Abstract
Metabolic changes are common features of many cancer cells and are frequently associated with the clinical outcome of patients with various cancers including hepatocellular carcinoma (HCC). Thus, aberrant metabolic pathways in cancer cells are attractive targets for cancer therapy. However, our understanding of cancer-specific regulatory mechanisms of cell metabolism is still very limited. We found that Tat activating regulatory DNA-binding protein (TARDBP) is a novel regulator of glycolysis in HCC cells. TARDBP regulates expression of the platelet isoform of phosphofructokinase (PFKP), the rate-limiting enzyme of glycolysis that catalyzes the irreversible conversion of fructose-6-phosphate to fructose-1,6-bisphosphate. Silencing of TARDBP expression in multiple HCC cell lines leads to impaired glucose metabolism and inhibition of in vitro and in vivo growth of HCC cells. Notably, the miR-520 family is an intermediate regulator of TARDBP-mediated regulation of glycolysis. Mechanistically, TARDBP suppressed expression of the miR-520 family, which in turn inhibited expression of PFKP. We further showed that expression of TARDBP is significantly associated with the overall survival of patients with HCC.
Conclusion
Our study provides new mechanistic insights into the regulation of glycolysis in HCC cells and reveals TARDBP as a potential therapeutic target for HCC.
Subject terms: TARDBP, PFKP, Glycolysis, miR-520, Hepatocellular carcinoma
INTRODUCTION
TARDBP was identified first as a transcription factor that binds to the human immunodeficiency virus transactivation response region1 and later as an RNA-binding protein linked to neurodegenerative diseases such as frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS).2–4 TARDBP is one of frequently mutated genes in sporadic and familial ALS, as well as in patients with FTLD, providing evidence of a direct link between TARDBP abnormalities and neurodegeneration.4 Although roles of TARDBP have been extensively studied in motor neuron linking to FTLD and ALS, recent reports suggested that TARDBP might play important roles in cellular metabolisms including glucose metabolism and lipid metabolism.5, 6 In addition, recent studies also suggested functional roles of TARDBP in human cancer.7–9 TARDBP expression is significantly altered in leukemia and TARDBP is significantly associated with susceptibility to Ewing sarcoma.8, 9 However, while link of TARDBP in human diseases have been confirmed by numerous reports, it is not clear how TARDBP contributes to diseases because very little is known about the molecular functions of TARDBP except for its roles in RNA metabolism.10
Most cancer cells including hepatocellular carcinoma (HCC) cells have very high demand for cellular metabolism to meet the need for new building blocks and energy required for cell growth.11–13 In particular, oncogenic transformation of cells is frequently associated with an increase in glycolytic flux, mainly caused by increased expression of glycolysis-regulating genes. MYC and HIF1A are best known transcriptional regulators controlling expression of glycolysis genes such as LDHA, HK2, PDK1 and GLUT1, whose expression levels are highly elevated in cancer cells.14, 15 However, because glycolysis is highly facilitated in cancer cells, more transcriptional regulators that actively promote glycolysis are expected to be involved.
In this study, we demonstrated that expression of TARDBP is significantly elevated in HCC and that it regulates expression of PFKP, rate-limiting enzyme for glycolysis, through negative-regulation of miR-520s. Thus, our study provides evidence that TARDBP is a novel transcriptional regulator of glycolysis in cancer and potential therapeutic target for treatment of patients with HCC.
Materials and methods
Additional Materials and Methods information can be found in the online Supporting Information (Supporting Materials).
shRNA, siRNA and miRNA mimics
shTARDBP (#38-TRCN0000016038 and #40-TRCN0000016040) and shControl (SHC002) clones were purchased from Sigma. The cell lines of interest were then infected with virus particles from the 293 cells. Virus-infected cells were selected for 3 days with 2μg/ml puromycin. siLuc, siTARDBP (SMARTpool)16 and miRNA mimics were purchased from Dharmacon. Cells were transfected with indicated siRNA or mimic-miRNA using Oligofectamine (Invitrogen) for 72 hrs and used for assays.
