This study shows that the small GTPase Rab11 is required for both autophagosome and endosome maturation. The authors demonstrate a novel cross-talk mechanism through which autophagy can orchestrate endosomal maturation events to ensure the required endolysosomal input for autophagosome maturation.
Abstract
During autophagy, double-membrane autophagosomes deliver sequestered cytoplasmic content to late endosomes and lysosomes for degradation. The molecular mechanism of autophagosome maturation is still poorly characterized. The small GTPase Rab11 regulates endosomal traffic and is thought to function at the level of recycling endosomes. We show that loss of Rab11 leads to accumulation of autophagosomes and late endosomes in Drosophila melanogaster. Rab11 translocates from recycling endosomes to autophagosomes in response to autophagy induction and physically interacts with Hook, a negative regulator of endosome maturation. Hook anchors endosomes to microtubules, and we show that Rab11 facilitates the fusion of endosomes and autophagosomes by removing Hook from mature late endosomes and inhibiting its homodimerization. Thus induction of autophagy appears to promote autophagic flux by increased convergence with the endosomal pathway.
INTRODUCTION
Macroautophagy (hereafter autophagy) is an evolutionarily conserved bulk degradation process of eukaryotic cells. The regulation of this process was originally described in yeast (Saccharomyces cerevisiae) but even has a key role in multicellular organisms as a cytoprotective response to stress and pathological conditions (Levine and Kroemer, 2008; Mizushima et al., 2008). Autophagy has the capacity to engulf large portions of the cytoplasm through the formation of double-membrane vesicles, called autophagosomes. These vesicles arise from preautophagosomal structures (PAS), which are defined sites of cytoplasm, marked by a subset of autophagy-related (Atg) proteins (Mizushima et al., 2011). Closed autophagosomes undergo a maturation process, as they subsequently fuse with endosomes and lysosomes.
On autophagy induction, the Atg1 kinase complex (ULK1/2 in mammals) localizes to the PAS (Chan and Tooze, 2009; Mizushima, 2010) and together with the class III phosphatidylinositol-3-kinase (Vps34) complex initiates the phagophore nucleation and expansion (Funderburk et al., 2010). After these events, the members of two ubiquitin-like conjugation systems are recruited to the phagophore membrane: the Atg5-12-16 complex and the phosphatidylethanolamine-conjugated Atg8a (LC3 in mammals; Geng and Klionsky, 2008). The lipid-conjugated form of Atg8a (Atg8a-II) is located on both sides of the membrane of the phagophore and autophagosomes as well. While the Atg8a located on the outer membrane is routed for recyclization, the other portion of Atg8a becomes trapped in the autolysosomal lumen and is degraded by lysosomal hydrolases. Thus Atg8a is a widely used marker of autophagic structures (Klionsky et al., 2012).
After completion, autophagosomes can fuse with lysosomes. However, there is also growing evidence about the convergence of the autophagic route with the earlier stages of endosomal pathways. Recent studies show that autophagosomes can fuse with different populations of both early (Tooze et al., 1990; Razi et al., 2009) and late (Lucocq and Walker, 1997; Köchl et al., 2006; Filimonenko et al., 2007) endosomes to form hybrid organelles called amphisomes (Gordon and Seglen, 1988).
In spite of numerous studies, many questions remained open concerning the spatial and temporal regulation of autophagosome maturation, amphisome formation, and the exact molecular mechanisms of the fusion events. As a recent review discussed, a cross-talk may exist between autophagic and endosomal processes, which likely plays an important role in the regulation of both degradative pathways (Lamb et al., 2013).
The members of the Rab small GTPase protein family are the main regulators of membrane trafficking and fusion events (Stenmark, 2009). The active, GTP-bound Rab proteins recruit several specific effectors to the membrane in which they are associated, thereby determining the membrane's identity, traffic, and fusion ability. Recent studies revealed a role for a subset of Rab proteins in autophagy (Chua et al., 2011). Rab11 was indicated to play an important role both at the early and late stages of autophagy. Recent studies found that Rab11 is involved in vesicle trafficking events from recycling endosomes (REs) to the phagophore during autophagosome formation (Longatti et al., 2012; Knævelsrud et al., 2013). Meanwhile, previous studies suggested a role for Rab11 in the maturation of autophagosomes (Fader et al., 2008; Richards et al., 2011).
RESULTS
Rab11 is required for amphisome formation
Rab11 was previously described as a potent regulator of autophagosome maturation in cultured mammalian cells (Fader et al., 2008). To better understand the role of Rab11 in autophagy, we examined the functions of this protein in Drosophila melanogaster. We analyzed whether the decrease in the level of functional Rab11 protein causes any defect upon autophagy. Using three independent RNA interference (RNAi) lines, we observed the accumulation of small-sized Atg8a-positive dots in Rab11-depleted cells under both fed and starved conditions (Figure 1A and Supplemental Figure S1, A, B, E, and F). Fat body cells overexpressing the GDP-locked dominant-negative form of Rab11 or Rab11 hypomorphic mutant clone cells showed the same phenotype (Figure S1, C, D, and F).
FIGURE 1:
Rab11 is required for autophagosome maturation. (A) Control and Rab11-depleted (green) fat body cells of L3 fed Drosophila larvae expressing mCherry-Atg8a (red). Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI, blue). The mCherry-Atg8a–positive dot per cell area ratio was calculated as described and compared between Rab11 RNAi and neighboring control cells using Wilcoxon's signed-rank test (n = 21, p < 0.0001). (B) Fat body cells of starved Rab11-depleted L3 starved larvae clonally expressing the mCherry-GFP-Atg8a autophagic flux marker. Nuclei were stained using DAPI (blue). Dot plot shows fluorescence intensity and colocalization profiles of mCherry and GFP channels of the main image. (C) Control and Rab11 RNAi (green) fat body cells of L3 fed larvae were stained with anti-p62 antibody (red). Nuclei were stained using DAPI (blue). P62-positive dot per cell area ratio was calculated and compared between the two groups using Wilcoxon's signed-rank test (n = 22, p < 0.0001). (D) Representative Western blot showing p62, Rab11, and α-tubulin levels (relative density) in fed and starved control and Rab11-depleted larvae. Asterisk marks aspecific band. (E) Western blots of the GFP-Atg8a processing assay in fed and starved control and Rab11-depleted larvae were measured as described and compared between groups using GLMM (n = 4, p < 0.0001). The differently colored sections of bars show the relative amounts of free GFP (green) and GFP-Atg8a (black) in the samples. (F) Ultrastructure of Rab11-depleted clone cells and neighboring control cells. Rab11-depletion results in the accumulation of immature (arrows) and abnormal (arrowheads) autophagosomes compared with the control cells. Nu, nuclei; M, mitochondria; L, lipid droplets. Scale bars: (A–C) 10 μm; (F) 1 μm. On box-and-whisker plots, bars (gray, control; green, Rab11 RNAi) show the data lying between the upper and lower quartiles; the median is indicated as a horizontal line within the box. Whiskers plot the smallest and largest observations. ***, p < 0.001.
