Abstract
Hox genes contain a homeobox encoding a 60-amino acid DNA binding sequence. The Hoxa-1 gene (Hox1.6, ERA1) encodes two alternatively spliced mRNAs that encode distinct proteins, one with the homeodomain (Hoxa1-993), and another protein lacking this domain (Hoxa1-399). The functions of Hoxa1-399 are unknown. We detected Hoxa1-993 and Hoxa1-399 by immunoprecipitation using Hoxa1 antibodies. To assess whether Hoxa1-399 functions in cellular differentiation we analyzed Hoxb1, a Hoxa1 target gene. Hoxa1-993 and its cofactor, Pbx1, bind to the Hoxb1 SOct-R3 promoter to transcriptionally activate a luciferase reporter. Results from F9 stem cells that stably express ectopic Hoxa1-399 (the F9-399 line) show that Hoxa1-399 reduces this transcriptional activation. Gel shift assays demonstrate that Hoxa1-399 reduces Hoxa1-993/Pbx1 binding to the Hoxb1 SOct-R3 region. GST-pull down experiments suggest that Hoxa1-399, Hoxa1-993, and Pbx1 form a trimer. However, the F9-399 line exhibits no differences in RA-induced proliferation arrest or endogenous Hoxb1, Pbx1, Hoxa5, Cyp26a1, GATA4, or Meis mRNA levels when compared to F9 wild type.
Keywords: Homeobox, Transcription Factor, Splice Variant, cell differentiation, F9 cells, retinoids, Hoxb1, Pbx, teratocarcinoma, stem cell, vitamin A
Hox genes encode a group of evolutionarily conserved transcription factors, originally identified in Drosophila melanogaster [Lewis, 1978], that are transiently expressed and are involved in establishing anterior to posterior pattern during vertebrate development [McGinnis and Krumlauf, 1992]. Four clusters of Hox genes exist in mouse and human, each cluster on a different chromosome, with related genes falling into 13 paralogous groups. Genes at the 3′ ends of the Hox chromosome clusters have a greater response to RA and are expressed in more anterior regions of the developing embryo as compared to genes at the 5′ ends, which are less responsive to RA and are expressed in more posterior regions [Gudas, 1994; Langston and Gudas, 1994; Means and Gudas, 1995; Simeone et al., 1990; Simeone et al., 1991]. Hox genes characteristically encode a 60-amino acid DNA-binding motif, the homeodomain, that allows Hox proteins to bind the promoter regions of their target genes to regulate their transcription [Langston and Gudas, 1994; Waskiewicz et al., 2001].
RA transcriptionally activates the Hoxa1 gene, the most 3′ gene of the Hox A cluster, in F9 teratocarcinoma stem cells [LaRosa and Gudas, 1988a]. The Hoxa1 gene encodes two alternatively spliced mRNAs that can direct the synthesis of two distinct proteins; one that contains the homeodomain (Hoxa1-993) and another truncated protein that lacks the DNA binding domain (Hoxa1-399). The resulting proteins are identical for the first 114 amino acids but have unique carboxy-termini [LaRosa and Gudas, 1988b]. The functions of the smaller, truncated Hoxa1 protein, which lacks the homeodomain, are not known and are the subject of the series of experiments reported here.
The expression of a truncated Hox protein lacking a homeodomain is not unique to Hoxa1. For example, alternatively spliced transcripts resulting in truncated Hox proteins which lack a homeodomain have been described for the murine Hoxa9, human HOXA1, murine Hoxb6, human HOXB6, and Xenopus XlHbox2 [Fujimoto et al., 1998; Hong et al., 1995; Shen et al., 1991; Wright et al., 1987]. The functions of these truncated Hox proteins that lack the homeodomain have not been studied in depth.
Hox proteins achieve DNA sequence specificity by binding to other DNA-binding proteins that act as cofactors to generate heterodimers that bind cooperative sites on a DNA target [Knoepfler et al., 1996]. Due to the increased size of the cooperative binding site, cofactor binding increases the affinity and specificity of the Hox protein binding to DNA [Chang et al., 1996; Shen et al., 1996]. These cofactors include the PBC and Meis classes of TALE (Three Amino Acid Loop Extension) homeodomain proteins [Moens and Selleri, 2006]. The PBC class is comprised of fly Extradenticle (Exd) and vertebrate Pbx, whereas the Meis family is made up of fly Homothorax (Hth) and vertebrate Meis and Prep [Moens and Selleri, 2006]. Hoxa1 has been shown to interact with Pbx1, a member of the PBC class, via a tryptophan-containing pentapeptide motif found N-terminal to the homeodomain. The protein sequence of Hoxa1-399 does not contain the pentapeptide motif [LaRosa and Gudas, 1988b], and therefore it would be expected that Hoxa1-399 does not bind Pbx1.
The Hoxa1 protein transcriptionally regulates genes involved in various cellular signaling and developmental pathways. Subtractive hybridization experiments comparing F9 Wt and F9 cells overexpressing Hoxa1-993 resulted in the identification of various putative Hoxa1 target genes, such as BMP-4, cadherin-6, HMG-1, SAP18, Gbx-2, Evx-2, and RBP-2 [Shen et al., 2000]. Microarray analyses of RA-treated Hoxa1−/− ES (Embryonic Stem) cells versus wild type ES cells indicate that Hoxa1 mediates the repression of genes involved in endodermal differentiation, such as Sox17, Gata6, Gata4, Dab2, and Lama1, while Hoxa1 promotes the expression of genes involved in ectodermal and mesodermal differentiation, such as Fgf5, Nnat, Wnt3a, BDNF, RhoB, Postn, and Col1a1 [Martinez-Ceballos et al., 2005; Martinez-Ceballos and Gudas, 2008]. Additionally, microarray analyses of forced expression or depletion of HOXA1 in human mammary carcinoma cells identified genes involved in the p44/42 MAP kinase activation pathway as downstream targets of HOXA1 [Mohankumar et al., 2007].
Two independent groups have generated Hoxa1 knockout mice [Chisaka et al., 1992; Lufkin et al., 1991]. Defects in these knockout mice stem from alterations in rhombomeres 4–7 [Mark et al., 1993], resulting in malformations of the inner ear, delayed hindbrain neural tube closure, and absence of certain cranial nerves and ganglia [Chisaka et al., 1992; Lufkin et al., 1991]. Recently, the genetic etiology of a congenital dysinnervation disorder, the human HOXA1 syndrome, in which early motoneuron development is disrupted, was described [Bosley et al., 2007; Engle, 2007]. The resulting phenotype includes deafness, horizontal gaze abnormalities, hypoventilation, vascular malformations of the internal carotid arteries and cardiac outflow tract, facial weakness, mental retardation, and autism spectrum disorder. The mutations in these families affect the synthesis of all HOXA1 transcripts, resulting in the complete loss of HOXA1 [Tischfield et al., 2005].
One of the best-studied Hoxa1 target genes is Hoxb1. The Hoxb1 gene contains multiple retinoic acid response elements (RAREs) downstream of the coding region which are required for the transcriptional activation of Hoxb1 in response to retinoids in cultured cells and in different areas of the developing embryo [Bel-Vialar et al., 2002; Huang et al., 2002; Huang et al., 1998; Langston et al., 1997; Marshall et al., 1996]. Genetic analysis of double mutant mice demonstrates that both Hoxa1 and Hoxb1 are necessary for establishing and maintaining rhombomere 4 identity [Studer et al., 1998]. Analysis of the Hoxb13′RAREallele and other targeted loss-of-function alleles from both Hoxa1 and Hoxb1 shows that Hoxb1 expression in rhombomere 4 is dependent on the early activation of both Hoxa1 and Hoxb1 by endogenous retinoids [Studer et al., 1998].
The Hoxa1 gene is a direct target of the retinoic acid receptors [Langston and Gudas, 1992; LaRosa and Gudas, 1988b]. In RA-treated F9 teratocarcinoma cells, Hoxa1-993 mRNA is rapidly transcriptionally activated (<8 hr), and at later times (72 hr) Hoxa1-399 mRNA transcripts accumulate as well [LaRosa and Gudas, 1988b]. Both transcripts have been detected in mouse embryos, where the two mRNAs are present in equal amounts early in development (E8.0), but the relative proportion of homeodomain protein producing message is decreased at later times (E9.0) [Murphy and Hill, 1991]. These data indicate that the expression pattern of Hoxa1 transcripts seen in F9 cells is similar to that seen during development, making F9 stem cells an ideal system for studying Hoxa1 signaling during cellular differentiation.
Here, we used stably transfected F9 cells that ectopically express the Hoxa1-399 transcript to determine its effects on the expression of known Hoxa1 target genes. Using several biochemical approaches, we provide evidence for a negative role for the Hoxa1-399 protein in the transcriptional regulation of the Hoxb1 gene.
MATERIALS AND METHODS
Plasmids
pERA-1-399S and pERA-1-993S
The pERA-1-399S and pERA-1-993S plasmids were previously constructed in our laboratory by inserting the EcoRI restriction fragment of Hoxa1-399 and Hoxa1-993 mouse cDNA, respectively, into the EcoRI restriction fragment of the pSG5 expression vector.
pMT-399S
The pMT-399S plasmid was previously constructed in our laboratory by inserting the EcoRI restriction fragment from plasmid pERA-399 that encodes the Hoxa1-399 cDNA into the EcoRI site of the expression vector pMT64AA. This fragment contains the full-length open reading frame of Hoxa1-399 but lacks a region of the 3′ untranslated region. pMT64AA contains a portion of the mouse metallothionein I promoter followed by an SV40 poly(A) adenylation sequence to allow induction of expression by zinc.
pAR3039-Hoxa1-993
The bacterial expression vector pAR3039-Hoxa1-993 was previously constructed in our laboratory by inserting the NdeI restriction fragment of PCR-amplified Hoxa1-993. The XhoI/BamHI fragment of pMT-993S was subsequently inserted.
pAR3039 –Hoxa1-399
The bacterial expression vector pAR3039-Hoxa1-399 was previously constructed in our laboratory by inserting the SacI/XhoI fragment of pMT-399S into pAR3039-993.
pCS2-Pbx1a
The pCS2-Pbx1a plasmid was a gift from the laboratory of Dr. Licia Selleri. The construct was generated by inserting the BamHI-ClaI fragment of Pbx1a mouse cDNA into the pCS2+ expression vector.
pGL3-wtARE
The pGL3-wtARE reporter construct was generated by annealing oligonucleotides corresponding to the SOct-R3 of the Hoxb1-ARE promoter region [5′ CAAGTGTCTTT GTCATGCTAATGATTGGGGGGTGATGGATGGGCGCTGC 3′] and inserting this into the KpnI/ BglII site of the pGL3-promoter vector (Promega).
pGEX-Hoxa1
The pGEX-Hoxa1-993 and pGEX-Hoxa1-399 were generated by inserting the corresponding cDNA restriction fragments into the EcoRI/ BamHI site of pGEX2T.
Protein detection
Radiolabeled, in vitro translated proteins were synthesized from pSV-993S and pSV-399S with [35S]-cysteine (1000 Ci/mmol) (cat # SJ232, GE Healthcare) using TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI, cat # L5020) according to the manufacturer’s protocol. Protein products (2 μL) were resolved by SDS-PAGE. The gel was treated with ENHANCE (cat # 6NE9701, Perkin Elmer, Boston, MA) according to the manufacturer’s protocol and visualized by Kodak BioMax films.