Detection of glucose uptake, lactate levels and cellular ATP levels
Glucose uptake and lactate level were measured using a Multiparameter Bioanalytical System (#YSI 7100; YSI Life Sciences). After SK-Hep1 cells were transduced with shRNA viruses, the media from cultured cells were used for measuring glucose or lactate levels. Amount of glucose taken up by cancer cells were measured by subtraction of remained glucose level in cultured media from total glucose level in uncultured media. Cellular ATP level was determined using an ATP Colorimetric Assay Kit (#K354-100; BioVision) and normalized to cell number.
Xenograft experiments
Female athymic nude mice (NCr-nu) were purchased from the NCI-Frederick Cancer Research and Development Center (Frederick, MD) and maintained as previously described.17 Tumor implantation, siRNA incorporation into dioleoyl phosphatidylcholine (DOPC) nanoliposomes and delivery in vivo were carried out as previously described.17, 18 For TARDBP silencing experiments, mice were randomized into one of the following treatment groups (n = 10 per group): control siRNA-DOPC (150 μg/kg i.v. twice weekly) and TARDBP siRNA-DOPC (150 μg/kg i.v. twice weekly). The control siRNA sequence used was UUCUCCGAACGUGUCACGU [dT][dT], and TARDBP siRNA sequences were the same as those used for the cell line experiments. Human HCC cells (SK-Hep1) were subcutaneously injected into mice (1.0 × 106 cells/animal) on day 0 and siRNA treatment was started on day 7. Five weeks later, mice were euthanized and subjected to necropsy, and tumors were harvested.
Results
TARDBP expression is elevated in HCC
Because recent studies suggested a potential link of TARDBP to cancer, we first examined the expression level of TARDBP in human liver tissues. Expression of TARDBP was significantly higher in tumors than in normal liver tissues surrounding the tumors (P = 1.0 × 10−14 by Student’s t-test, Fig. 1A), indicating potential roles of TARDBP in HCC. Consistent with the gene expression data from patient tissues, expression of TARDBP was detected in all HCC cell lines examined (Fig. 1B).
Fig. 1. TARDBP expression in HCC cells.
(A) Microarray data from Fudan University Liver Cancer Institute (FULCI) cohorts40 were used to examine expression level of TARDBP in surrounding and tumor tissues. (B) Western blot analysis of 7 HCC cell lines with anti-TARDBP and anti-α-tubulin antibodies. (C, D) MTT and clonogenicity assay after siRNAs transfection in SK-Hep1. (E) Western blot analysis of fractionated cell lysates (nucleus and cytoplasmic parts) from SK-Hep1 and HUH7 with indicated antibodies. Data represent mean ± standard deviation (s.d.) from three independent replicates (*P < 0.05, **P < 0.01, ***P < 0.005 by Student’s t-test).
We next depleted expression of TARDBP with specific siRNAs to TARDBP to test whether TARDBP plays significant roles in the growth of HCC cells. Silencing of TARDBP expression with specific siRNAs significantly attenuated growth of SK-Hep1 and HUH7 cells (Fig. 1C and Supporting Fig. 1A), strongly suggesting that TARDBP is necessary for growth and survival of HCC cells. Consistent with cell growth assay, the colony formation is also significantly reduced upon depletion of TARDBP with specific siRNAs (Fig. 1D). Similar levels of growth inhibition upon silencing of TARDBP expression were observed in additional HCC cells (SNU-449 and Hep3B) (Supporting Fig. 1A and B).
In agreement with previous reports,1, 2 cell fractionation showed that TARDBP is predominantly localized in the nucleus of SK-Hep1 cells (Fig 1E and Supporting Fig. 1C), suggesting that its biological roles in cancer cell growth might be mediated by its roles as a transcription factor or regulator of RNA processing.