Accumulation of autophagic structures can be caused both by increased induction of autophagy and by failures in the autophagosome maturation. For monitoring autophagic flux, we used tandem-tagged mCherry–green fluorescent protein–Atg8a reporter (mCherry-GFP-Atg8a; Kimura et al., 2007). In Rab11 RNAi cells, we observed double-labeled immature autophagic structures, suggesting depletion of Rab11 blocks the autophagic flux (Figure 1B). Furthermore, our Western blot and immunostaining experiments showed the accumulation of p62, a well-known target of autophagic degradation due to Rab11 silencing (Figure 1, C and D). Rab11 depletion resulted in the accumulation of GFP-Atg8a–positive puncta both in presence and in absence of chloroquine (CQ), an inhibitor of lysosomal acidification. However, CQ treatment did not cause a further increase in the number of autophagic structures (Figure S1, G and H). The GFP-Atg8 processing assay is also suitable for investigating autophagic degradation (Klionsky et al., 2012). Using this tool, we found significantly less free GFP in Rab11-depleted larvae compared with control larvae (Figures 1E and S1I). Moreover, our ultrastructural studies also suggest that Rab11 RNAi leads to the accumulation of abnormal autophagic vacuoles (Figure 1F).
Surprisingly, when we examined the influence of Rab11 depletion on the pattern of LysoTracker Red (LTR), a marker of acidic structures, we observed an accumulation of LTR-positive dots under both fed and starved conditions, compared with control cells (Figures 2A and S2, A and D). However, we found a decrease in the colocalization frequency of GFP-Atg8a with LTR (from 43 ± 1.6 to 8 ± 0.8%), indicating that a drop in Rab11 protein levels results in a reduced number of late autophagic structures (Figure S2, E and F). Next we tested whether LTR colocalizes with any marker of the endolysosomal system. Using Lamp1-GFP, a marker of late endosomes and lysosomes (Schwake et al., 2013), we observed an increased number of Lamp1-positive dots and 90 ± 1.4% of LTR-positive dots colocalized with them in Rab11 RNAi cells (Figures 2A and S2, B–D). Moreover, we found the accumulation of the late endosomal marker Rab7 (Wang et al., 2011), but not that of the early endosomal marker Rabenosyn-5 (de Renzis et al., 2002; Figures 2B and S2G).
FIGURE 2:
Rab11 is required for amphisome formation. (A) Fat body cells of Rab11-depleted fed L3 larvae clonally expressing Lamp1-GFP (green) were stained with LTR (red). Nuclei were stained using DAPI (blue). The LTR-positive dot per cell area ratio was calculated and compared between clone and neighboring control cells using Wilcoxon's signed-rank test (n = 21, p < 0.0001). Lamp1-GFP–positive dot per cell area ratio was also calculated and compared between control (see Figure S2C) and Rab11-depleted groups using the Mann-Whitney test (nctrl = 34, nRNAi = 27, p < 0.0001). (B) Control and green, Rab11-depleted fat body cells of fed L3 larvae were stained with anti-Rab7 antibody (red). Nuclei were stained using DAPI (blue). Dot per cell area ratio was calculated and compared between the groups using Wilcoxon's signed-rank test (Rab7: n = 28, p < 0.0001; Rbsn5: n = 15, p = 0.8647). (C–C′) Colocalization between GFP-Atg8a (green) and TRA (red) decreased due to Rab11 depletion. Nuclei were stained with DAPI (blue). Colocalization between GFP-Atg8a and TRA, mCh-Atg8a and anti-Rab7, and mCh-Atg8a and Lamp1-GFP was quantified and compared between the groups using the Mann-Whitney test (Atg8-TRA: nctrl = 100, nRNAi = 190, p = 0.0002; Atg8-Rab7: nctrl = 41, nRNAi = 39, p = 0.0008; Atg8-Lamp1: nctrl = 57, nRNAi = 57, p < 0.0001). On box-and-whisker plots, bars (gray, control; green, Rab11 RNAi) show the data lying between the upper and lower quartiles; the median is indicated as a horizontal line within the box. Whiskers plot the smallest and largest observations. NS, p > 0.05; ***, p < 0.001. Scale bars: 10 μm.
Because our results showed the accumulation of both autophagosomes and late endosomes, we tested whether Rab11 depletion affects amphisome formation. Colocalization of Atg8a with different endosomal markers was studied both in the presence and absence of Rab11 during starvation-induced autophagy. We found a significant decrease in colocalization of the endocytic tracer Texas Red Avidin (TRA) with GFP-Atg8a due to Rab11-depletion (from 43 ± 0.9 to 8 ± 0.2%; Figure 2, C and C′). Silencing of Rab11 decreased the colocalization of mCherry-Atg8a with the endosomal markers Rab7 (from 20±1.6% to 1.8±0.3%) and Lamp1 (from 75 ± 1.7 to 32 ± 1.6%) as well (Figures 2C and S2, H and I). Supporting these findings, our electron microscopic analysis showed acidic late endosomes often located in proximity to autophagosomes, but we could not detect any acidic autophagic vacuoles (Figure S2J).
These findings suggest that Rab11 is required for amphisome formation and lack of Rab11 results in the accumulation of abnormal autophagosomes and late endosomes.
