 103N mutation at study entry and primary study endpoints (P = .009; Table 3). There was also no association between the detection of the 181C mutation at baseline and study endpoints among women without prior sdNVP exposure, (21% [3/14] with 181C at entry failed vs 18% [36/203] without 181C; P = .72; Table 3 and Figure 3). Of note, a similar number of women without and with prior sdNVP exposure had baseline 181C mutants (14 and 13, respectively), but only women with prior sdNVP exposure and 181C mutants were significantly more likely to reach a primary endpoint. Specifically, in trial 1, 77% (10/13) of women with 181C developed a primary endpoint compared with 19% (18/96) without 181C (P ≤ .001) [20]. This difference in association between women without vs with sdNVP exposure was significant (interaction P = .013). Further, women in trial 1 with 181C were 7 times more likely to experience an endpoint than women without this mutation (hazard ratio [HR], 7.1; 95% confidence interval [CI], 3.0–16.8). In marked contrast, women in trial 2 with the 181C mutation were no more likely to experience an endpoint than those without the mutation (HR, 1.3; 95% CI, .4–4.2). The 190A mutation was not associated with study endpoints in either trial (Table 3).
Table 3.
Significant Risk of Endpoints Associated With 103N and 181C Mutations Among Women With Prior Single-Dose Nevirapine (sdNVP) Exposure And Not In Women With No sdNVP Exposure
| Prior sdNVP Exposure | Mutation | No. of Subjects | Primary Endpoint | P Valuea |
|---|---|---|---|---|
| No | No 103N | 202 | 18% | 1.00 |
| 103N | 17 | 18% | ||
| Yes | No 103N | 69 | 17% | .009 |
| 103N | 45 | 40% | ||
| No | No 181C | 203 | 18% | .72 |
| 181C | 14 | 21% | ||
| Yes | No 181C | 96 | 19% | <.001 |
| 181C | 13 | 77% | ||
| No | No 190A | 205 | 17% | .061 |
| 190A | 13 | 39% | ||
| Yes | No 190A | 100 | 25% | .69 |
| 190A | 9 | 33% |
Abbreviation: sdNVP, single-dose nevirapine.
a Fisher exact test.
Figure 3.
Kaplan-Meier plot showing the proportion of women identified with 181C at entry in trial 2 (A, women without single-dose nevirapine [sdNVP] exposure) and trial 1 (B, women with sdNVP exposure) reaching a study endpoint of virologic failure (VF) or death and compared to women with no mutation. The median mutant frequency for trial 1 was 0.6% and for trial 2 was 0.7% (P = .59); the median mutant copy number was 1297/mL and 1007/mL (P = .25) and the number of women in each trial identified with 181C was 14 and 13, respectively. P values from proportional hazards models.
Comparison of Failure Genotypes
We next analyzed the relationship between minor mutations detected at baseline by ASP and mutations detected at VF by standard genotyping. Among women without sdNVP exposure, 13 of the 29 women (45%) experiencing failure had NVP resistance mutations detected at failure, whereas among women with sdNVP exposure experiencing failure, 21 of the 26 women (81%) had NVP resistance mutations detected at failure (P = .012). In addition, only 2 of the 13 women (15%) without sdNVP exposure who had NVP resistance mutations at failure had the same mutations detected by ASP at entry, whereas 12 of the 21 (57%) sdNVP-exposed women with NVP resistance mutations at failure had the same mutations identified by ASP at study entry (P = .030). We also compared the genotypes for nucleoside reverse transcriptase inhibitor (NRTI) resistance at VF. There were higher rates of NRTI resistance at VF in the sdNVP-exposed women: 15 of 26 (58%), compared with 8 of 34 women (24%) in sdNVP-unexposed women (P = .009).
DISCUSSION
In the AIDS Clinical Trials Group (ACTG) OCTANE/A5208 trial of initial cART in women with prior exposure to sdNVP (trial 1), the primary study endpoint of VF or death was significantly associated with NVP resistance detected at study entry by ASP in women randomized to a NVP-based cART regimen [20]. The finding of an association between low-frequency drug-resistant variants and VF of cART is consistent with other reports [31, 32]. By contrast, we now show in the parallel ACTG OCTANE/A5208 trial 2 of initial cART in women never exposed to sdNVP and randomized to a NVP-based cART regimen that the primary study endpoint of VF or death was not associated with NVP resistance detected by ASP at study entry.