TARDBP regulates glycolysis in HCC cells
To investigate downstream targets of TARDBP that could regulate cell growth, we carried out microarray experiments after depleting TARDBP in SK-Hep1 cells (Fig. 2A). As expected, silencing of TARDBP expression led to down-regulation of genes involved in cell growth (i.e, CDK6, RANBP1 and CENPE). Surprisingly, a large number of the down-regulated genes are directly involved in glucose transport and glycolysis (i.e., SLC2A1, PFKP, PFKFB4, PGK1 and ENO2), strongly suggesting potential roles of TARDBP in regulating glucose metabolism. Notably, expression of PFKP, among many glycolysis-related genes, was most significantly altered by TARDBP (P = 7.5 × 10−5 by Student’s t-test, 4.7-fold). Expression of PFKP and other glycolysis-related genes was also significantly down-regulated by depleting TARDBP in two additional HCC cell lines (FOCUS and HUH7) when their expression was assessed by quantitative real-time polymerase chain reaction (qRT-PCR) (Fig. 2B and C). These results strongly suggested conserved and universal roles of TARDBP in glucose metabolism in HCC cells, particularly through regulation of PFKP.
Fig. 2. PFKP expression measurement by silencing TARDBP.
(A) Gene expression data from SK-Hep1 cells transfected with siLuc or siTARDBP. A total of 1019 genes were identified as significantly changed by silencing of TARDBP expression (P < 0.001). Data are presented in matrix format; each row represents an individual gene, and each column represents a transfected SK-Hep1 cell sample. In the matrix, red and green reflect relatively high and low expression levels of genes, respectively, as indicated in the scale bar (a log2-transformed scale). Genes involved in glycolysis are highlighted in red text. (B, C) qRT-PCR experiments with mRNAs from the indicated cancer cell lines after transfection with siLuc or siTARDBP. Data represent mean ± s.d. from three independent replicates. (D) Western blot analysis of the three isoforms of the glycolysis enzyme PFK in SK-Hep1 cells transfected with indicated siRNAs. (E) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) experiments with mRNAs from SK-Hep1 cell line after transfection with siLuc or siTARDBP (*P < 0.05, **P < 0.01, ***P < 0.005 by Student’s t-test).
Phosphofructokinase (PFK) is a key regulatory enzyme in glycolysis that catalyzes the irreversible conversion of fructose-6-phosphate to fructose-1,6-bisphosphate. Humans have three PFK isoforms: liver (PFKL), muscle (PFKM) and platelet (PFKP).19, 20 Thus, we examined whether TARDBP regulates expression of the other PFK isoforms in addition to PFKP. The results showed that protein expression of PFKL and PFKM was not altered by silencing of TARDBP expression, while all three siRNAs specific to TARDBP successfully down-regulated expression of PFKP in SK-Hep1 cells (Fig. 2D). Consistent with western blot experiments, qRT-PCR experiments showed that TARDBP regulates expression of only PFKP in SK-Hep1 cells (Fig. 2E).
Since TARBDP regulated expression of many glycolysis genes including PFKP in multiple HCC cells, we determined effect of depletion of TARDBP in metabolic response. Glucose uptake of SK-Hep1 cells was significantly reduced by silencing of TARDBP expression (Fig. 3A and B). Furthermore, silencing of TARDBP expression resulted in a decrease in lactate production and ATP levels indicating a decrease of glycolysis (Fig. 3B). Thus, our findings strongly support the proposed roles of TARDBP in HCC cell growth through regulation of glucose and energy metabolism.
Fig. 3. Glucose metabolite measurement in HCC cell.

(A) Western blot analysis for SK-Hep1 cells stably transduced with TARDBP (#38 and #40) and control short hairpin RNAs. (B) Analysis of metabolites in SK-Hep1 cells stably transduced with the indicated short hairpin RNAs. Glucose uptake and lactate level were measured from media of cell culture. Intracellular ATP levels were measured at 72 hrs after lentiviral infection. Data represent mean ± s.d. from three independent replicates (*P < 0.05, **P < 0.01, ***P < 0.005 by Student’s t-test).