Rab11 localizes to autophagosomes
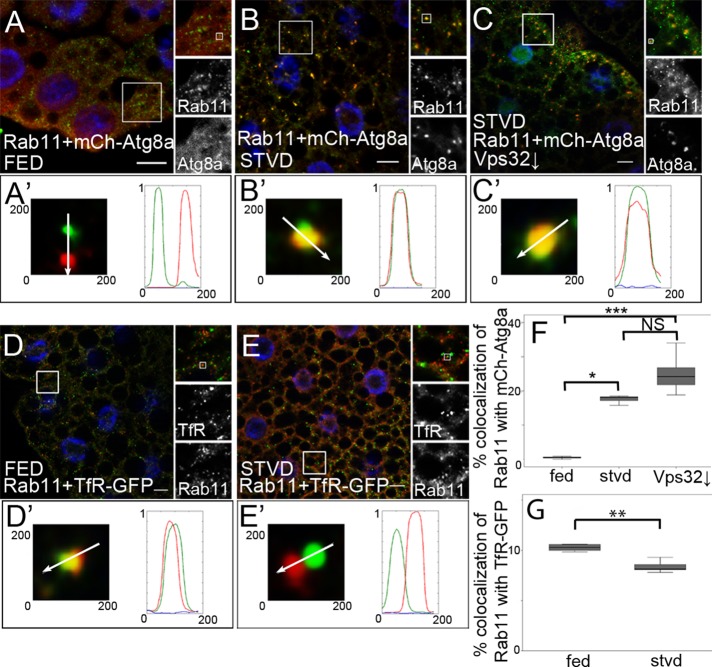
As our results suggest, Rab11 presumably plays an important role in the fusion of autophagosomes with late endosomes. Therefore our aim was to verify whether Rab11 indeed localizes to autophagic structures. At first, we investigated the colocalization of Rab11 with Atg8a. We found that 40 ± 0.4% of ectopically expressed Rab11-GFP–positive puncta colocalized with mCherry-Atg8a under starved conditions (Figure S3A). Supporting this result, mCherry-Atg8a was also found to colocalize with endogenous Rab11 (Figure 3B).
FIGURE 3:
Rab11 localizes to autophagosomes. Fat body cells of fed (A) and starved (B) L3 larvae expressing mCherry-Atg8a (red) were stained with anti-Rab11 antibody (green). Nuclei were stained using DAPI (blue). (C) Fat body cells of starved larvae expressing mCherry-Atg8a (red) were stained with anti-Rab11 antibody (green). Vps32 is depleted (↓) in GFP-expressing cells. Nuclei were stained using DAPI (blue). Fat body cells of a fed (D) and starved (E) L3 larva expressing TfR-GFP (green) were stained with anti-Rab11 antibody (red). Nuclei were stained using DAPI (blue). (A′–E′) show the indicated single dots from their main images. Arrows indicate the direction of scanning intensity. Diagrams show the intensity of red, green, and blue channels. (F) Autophagy induction increases the colocalization of Rab11 with mCherry-Atg8a. Colocalization was calculated and compared between the groups using the Kruskal-Wallis test (nfed = 72, nstvd = 143, nVps32 = 20, p < 0.0001; fed-starved: p = 0.0140; fed-Vsp32↓: p < 0.0001; starved-Vps32 ↓: p = 0.1840). (G) Colocalization of Rab11 with TfR-GFP was calculated and compared between cell groups described in (D and E) using the Mann-Whitney test (nfed = 35, nstvd = 34, p < 0.0017). On box-and-whisker plots, bars show the data lying between the upper and lower quartiles; the median is indicated as a horizontal line within the box. Whiskers plot the smallest and largest observations. NS, p > 0.05; *, p < 0.05; **, p < 0.001; ***, p < 0.001. Scale bars: 10 μm.
These findings raised the question of whether there is an autophagy induction–dependent change in the colocalization of endogenous Rab11 with mCherry-Atg8a. We observed that only 0.46 ± 0.1% of Rab11-positive dots colocalized with mCherry-Atg8a under fed conditions, while amino acid starvation significantly increased the colocalization frequency to 18 ± 1.5% (Figure 3, A, B, and F). However, autophagy induction did not affect the number of Rab11-positive structures in fat body cells (Figure S3B). This suggests that Rab11, besides being found in REs, can be also found on autophagic structures.
Vps32/Snf7 is a member of the ESCRT-III complex, playing a key role in the maturation of endosomes (Williams and Urbé, 2007; Hanson et al., 2008; Raiborg and Stenmark, 2009). It was previously described that ESCRT proteins are required for amphisome formation and that immature autophagosomes accumulate due to the lack of ESCRT components (Filimonenko et al., 2007; Lee et al., 2007). We detected a moderate increase in the frequency of colocalization between Rab11 and mCherry-Atg8a in Vps32-silenced cells. This observation also provides further evidence that Rab11 localizes to immature autophagosomes (Figure 3, C and F).
Rab11 is a widely used marker of REs (Hsu and Prekeris, 2010). Thus we examined whether autophagy induction affects the colocalization of Rab11 with GFP-labeled transferrin receptor (TfR), a well-known cargo of the Rab11-dependent recycling pathway (Maxfield and McGraw, 2004). Starvation-induced autophagy caused significant decrease in colocalization of Rab11 with TfR compared with control cells, while the amount of TfR-positive dots did not change (Figures 3, D, E, and G, and S3C).
These results suggest that, upon autophagy induction, Rab11 translocates from REs to autophagosomes.
Rab11 supports endosome maturation by regulating Hook
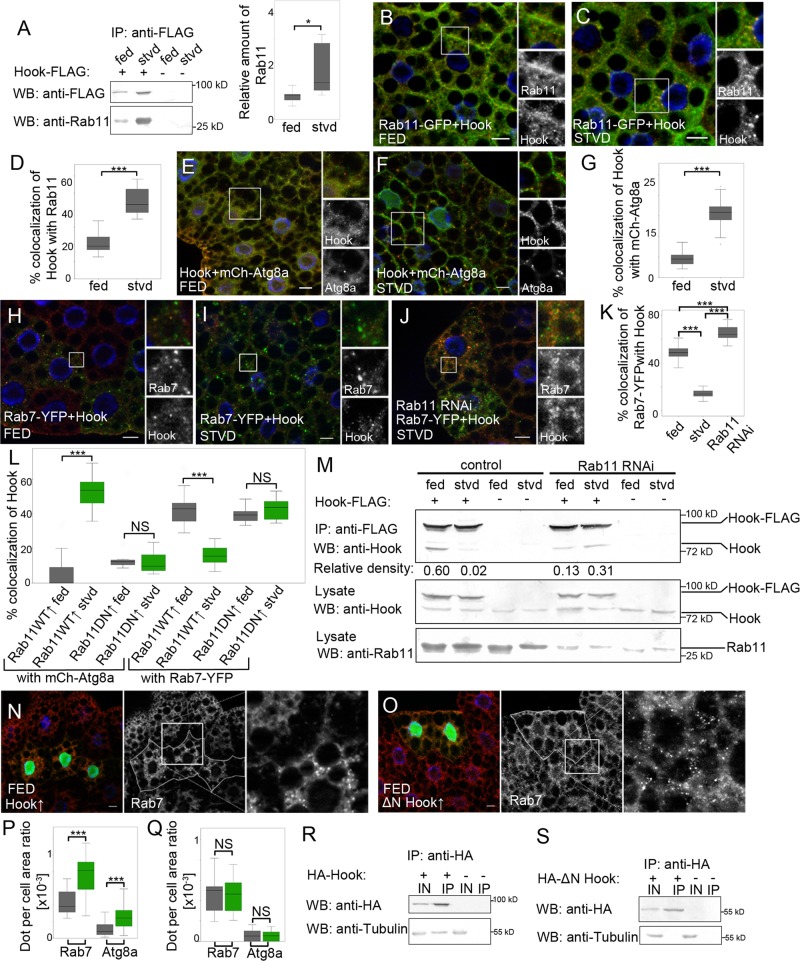
A recent study in cultured HEK293T cells showed that Rab11 can bind to Hook, a protein playing a role in the maturation of endosomes (Luiro et al., 2004). We also found a strong interaction between transgenic Hook and Rab11 in cultured Drosophila D.Mel-2 cells, while Rab11 did not bind to another late endosomal protein, Lamp1 (Figure S3, D and D′). Confirming these results, we could detect in vivo interaction of Hook-FLAG with endogenous Rab11 (Figure 4A), and we found that Rab11 interacts with Hook in a GTP-dependent manner (Figure S3E). Moreover, the strength of this interaction increased due to autophagy induction by starvation (Figure 4A). Finally, our coimmunoprecipitation experiments showed that Rab11 binds to the central coiled-coil domain of Hook, which was previously found to be responsible for homodimerization (Krämer and Phistry, 1999; Figure S3, F–K).