BL21 bacteria were transformed with pAR3039-Hoxa1-993 and pAR3039-Hoxa1-399 plasmids. A single colony was inoculated, grown in LB, and induced with isopropyl-β-D-thiogalactopyranoside to express Hoxa1-993 and Hoxa1-399 proteins. Bacterial lysates were resolved by SDS-PAGE. Hoxa1-993 and Hoxa1-399 proteins were detected by Western analysis, using standard protocols as previously described [Goliger and Gudas, 1992], with the Hoxa1 primary antibodies generated in our laboratory.
Generation of the mouse Hoxa1 antibodies
Antisera 425 - A N-terminal peptide of mouse Hoxa1 (-M D N A R M N S F L E Y P I L G S-) was custom synthesized (Peninsula Labs, Belmont, CA). This peptide is located in the most N-terminal region of Hoxa1, which is common to both Hoxa1-993 and Hoxa1-399, and therefore the antibody that is generated using this peptide will recognize both proteins. The peptide was coupled to keyhole limpet hemocyanin (KLH) and injected into guinea pigs, using standard methods, to generate polyclonal antisera to the mouse Hoxa1 peptide (Pocono Rabbit Farm and Laboratory, Inc. Canadensis, PA). Thereafter, animals with positive sera were bled weekly and boosted with 25 μg antigen in Incomplete Freund’s Adjuvant (IFA) monthly. The IgG fraction was obtained by passing crude sera through a DEAE Affi-gel blue (Bio-Rad) column. This was followed by affinity chromatography of the IgG fraction on resins prepared by carbodiimide coupling of the peptide to sepharose gel (Pharmacia Sepharose 4B). The two-step purification steps were monitored by dot-blot assays, with peptide spotted onto nitrocellulose discs. After incubation with column fractions, the discs were processed with 1×104–3×104 cpm of 125I-protein A (New England Nuclear, Boston, MA). Positive IgG fractions were detected by radiography. Antisera 6466 – This antibody was generated by injecting a custom synthesized N-terminal peptide consisting of amino acids 18–32 (-C-S-A-R-A-Y-P-S-D-H-G-I-T-T-F-) of mouse Hoxa1 into rabbits (Pocono Rabbit Farm and Laboratory, Inc. Canadensis, PA). This region of Hoxa1 is common to both Hoxa1-993 and Hoxa1-399 and therefore the antibody that is generated using this peptide will recognize both proteins. This antisera was purified in a similar manner as antisera 425. Antisera 397 & 398 - These antibodies were generated by injecting a custom synthesized C-terminal peptide consisting of the last 17 amino acids of Hoxa1-399 (-R-S-L-S-F-P-C-F-R-D-V-FS-S-A-D-L) into two guinea pigs (Pocono Rabbit Farm and Laboratory, Inc. Canadensis, PA). This stretch of amino acids is unique to Hoxa1-399 and therefore the antibody that is generated using this peptide will recognize only Hoxa1-399. These antibodies were purified as described above.
Generation of F9-399-83 and F9-399-72 cell lines
F9 murine teratocarcinoma cells were co-transfected with the pMT-399S and neomycin resistance (pSV2-neo) plasmids through electroporation. Two independent clones were selected and screened to generate the EW-F9-MT-Hoxa1-399S-83 and EW-F9-MT-Hoxa1-399S-72 stable cell lines (abbreviated F9-399-83 and F9-399-72, respectively). These cells, in response to zinc, express an exogenous Hoxa1-399 mRNA that is 1.5 kB because a region of the 3′ untranslated region was truncated in the pMT-399S construct (Table 1). A similar cell line, F9-993-10, was previously generated [Goliger and Gudas, 1992] to overexpress Hoxa1-993 in response to zinc.
Table 1.
Hoxa1 Transcript Sizes in F9 Wt and Stably Transfected Cell Lines (kb)
| RA-inducible transcripts | zinc-inducible transcripts | |||
|---|---|---|---|---|
|
| ||||
| Endogenous Hoxa1-993 | Endogenous Hoxa1-399 | Exogenous Hoxa1-399 | Exogenous Hoxa1-993 | |
| F9 Wt | 2.2 | 2.0 | - | - |
| EW-F9-399-83 | 2.2 | 2.0 | 1.5 | - |
| EW-F9-399-72 | 2.2 | 2.0 | 1.5 | - |
| JG-F9-993-10 | 2.2 | 2.0 | - | 1.5 |
Cell Culture
COS cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% bovine calf serum (Invitrogen). The cells were maintained at 10% CO2 at 37°C. F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cells were grown on gelatinized plates in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% bovine calf serum and 2 mM glutamine. The cells were maintained at 10% CO2 at 37°C.
Cell Proliferation studies
F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cells were plated on 12-wellplates at a density of 1 × 104 cells/well. The day after plating, cells were treated in triplicate wells with 100 μM ZnCl2, with or without 1 μM RA, and were again treated after 48 hrs. After various times in culture, cells were trypsinized and counted using an electronic particle counter (model: Coulter Z1; Beckman Coulter, Inc., Fullerton, CA). All cells were treated with ZnCl2 alone to ensure maximum expression of the exogenous Hoxa1. Error bars are indicative of Standard Error of the Mean (S.E.M.). Results are plotted as percentage of cell count of cells treated with RA and zinc compared to cells treated with zinc alone.
Northern Analysis
All experiments were done in the presence of 100 μM zinc chloride to ensure maximum expression of exogenous Hoxa1. Experiments done in the absence of zinc are not discussed as the metallothionein promoter driving the expression of exogenous Hoxa1 has been found to be active in the absence of zinc (Fig. 1b), presumably due to trace amounts of zinc present in the cell culture media [Goliger and Gudas, 1992]. F9 Wt, F9-399-83, F9-399-72, and F9-93-10 cells were treated with 100 μM zinc chloride with or without 1 μM all-trans-retinoic acid (RA) (cat #R2625, Sigma, St. Louis, MO) for 24, 48, and 72 hours. After treatment, cells were harvested and total RNA was isolated using RNA Stat60 (Tel-Test, Inc.) according to the manufacturer’s instructions. RNA samples were separated on 1% agarose, 2.2 M formaldehyde gels, transferred to nylon membranes, and UV crosslinked using UV Stratalinker 2400 (Stratagene, La Jolla, CA). Membranes were incubated with radiolabeled probes in standard hybridization buffer (5X SSC, 5 mM EDTA, 50 mM phosphate buffer, 5X Denhardt’s solution, 0.1% SDS, 10% dextran sulfate, 100μg/ul denatured salmon sperm, 50% formaldehyde) for 24 hours at 42°C. After hybridization, membranes were washed two times for 15 min each at 42°C in low stringency wash (2x SSC, 0.1% SDS) followed by one high stringency wash (0.2x SSC, 0.1% SDS) for 30 minutes at 60°C. After washing, the membranes were exposed to Kodak BioMax films or PhosphorImager analysis (Amersham Biosciences, Piscataway, NJ).
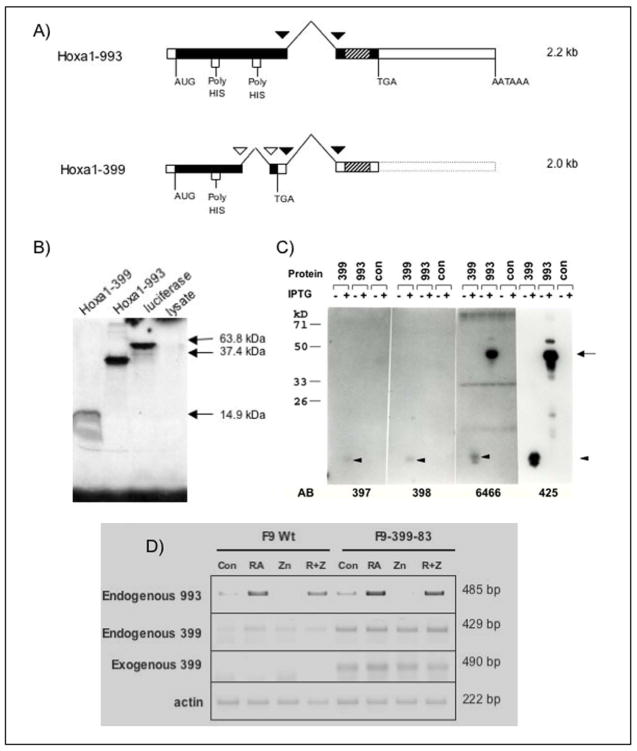
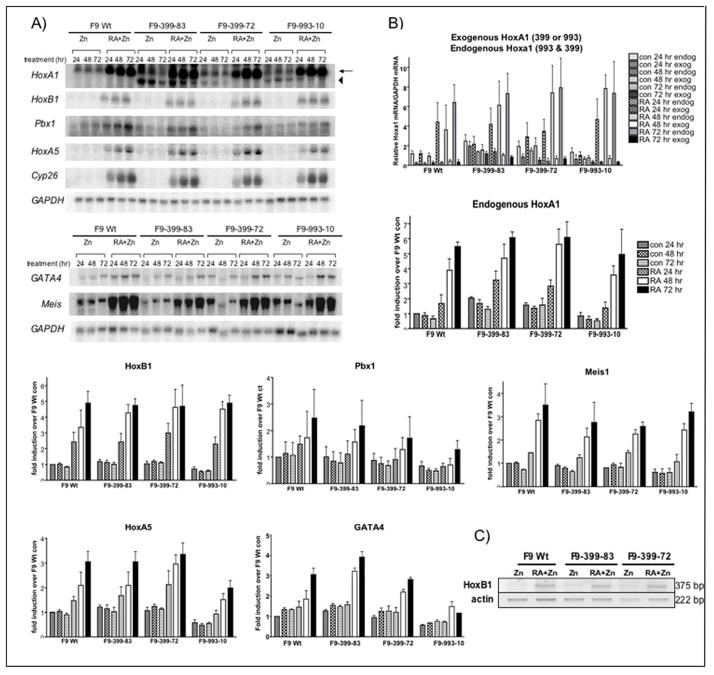
Figure 1. Splicing patterns of Hoxa1-993 and Hoxa1-399 cDNAs.