miR-520s down-regulates PFKP expression
We next attempted to determine the molecular mechanism of how TARDBP regulates PFKP expression. Given that the best known function of TARDBP is RNA processing as an RNA-binding protein,21 we examined whether TARDBP directly interacts with mRNA of PFKP. However, analysis of RNA immunoprecipitation data with anti-TARDBP antibody 21 failed to demonstrate interaction of TARDBP with PFKP mRNA (Supporting Fig. 2), suggesting that TARDBP likely regulates PFKP by other mechanisms. Because TARDBP positively regulates expression of PFKP and also functions as a transcription repressor,22 we hypothesized that PFKP could be negatively regulated by intermediate regulators that are in turn directly suppressed by TARDBP. Recent studies showed that TARDBP is involved in regulation of miRNAs,23, 24 suggesting that miRNAs might be good candidates for intermediaries between TARDBP and PFKP. To identify such intermediary regulators, we explored target miRNAs that can suppress PFKP based on sequence alignment (Fig. 4A). Sequence analysis with the starBase database25 revealed that 26 miRNAs contain direct binding sequences for the PFKP 3′UTR (Supporting Table 1). Interestingly, three all independent prediction programs (target Scan, picTar and miRanda) predicted miR-520 and miR-302 family as major regulatory miRNAs for PFKP.26 Because previous studies showed that miR-520b and miR-520e can inhibit cancer cell growth,27–29 we next tested if inhibition of cell growth by miR-520 is mediated by regulation of PFKP expression. When SK-Hep1 cells were treated miR-520a-3p, miR-520b, and miR-520e (here after miR-520a/b/e), expression of PFKP was significantly down-regulated (Fig. 4B), suggesting that PFKP might be a direct target of miR-520a/b/e. However, expression of other glycolysis genes were not significantly altered by miR-520a/b/e (Supporting Fig. 3), suggesting that these miRNAs regulate glycolysis mainly through inhibition of PFKP. To determine whether miR-520a/b/e directly targets the 3′UTR of PFKP mRNA, we assessed a luciferase reporter vector containing the 3′UTR sequence of PFKP, including the predicted binding site for miR-520a/b/e in SK-Hep1 cells. Luciferase activity was significantly inhibited by the PFKP 3′UTR sequence when only miR-520a/b/e were co-transfected (Fig. 4C). However, luciferase activity was not inhibited by a mutant 3′UTR sequence (Fig. 4D and E), strongly demonstrating that miR-520a/b/e directly targets the 3′UTR sequence of PFKP mRNA and inhibits the expression of PFKP.
Fig. 4. PFKP gene expression by miR-520.
(A) Schematic diagram of strategy for identification miRNAs targeting PFKP. (B) Western blot analysis of SK-Hep1 cells after transfecting with miR-520a-3p, miR-520b and miR-520e. β-actin was used as an internal control. (C) Firefly luciferase assay with a luciferase reporter vector containing the wild-type 3′UTR sequence of PFKP in SK-Hep1 cells. Luciferase activities were measured after transfecting the indicated miRNAs. Data represent means ± s.d. from three independent replicates. (D) Sequence alignment within miR-520a/b/e and between miR-520b and the 3′UTR of human PFKP. Seed sequences are highlighted in the gray box. (E) Firefly luciferase assay with luciferase reporter vector containing a mutant 3′UTR sequence of PFKP in SK-Hep1 cells. Luciferase activities were measured after transfecting the indicated miRNAs (*P < 0.05, **P < 0.01 by Student’s t-test).
TARDBP regulates PFKP by suppressing miR-520s
Since expression of PFKP was down-modulated by miR-520s, we next tested whether miR-520a/b/e are regulated by TARDBP by measuring expression of these miRNAs by qRT-PCR after silencing of TARDBP in SK-Hep1 and SNU449. The results showed that expression of miR-520a/b/e was significantly increased when TARDBP was silenced (Fig 5A), with expression of miR-520b strongly induced by silencing TARDBP in two HCC cell lines. TARDBP is a DNA-binding protein that binds to the GTGTGT sequence in target promoter regions.22 Thus, we examined whether TARDBP can directly bind to the miR-520b promoter. Indeed, a chromatin immunoprecipitation (ChIP) assay demonstrated that TARDBP directly bound to the miR-520b promoter region, specifically in GT-rich regions (Fig 5B), suggesting that miR-520b is indeed a direct downstream target of TARDBP.