FIGURE 4:
Rab11 facilitates endosome maturation by regulating Hook localization. (A) Lysates of Hook-FLAG–expressing and control fed and starved L3 Drosophila larvae were incubated with anti-FLAG antibody–conjugated agarose beads, and bound proteins were detected by Western blotting. The relative density of Rab11 bands compared with Hook-FLAG bands was measured as described and compared between fed and starved groups using Wilcoxon's signed-rank test (n = 5, p = 0.0431). Rab11-GFP–expressing (green) fat body cells of fed (B) and starved (C) L3 larvae were stained with anti-Hook antibody (red). Nuclei were stained using DAPI (blue). (D) Colocalization of Hook with Rab11 was calculated and compared between fed and starved groups using the Mann-Whitney test (nfed = 132, nstvd = 64, p < 0.0001). Fat body cells of fed (E) and starved (F) larvae expressing mCherry-Atg8a (red) were stained with anti-Hook antibody (green). Nuclei were stained using DAPI (blue). (G) Colocalization of Hook with mCherry-Atg8a was calculated and compared between two groups using the Mann-Whitney test (nfed = 20, nstvd = 20, p < 0.0001). Fat body cells of fed (H) and starved (I) Rab7-YFP–expressing (green) larvae were stained with anti-Hook antibody (red). Nuclei were stained with DAPI (blue). (J) Starved Rab7-YFP–expressing (green) Rab11 RNAi cells (marked by GFP expression) were stained with anti-Hook antibody (red). Nuclei were stained with DAPI (blue). (K) Colocalization of Rab7-YFP with Hook was calculated and compared between the groups using the Kruskal-Wallis test (p < 0.0001; nfed = 40, nstvd = 35, nRNAi = 24, in all pairwise comparisons p < 0.0001). (L) Colocalization of Hook with mCh-Atg8a or Rab7-YFP was measured in the fat body cells of fed and starved larvae overexpressing wild-type (Rab11WT↑) or GDP-locked form (Rab11DN↑) of Rab11. Data were compared between fed and starved groups using the Mann-Whitney test (Hook-Atg8a: Rab11WT↑: nfed = 33, nstvd = 30, p < 0.0001; Rab11DN↑: nfed = 10, nstvd = 14, p = 0.7252; Hook-Rab7: Rab11WT↑: nfed = 49, nstvd = 36, p < 0.0001; Rab11DN↑: nfed = 13, nstvd = 16, p = 0.2729). Green bars, starved groups; gray bars, fed groups. (M) Lysates of Hook-FLAG expressing and lacking fed and starved control and Rab11 RNAi L3 Drosophila larvae were incubated with anti-FLAG antibody–conjugated agarose beads. Bound proteins and Rab11, Hook, and Hook-FLAG levels of lysates were detected by Western blotting. (N) Control and full-length Hook overexpressing (green) fat body cells of fed L3 larvae were stained with anti-Rab7 antibody (red). Nuclei were stained using DAPI (blue). (O) Control and ΔN-Hook overexpressing (green) fat body cells of fed L3 larvae were stained with anti-Rab7 antibody (red). Nuclei were stained with DAPI (blue). (P) Rab7 and mCh-Atg8a–positive dot per cell area ratio was calculated and compared between the groups indicated on (N) image using Wilcoxon's signed-rank test (Rab7: n = 18, p = 0.0002; Atg8: n = 29, p < 0.0001). Green bars, Rab11 RNAi; gray bars, control groups. (Q) Rab7 and mCh-Atg8a–positive dot per cell area ratio was calculated and compared between the groups indicated on (O) image using Wilcoxon's signed-rank test (Rab7: n = 22, p = 0.8838; Atg8: n = 20, p = 0.4955). Green bars, Rab11 RNAi; gray bars, control groups. D.Mel-2 cells were transfected with HA-tagged full-length Hook (R) or ΔN-Hook (S) constructs. Lysates were incubated with anti-HA antibody–conjugated agarose beads, and bound proteins were detected by Western blotting. IN, 1% input; IP, anti-HA bound protein samples. On box-and-whisker plots, bars show the data lying between the upper and lower quartiles; the median is indicated as a horizontal line within the box. Whiskers plot the smallest and largest observations. NS, p > 0.05; *, p < 0.05; ***, p < 0.001. Scale bars: 10 μm.
Furthermore, we found a 21 ± 1.5% colocalization ratio between Hook and Rab11 in fat body cells of fed larvae. This ratio increased to 45 ± 2.2% upon autophagy induction by starvation (Figure 4, B–D), while the number of Hook-positive structures and the level of Hook protein remained unaffected (Figure S4, A and B).