(A) Due to alternative splicing, two transcripts of Hoxa1 (previously Era-1) exist. Both transcripts share common splice sites, indicated by black triangles. However, Hoxa1-399 undergoes an additional splicing event, indicated by white triangles, causing a shift in the reading frame. This results in a truncated protein which lacks the homeodomain and a portion of the poly-His region that is present in Hoxa1-993. Hatched rectangles indicate the homeobox. (B) In vitro translated Hoxa1-993 and Hoxa1-399 detected by SDS-PAGE and subsequent radiography. Hoxa1-993 and Hoxa1-399 electrophoresed at the expected sizes of ~36 kDa and ~15 kDa, respectively. A luciferase expression construct was used as a positive control for the in vitro transcription and translation reaction. The luciferase protein electrophoresed at the expected size of 61 kDa. The rabbit reticulocyte lysate used for the in vitro transcription reaction was included as a negative control. (C) Hoxa1-993 and Hoxa1-399 proteins detected by western blot from IPTG-induced recombinant protein expressed in bacteria. The empty bacterial expression vector was included as a negative control. The primary antibodies used in these experiment were affinity-purified, polyclonal guinea pig or rabbit anti-mouse Hoxa1 antibodies. Antisera 6466 and 425 are specific to two different regions of the N-terminus of the Hoxa1 protein and recognize both Hoxa1-993 and Hoxa1-399. Antisera 397 and 398 are specific to the C-terminus of Hoxa1-399 and recognize Hoxa1-399 only. The proteins were immunoprecipitated with these antibodies and then electrophoresed. These experiments were performed at least two times with similar results. Arrow, expression of Hoxa1-993 protein; Arrowhead, expression of Hoxa1-399 protein. (D) RT-PCR analysis of exogenous and endogenous Hoxa1 transcripts in F9 Wt and F9-399-83 cells after 48 hr treatment with 1 μM RA and/or 100 μM ZnCl2. These assays were performed three times with similar results obtained.
Results of Northern blot assays were normalized to GAPDH expression as this gene is constitutively expressed in these cell lines. Data analysis was performed by the computer software program NIH Image 1.62 or Imagequant and graphed as fold induction over results in F9 Wt zinc treatment at 24 hr except exogenous Hoxa1 which was graphed as exogenous Hoxa1 mRNA levels over GAPDH mRNA levels. Statistical analysis was performed by T-test using GraphPad Prism 4.0. For the statistical analysis of northern blot assays, normalized values of stably transfected F9 samples were compared to normalized values of F9 Wt cells at the same time point and respective treatment. Significance indicates a p value of <0.05.
The following cDNA probes were used in the Northern blot assays: Hoxa1 cDNA was isolated as a 1-kb fragment, initially isolated as ERA-1-993, inserted into the EcoRI site of the pSG5 vector. Hoxb1 was excised as a 435-bp EcoRI-HindIII fragment from pGEM72f-Hoxb1. For Pbx1, a 750-bp EcoRI-NcoI fragment was isolated from pBS-Pbx1 (obtained from Drs. Dwaine Wright and Mark Kamps, UC San Diego, San Diego, CA). Meis1 cDNA was excised as a 1.2-kb EcoRI fragment from pMSCV-Meis1a (gift from Drs. Scott Steelman and Arthur M. Burchberg). Hoxa5 cDNA was isolated as a 637-bp fragment from the EcoRI-BglII site of the pUC18-Hoxa5 vector (obtained from Dr. W.F. Odenwald). GATA4 cDNA was isolated as a 1.9-kb fragment excised from pBluescript (obtained from Dr. Stuart H. Orkin). Hoxc9 was isolated from the EcoRI site of the pCRII vector. For GAPDH, a 280-bp fragment was excised from the PstI site of pGEM5.
Semi-quantitative RT-PCR
F9 Wt, F9-399-83, F9-399-72 cells were treated with 1 μM RA and/or 100 μM zinc chloride for 48 or 72 hours. Total RNA was isolated from the cells using RNA Stat 60. RNA (5μg) was reverse transcribed using Superscript III (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and random primed using 1μg random hexanucleotides. The cDNA product was diluted 1:10 with sterile, ultrapure water. The diluted cDNA (2 μL) was amplified in the linear range by PCR using the primer pairs listed in Table 2. For the exogenous Hoxa1-399, 30 cycles with an annealing temperature of 60°C were performed. For endogenous Hoxa1-993, endogenous Hoxa1-399, Hoxb1, and β-actin, touch-down protocols were employed that consisted of 19 cycles with a decreasing annealing temperature of 70 to 60°C, followed by 12, 10, 10, and 2 cycles, respectively, with an annealing temperature of 60°C.
Table 2.
Primer Pairs used for Hoxa1 and Hoxb1 transcript detection
| Endogenous Hoxa1-993 | 5′-TGG AGG AAG TGA GAA AGT TGG C -3′ (forward) 5′-ATG GGA GTC GAG AGG TTT CC -3′ (backward) |
| Endogenous Hoxa1-399 | 5′-TGG AGG AAG TGA GAA AGT TGG C -3′ (forward) 5′-TTC TTG GTG GGT CTG CTT CC -3′ (backward) |
| Exogenous Hoxa1-399 | 5′-TCT CCT CAC TTA CTC CGT AGC TCC -3′ (forward) 5′-TTC TTG GTG GGT CTG CTT CC -3′ (backward) |
| Endog Hoxa1 (spans intron) | 5′-TGG AGG AAG TGA GAA AGT TGG C -3′ (forward) 5′-AGA AGA CGT CTC TGA AGC AGG-3′ (backward) |
| Endog Hoxa1-993 (spans intron) | 5′-TAA CTC CTT ATC CCC TCT CCA C-3′ (forward) 5′-TTC TCC AGC TCT GTG AGC TGC TTG GTG G-3′ (backward) |
| β-actin | 5′-AAG TGT GAC GTT GAC ATC CG -3′ (forward) 5′-GAT CCA CAT CTG CTG GAA GG -3′ (backward) |
| Hoxb1 | 5′-CTC CGC CGC AGC CCC CAT AC –3′ (forward) 5′-TCG CTC GCG TTT CTT CTG CTT CAT –3′ (backward) |
| 36B4 | 5′-AGA ACA ACC CAG CTC TGG AGA AA –3′ (forward) 5′-ACA CCC TCC AGA AAG CGA CAG T –3′ (backward) |
Transient Transfections and Reporter Assays
F9 Wt and F9-399-83 cells (1×106) were plated on gelatinized 100 mm tissue culture plates for transfection by the calcium phosphate method. pGL3-wtARE or pGL3-promoter (8 μg) (Promega, Madison, WI), 4 μg pSV-Hoxa1-993, 4 μg pCS2-Pbx1a, and 1μg Renilla/Luciferase (as an internal control) constructs were transfected. pSG5 was used in appropriate amounts when necessary to normalize for the amount of DNA used in each experimental condition. Briefly, DNA in a total volume of 420 μl 1 mM Tris, pH 7.4, was mixed with 60 μl 2M CaCl2 by gentle vortexing. The DNA CaCl2 mixture was then added to an equal volume of 2x Hepes Buffered Saline [280 mM NaCl, 50 mM Hepes, 1.5 M Na2HPO4], pH 7.05, by bubbling, and incubated at room temperature for 30 minutes before being added dropwise to the cells. After 12–15 hours, the cells were washed twice with TBS [25 mM Tris, 137 mM NaCl, 5 mM KCl, 0.7mM CaCl2, 0.5 mM MgCl2, 0.6 mM Na2HPO4], pH 7.5, and incubated in regular formulated media for 24 hours before being harvested.
For transient transfection of COS cells, the DEAE-dextran method was used. DNA amounts were the same as those used for transient transfection of F9 cells (described above). Cells (2 ×106) were plated in 100 mm dishes 24 hrs hours prior to transfection. DNA was brought to a final volume of 1.9 ml in PBS and mixed with 100 μl of 10 mg/ml DEAE-dextran. Cells were washed twice with PBS before the DNA-DEAE mixture was added dropwise. After incubating for 30 minutes, 8 ml media containing 80 μM chloroquine was added to the cells. The cells were then incubated for an additional 2.5 hrs. The media containing chloroquine was removed and the cells were then incubated at room temperature with media containing 10% DMSO for 2.5 minutes. At that time, the media containing DMSO was removed, regular formulated media was added, and the cells were incubated for an additional 24 hours before harvesting. Semi-quantitative RT-PCR analysis was performed to confirm the efficiency of the transfection of COS cells. Total RNA was isolated and reverse transcribed as described above (see Semi-quantitative RT-PCR). cDNA (2 μl) was amplified by PCR using the primer pairs listed in Table 3. For all genes examined, 20 cycles with an annealing temperature of 60°C were performed.
Table 3.
Primer Pairs used for transiently transfected COS
| Hoxa1-993 | 5′-CCC CTC TGA CCA TGG GAT TAC AAC TT -3′ (forward) 5′-ATG GGA GTC GAG AGG TTT CC -3′ (backward) |
| Hoxa1-399 | 5′-CCC CTC TGA CCA TGG GAT TAC AAC TT -3′ (forward) 5′-TTC TTG GTG GGT CTG CTT CC -3′ (backward) |
| Pbx1 | 5′-GGA CAT CGG GGA CAT TTT ACA GCA -3′ (forward) 5′-GTA GTA GCA TCC TGC CAA CCT CCA TT -3′ (backward) |
Luciferase assays were conducted using the Dual Luciferase Reporter Assay kit (Promega, Madison, WI, cat # E1960) according to the manufacturer’s instructions. Luciferase assays were monitored using the TD-20/20 Luminometer (Turner Designs). Results of the luciferase assays were normalized to values obtained for pGL3-wtARE transfected alone into F9 Wt or COS cells. Statistical analysis was performed by 2-way analysis of variance using GraphPad Prism 4.0. Normalized values of F9-399-83 cells were compared to normalized values of F9 Wt cells at the same time experimental condition. Significance indicates a p value of <0.05
GST-Pull Down Assays
Radiolabeled, in vitro translated proteins were synthesized from pSV-993S, pSV-399S, and pCS2-Pbx1a plasmids with [35S]-cysteine (1000 Ci/mmol) (cat # SJ232, GE Healthcare) using TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI, cat # L5020) according to the manufacturer’s protocol. BL21 bacteria were transformed with pGEX-Hoxa1 constructs. A single colony was inoculated and grown in LB for approximately 16 hours at 37°C. The culture was then diluted 1:20 into one liter of LB. When the OD600 reached 0.6, 10 ml of 50 mM isopropyl-β-D-thiogalactopyranoside was added to the culture for 2–4 hours to induce expression of fusion proteins. The culture was then centrifuged at 6000 rpm at 4°C for 5 minutes and subsequently resuspended in lysis buffer [PBS, 1 mM phenylmethylsulfonyl fluoride, 0.5 μg/ul leupeptin, 1mM DTT], except GST-399, which was supplemented with 0.5% sarkosyl (Sigma) to increase protein solubility. Suspensions were sonicated 3 times for 10 seconds on ice at output level 3 (Branson Sonifier 150) and microcentrifuged at 14,000 rpm for 5 minutes at 4°C to isolate soluble proteins. The supernatants (soluble protein) were then collected. To increase binding of solubilized proteins to glutathione agarose, 0.3% and 4% Triton-X 100 was added to GST-993 and GST-399 lysates, respectively, and rocked for 30 minutes at 4°C. Recombinant GST, GST-993 and GST-399 were immobilized on 50 μl bed volume of glutathione Sepharose 4B beads (Amersham Biosciences, cat # 17-0756-01) by rocking for 1 hr at 4°C. After washing 3 times with PBST and equilibration in GST binding buffer (20 mM Tris pH 7.9, 100 mM NaCl, 0.1% Tween, 10% glycerol), the amount of fusion protein bound to glutathione beads was determined by comparison with BSA protein concentration standards by SDS-PAGE and subsequent coomasie blue staining.