Fig. 5. The miR-520 family is a key intermediate regulator of TARDBP-mediated PFKP expression.
(A) qRT-PCR assay with RNAs from the indicated cell lines after transfection with indicated siRNA. Expression of three miRNAs (miR-520a-3p, miR-520b and miR-520e) was measured with specific primers. Data represent mean ± s.d. from three independent replicates (*P < 0.05, **P < 0.01 by Student’s t-test). (B) ChIP assay with anti-TARDBP antibody in SK-Hep1 cells. The left panel shows a schematic representation of miR-520b promoter regions for the ChIP assay. Recruitment of TARDBP to the miR-520b promoters was analyzed using primers specific to the indicated promoter region. IgG was used as an internal control. (C) SK-Hep1 cells were transfected with indicated cDNA and miRNA mimics. The cells were used for western blot with indicated antibody. (D, E) After SK-Hep1 were infected with shTARDBP (or shControl), the cells were transiently transfected with indicated cDNA and were used for cell proliferation assay (MTT) (D) or measuring the indicated metabolites (E).
Our results strongly suggested that growth inhibition following depletion of TARDBP might be mediated by miR-520a/b/e. To test this hypothesis, we introduced miR-520a/b/e into two HCC cell lines and observed that cell growth was significantly reduced (Supporting Fig. 4). To further test whether growth inhibition is mediated by down-regulation of PFKP, we introduced exogenous myc-tagged PFKP after silencing TARDBP in SK-Hep1 cells. Because myc-tagged PFKP lacks 3′ UTR sequence, its expression is not inhibited by miR-520s (Fig. 5C). Growth inhibition by silencing TARDBP was rescued by exogenous PFKP (Fig. 5D). Furthermore, glucose uptake, lactate production, and ATP level were significantly increased by exogenous PFKP (Fig. 5E). When induced miR-520b in TARDBP-silenced SK-Hep1 was inhibited by specific antisense miR-520b inhibitor, expression of PFKP is recovered (Supporting Fig. 5), demonstrating that regulation of PFKP by TARDBP is mediated through miR-520s. Taken together, our data suggested that PFKP is main regulatory target for TARDBP-miR-520s mediated regulation of cell growth. These observations agree very well with the finding of previous studies showing that miR-520b and miR-520e might function as negative regulators of cell proliferation.27, 30
Clinical relevance of TARDBP, PFKP, and miR-520s expression in cancer
Our data suggested functional roles of TARDBP and PFKP as positive regulators and miR-520s as negative regulator of cell proliferation. This view is strongly supported by expression patterns of these genes in the NCI-60 cancer cell lines. Expression of both TARDBP and PFKP was very high in the vast majority of the 60 cancer cell lines (Supporting Fig. 6A). In contrast, miR-520a/b/e were barely expressed, strongly supporting its role as negative regulator of cell growth. Expression of PFKP was the highest among PFK isoforms in NCI-60 cell lines (Supporting Fig. 6B), further supporting that cancer-specific expression of PFKP is regulated by miR-520a/b/e and TARDBP.