Our further experiments showed that 4 ± 0.4% of Hook-positive puncta colocalizes with mCherry-Atg8a under fed conditions, and this ratio increased to 16 ± 1% after autophagy induction (Figure 4, E–G). Parallel with these results, we could observe a decrease (from 48 ± 0.9 to 13 ± 0.6%) in colocalization between Hook and the late endosomal marker Rab7 after autophagy induction (Figure 4, H, I, and K). Meanwhile, no changes were detected in the number of Rab7-positive structures (Figure S4C). Similarly, induction of autophagy did not significantly increase the colocalization between Rab11 and Rab7 (Figure S4, D–F). This suggests that the increased colocalization of Hook with mCherry-Atg8a is not due to the increased convergence of autophagic and endosomal pathways. Furthermore, we could not detect any changes in the frequency of Hook-Rab7 colocalization upon amino acid starvation in cells lacking Atg1 protein, which is required for autophagy induction (Chan and Tooze, 2009; Figure S4, G–I). These results suggest that autophagy induction by starvation results in the translocation of Hook from Rab7-positive endosomes to autophagic structures.
We investigated whether Rab11 is required for the altered Hook localization. We found that silencing of Rab11 in fed larvae resulted in the accumulation of Hook on Rab7-positive late endosomes, indicated by an increased colocalization (65 ± 1.3% compared with 48 ± 0.9% in control cells; Figure 4, J and K). Moreover, we could not detect any changes in the frequency of colocalization between Hook and mCherry-Atg8a upon autophagy induction in Rab11-depleted cells (Figure S4, J–L). In addition to these results in fat body cells overexpressing wild-type Rab11, we observed an increase in the colocalization of Hook with mCherry-Atg8a (from 4 ± 4 to 52 ± 8.1%) and a decrease in the frequency of Hook-Rab7 colocalization (from 42 ± 7.9 to 16 ± 5.1%) due to amino acid starvation, whereas overexpression of GDP-locked Rab11 did not result in any changes in the colocalization of Hook with mCherry-Atg8a or Rab7–yellow fluorescent protein (Rab7-YFP; Figures 4L and S4, M–T).
Next we examined whether Rab11 has any effect on heterodimerization of endogenous Hook with transgenic Hook-FLAG. Our coimmunoprecipitation studies showed that starvation resulted in a decrease in heterodimer formation of Hook with Hook-FLAG in the presence of Rab11. However, we could not detect any changes in Hook heterodimerization in Rab11 RNAi larvae (Figure 4M).
A previous study showed that Hook has a negative regulatory role in the process of endosome maturation (Narayanan et al., 2000). Supporting this result, we found that overexpression of full-length Hook in fat body cells results in the accumulation of Rab7-positive structures (Figure 4, N and P). Moreover, we observed a moderate accumulation of mCherry-Atg8a–positive puncta in cells overexpressing Hook (Figures 4P and S4U).
It was previously described that the N-terminal domain of Hook is responsible for microtubule binding (Krämer and Phistry, 1999). We also could detect the interaction of the Hook N-terminus with α-tubulin (Figure 4, R and S). Furthermore, we found that overexpression of N-terminally truncated Hook in fat body cells does not result in accumulation of autophagic or Rab7-positive endosomal structures (Figures 4, O and Q, and S4V).
All of these data suggest that autophagy induction results in a Rab11-dependent translocation of Hook from late endosomes to autophagic structures. Through their interaction, Rab11 can inhibit the negative regulatory role of Hook on late endosomal development; therefore the presence of Rab11 allows the endosomes to mature.
DISCUSSION
Former work suggested a role for Rab11 in the maturation process of autophagosomes (Fader et al., 2008; Richards et al., 2011). In this in vivo study, we identified novel key functions of Rab11 in this process in Drosophila melanogaster. Loss of Rab11 leads to the accumulation of abnormal autophagosomes and acidic late endosomes, which is probably a consequence of impaired amphisome formation.
Furthermore, we observed autophagy induction–dependent translocation of Rab11 from TfR-positive REs to autophagosomes. This finding supports previous reports, which revealed REs as a possible membrane source for autophagosome formation and a role for Rab11 in vesicle trafficking from REs to the sites of autophagosome formation in cultured mammalian cells (Longatti et al., 2012; Puri et al., 2013). In line with these findings, a recent study of Knævelsrud and colleagues showed that Rab11 participates in membrane traffic from REs to the autophagosome precursors together with the Drosophila homologue of SNX18 (Knævelsrud et al., 2013). This study suggests that Rab11 has a similar role in Drosophila melanogaster and a role in providing an RE-derived membrane source for phagophore expansion. However, we did not observe any defect in autophagosome formation in the absence of functional Rab11 protein.
The molecular mechanism of autophagosome formation is a poorly understood process. Recently several organelles were implicated as possible membrane sources for autophagosomes (Mari et al., 2011). Many studies suggest that the possible origin of the autophagosomal membrane might vary, even in different mammalian cell types (Hayashi-Nishino et al., 2009; Hailey et al., 2010; Ravikumar et al., 2010; Yamamoto et al., 2012; Hamasaki et al., 2013), and this question needs further investigation in Drosophila melanogaster as well. Presumably, although REs contribute to the formation of autophagic membrane in flies, they are not likely to be the main membrane source. This could explain why the loss of Rab11 did not block autophagosome formation in our experiments.
We demonstrated the interaction of both transgenic and endogenous Rab11 with Hook protein in Drosophila cell culture and even in the whole organism. These results confirm a former study that showed the same phenomenon using COS-1 cells cotransfected with tagged Hook and Rab11 constructs (Luiro et al., 2004). Furthermore, we found an autophagy induction–dependent increase in the strength of this physical interaction.
In addition to these findings, our results provide mechanistic insights into the maturation of endosomes and autophagosomes: the interaction between Rab11 and Hook is crucial for the maturation of these structures. Autophagy induction by amino acid starvation increased the colocalization of Rab11 with Hook and Hook with Atg8a. In line with that, we found a significant decrease in Hook-Rab7 colocalization during starvation-induced autophagy. Our data suggest a Rab11-dependent translocation of Hook from Rab7-labeled late endosomes to autophagic structures. This idea is supported by the finding that lack of Rab11 causes the accumulation of Hook on Rab7-positive late endosomes.