GST-fusion proteins (10 μg) bound to glutathione beads were incubated with 10 μl in vitro translated Hoxa1-993, Hoxa1-399, or Pbx1a and rocked for 1 hour at 4°C in GST binding buffer. After washing three times at 4°Cby rocking with PBST, thebound proteins were resolved by SDS-PAGE. The gel was treated with ENHANCE (cat # 6NE9701, Perkin Elmer, Boston, MA) according to the manufacturer’s protocol and visualized by phosphorimager and autoradiography. Data analysis was performed by the computer software programs NIH Image 1.62 or ImageQuant v1.11 and graphed as percentage of protein bound compared to input controls. Statistical analysis was performed by unpaired T-test using GraphPad Prism 4.0.
Electrophoretic Mobility Shift Assays
Oligonucleotides used in EMSA were obtained commercially fromMWG Biotech (USA) and correspond to the R3 region of the Hoxb1 ARE [5′-GGGGGGTGATGGATGGGC GCTGC-3′] [Ferretti et al., 2005]. Oligonucleotides were resuspended and diluted in Tris-EDTA. Double-stranded oligonucleotides wereprepared by mixing equal molar ratios of single-stranded oligonucleotides. The mixture was heated at 90°C for 5 min and slowly cooled to room temperature.
In vitro translated proteins were synthesized from 2 μg pSV-993S, pSV-399S, and pCS2-Pbx1a plasmids using the TNT Quick Coupled Reticulocyte Lysate system (Promega) according to the manufacturer’s protocol. [35]S-cysteine- labeled proteins were prepared in parallel, run on a 15% acrylamide gel, and analyzed by phosphorimager and autoradiography to determine translation efficiency. The amounts of proteins were normalized for the amount of cysteine of each protein. Equal amounts of proteins (1 μl of Hoxa1-993, 1 μl Pbx1 and 6 or 12 μl Hoxa1-399) were incubated on ice with 40,000 cpm [32P]-labeled oligonucleotide [5′ GGGGGTGATGGATG GGCGCTGC 3′] in binding buffer (10 mM Tris pH 7.5, 75mM NaCl, 1 mM EDTA, 6% glycerol, 3 mM spermidine, 1 mM DTT, 0.5 mM PMSF, 1 μg poly dI-dC) in a total volume of 20 μl for 30 minutes. The amount of reticulocyte lysate added to each reaction was adjusted in order to normalize for the amount of translated protein products in each experiment. The reaction mixture was separated on a 6% non-denaturing polyacrylamide gel in 0.5x Tris-buffered EDTA. After gel drying, the DNA-bound proteins were detected by phosphorimager and autoradiography. Quantitation was performed using ImageQuant v1.11. Statistical analysis was performed using the software GraphPad Prism v4.0 by an unpaired T-test comparing band intensity of the band corresponding to the 993/Pbx1 complex in the absence of 399 (Fig. 3, lane 2) to all other experimental conditions. Error bars are indicative of the S.E.M.
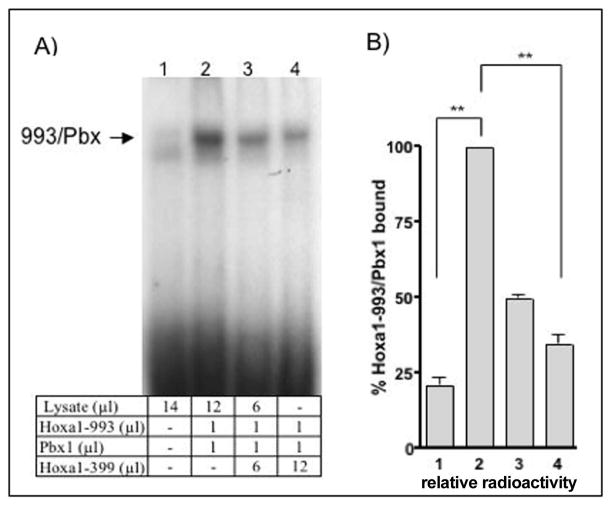
Figure 3. Hoxa1-399 inhibits the formation of a DNA-bound Hoxa1-993/Pbx.
(A) EMSA was performed using equal units of in vitro translated Hoxa1-993, Pbx1 and increasing amounts of Hoxa1-399 incubated with 32P-labeled DNA corresponding to the HoxB1-ARE-R3 region of the HoxB1 promoter. Lysate lane (1) indicates endogenous protein present in the rabbit reticulocyte lysate used for the in vitro transcription. (B) Quantitation of EMSA band intensities depicted in (a). Data was normalized to the intensity of the band corresponding to the 993/Pbx complex (lane 2). Error bars are indicative of S.E.M. Data representative of three independent experiments except lane 3 where n=2. ** very significant; indicates a p value of <0.01, when compared to intensity of 993/Pbx1 band.
RESULTS
Differential splicing of Hoxa1
RA transcriptionally activates the Hoxa1 gene, the most 3′ gene of the Hox A homeobox gene cluster, in F9 teratocarcinoma stem cells [LaRosa and Gudas, 1988a]. The Hoxa1 gene encodes two alternatively spliced mRNAs which can direct the synthesis of two distinct proteins, one that contains the homeodomain (Hoxa1-993), and a second, truncated protein that lacks the DNA binding domain (Hoxa1-399). Using this nomenclature, the numbers −993 and −399 correspond to the amino acid lengths of the respective open reading frames. The splicing event causes a shift in the reading frame, resulting in an earlier TGA stop codon in the Hoxa1-399 transcript. Therefore, the translation of the Hoxa1-399 transcript results in a truncated protein that lacks the homeodomain (Fig 1a). The Hoxa1-993 transcript encodes a protein of 36 kDa, while the truncated Hoxa1-399 transcript encodes a15 kDa protein. The two proteins are identical for the first 114 amino acids but have unique caroboxy-termini [LaRosa and Gudas, 1988b]. The functions of the smaller Hoxa1 protein, which lacks the homeodomain, are not known.
Hoxa1 protein expression
Hoxa1-993 and Hoxa1-399 were in vitro transcribed and translated using the pSV-993S and pSV-399S plasmids, respectively, as described in Materials and Methods. The cDNA encoded by each of these plasmids contains the full-length coding sequence for each protein. The apparent molecular masses of the mouse Hoxa1 proteins, determined by acrylamide gel electrophoresis, are similar to the calculated molecular masses [LaRosa and Gudas, 1988b]. In vitro translated Hoxa1-993 and Hoxa1-399 electrophoresed at the expected sizes of 36 kDa and 15 kDa, respectively (Fig. 1b). The molecular masses of both Hoxa1-993 and Hoxa1-399 were also confirmed by bacterial expression of the proteins, followed by immunoprecipitation using affinity purified, polyclonal guinea pig or rabbit anti-mouse Hoxa1 antibodies (Fig. 1c).
Generation of F9 cells which stably express Hoxa1-399 or Hoxa1-993
To determine the effects of increased expression of Hoxa1-399 on the expression of known Hoxa1 target genes, F9 Wt teratocarcinoma stem cells were stably transfected by electroporation with pMT-399S to express ectopically Hoxa1-399 under the regulation of the metallothionein promoter. We selected the metallothionein promoter for inducible expression because we wanted to be able to compare directly these lines with the F9-993-10 line, which we had generated earlier (see below). These cell lines, referred to as F9-399-83 and F9-399-72, ectopically express Hoxa1-399 upon the addition of zinc chloride to the cell culture media. A similar cell line, F9-993-10, was previously generated by electroporation of pMT-993S into F9 Wt cells to express Hoxa1-993 ectopically in response to zinc chloride [Goliger and Gudas, 1992]. We previously showed that 100 μM zinc chloride had no effects on cell proliferation rates or on RA-regulated gene expression [Goliger and Gudas, 1992].
The exogenous Hoxa1 message that is ectopically expressed in the F9-399-83, F9-399-72, and F9-993-10 cell lines is shorter than the endogenous Hoxa1 transcript because a portion of the Hoxa1 3′ UTR was removed. The endogenous Hoxa1-993 and Hoxa1-399 messages are 2.2 kb and 2.0 kb, respectively, while the exogenous Hoxa1-399 and Hoxa1-993 messages are both 1.5 kb (Table 1). By Northern blot analysis, the endogenous Hoxa1 transcripts (Hoxa1-993 and Hoxa1-399) versus the exogenous Hoxa1 transcripts can be distinguished and separated on the basis of their different sizes. By RT-PCR, all four different transcripts can also be easily distinguished. RT-PCR analysis of the F9 Wt and the F9-399-83 cell lines, treated for 48 hr with RA and/or ZnCl2, showed exogenous Hoxa1-399 transcripts in the F9-399-83 cell line, regardless of treatment, even in the absence of zinc (Fig. 1d). The expression of the exogenous transcript in the absence of zinc may have resulted from the activity of the metallothionein promoter under these culture conditions [Goliger and Gudas, 1992; Kim and Wold, 1985; Pecorino et al., 1988]. Endogenous Hoxa1-993 mRNA was similarly induced in the F9 Wt and F9-399-83 cell lines in response to RA with and without zinc treatment (Fig. 1d). Interestingly, RT-PCR analysis of RA and/or zinc treated F9 Wt and F9-399-83 cells indicated that ectopic expression of exogenous Hoxa1-399 in F9-399-83 cells increases the expression of endogenous Hoxa1-399 compared to that in F9 Wt cells (Fig. 1d). By RT-PCR analysis, the mRNA levels of endogenous Hoxa1-399 and exogenous Hoxa1-399 are quite similar after a 48 hr treatment of RA and/or zinc. These data were confirmed in both F9-399 cell lines using different primer pairs that spanned the Hoxa1-399 intron (Table 2), verifying that the increased expression of endogenous Hoxa1-399 in F9-399-83 cells did not result from genomic DNA contamination of the RNA samples (data not shown).
Hoxa1-399 inhibits a luciferase reporter driven by the Hoxb1 SOct-R3 motif in F9 cells
Hoxa1-993 has been shown to play a vital role in the transcriptional activation of Hoxb1 at a specific region of the Hoxb1 promoter. Three evolutionarily conserved, related sequence motifs (R1, R2, and R3) have been identified in the promoter region of the Hoxb1 gene that are necessary for proper Hoxb1 transcription in the developing hindbrain [Popperl et al., 1995]. These conserved elements have evolved to form an auto-regulatory loop (b1-ARE) for Hoxb1 expression and are made up of three consensus Hox/Pbx sites [Studer et al., 1998]. On the b1-ARE, and specifically in the R3 region within it, only a subset of Hox proteins (Hoxa1, Hoxb1, and Hoxb2) can bind Pbx1 to activate transcription [Di Rocco et al., 1997]. Additionally, it has been demonstrated that in vivo, Hoxa1 is the primary mediator responsible for the RA-induced expression of Hoxb1 through the b1-ARE [Di Rocco et al., 2001]. Other published studies have shown that mutation of the SOct motif within the b1-ARE significantly suppresses the transcriptional activation of Hoxb1 by Hoxa1/Pbx1 [Di Rocco et al., 2001] (Fig. 2a).