The different clinical significance of low-frequency drug-resistant variants in women with and without prior exposure to sdNVP is clearly illustrated by analyses of the Y181C mutation. Although the distribution of mutant frequencies of the Y181C mutation in baseline samples was very similar between trial 1 and trial 2 (Supplementary Figure 1), Y181C was strongly associated with study endpoints in women exposed to sdNVP (trial 1) but not among women without prior exposure (trial 2). The reason for this important difference in outcome is not clear. Errors in mutant detection (false positive) are an unlikely explanation. The same ASP detection method was used for both trials and performed by the same operator who was blinded to study outcome. Specifically, in trial 1, 10 of 13 women (77%) with 181C mutations detected subsequently failed ART, indicating that most 181C mutants detected were associated with VF. In trial 2, a very similar number of women (n = 14) had 181C detected and the frequency of 181C mutants within the virus population was not different between the 2 trials (Supplementary Figure 1), yet only 3 (21%) of women in trial 2 with 181C failed cART, which was not more often than women without Y181C detected (18%). Thus, the difference between the outcomes of women with 181C is not related to frequency of mutant detection or the mutant frequency within the virus population, but rather to the history of prior exposure to sdNVP.
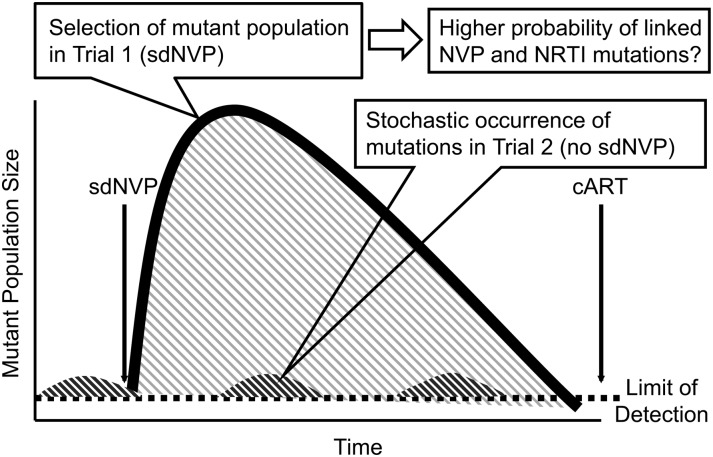
These findings suggest that the 181C mutation alone is not sufficient to cause failure of cART, and that more mutations are required. The model shown in Figure 4 proposes that linked resistance mutations conferring resistance to >1 component of cART are a potential explanation for the different outcomes associated with low-frequency resistant mutants in NVP exposed vs nonexposed women. With sdNVP exposure (trial 1), stochastically appearing NVP-resistant variants present at the time of exposure are selected and deterministically undergo a population outgrowth containing many cycles of replication to become the dominant circulating variant, during which time additional mutations may accumulate on the same genome (such as other NVP-resistance mutations or NRTI mutations). Without prior sdNVP exposure (trial 2) and thus no selection, mutants that stochastically appear would not expand to a large replicating virus population and thus would unlikely accumulate additional linked resistance mutations. With initiation of cART, variants with linked drug-resistance mutations that arose following sdNVP selection (trial 1) would be more likely to cause treatment failure than single drug-resistant variants that arise stochastically without sdNVP selection (trial 2). In the latter instance, single-drug-resistant variants would likely be suppressed by cART because it confers resistance to only 1 drug of the combination. In support of this model, the population genotype results at the time of failure show that the majority of women in trial 1 failed with NVP-resistance mutations that were identified by ASP at entry in association with NRTI resistance mutations detected at VF. These data are compatible with reports of reduced treatment efficacy in patients with prior experience to cART [13, 33, 34]. By contrast, in trial 2, only a small minority of the genotypes at failure (15%) contained NVP-resistance mutations that were detected at study entry by ASP. Nevertheless, additional analytical methods are needed to detect linked, low-frequency mutations to test the validity of the proposed model.
Figure 4.
Model proposed to explain differing impact of preexisting nevirapine (NVP)–resistant mutations in trial 1 (women with single-dose NVP [sdNVP] exposure) and trial 2 (women without sdNVP exposure). The greater area under the curve shaded in gray represents the greater expansion of mutant populations under drug selective pressure in women with sdNVP exposure providing more opportunity for linkage of other resistance mutations during ongoing cycles of replication. The smaller area under the curve shown in dark gray represents the smaller proportions of mutations occurring stochastically in women without sdNVP exposure, providing fewer opportunities for linkage to other resistance mutations on the same genome. Abbreviations: cART, combination antiretroviral therapy; NRTI, nucleoside reverse transcriptase inhibitor.