Previously Hook was found to be a negative regulator of endosome maturation in Drosophila (Narayanan et al., 2000) and the Hook-like protein Acinus was described as an important regulator of endosome maturation and autophagic processes (Haberman et al., 2010). In line with these findings, we found that overexpression of full-length Hook protein mimics the effects of Rab11 depletion on the maturation of autophagosomes and endosomes. Moreover, we show that the N-terminal microtubule-binding domain of Hook is required for its negative regulatory role. Our latter results reveal a key mechanistic role for Rab11 in removal of Hook from late endosomes, which allows termination of endosome maturation and subsequent fusion with lysosomes. As was demonstrated in previous work, mature late endosomes can undergo fusion with nascent autophagosomes and promote their maturation (Lucocq and Walker, 1997; Köchl et al., 2006; Filimonenko et al., 2007). On the basis of these findings, we developed a model representing a mode of cross-talk between the autophagic and endosomal pathways (Figure 5). On starvation, the enhanced autophagic activity requires an increased input from the endolysosomal system. For this purpose, Rab11 removes Hook from the late endosomes, thereby allowing subsequent fusion of these compartments with immature autophagosomes as well.
FIGURE 5:
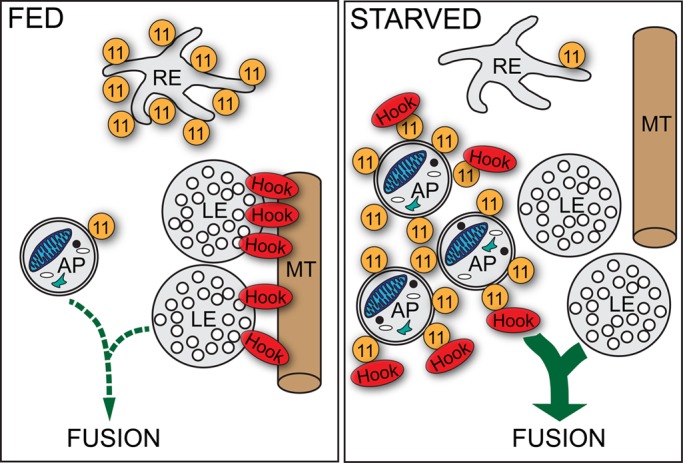
Model for the role of Rab11-Hook interaction in autophagosome maturation. Under fed conditions, Rab11 (11) localizes to REs, and Hook anchors late endosomes (LE) to microtubules (MT), thereby suppressing endosome maturation and fusion events. Autophagy induction by starvation results in the translocation of Rab11 from REs to autophagosomes (AP). Meanwhile, Rab11 removes Hook from LEs, allowing subsequent fusion of LEs with APs.
Why Hook does not inhibit the maturation of autophagosomes once it localizes to these organelles is still an open question. We mapped the Hook interaction surface with Rab11 to residues 180–530, which represents the coiled-coil domain of Hook. This domain was previously found to be responsible for homodimerization of Hook (Krämer and Phistry, 1999). Conceivably, homodimerization and Rab11 binding are distinct and separable functions of the Hook coiled-coil domain, as was shown in the case of the FIP2 coiled-coil domain, which also interacts with Rab11 (Wei et al., 2006). Our further experiments showed that Rab11 binding prevents Hook from homodimerization. Thus our results suggest that Rab11 promotes autophagic flux by decreasing Hook homodimerization and inhibiting its negative regulatory role on endosome maturation.
MATERIALS AND METHODS
Drosophila stocks and culture
Fly stocks used in this study (derived from the Vienna Drosophila RNAi Center [referred to as VDRC; Vienna, Austria] and Bloomington Drosophila Stock Center [referred to as BDSC; Bloomington, IN]) are listed in Supplemental Table S1. UAS-Hook-FLAG, UAS–ΔN-Hook–hemagglutinin (UAS-ΔN-Hook-HA), and UAS-Rab11-HA stocks were generated by BestGene, using constructs described below. Flies were raised on standard yeast/cornmeal/agar media at 25°C, 50% humidity and a 12-h light/12-h dark daily cycle, under uncrowded conditions. During amino acid starvation, well-fed 72–86 h AEL (after egg laying) larvae were floated in 20% sucrose solution in a microfuge tube for 3 h at room temperature.
Cell culture and constructs
Drosophila D.Mel-2 cells were cultured in Express Five Serum-Free Medium (Invitrogen). Transfections were performed with TransIT-2020 reagent (Mirus) according to the manufacturer's recommendations. HA2-679hook, HA248-679hook (ΔN-Hook), and hook1-551MYC (ΔC-Hook) constructs controlled by inducible metallothionein promoter (hereafter pMT) were kind gifts of Helmut Krämer (Krämer and Phistry, 1999). Full-length Hook and the coiled-coil domain, N-terminus, and C-terminus of Hook, Rab11, and Lamp1 were amplified from the Drosophila cDNA clones LD05265, GM06568, and RE72002 (provided by Drosophila Genomics Resource Center [DGRC]). UAS constructs were cotransfected into Drosophila D.Mel-2 cells with pMT-GAL4 vector (provided by DGRC, ID 1042). Cells were transfected with 2500 ng of plasmids in each case, and protein expression was induced 24–48 h later by overnight incubation with 1 mM CuSO4. Primers for amplification were as follows: full-length Hook, 5′-ATGTCCGCGCCCAAGAACGA-3′ and 5′-ATCCCTTTGATTTCATTGCA-3′; Rab11, 5′-ATGGGTGCAAGAGAAGACGA-3′ and 5′-ATCCCTGACAGCACTGTTTG-3′; Lamp1, 5′-ATGTTCGCCAACAAATTGTT-3′ and 5′-ATCCGAAGCTCATGTAACCG-3′. All these PCR products were inserted into XmnI/EcoRV-digested pENTR1A Gateway vector (Invitrogen). Entry clones were recombined with the destination vector pTWF in case of C-terminally FLAG-tagged proteins or pTWH in case of C-terminally HA-tagged protein using LR Clonase II Enzyme Mix (Invitrogen). For mapping, the Rab11-binding site Hook coiled-coil domain (180–530 aa), N-terminus, and C-terminus of Hook were amplified from the LD05265 clone using the following primers: coiled-coil domain, 5′-CTGGACAGCAAGGCAGTTCAG-3′ and 5′-CTATTGTTTACCAAACTCGCTCTC-3′; N-terminus of Hook, 5′-TCCGCGCCCAAGAACGAGATG-3′ and 5′-TTGTTTACCAAACTCGCTCTC-3′; C-terminus of Hook, 5′-ATCAAACAGTTAATGGAGCTA-3′ and 5′-CTTTGATTTCATTGCACTTAG-3′. All of these oligonucleotides included a 5′ NotI restriction site and a 3′ Acc65I restriction site. Resulting products were inserted into NotI/Acc65I-digested UAS-3×HA vector. For all PCR amplifications, we used Phusion DNA Polymerase (Finnzymes). For the UAS-ΔN-Hook construct, HA248-679hook construct from Helmut Krämer was cloned into pENTR1A Gateway vector (Invitrogen) using Acc65I and NotI restriction sites. Entry clones were recombined with the destination vector pTWH using LR Clonase II Enzyme Mix (Invitrogen).