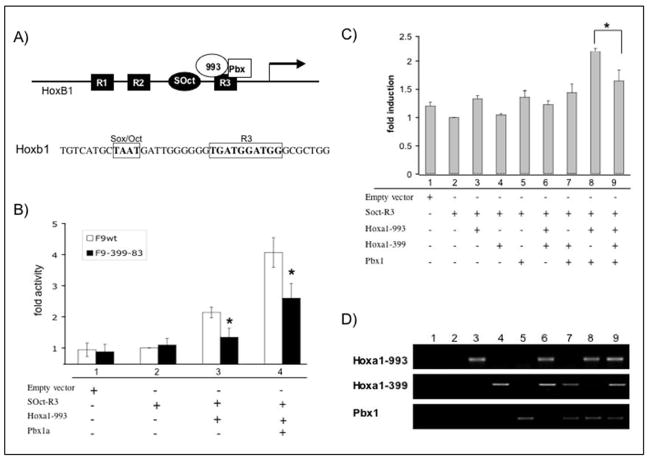
Figure 2. Luciferase activity driven by the SOct-R3 region of the HoxB1 promoter is reduced by Hoxa1-399.
(A) (top) Diagram illustrating the region of the Hoxb1 promoter that is known to be regulated by Hoxa1. The Hoxb1 auto-regulatory element is known to contain three related sequence motifs, R1, R2, and R3, and a Sox/Oct binding site (SOct). Hoxa1/Pbx1 bind the R3 region to aid in the establishment of Hoxb1 expression in rhombomere 4. (bottom) Sequence of Hoxb1 promoter containing the Sox/Oct and R3 regions. (B) Fold induction of luciferase activity assayed from transiently transfected F9 Wt and F9-399-83 cells. Cells were transfected with 8 μg reporter construct together with 4 μg expression vectors for Hoxa1-993 and Pbx1a where indicated. In all experiments, 1μg renilla/luciferase was co-transfected as internal control. Bars indicate SEM of four independent experiments. Significance * indicates a p value of <0.05 comparing normalized values of F9-399 cells to normalized values of F9-Wt cells at the same time experimental condition. (C) Fold induction of luciferase activity assayed from transiently transfected COS cells. Cells were transfected with 8 μg reporter construct together with 4 μg expression vectors for Hoxa1-993, Hoxa1-399, and Pbx1a where indicated. In all experiments, 1 μg renilla/luciferase was co-transfected as internal control. Bars indicate S.E.M. of four independent experiments. Significance * indicates a p value of <0.05 when comparing fold induction of cells transfected with reporter, Pbx1, and Hoxa1-993 versus cells that were transfected with Hoxa1-399 as well (lanes 8 versus 9). (D) RT-PCR analysis of transfected COS cells described in (c).
We next wanted to determine if Hoxa1-399 could play a role in the transcriptional regulation of Hoxa1 target genes in the presence of Hoxa1-993. We used the transcriptional activation of a small portion of the Hoxb1 promoter region as a model system. To address this question, a reporter construct was generated, named pGL3-wtARE, which contains the SOct-R3 region (TAATGATTGGGGGGTGATGGATGG) (Fig. 2a) of the Hoxb1 promoter driving the expression of a luciferase reporter gene. As endogenous Hoxa1-399 mRNA is expressed upon RA treatment in the F9 Wt and stably transfected F9-399 cell lines and could potentially mask any effects of exogenous Hoxa1-399, this reporter construct was transiently transfected into F9 Wt and F9-399-83 cells in the absence of RA, together with expression constructs for Hoxa1-993 (pSV993) and Pbx1 (pCS2-Pbx1). Both cell lines show similar induction of luciferase activity when transiently transfected with the empty vector or the pGL3-wtARE reporter construct (Fig. 2b, lanes 1 and 2). Transient transfection of Pbx1, together with the reporter construct, showed no significant induction of luciferase activity (data not shown). However, when Hoxa1-993 was transiently transfected into the cells, together with the reporter construct, F9 Wt cells exhibited a 2.1-fold +/− 0.1 induction of luciferase activity. F9-399-83 cells showed a 37% (p=0.0221) (1.4-fold +/− 0.2) lower activation of the luciferase reporter gene compared to Wt cells (Fig. 2b, lane 3). In the presence of both Hoxa1-993 and Pbx1, the Wt cells induced luciferase activity by 4.1-fold +/− 0.4, while F9-399-83 cells induced luciferase activity by 2.6-fold +/− 0.4 (Fig. 2b, lane 4), indicating a 36% (p=0.0279) inhibition of the transcriptional activation of luciferase in cells that ectopically express Hoxa1-399. The results from these experiments indicate that transactivation of luciferase through the Soc-R3 region of the Hoxb1 promoter is lower in F9 cells that ectopically express Hoxa1-399.
Testing of the Hoxb1 SOct-R3 site in Transfected COS cells
Since the reporter assays were performed in two different cell lines (F9 Wt and F9-399), the transient transfections and reporter assays were repeated in COS cells for further confirmation. COS cells were transiently transfected with the same SOct-R3 reporter construct together with expression vectors for Hoxa1-993, Pbx1, and an additional construct to overexpress Hoxa1-339 (pSV399). RT-PCR analysis of transiently transfected cells confirmed the proper expression of transfected constructs in COS cells (Fig. 2d). Expression of Hoxa1-399 in COS cells inhibited luciferase reporter activity (1.7-fold induction +/− 0.2) by 25% (p=0.0393) as compared to cells that were transiently transfected with expression constructs for Hoxa1-993 and Pbx1 alone (2.3-fold induction +/− 0.1) (Fig. 2c, compare lanes 8 and 9). These results were similar to the results in F9 Wt and F9-399 cells and they show that ectopic expression of Hoxa1-399 inhibits the transactivation ability of Hoxa1-993 and Pbx1 through the SOct-R3 element in the Hoxb1 promoter.
Lower levels of DNA-bound Hoxa1-993/Pbx1 is detected in the presence of Hoxa1-399
We next sought to determine the mechanism by which Hoxa1-399 inhibits the transactivation ability of Hoxa1-993 and Pbx1 through an element in the Hoxb1 promoter. Because the Hoxa1/Pbx1 dimer is known to bind directly to the Hoxb1 promoter, we wanted to determine if Hoxa1-399 could affect this association by the use of electrophoretic mobility shift assays (EMSA). In vitro translated Hoxa1-993 and Pbx1 were incubated with [32P]-labeled oligonucleotides corresponding to the R3 region (TGATGGATGG) (Fig. 2a) of the Hoxb1 promoter. As expected, a protein-bound DNA complex was detected when Hoxa1-993 and Pbx1 were incubated with the DNA (Fig. 3a, lane 2). However, a lower level of the protein complex bound to the R3-ARE was seen in the presence of Hoxa1-399. As increasing amounts of in vitro translated Hoxa1-399 were added to the reaction, a corresponding decrease in the amount of protein bound to DNA resulted (Fig. 3, lanes 2–4). The amount of Hoxa1-993/Pbx1 bound to DNA was analyzed by radiography (Fig. 3b). When equal molar amounts of Hoxa1-993, Pbx1, and Hoxa1-399 proteins were incubated with DNA, the intensity of the band generated by Hoxa1-993/Pbx1 bound to DNA was 34.8% +/− 3.8 (p=0.0022) of the band formed in the absence of Hoxa1-399. In the presence of one half of the molar amount of Hoxa1-399 added to the reaction, the intensity of the band generated by Hoxa1-993/Pbx1 bound to DNA was 48.9% +/− 1.13 (p<0.0001) of the band formed in the absence of Hoxa1-399. These results suggest that in the presence of Hoxa-399, a significantly lower amount of Hoxa1-993/Pbx1 bound to the R3 region.
Hoxa1-399 and Hoxa1-993 interact in vitro
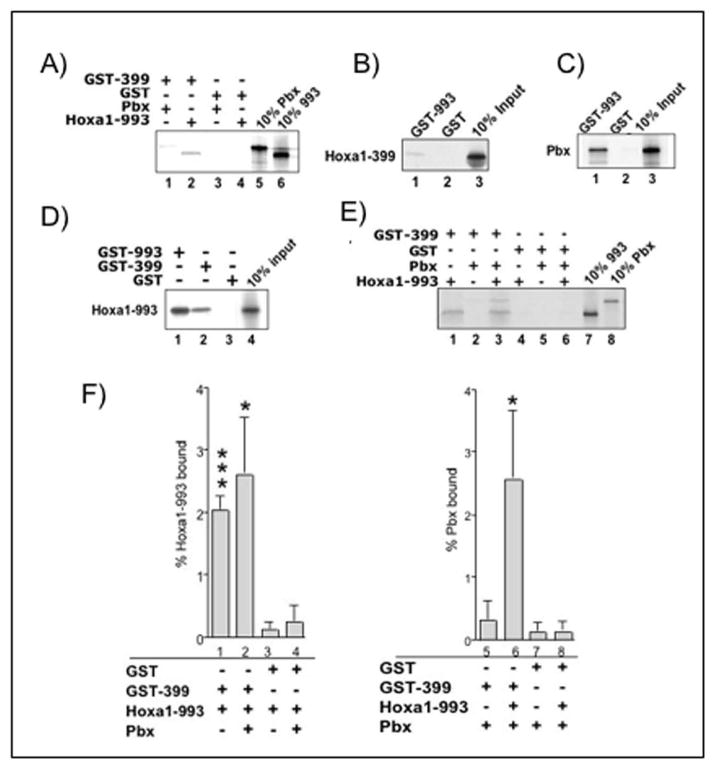
Through EMSA, we demonstrated that Hoxa-399 inhibits the binding of Hoxa1-993/Pbx1 to the R3 region in vitro. To further explore how this occurs, GST pull-down assays were performed to determine if Hoxa1-993 and Hoxa1-399 interact in vitro. GST-Hoxa1-399 (GST-399) was incubated with [35S]-labeled full-length Hoxa1-993. When compared to the amount of GST (negative control) that bound Hoxa1-993 (0.13% +/− 0.1 of input), more binding was detected between GST-399 and Hoxa1-993 (2% +/−0.3 of input, p=0.0008) (Fig. 4a, compare lanes 2 and 4, Fig. 4d, compare lanes 2 and 3, Fig. 4e, compare lanes 1 and 4, Fig. 4f, compare lanes 1 and 3). To confirm this interaction, the reverse experiment was performed using GST-Hoxa1-993 (GST-993) incubated with [35S]-labeled Hoxa1-399. As expected, GST-993 was able to bind Hoxa1-399, while GST alone was not able to bind, as GST-993 bound 8-fold more Hoxa1-399 that GST alone (Fig. 4b). These results demonstrate that Hoxa1-399 can directly interact with Hoxa1-993. Because these findings indicate a heterodimerization of Hoxa1-993 and -399, we next repeated similar GST pull-down experiments to determine if Hoxa1 proteins can homodimerize. Experiments performed using GST-993 incubated with [35S]-labeled Hoxa1-993 showed significant binding, indicating a homodimerization of Hoxa1-993 (Fig. 4d, lane 1). A similar experiment was done with GST-399 and radiolabeled Hoxa1-399, but no binding was detected (data not shown).
Figure 4. Hoxa1-993 and Hoxa1-399 form dimers in vitro.