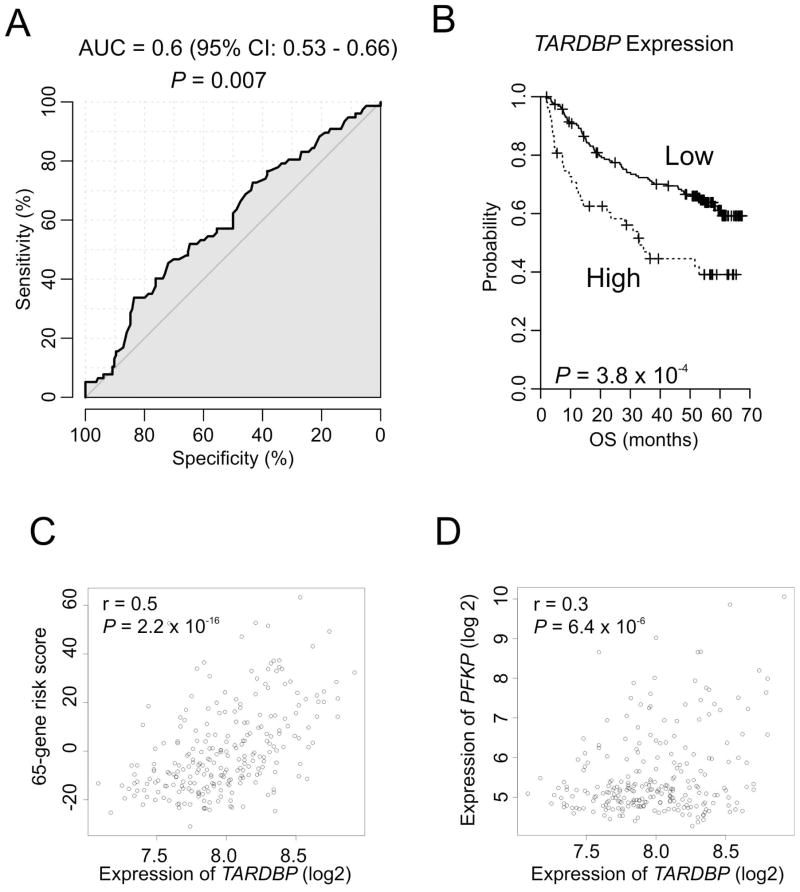
We next assessed the clinical relevance of TARDBP in HCC. Expression of TARDBP is significantly associated with prognosis when estimated by receiver operating characteristic analysis. Areas under curve of TARDBP expression over 3-year overall survival is 0.6 (95% CI, 0.53 – 0.66, P = 0.007) (Fig. 6A). When patients were stratified according to expression level of TARDBP, patients with high TARDBP expression showed significantly shorter survival (P = 3.8 × 10−4, Fig. 6B). Association of TARDBP with prognosis is further supported by its significant correlation with 65-gene risk score (r = 0.5; P = 2.2 × 10−16) (Fig. 6C) that was previously developed for prediction of recurrence.31 Significant positive correlation between expression of TARDBP and PFKP in HCC patients is also concordant with their roles as positive regulator for cell growth (Fig. 6D).
Fig. 6. Clinical relevance of TARDBP in patients with HCC.
(A) Prognostic significance of TARDBP estimated by AUC from receiver operating characteristics analysis for 3-year OS. AUC: area under curve, CI: confident internal of AUC. (B) Kaplan–Meier plots of overall survival of HCC patients. Patients were stratified according to expression level of TARDBP. P-values were calculated with the log-rank test. (C) Scatter plots between 65-gene risk score and expression level of TARDBP in HCC (D) Scatter plots between expression level of TARDBP and PFKP in HCC.
Therapeutic efficacy of PFKP in a mouse model
The critical roles of TARDBP and its downstream targets, the miR-520 family, in cell growth and the significant correlation of TARDBP with patient survival strongly suggested that TARDBP and its downstream targets would be potential therapeutic targets for cancer treatment. To test this, we carried out a mouse xenograft experiment with SK-Hep1 cells and siRNA specific to TARDBP. Compared with treatment with control siRNA, treatment with siTARDBP resulted in significant reduction in tumor weight (Fig. 7A), recapitulating the effects of silencing TARDBP in vitro. Efficient silencing of TARDBP by siRNA was confirmed by immunostaining of TARDBP and its downstream target PFKP and further validated by qRT-PCR (Fig. 7B and C and Supporting Fig. 7). As expected, cell proliferation, as examined by Ki67 immunostaining, was significantly decreased in tumors treated with siTARDBP (Fig. 7B). In addition, lactate and ATP levels were also significantly decreased (Fig. 7C) and expression of miR-520b and miR-520e (Fig. 7D) was significantly increased in siTARDBP treated mice compared to control. These results clearly demonstrate the importance of TARDBP in tumor growth and the potential of TARDBP as a therapeutic target.