Immunoprecipitation
Transfected cells were harvested, pelleted (1000 × g for 3 min), and washed twice with phosphate-buffered saline (PBS). All steps of the following procedure were performed on ice or at 4°C. Cells were lysed in 500 μl lysis buffer (0.5% Triton-X100, 150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 7.5, and complete protease inhibitor cocktail [Sigma-Aldrich]). Lysate was cleared by centrifugation at 10,000 × g for 10 min in an Eppendorf 5430R microcentrifuge. Before incubation, beads were washed three times with lysis buffer. Two hours later, beads were pelleted (8000 × g for 30 s), thoroughly washed five times with lysis buffer, and finally boiled in 30 μl Laemmli sample buffer (Sigma-Aldrich) for 5 min. Cell lysates were incubated with 30 μl anti-FLAG– or anti-HA–conjugated agarose beads (Sigma-Aldrich) for 2 h at 4°C. For immunoprecipitation studies carried out in whole organisms, expression of Hook-FLAG was induced by heat shocking well-fed L3 larvae for 1 h at 37°C. After this procedure, larvae were kept at 25°C for a 2-h recovery incubation, which was followed by 2 h of starvation. Larvae (100 mg) were washed twice and homogenized in 1 ml RIPA buffer (Sigma-Aldrich) containing protease inhibitor cocktail (Sigma-Aldrich). Detritus was pelleted (10,000 × g for 10 min), and the middle fraction was incubated with 30 μl anti-FLAG–conjugated agarose beads (Sigma-Aldrich) for 2 h at 4°C. Beads were collected by centrifugation for 30 s at 8000 × g, washed extensively, and boiled in 30 μl Laemmli sample buffer. Bound proteins were detected by Western blotting.
Western blots and immunostainings
Larvae (10–15 mg) were washed twice with PBS, and each mg of larvae was homogenized in 20 μl Laemmli sample buffer (Sigma-Aldrich). Homogenate was boiled for 5 min and pelleted at 10,000 × g for 10 min at 4°C, and the middle fraction was collected. Protein samples were separated on 8–12% polyacrylamide gel and were transferred to nitrocellulose membrane (Bio-Rad). After incubation in blocking solution (3% bovine serum albumin [BSA] in Tween-20/TBS [TBST]) for 1 h at room temperature, membranes were incubated with primary antibody in antibody solution (1% BSA in TBST) for 1 h at room temperature; this was followed by three 10-min washes in TBST. Signals were detected using alkaline phosphatase–coupled secondary antibodies diluted in antibody solution. Finally, membranes were developed by BCIP/NBT solution (Sigma-Aldrich). For immunostaining studies, we used larvae in the nutrient conditions and at the ages indicated above. The larval cuticle was opened, and tissue was fixed with 4% formaldehyde freshly depolymerized from paraformaldehyde in PBS overnight at 4°C. After fixation, larvae were washed three times for 30 min each time in PBST (0.1% Triton X-100 in PBS) and incubated in blocking solution (5% normal goat serum in PBST) for 2 h at room temperature. Larvae were incubated in primary antibody in PBST overnight at 4°C; this was followed by three 30-min washes in PBST. Afterward, larvae were incubated with fluorescent dye–conjugated secondary antibody; this was followed by three 30-min washes. Finally, fat bodies were dissected and analyzed by microscopy.
Antibodies
Antibodies used in this study are listed in Table S2. The EGFP gene was amplified from the pIRES2-EGFP vector (Clontech) and directly cloned into the pQE-UA bacterial expression vector (Qiagen). Recombinant protein encoding the full-length EGFP fused to an N-terminal hexahistidine tag was expressed in Escherichia coli M15 cells. Protein purification was performed using the QIAexpressionist kit (Qiagen). Mice were immunized with the fusion protein, and the resulting polyclonal antisera were used for further investigation.
Histology and imaging
Fat bodies from well-fed or amino acid–starved larvae were dissected in PBS and incubated for 2 min in 50 nM LTR (Invitrogen) and 1 μM Hoechst in PBS at room temperature. Stained fat bodies were washed with PBS, covered, and immediately analyzed by microscopy. For TRA uptake assay, larvae were dissected in M3 medium containing Insect Medium Supplement (Sigma-Aldrich). A 30-min incubation in pulse solution (TRA [Invitrogen] 1:25 in 5% fetal calf serum [FCS] in M3/IMS medium) was followed by a 10-min chase (5% FCS in M3/IMS medium). Larvae were washed three times with 5% BSA in PBS at 4°C and fixed in 4% paraformaldehyde solution. Fixed larvae were washed with PBS, and nuclei were stained with 1 μM Hoechst in PBS. Prepared tissues were covered in a solution containing 75% glycerol and 25% PBS. Primary images were edited using Adobe Photoshop CS5 software: area of interest was cropped, and brightness and contrast adjustments were modified, if it was necessary. To capture images, we used a Zeiss Axio Imager Z1 microscope equipped with an ApoTome unit and a Plan-Neofluar 40 × 0.75 numerical aperture objective using AxioCam MRm camera with AxioVision 4.82 software.