(A,B,D) Hoxa1-399 binds Hoxa1-993 but not Pbx1a. GST, GST-Hoxa1-993, and GST-Hoxa1-399 fusion proteins (10 μg) bound to glutathione beads were incubated with radiolabeled Hoxa1-993, Hoxa1-399, or Pbx1a (10 μl). (C) GST-993 binds Pbx1a as previously reported. GST and GST-Hoxa1-993 fusion protein bound to glutathione beads were incubated with radiolabeled Pbx1a. (E) GST-Hoxa1-399 forms a trimer with Pbx1a and Hoxa1-993 by binding to Hoxa1-993. GST and GST-Hoxa1-399 fusion proteins bound to glutathione beads were incubated with radiolabel Hoxa1-993 and Pbx1a. Similar results were seen in at least two independent experiments. (F) quantitation of GST pull down experiment shown in e. Error bars are indicative of S.E.M. Data are representative of three independent experiments. *** very significant; indicates a p value of <0.001, * significant; indicates a p value of <0.05 when comparing % bound to GST-399 versus GST under the same experimental condition.
Hoxa1-399 interacts with the Hoxa1-993/Pbx1 complex by binding to Hoxa1-993
Hoxa1-993 has been shown to interact with Pbx1 via a tryptophan-containing pentapeptide motif found N-terminal to the homeodomain [Phelan et al., 1995]. Hoxa1-399 does not contain the pentapeptide motif [LaRosa and Gudas, 1988b], and therefore presumably does not bind to Pbx1. We tested whether Hoxa1-399 can interact with Pbx1 via GST pull-down assays using GST-399 incubated with [35S]-labeled Pbx1. As expected, no significant binding was detected between GST-399 and Pbx1 (0.32% +/−0.4 of input), and no binding was detected between Pbx1 and GST alone (0.13% +/−0.2 of input) (Fig. 4a, compare lanes 1 and 3, Fig. 4e, compare lanes 2 and 5, Fig. 4f, compare lanes 5 and 7). However, in the presence of both radiolabeled Hoxa1-993 and Pbx1, GST-399 was able to bring down both proteins (Fig. 4e, lane 3). GST-399 bound 8-fold more Pbx1 in the presence of Hoxa1-993 (p=0.06) (2.6% +/− 1.3 of input) (Fig. 4f, compare lanes 5 and 6). The presence of both Hoxa1-993 and Pbx1 had no significant effect on the affinity of GST-399 for Hoxa1-993 (p=0.2896) (Fig. 4f, compare lanes 1 and 2). The presence of both proteins also had no effect on the affinity of GST for Hoxa1-993 (0.26% +/− 0.2 of input) (p=0.3422) (Fig. 4f, compare lanes 3 and 4) or for Pbx1 (p=0.4770) (Fig. 4f, compare lanes 7 and 8). These results indicate that in vitro, Hoxa1-399 forms a trimer with Hoxa1-993 and Pbx1 by binding to Hoxa1-993.
Effects of ectopic expression of exogenous Hoxa1 on the expression of Hoxa1 and/or RA target genes
We next analyzed the effects of RA treatment on F9 cells that ectopically express exogenous Hoxa1-399 or Hoxa1-993. The expression of endogenous (399 and 993) versus exogenous (399 or 993) Hoxa1 mRNAs was examined by Northern analysis. F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cell lines were treated with 100 μM ZnCl2 in the presence or absence of 1 μM RA for 24, 48, and 72 hours. Exogenous Hoxa1-399 message was expressed in both F9-399 cell lines in response to zinc, with or without RA (Fig. 5a). Similarly, the exogenous Hoxa1-993 transcript was detected in F9-993-10 cells in response to zinc, with or without RA (Fig. 5a). F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cells all exhibited similar endogenous Hoxa1 (399 and 993) mRNA levels in response to a 72 hr treatment with RA as compared to F9 Wt cells. F9 Wt cells exhibited a 5.5-fold +/− 0.3 increase, the F9-399-83 cells showed a 6.1–fold +/−0.5 increase, the F9-399-72 cell line exhibited a 6.1–fold +/− 1.3 increase, and the F9-993-10 cell line exhibited a 5.0-fold +/− 1.7 increase in endogenous Hoxa1 (399 and 993) message in response to RA (Fig. 5a,b). Statistical analysis revealed no significant differences among F9 Wt and the two independently isolated, stably transfected cell lines (p values ranged from 0.1318–0.3833) for RA-induced Hoxa1 mRNA.
Figure 5. Characterization of F9 cell lines overexpressing Hoxa1-399 and Hoxa1-993.
(A) Total RNA from F9 Wt and the stably transfected cell lines F9-399-83, F9-399-72, and F9-993-10 treated with 100 μM ZnCl2 with or without 1 μM RA for the indicated times were subjected to Northern blot analysis and probed with various Hoxa1 and/or RA target genes. One representative is shown, although the experiments were performed three times (except where indicated) with different RNA preparations with similar results. Arrow, expression of the total endogenous Hoxa1. Arrowhead, expression of exogenous Hoxa1-399 mRNA. (B) Quantitation of the mRNA levels of Hoxa1 and/or RA targets by phosphorimager analysis from three independent experiments (except Pbx1 and Meis1 where n=2). Northern blots were normalized to GAPDH mRNA levels, and the data are shown as fold increase over the relative mRNA levels of F9 Wt zinc-treatment for 24 hrs. *** very significant; indicates a p value of <0.001, ** very significant; indicates a p value of <0.01,* significant; indicates a p value of <0.05 when comparing relative mRNA levels to F9 Wt under the same experimental condition. (C) RT-PCR analysis of the Hoxb1 transcript in F9 Wt, F9-399-83, and F9-399-72 cells after 72 hr treatment with 100 μM ZnCl2 with or without 1 μM RA.
Next, we wanted to determine if ectopically expressed Hoxa1-399 or Hoxa1-993 affects the mRNA levels of Hoxa1 target genes in response to RA. The F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cell lines were cultured in the presence of 100 μM ZnCl2 and 1 μM RA for 24, 48, or 72 hours. RNA was isolated and analyzed by Northern blot. The levels of RA-induced Hoxb1 mRNAs were similar at all time points in the transfected F9 cells as compared to F9 Wt, as determined by Northern analysis (Fig 5a,b). Quantitation of Northern blots of three separate, individual experiments showed that Hoxb1 mRNA levels were 2.4-fold +/−0.7 higher in F9 Wt after 24 hour treatment of RA plus zinc, while the F9-399-83 and F9-399-72 cell lines showed a 2.4-fold +/−0.7 (p= 0.4987) and 2.3-fold +/− 0.7 (p= 0.2639) increase, respectively, and F9-993-10 cells exhibited a 2.3-fold +/− 0.5 (p=0.4396) increase. At 72 hours of treatment with RA+zinc, F9 Wt cells showed a 4.9-fold +/− 0.9 induction while F9-399-83 and F9-399-72 cells exhibited a 4.7–fold +/− 0.5 (p=0.4309) and 4.7-fold +/− 1.6 (p=0.4485) induction, respectively, and F9-993-10 cells showed a 4.9-fold +/− 0.6 (p=0.4994) induction. These results indicate that ectopic expression of Hoxa1-399 or Hoxa1-993 had no effect on the levels of Hoxb1 mRNA, with or without RA treatment. These results for ectopic expression of Hoxa1-399 on Hoxb1 mRNA expression were confirmed by RT-PCR (Fig. 5c) and quantitative real-time PCR (data not shown).
We also analyzed the effects of ectopic expression of Hoxa1-399 or Hoxa1-993 on the mRNA levels of other RA-regulated genes (Pbx1, Hoxa5, Meis, Cyp26) and genes previously identified as Hoxa1 targets (Hoxc9, GATA4) [Martinez-Ceballos et al., 2005]. Results from Northern blot experiments indicated that ectopic expression of Hoxa1-399 or Hoxa1-993 in F9 cells had no effect on the mRNA levels of these target genes (Fig. 5a,b).
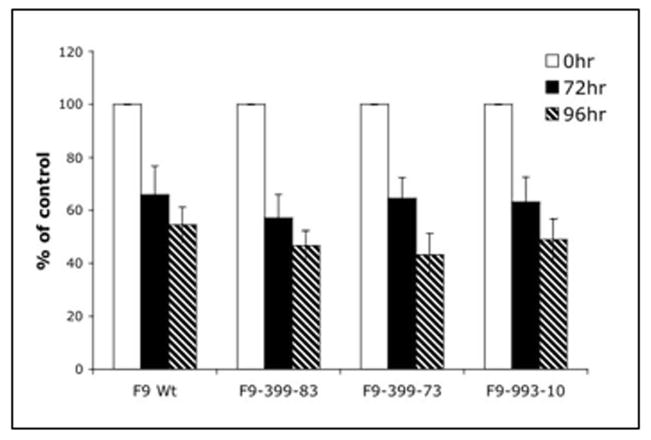
Characterization of the functions of Hoxa1-399 and Hoxa1-993 in F9 cells: Cell Proliferation
ES cells that lack a functional Hoxa1 are somewhat resistant to the antiproliferative effects of RA [Martinez-Ceballos et al., 2005]. Therefore, we next performed cell proliferation studies to determine the effects of ectopic expression of Hoxa1-399 or Hoxa1-993 on the anti-proliferative properties of RA. For the proliferation analyses, F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cells were treated with 100 μM ZnCl2 with or without 1 μM RA for 0, 72, and 96 hours. The cell numbers of F9 Wt cells treated with RA and zinc for 72 hours was 66% +/− 10.8 of control, untreated cells (Fig. 6). In comparison, the cell numbers of F9-399-83 and F9-399-72 cells treated with RA were 57% +/− 8.8 (p=0.4987) and 65% +/− 7.8 (p=0.2639) of control, untreated cells, respectively. Similarly, the cell numbers of F9-993-10 cells treated with RA were 63% +/− 9.4 (p=0.4396) of control. After 96 hours of RA treatment, no significant differences were seen among F9 Wt and the two stably transfected F9 cell lines as the cell numbers of F9 Wt, F9-399-83, F9-399-72, and F9-993-10 cells were 57% +/− 6.7, 47% +/− 5.7 (p=0.4309), 43% +/− 7.8 (p=0.4485), and 49% +/− 7.7 (p=0.4994), respectively, of the control cell numbers. Therefore, ectopic expression of exogenous Hoxa1-399 or Hoxa1-993 does not alter the antiproliferative effect of RA.
Figure 6. Inhibition of cell proliferation in response to RA treatment in F9 Wt and stably transfected cells.
Cells were plated on 12-well dishes at a density of 1×104 cells/well. The day after plating, 0 hr cells were harvested and counted. Cells were treated with 100 μM ZnCl2 with or without 1μM RA at 0 and 48 hr and harvested at the indicated times. The results are plotted as the percentage of cell proliferation versus RA treatment over time. Data points are the means of triplicate samples. The experiments were performed three times, starting with fresh cells. Error bars are indicative of SEM.