Fig. 7. Therapeutic potential of TARDBP.
(A) Growth of SK-Hep1 tumors in mice treated with TARDBP siRNA-DOPC or control siRNA-DOPC. Tumor volume and weight were measured at day 33. (B) Immunostaining of Ki-67, TARDBP and PFKP in SK-Hep1 tumor tissues treated with TARDBP siRNA or control siRNA. (C) qRT-PCR assay with RNA and metabolites measured from SK-Hep1 tumor tissues treated with TARDBP siRNA or control siRNA. (D) qRT-PCR assay with miRNAs from SK-Hep1 tumor tissues treated with TARDBP siRNA or control siRNA. Data represent mean ± s.d. from three independent replicates (*P < 0.05, **P < 0.01, ***P < 0.005 by Student’s t-test). (E) Schematic diagram of the regulatory pathway from TARDBP to glycolysis.
DISCUSSION
In the current work, we have presented a mechanistic link from TARDBP to PFKP, the rate-limiting enzyme of glycolysis, and we also have provided evidence suggesting that this pathway is associated with poor prognosis of HCC. A notable finding was the identification of the miR-520 family as an intermediary regulator of this pathway.
While TARDBP was originally identified as a transcription repressor binding to the HIV transactivation response region,1 down-stream targets and molecular mechanisms related to its transcription repressor activity has not been properly explored. Our data demonstrated that TARDBP function as a transcription repressor and its molecular activity as transcription repressor plays key roles in regulation of glycolysis in cancer cells. TARDBP strongly represses expression of miR-520 family as evidenced by significant increase in their intracellular levels upon TARDBP depletion. This notion was strengthen by a ChIP assay demonstrating that TARDBP directly bound to GT-rich regions in the miR-520b promoter. In addition to regulation of miR-520s in transcriptional level, TARDBP may regulate miR-520s post-transcriptionally because the involvement of TARDBP in miRNA processing has been observed in other systems.23, 24 Our data are in good agreement with previous studies demonstrating repression of the spermatid-specific gene SP-10 expression by TARDBP.22, 32 Thus, our study rediscovered previously recognized molecular function of TARDBP and updated molecular mechanisms and biological roles of TARDBP especially in cellular metabolism with use of miRNAs as intermediary regulators.
There are growing evidences supporting that miRNAs play important roles in regulation of cellular metabolism.33, 34 Recent studies revealed that miRNAs such as miR-124, miR-137, miR-340 miR-143 and miR-155 regulate glycolysis by directly targeting the 3′ UTR region of HK2 and PKM2.35–37 With prediction analysis based on sequence, we discovered that miR-520a/b/e are major regulators of glycolysis by directly targeting the 3′UTR of PFKP mRNA. We further demonstrated that TARDBP-mediated suppression of miR-520a/b/e is important for growth and survival of HCC cells.
Importantly, analyses of gene expression patterns from multiple cancer lineages provide evidences supporting our findings related to molecular functions and mechanisms of TARDBP-mediated PFKP regulation. Expression of TARDBP is significantly higher in tumors than normal tissues. We also have found inversed correlation of expression patterns among TARDBP, PFKP and miR-520s. Expression of TARDBP and PFKP is significantly high in vast majority of cancer cell lines while expression of miR-520s is very low (Supporting Fig. 4A). This is in good agreement with mechanism postulating that TARDBP suppresses expression of miR-520s that directly inhibit PFKP to maintain increased glycolysis in cancer cells (Fig. 7E).
In conclusion, we found that novel roles of TARDBP linked to glycolysis via PFKP in HCC. Deregulation of cellular metabolism is one of the hallmarks of cancer cells,38 and altered components of the metabolic pathway represent attractive therapeutic targets.13, 39 Thus, the identification of the TARDBP-miR-520-PFKP axis regulating glycolysis and ATP production elicits a potential new approach to target the tumor-specific metabolic pathway.