Quantification and statistical analysis
We determined the number of dots in the cells of interest using ImageJ software. These data were weighted for the cell area to exclude unwanted biases, which could derive from variations in age or size of larvae. Cell area was measured using ImageJ software. In this way, we generated dot per cell area ratio data. For colocalization analyses, the number of double- and single-labeled dots was measured using the Red and Green Puncta Colocalization Macro of ImageJ software. We calculated their percentage ([double-labeled/single-labeled dots] × 100). When using a clonal analysis system during a study, we tried to choose the nearest neighbor of the clone cell in the same location (inside or on the periphery of the tissue) for control. If that was not possible, we took the next neighboring cell in a clockwise direction. When analyzing whole fat bodies, we measured the number of dots in 8–10 cells per larva and calculated an average value for each animal. Blots were quantified using ImageJ software. Density of P62, Rab11, GFP-Atg8, and free GFP bands were measured using ImageJ and weighted with the relative protein amount of the protein samples (density of tubulin bands). Statistical analysis of the GFP-Atg8a processing assay was performed using general linear mixed models (GLMM). We applied linear modeling for analysis, but the normal distribution is a strong requirement for this process, so we applied square-root transformation to normalize data distribution. Three main effects were investigated: two groups (Rab11 RNAi and control), two measurements (relative density of GFP-Atg8a and free GFP bands), and two treatments (fed and starved), respectively the two- and three-way interaction of these. The model showed the following results: there is significant difference between the two groups (F(1,24) = 37.426; p < 0.001) and the two measurements (F(1,24) = 7.336; p = 0.012), and these two are in significant interaction with each other (F(1,24) = 8.552; p = 0.007). The different treatments do not result in significant difference (F(1,24) = 0.235; p = 0.632). The post hoc test shows there is less free GFP in Rab11 RNAi larvae (t(24) = 5.603; p < 0.001), while there is no difference (t(24) = −0.125; p = 0.902) in the control. Density of Rab11 and Hook bands was measured and weighed with the density of Hook-FLAG bands. Statistical analysis was performed using the indicated tests in IBM SPSS Statistics software. Box-and-whisker plot figures were generated using the same software. On box-and-whisker plots, bars show the data lying between the upper and lower quartiles; the median is indicated as a horizontal line within the box. Whiskers plot the smallest and largest observations, while dots and asterisks indicate outliers. A p value of less than 0.05 was considered to be significant; * means p < 0.05; ** means p < 0.01; *** means p < 0.001. For details and results of statistical analyses, see Table S3. Analysis of colocalizing dots was performed using Matlab (R2012b) software.
Ultrastructural analysis
Transmission electron microscopy.
Larvae were dissected in ice-cold PBS, and fat bodies were fixed in a solution containing 2% formaldehyde, 0.5% glutaraldehyde, 3 mM CaCl2, and 1% sucrose in 0.1 M Na-cacodylate (pH 7.4; 30 min at room temperature); postfixed in 0.5% osmium tetroxide (45 min at room temperature) and in half-saturated aqueous uranyl acetate (30 min at room temperature); dehydrated in graded series of ethanol; embedded in LR White according to the manufacturer's instructions; and cured for 36 h at 60°C. Ultrathin sections were stained with lead citrate for 5 min. Grids were analyzed in a JEOL JEM 1011 transmission electron microscope operating at 60 kV. Images were taken using an Olympus Morada 11 megapixel camera and iTEM software (Olympus). All reagents and materials used for electron microscopy were obtained from Sigma-Aldrich.
Acid phosphatase cytochemistry.
Fat body lobes were fixed as described above and washed in 0.05M Na-acetate buffer (pH 5.0; three times for 5 min each time at room temperature), incubated in Gömöri's medium (5 mM β-glycerophosphate and 4 mM lead nitrate dissolved in 0.05M acetate buffer) for 30 min at room temperature, and then washed in acetate buffer (three times for 5 min each time) and processed for electron microscopy as described above. Substrate-free medium was used for control experiments.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Hungarian Scientific Research Funds (NK78012) provided to M.S. The European Union and the European Social Fund have provided financial support to this project under grant agreement no. TAMOP 4.2.1./B-09/KMR-2010-0003. T.F. was supported by the Hungarian Academy of Sciences (F01/031). We address special thanks to Helmut Krämer for the indicated antibodies, stocks, and constructs. We are thankful to János Elek for the colocalization analysis script in Matlab software.
Abbreviations used:
- Atg
autophagy-related gene
- BDSC
Bloomington Drosophila Stock Center
- BSA
bovine serum albumin
- CQ
chloroquine
- DAPI
4′,6-diamidino-2-phenylindole
- DGRC
Drosophila Genomics Resource Center
- GFP
green fluorescent protein
- HA
hemagglutinin
- LTR
LysoTracker Red
- PAS
preautophagosomal structures
- PBS
phosphate-buffered saline
- pMT
metallothionein promoter
- RE
recycling endosome
- RNAi
RNA interference
- TBST
Tween-20/TBS
- TfR
transferrin receptor
- TRA
Texas Red Avidin
- VDRC
Vienna Drosophila RNAi Center
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0574) on December 19, 2013.
The authors declare they have no conflict of interest.
REFERENCES
- Chan EYW, Tooze SA. Evolution of Atg1 function and regulation. Autophagy. 2009;5:758–765. doi: 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- Chua CEL, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci. 2011;68:3349–3358. doi: 10.1007/s00018-011-0748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S, Sönnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- Fader CM, Sánchez DG, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EMC, Isaacs AM, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. [DOI] [PubMed] [Google Scholar]
- Haberman AS, Akbar MA, Ray S, Krämer H. Drosophila acinus encodes a novel regulator of endocytic and autophagic trafficking. Development. 2010;137:2157–2166. doi: 10.1242/dev.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, et al. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–534. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knævelsrud H, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl R, Hu XW, Chan EYW, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Krämer H, Phistry M. Genetic analysis of hook, a gene required for endocytic trafficking in Drosophila. Genetics. 1999;151:675–684. doi: 10.1093/genetics/151.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: shared machinery for degradation. Bioessays. 2013;35:34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- Lee J-A, Beigneux A, Ahmad ST, Young SG, Gao F-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq J, Walker D. Evidence for fusion between multilamellar endosomes and autophagosomes in HeLa cells. Eur J Cell Biol. 1997;72:307–313. [PubMed] [Google Scholar]
- Luiro K, Yliannala K, Ahtiainen L, Maunu H, Järvelä I, Kyttälä A, Jalanko A. Interconnections of CLN3, Hook1 and Rab proteins link Batten disease to defects in the endocytic pathway. Hum Mol Genet. 2004;13:3017–3027. doi: 10.1093/hmg/ddh321. [DOI] [PubMed] [Google Scholar]
- Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Krämer H, Ramaswami M. Drosophila endosomal proteins hook and deep orange regulate synapse size but not synaptic vesicle recycling. J Neurobiol. 2000;45:105–119. doi: 10.1002/1097-4695(20001105)45:2<105::aid-neu5>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards P, et al. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington's disease. Cell Death Differ. 2011;18:191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwake M, Schröder B, Saftig P. Lysosomal membrane proteins and their central role in physiology. Traffic. 2013;14:739–748. doi: 10.1111/tra.12056. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol. 1990;111:329–345. doi: 10.1083/jcb.111.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal. 2011;23:516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Wei J, Fain S, Harrison C, Feig LA, Baleja JD. Molecular dissection of Rab11 binding from coiled-coil formation in the Rab11-FIP2 C-terminal domain. Biochemistry. 2006;45:6826–6834. doi: 10.1021/bi052655o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.