DISCUSSION
The expression of a truncated Hox protein lacking a homeodomain is not unique to Hoxa1. For example, an alternatively spliced transcript for Hoxa9 has been detected by RT-PCR. The alternative splicing causes a frameshift, resulting in the translation of a truncated protein that lacks the homeodomain [Fujimoto et al., 1998]. Differentially spliced transcripts resulting in Hox proteins that lack homeodomains have also been described for human HOXA1, murine Hoxb6, human HOXB6, and Xenopus XlHbox2 [Fujimoto et al., 1998; Hong et al., 1995; Shen et al., 1991; Wright et al., 1987]. The functions of these truncated Hox proteins that lack homeodomains have not been studied in depth. However, the truncated HOXB6 protein is expressed in undifferentiated keratinocytes and the full-length protein is induced by differentiation [Komuves et al., 2000]. Moreover, full-length HOXB6 is localized to the nucleus, while the truncated HOXB6 protein is cytoplasmic [Komuves et al., 2000]. The same protein localization patterns have been described for HoxA9 [Dintilhac et al., 2004]. Additionally, the Drosophila melanogaster homothorax (hth) gene also encodes two protein isoforms, one that contains the homeodomain and another truncated protein that does not. These two proteins carry out distinct functions during development [Noro et al., 2006]. Both proteins are capable of forming functional hth/Exd complexes (vertebrate ortholog of Meis1/Pbx complex), but with putative distinct transcriptional properties [Noro et al., 2006]. These data indicate that truncated Hox proteins that lack homeodomains may function in regulating differentiation. Previously Hoxa1-993 was shown to be required for proper differentiation of ES cells [Martinez-Ceballos et al., 2005] and F9 cells [Shen et al., 2000]. In this study, we sought to determine the role of Hoxa1-399 in the differentiation of F9 stem cells induced by RA.
Protein expression of Hoxa1-993 and Hoxa1-399
The molecular masses of Hoxa1-993 and Hoxa1-399 were confirmed by in vitro translation and immunoprecipitation (Fig. 1). Though we could detect the proteins when bacterial expression vectors were employed (Fig. 1), we were unable to detect the endogenous proteins in F9 Wt or the stably transfected cell lines using the antibodies that we generated against the specific isoforms of Hoxa1 (data not shown). Previously, our laboratory had detected Hoxa1-993 in F9 cells that overexpress Hoxa1-993 [Goliger and Gudas, 1992]. The endogenous Hoxa1-993 protein has also been detected in MCF7 cells [Mohankumar et al., 2007; Zhang et al., 2006; Zhang et al., 2003].
Alternative splicing of Hoxa1 and cell differentiation
Nothing is known about the mechanisms governing the alternative splicing of the Hoxa1 mRNA. Hoxa-399 was not detected in untreated COS cells that were transiently transfected for 24 hours with an expression vector for Hoxa1-993 (Fig. 2d). These experiments suggest that the splicing of Hoxa1 mRNA is dependent on RA treatment, 24 hours of Hoxa1-993 expression is not sufficient for splicing to occur, and/or that COS cells do not express the factors that are necessary for the splicing of Hoxa1. An unspliced transcript of Hoxb6 which encodes a truncated protein lacking a homeodomain is expressed earlier during differentiation and a spliced transcript, which encodes the full-length protein, at later stages of differentiation [Komuves et al., 2000; Shen et al., 1991]. These data support the concept that the splicing of Hox transcripts is dependent and regulated by factors that are involved in cellular differentiation.
Transcriptional activation through the SOct-R3 region of the Hoxb1 promoter is reduced in F9 cells that ectopically express Hoxa1-399 in the absence of RA
We performed luciferase assays using the Soct-R3 region of the HoxB1 promoter to drive the expression of the reporter gene in F9 Wt and F9-399 cells. F9-399 cells exhibited a reduced transcriptional activation of Hoxb1 compared to F9 Wt cells (Fig. 2). It should be highlighted that these experiments were performed in the absence of RA. Transiently transfected F9 Wt and F9-399 cells that were treated with RA for 24 hrs exhibited a decreased transcriptional activation of a reporter gene driven by the SOct-R3 region of the Hoxb1 promoter in comparison to untreated cells (data not shown). As RA induces Hoxa1 and Pbx1 expression in teratocarcinoma cells [Knoepfler and Kamps, 1997; Qin et al., 2004], it would be expected that the Hoxa1/Pbx1 dimer would thereby bind the R3 region to enhance reporter activity. However, in the early phases of development, Hoxa1 and Hoxb1 are induced by endogenous retinoids, functioning at the RAREs found 3′ of both genes, to establish initial rhombomere 4 identity by triggering the Hoxb1-ARE [Popperl et al., 1995; Studer et al., 1998; Studer et al., 1996]. Later in development, rhombomere 4 identity is maintained as Hoxb1 participates in an auto-regulatory feedback loop to maintain its expression through the b1-ARE [Studer et al., 1998; Studer et al., 1996]. Thus after the addition of RA to the transiently transfected F9 Wt and F9-399 cells, the Soct-R3 region of the Hoxb1 promoter may not have a major role in the transcriptional activation of Hoxb1 in comparison to the RARE.
Lower levels of DNA-bound Hoxa1-993/Pbx1 are detected in the presence of Hoxa1-399
Through the use of EMSA, we have detected lower levels of DNA-bound Hoxa1-993/Pbx1 in the presence of Hoxa1-399. As our EMSA experiments were performed by co-incubating in vitro translated Hoxa1-993 and Pbx1, it cannot be determined under these conditions if Hoxa1-993 or Pbx1 were bound to DNA as a monomer. Therefore, the possibility remains that the presence of Hoxa1-399 resulted in lower levels of DNA-bound Hoxa1-993 or Pbx1. However, other groups have previously shown that Hoxa1-993 has intrinsically weak DNA-binding activity [Phelan et al., 1995; Phelan et al., 1994] and that in vitro translated full-length Pbx1 alone produces no measurable DNA shift by EMSA [Phelan et al., 1995]. Therefore, the band that we detect in the EMSA when Hoxa1-993 and Pbx1 are co-incubated with DNA (Fig. 3, lanes 2–4) is most likely generated by a DNA-bound Hoxa1-993/Pbx1 heterodimer. The EMSA experiments do not differentiate between inhibition of the binding of Hoxa1-993/Pbx1 to DNA by Hoxa1-399 and promotion of the dissociation of Hoxa1-993/Pbx1 from DNA.
We were unable to detect a Hoxa1-993/Pbx1/DNA complex when performing the EMSA experiments using endogenous proteins from F9 Wt and F9-399-83 cell extracts. We were also unable to produce supershifts in the EMSA experiments using the antibodies that we generated against the specific isoforms of Hoxa1 (data not shown). To date, gel-shift experiments involving Hoxa1 have been performed using in vitro translated proteins or epitope-tagged bacterially expressed proteins [Abramovich et al., 2000; Chen and Ruley, 1998; Di Rocco et al., 1997; Green et al., 1998; Huang et al., 2005; Phelan and Featherstone, 1997; Phelan et al., 1995]. This may be because of very low endogenous protein levels or limited stability of the endogenous proteins.
Interactions among Hoxa1-399, Hoxa1-993, and Pbx1
Although no significant binding is detected between GST-Hoxa1-399 and Pbx1 (Fig. 4E), the binding of Pbx1 to GST-Hoxa1-399 increases by 8-fold in the presence of Hoxa1-993 (Fig. 4). It is likely that Hoxa1-399, by binding to Hoxa1-993 (which in turn is bound to Pbx1), forms a trimer with Hoxa1-993 and Pbx1, and therefore, that Hoxa1-399 does not directly bind to Pbx1. This is most likely the case since Hoxa1-399 does not contain the pentapeptide that is essential for Pbx/Hox binding [Phelan et al., 1995]. Another possibility is that Hoxa1-399 binds to Pbx1through an alternative motif that is only exposed when Pbx1 is bound to Hoxa1-993. Although most Hox proteins bind Pbx through the conserved YPWM motif N-terminal to the homeodomain [Chang et al., 1995; Johnson et al., 1995; Knoepfler and Kamps, 1995; Neuteboom et al., 1995; Phelan et al., 1995], there are a few exceptions. For instance, Hoxa10 binds through an alternative tryptophan-containing motif [Chang et al., 1996]. However, the Hoxa1-399 amino acid sequence does not contain these alternative motifs either.
Hoxa1-993 can form homodimers
In addition to demonstrating that Hoxa1-993 and Hoxa1-399 can heterodimerize, our GST pull-down assays indicate that Hoxa1-993 can homodimerize (Fig. 4). These data are consistent with several other studies that report Hoxa1/Hoxa1 bound to DNA by EMSA [Green et al., 1998; Phelan et al., 1995].
There are several examples of homeodomain-containing proteins, such as Pbx3 [Neuteboom and Murre, 1997] and yeast α2 [Goutte and Johnson, 1993], that have the ability to bind DNA as homodimers. The yeast α2 protein has also been found to have different affinity and specificity to DNA as a homodimer versus as a heterodimer [Goutte and Johnson, 1993]. The hetero- versus homo-dimerization of Hoxa1-993 may modulate the DNA binding properties of the protein.
Hoxa1-993 mRNA encodes a protein estimated to be 36 kDa, whereas the truncated Hoxa1-399 transcript encodes a 15 kDa protein. The two proteins are identical for the first 114 amino acids but have unique caroboxy-termini [LaRosa and Gudas, 1988b]. Because Hoxa1-993 is able to bind itself (Fig. 4) and Hoxa1-399 is not (data not shown), we hypothesize that Hoxa1-993 homodimerizes through its unique COOH-terminal which encompasses 50% of the Hoxa1-993 protein. Additionally, there are two poly-histidine regions made up of 5 and 11 histidines encoded within the Hoxa1 protein. The second poly-histidine region, made up of 5 histidines, is spliced out of Hoxa1-399. The functions of these polyhistidine regions are not known. However, COS cells transfected with human HOXA1 containing expansions in the second polyhistidine region show increased cell aggregation and cell death [Paraguison et al., 2005]. Interaction assays using GST fusion proteins of truncated Hoxa1-993 incubated with a radiolabeled full-length Hoxa1-993 protein could be performed to determine which regions of the protein are involved in dimerization. These GST fusion proteins of truncated domains of Hoxa1-993 could also be used in experiments with radiolabeled Hoxa1-399 to determine the amino acid sequences responsible for the heterodimerization.
Ectopic expression of Hoxa1-399 in F9 cells does not alter the expression levels of putative Hoxa1 targets in response to RA
Despite the extensive data presented here supporting a negative role for Hoxa1-399 in the transcriptional activation of the Hoxb1 promoter (Fig. 2), ectopic expression of Hoxa1-399 in F9 cells did not affect the mRNA levels of Hoxb1 or other Hoxa1 targets after RA addition (Fig. 5). There could be for several reasons for this. First, the endogenous Hoxa1-399 is expressed in response to RA in F9 cells and therefore may mask any effects of ectopically expressed Hoxa1-399. Second, the transcriptional regulation of the Hoxb1 gene is complex. Within the vertebrate embryo, multiple retinoic acid response elements cooperate with positive and negative enhancers to specify Hoxb1 expression during pattern formation [Huang et al., 2002; Huang et al., 1998; Langston and Gudas, 1994; Langston et al., 1997; Marshall et al., 1996]. Other researchers have previously identified several Hoxb1 promoter regions that have a negative effect on transcription via multiple Prep1-Pbx1 and Pbx1-Hoxb1 binding sites functioning together to modulate and spatially restrict Hoxb1 expression in rhombomere 4 [Ferretti et al., 2005]. The experiments performed in this report focused only on the R3 region.