Supplementary Material
Acknowledgments
This research is supported in part by 2011 and 2012 cycle of MD Anderson Sister Institute Network Fund (J-S.L.), 5U54 CA112970-08 (G.B.M.), 5P01CA099031-07 (G.B.M.), P30 CA016672 (G.B.M.), and CA016672 (MD Anderson Cancer Center Support Grant) from National Institutes of Health
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Supporting Information is available at Hepatology Online.
References
- 1.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauffenberger A, Vaccaro A, Aulas A, Velde CV, Parker JA. Glucose delays age-dependent proteotoxicity. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci U S A. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teittinen KJ, Karkkainen P, Salonen J, Ronnholm G, Korkeamaki H, Vihinen M, Kalkkinen N, et al. Nucleolar proteins with altered expression in leukemic cell lines. Leuk Res. 2012;36:232–236. doi: 10.1016/j.leukres.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 9.Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, Oberlin O, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–327. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 10.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiesel FC, Voigt A, Weber SS, Van den Haute C, Waldenmaier A, Gorner K, Walter M, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaker PH, Han LY, Kamat AA, Arevalo JM, Takahashi R, Lu C, Jennings NB, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat Med. 2006;12:939–944. doi: 10.1038/nm1447. [DOI] [PubMed] [Google Scholar]
- 18.Park YY, Jung SY, Jennings N, Rodriguez-Aguayo C, Peng G, Lee SR, Kim SB, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Martinez C, Estevez AM, Aragon JJ. Phosphofructokinase C isozyme from ascites tumor cells: cloning, expression, and properties. Biochem Biophys Res Commun. 2000;271:635–640. doi: 10.1006/bbrc.2000.2681. [DOI] [PubMed] [Google Scholar]
- 20.Sola-Penna M, Da Silva D, Coelho WS, Marinho-Carvalho MM, Zancan P. Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life. 2010;62:791–796. doi: 10.1002/iub.393. [DOI] [PubMed] [Google Scholar]
- 21.Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287:15635–15647. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalmansingh AS, Urekar CJ, Reddi PP. TDP-43 is a transcriptional repressor: the testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J Biol Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratti E, Baralle FE. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- 24.Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L, Lu Z. Global expression analysis of miRNA gene cluster and family based on isomiRs from deep sequencing data. Comput Biol Chem. 2010;34:165–171. doi: 10.1016/j.compbiolchem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Li BA. A novel tumor suppressor miRNA miR-520e contributes to suppression of hepatoma. Acta Pharmacol Sin. 2012;33:3–4. doi: 10.1038/aps.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-kappaB-inducing kinase (NIK) Oncogene. 2012;31:3607–3620. doi: 10.1038/onc.2011.523. [DOI] [PubMed] [Google Scholar]
- 29.Cui W, Zhang Y, Hu N, Shan C, Zhang S, Zhang W, Zhang X, et al. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3′UTR of CD46. Cancer Biol Ther. 2010;10:232–241. doi: 10.4161/cbt.10.3.12277. [DOI] [PubMed] [Google Scholar]
- 30.Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, Bai X, et al. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem. 2011;286:13714–13722. doi: 10.1074/jbc.M110.204131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi PP. cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 33.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh PK, Mehla K, Hollingsworth MA, Johnson KR. Regulation of Aerobic Glycolysis by microRNAs in Cancer. Mol Cell Pharmacol. 2011;3:125–134. [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, Li B, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–1998. doi: 10.1038/emboj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2012 doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 37.Gao P, Sun L, He X, Cao Y, Zhang H. MicroRNAs and the Warburg Effect: New Players in an Old Arena. Curr Gene Ther. 2012;12:285–291. doi: 10.2174/156652312802083620. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Dang CV, Hamaker M, Sun P, Le A, Gao P. Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl) 2011;89:205–212. doi: 10.1007/s00109-011-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.