The literature contains contrasting reports of low-frequency drug-resistant mutations being associated [12–15, 20] or not associated [16–19] with failure of cART. There are several possible explanations for these discordances. Prior exposure to antiretrovirals, as illustrated by our findings with sdNVP, may explain some of the reported differences. Geographic differences in the frequency of transmitted resistance may also be important. In developed countries, where access to cART is widespread, low-frequency variants may arise from transmitted resistance. Initially, the transmitted drug-resistant variant would be a dominant replicating species, resulting in the higher likelihood of additional resistance mutations accumulating on the same genome that would persist even as the total mutant population declined to low levels and could lead to failure of cART after its initiation. A pooled analysis described by Li et al [35] revealed that low-frequency variants in naive individuals from developed countries were significantly associated with increased risk of VF. By contrast, in the OCTANE/A5208 trial 2, there was no association between low-frequency NVP resistant variants and VF. At the time of the OCTANE studies in Africa, transmission of drug resistance was not likely because access to cART was limited; hence, low-frequency variants in this cohort were more likely to have arisen stochastically rather than from transmission. Additionally, the most common HIV-1 subtype in sub-Saharan Africa and in both OCTANE trials is subtype C, whereas subtype B was likely dominant in the report by Li et al [35]. It is possible that the significance of low-frequency mutations differs by HIV-1 subtype. Further research is needed to clarify the contribution of HIV-1 subtype, transmitted drug resistance, and linked resistance mutations to the clinical significance of low-frequency variants.
In summary, our analysis shows that the risk of treatment failure of NVP-containing ART associated with detection of minor NVP-resistant variants differs significantly between women who have been and have not been exposed to sdNVP, indicating that antiretroviral exposure history is critical for assessing the significance of low-frequency variants. In the absence of accurate antiretroviral exposure history, the implications of mutant detection in African women with subtype C infection are uncertain. These findings significantly change our understanding of the clinical impact of preexisting drug-resistant mutations and reveal the importance of developing new methods for distinguishing minority variants that may compromise future treatment from those that may be clinically insignificant.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. This study would not have been possible without the women who participated in the study or the 10 study sites in Africa and key personnel who implemented the study: Ms Elizabeth Dangaiso, Mohammed S. Rassool, MD, Josephine Tsotsotetsi, KMRI/Walter Reed Project Clinical Research Center, Charity Potani, Regina Mwausegha, Dr Fatima Laher, Reinet Hen-Boisen, Kipruto Kirwa, Agnes Nzioka, Dr Margaret Chibowa, Dr Jeffrey Stringer, Kagiso Sebina, Kinuthia Mburu, Tebogo Kakhu (Gaborone Unit), Banno Moorad (Molepolole Unit), Cissy Kityo and Sandra Rwambuya, Drs Farida Amod, Umesh Lalloo, and Sandy Pillay.
We also thank the following members of the A5208 ACTG Study Team, without whom this study would not have been possible: Beth Zwickl, Cissy Kityo Mutuluuza, Christine Kaseba, Charles C. Maponga, Heather Watts, Daniel Kuritzkes, Thomas B. Campbell, Lynn Kidd-Freeman, Monica Carten, Jane Hitti, Mary Marovich, Peter N. Mugyenyi, Sandra Rwambuya, Ian M. Sanne, Beverly Putnam, Cheryl Marcus, Carolyn Wester, Robin DiFrancesco, Elias Halvas, Annie Beddison, Sandra Lehrman, Francesca Aweeka, Betty Dong, Peter Ndhleni Ziba, Michael S. Saag, William C. Holmes, and Scott M. Hammer. In addition, we thank Abbvie's support of ACTG 5208 for the drug supply.
We also thank the following companies for their generous donation of the study drugs: Gilead, Boehringer-Ingelheim, Abbott, GlaxoSmithKline, and Bristol-Myers Squibb. We also thank Frank Maldarelli for helpful discussions and comments and Luke Smith for helping with the formatting of the tables.
Disclaimer. Gilead, Boehringer-Ingelheim, Abbott, GlaxoSmithKline, and Bristol-Myers Squibb were given the opportunity to check the data used in this manuscript for factual accuracy only.
Financial support. This work was supported by intramural funding from the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Division of AIDS (University of Pittsburgh CTU Grant 1U01 AI069494-01), a Virology Support Laboratory subcontract (204VC009) of the ACTG Central Group (grant 1 U01AI068636-01) and ACTG Statistical and Data Management Center (1 U01-AI068634), and the National Cancer Institute (SAIC contract 25XS119). J. M. C was the recipient of a research professorship from the American Cancer Society, with support from the FM Kirby Foundation.
Potential conflicts of interest. M. D. H. reports having previously been a paid data and safety monitoring committee member for Boehringer Ingelheim, Medicines Development, Pfizer, and Tibotec. J. W. M. is a consultant for Gilead Sciences and has share options in RFS Pharmaceuticals. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–9. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Kearney M, Palmer S, Maldarelli F, et al. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gianella S, Richman DD. Minority variants of drug-resistant HIV. J Infect Dis. 2010;202:657–66. doi: 10.1086/655397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richman DD, Havlir D, Corbeil J, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–6. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayers DL, Japour AJ, Arduino JM, et al. Dideoxynucleoside resistance emerges with prolonged zidovudine monotherapy. The RV43 Study Group. Antimicrob Agents Chemother. 1994;38:307–14. doi: 10.1128/aac.38.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JA, Li JF, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 7.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A. 2006;103:7094–9. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korn K, Reil H, Walter H, Schmidt B. Quality control trial for human immunodeficiency virus type 1 drug resistance testing using clinical samples reveals problems with detecting minority species and interpretation of test results. J Clin Microbiol. 2003;41:3559–65. doi: 10.1128/JCM.41.8.3559-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society–USA panel. Clin Infect Dis. 2008;47:266–85. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 10.Johnson VA, Calvez V, Gunthard HF, et al. 2011 update of the drug resistance mutations in HIV-1. Top Antiviral Medicine. 2011;19:156–64. [PMC free article] [PubMed] [Google Scholar]
- 11.Lockman S, Hughes MD, McIntyre J, et al. Antiretroviral therapies in women after single-dose nevirapine exposure. N Engl J Med. 2010;363:1499–509. doi: 10.1056/NEJMoa0906626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis. 2010;201:672–80. doi: 10.1086/650542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–71. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, Paredes R, Ribaudo HJ, et al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS. 2012;26:185–92. doi: 10.1097/QAD.0b013e32834e9d7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakobsen MR, Tolstrup M, Sogaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010;50:566–73. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- 17.Metzner KJ, Rauch P, von Wyl V, et al. Efficient suppression of minority drug-resistant HIV type 1 (HIV-1) variants present at primary HIV-1 infection by ritonavir-boosted protease inhibitor-containing antiretroviral therapy. J Infect Dis. 2010;201:1063–71. doi: 10.1086/651136. [DOI] [PubMed] [Google Scholar]
- 18.Metzner KJ, Rauch P, Braun P, et al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol. 2010;50:156–61. doi: 10.1016/j.jcv.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Messiaen P, Verhofstede C, Vandenbroucke I, et al. Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology. 2012;426:7–11. doi: 10.1016/j.virol.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A. 2011;108:9202–7. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lockman S, Hughes M, Sawe F, et al. Nevirapine- versus lopinavir/ritonavir-based initial therapy for HIV-1 infection among women in Africa: a randomized trial. PLoS Med. 2012;9:e1001236. doi: 10.1371/journal.pmed.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer S, Boltz V, Maldarelli F, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. Aids. 2006;20:701–10. doi: 10.1097/01.aids.0000216370.69066.7f. [DOI] [PubMed] [Google Scholar]
- 23.Boltz VF, Maldarelli F, Martinson N, et al. Optimization of allele-specific PCR using patient-specific HIV consensus sequences for primer design. J Virol Methods. 2010;164:122–6. doi: 10.1016/j.jviromet.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok S, Kellogg DE, McKinney N, et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 26.Hance AJ, Lemiale V, Izopet J, et al. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J Virol. 2001;75:6410–7. doi: 10.1128/JVI.75.14.6410-6417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halvas EK, Aldrovandi GM, Balfe P, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol. 2006;44:2612–4. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambrose Z, Palmer S, Boltz VF, et al. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J Virol. 2007;81:12145–55. doi: 10.1128/JVI.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer S, Boltz VF, Chow JY, et al. Short-course Combivir after single-dose nevirapine reduces but does not eliminate the emergence of nevirapine resistance in women. Antivir Ther. 2011;17:327–36. doi: 10.3851/IMP1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boltz VF, Ambrose Z, Kearney MF, et al. Ultrasensitive allele-specific PCR reveals rare preexisting drug resistant variants and a large replicating virus population in macaques infected with RT-SHIV. J Virol. 2012;86:12525–30. doi: 10.1128/JVI.01963-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–47. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 32.Jourdain G, Wagner TA, Ngo-Giang-Huong N, et al. Association between detection of HIV-1 DNA resistance mutations by a sensitive assay at initiation of antiretroviral therapy and virologic failure. Clin Infect Dis. 2010;50:1397–404. doi: 10.1086/652148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecossier D, Shulman NS, Morand-Joubert L, et al. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr. 2005;38:37–42. doi: 10.1097/00126334-200501010-00007. [DOI] [PubMed] [Google Scholar]
- 34.Svarovskaia ES, Margot NA, Bae AS, et al. Low-level K65R mutation in HIV-1 reverse transcriptase of treatment-experienced patients exposed to abacavir or didanosine. J Acquir Immune Defic Syndr. 2007;46:174–80. doi: 10.1097/QAI.0b013e31814258c0. [DOI] [PubMed] [Google Scholar]
- 35.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–35. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.