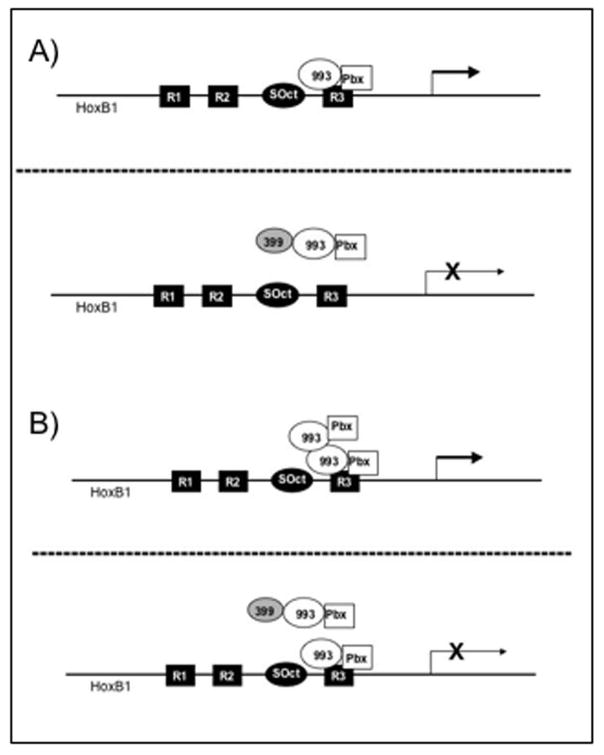
Based on these results, we present two models for the mechanism by which Hoxa1-399 plays a role in the negative regulation of Hoxb1 at a specific region of the promoter, the SOct-R3 motif (Fig. 7). Hoxa1-993 is expressed and forms a dimer with Pbx1a that binds directly to the R3 region of the Hoxb1 promoter, resulting in transcription. However, in the presence of Hoxa1-399, the protein forms a trimer with Hoxa1-993 and Pbx1, possibly inducing a conformational change in the 993/Pbx1 complex that inhibits the Hoxa1-993/Pbx1 dimer from binding to DNA or decreases the binding affinity. This event causes a reduction in the transcriptional activation of Hoxb1 (Fig. 7a). As a result of the in vitro homodimerization that we report, it is also possible that there are two Hoxa1-993/Pbx1 dimers present at the R3 region (Fig. 7b). The Hoxa1-399 protein could bind one of the Hoxa1-993/Pbx1 dimers, resulting in the loss of one of the 993/Pbx1 complexes from the DNA; a second, functional 993/Pbx1 complex would still be bound to the promoter. This could result in a reduction in the transcriptional activation of Hoxb1. The possibility also exists that Hoxa1-399 binds Hoxa1-993 to not only decrease the affinity of Hoxa1-993 to the R3 region, but also to mask the activation domain of Hoxa1-993. As previously mentioned, Hox genes are transiently and spatially expressed during embryogenesis. Therefore, Hoxa1-399 may participate in a mechanism to inhibit Hoxb1 gene expression and the expression of its targets at specific times during development and/or Hoxa1-399 may play a role in the quantitative regulation of Hoxb1 gene expression during cellular differentiation.
Figure 7. Proposed model for the role of Hoxa1-399 at the R3 region of the HoxB1 promoter.
(A) Hoxa1-993 forms a complex with Pbx1a. The dimer then binds directly to the R3 region of the B1 promoter to induce its expression (top panel). Later, Hoxa1-399 is expressed. The protein binds Hoxa1-993 to form a trimer with Pbx1a, inhibiting the 993/Pbx1a complex from binding to DNA. This reduces the transcriptional activation of HoxB1 (bottom panel). (B) Due to the in vitro homodimerization that we report, it is possible that there are two Hoxa1-993/Pbx1 multimers present are the R3 (top panel). Later, Hoxa1-399 is expressed. The protein binds one of the Hoxa1-993 proteins to form a trimer with Pbx1a, inhibiting one of the 993/Pbx1a complexes from binding to DNA. This reduces the transcriptional activation of HoxB1 (bottom panel).
Improper regulation of HOXA1 gene expression has been implicated in various types of cancers [Abe et al., 2006; Calvo et al., 2000; Hassan et al., 2006; Hung et al., 2003; Maeda et al., 2005]. For example, Hoxa1 has been detected in carcinomas of the mouse mammary gland, but not in normal mammary tissue [Zhang et al., 2003]. The human gene is expressed in a variety of breast cancer lesions [Chariot and Castronovo, 1996]. In MCF7 human breast cancer cells, a third alternatively spliced transcript, which also lacks the homeodomain, has been described [Chariot et al., 1995]. All three transcripts are induced by RA and progestins, another differentiating agent, in these cells [Chariot and Castronovo, 1996; Chariot et al., 1995]. Moreover, the HOXA1 mRNA encoding the homeodomain-containing protein has a higher level of expression than HOXA1 mRNA encoding homeodomain-less proteins in human breast cancer lesions [Chariot and Castronovo, 1996]. These data support the idea that the regulation of both the full-length and truncated Hoxa1 proteins is potentially important for cellular differentiation, particularly breast epithelial cell differentiation.
Acknowledgments
We thank Elana Wilen for generating the F9-399 stably transfected cell lines. We also credit Allison Ait-Aouane for the preliminary bacterial protein expression work. We are grateful to other members of the Gudas laboratory, especially Robert Gillespie, for helpful discussions and critical reading of this manuscript. We are especially thankful to Christopher Kelly for assistance in the preparation of this manuscript for publication.
Contact grant sponsor: NIH
Contact grant numbers: NCI RO1CA43796 and UR 5U19 HD03546 to LJ Gudas.
References
- Abe M, Hamada J, Takahashi O, Takahashi Y, Tada M, Miyamoto M, Morikawa T, Kondo S, Moriuchi T. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006;15:797–802. [PubMed] [Google Scholar]
- Abramovich C, Shen WF, Pineault N, Imren S, Montpetit B, Largman C, Humphries RK. Functional cloning and characterization of a novel nonhomeodomain protein that inhibits the binding of PBX1-HOX complexes to DNA. J Biol Chem. 2000;275:26172–7. doi: 10.1074/jbc.M001323200. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–15. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- Bosley TM, Salih MA, Alorainy IA, Oystreck DT, Nester M, Abu-Amero KK, Tischfield MA, Engle EC. Clinical characterization of the HOXA1 syndrome BSAS variant. Neurology. 2007;69:1245–53. doi: 10.1212/01.wnl.0000276947.59704.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, Rosell R, Gemmill RM, Drabkin HA. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A. 2000;97:12776–81. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–45. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–74. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Chariot A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Commun. 1996;222:292–7. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- Chariot A, Moreau L, Senterre G, Sobel ME, Castronovo V. Retinoic acid induces three newly cloned HOXA1 transcripts in MCF7 breast cancer cells. Biochem Biophys Res Commun. 1995;215:713–20. doi: 10.1006/bbrc.1995.2522. [DOI] [PubMed] [Google Scholar]
- Chen J, Ruley HE. An enhancer element in the EphA2 (Eck) gene sufficient for rhombomere-specific expression is activated by HOXA1 and HOXB1 homeobox proteins. J Biol Chem. 1998;273:24670–5. doi: 10.1074/jbc.273.38.24670. [DOI] [PubMed] [Google Scholar]
- Chisaka O, Musci TS, Capecchi MR. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–20. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- Di Rocco G, Gavalas A, Popperl H, Krumlauf R, Mavilio F, Zappavigna V. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J Biol Chem. 2001;276:20506–15. doi: 10.1074/jbc.M011175200. [DOI] [PubMed] [Google Scholar]
- Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. Embo J. 1997;16:3644–54. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintilhac A, Bihan R, Guerrier D, Deschamps S, Pellerin I. A conserved non-homeodomain Hoxa9 isoform interacting with CBP is co-expressed with the ‘typical’ Hoxa9 protein during embryogenesis. Gene Expr Patterns. 2004;4:215–22. doi: 10.1016/j.modgep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Engle EC. Oculomotility disorders arising from disruptions in brainstem motor neuron development. Arch Neurol. 2007;64:633–7. doi: 10.1001/archneur.64.5.633. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol. 2005;25:8541–52. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Araki K, Chisaka O, Araki M, Takagi K, Yamamura K. Analysis of the murine Hoxa-9 cDNA: an alternatively spliced transcript encodes a truncated protein lacking the homeodomain. Gene. 1998;209:77–85. doi: 10.1016/s0378-1119(98)00014-6. [DOI] [PubMed] [Google Scholar]
- Goliger JA, Gudas LJ. Mouse F9 teratocarcinoma stem cells expressing the stably transfected homeobox gene Hox 1.6 exhibit an altered morphology. Gene Expr. 1992;2:147–60. [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Johnson AD. Yeast a1 and alpha 2 homeodomain proteins form a DNA-binding activity with properties distinct from those of either protein. J Mol Biol. 1993;233:359–71. doi: 10.1006/jmbi.1993.1517. [DOI] [PubMed] [Google Scholar]
- Green NC, Rambaldi I, Teakles J, Featherstone MS. A conserved C-terminal domain in PBX increases DNA binding by the PBX homeodomain and is not a primary site of contact for the YPWM motif of HOXA1. J Biol Chem. 1998;273:13273–9. doi: 10.1074/jbc.273.21.13273. [DOI] [PubMed] [Google Scholar]
- Gudas LJ. Retinoids and vertebrate development. J Biol Chem. 1994;269:15399–402. [PubMed] [Google Scholar]
- Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, Zhang X, Kashiwazaki H, Yamazaki Y, Inoue N, Moriuchi T. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. 2006;16:217–24. doi: 10.3727/000000006783981080. [DOI] [PubMed] [Google Scholar]
- Hong YS, Kim SY, Bhattacharya A, Pratt DR, Hong WK, Tainsky MA. Structure and function of the HOX A1 human homeobox gene cDNA. Gene. 1995;159:209–14. doi: 10.1016/0378-1119(95)92712-g. [DOI] [PubMed] [Google Scholar]
- Huang D, Chen SW, Gudas LJ. Analysis of two distinct retinoic acid response elements in the homeobox gene Hoxb1 in transgenic mice. Dev Dyn. 2002;223:353–70. doi: 10.1002/dvdy.10057. [DOI] [PubMed] [Google Scholar]
- Huang D, Chen SW, Langston AW, Gudas LJ. A conserved retinoic acid responsive element in the murine Hoxb-1 gene is required for expression in the developing gut. Development. 1998;125:3235–46. doi: 10.1242/dev.125.16.3235. [DOI] [PubMed] [Google Scholar]
- Huang H, Rastegar M, Bodner C, Goh SL, Rambaldi I, Featherstone M. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J Biol Chem. 2005;280:10119–27. doi: 10.1074/jbc.M413963200. [DOI] [PubMed] [Google Scholar]
- Hung YC, Ueda M, Terai Y, Kumagai K, Ueki K, Kanda K, Yamaguchi H, Akise D, Ueki M. Homeobox gene expression and mutation in cervical carcinoma cells. Cancer Sci. 2003;94:437–41. doi: 10.1111/j.1349-7006.2003.tb01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Parker E, Krasnow MA. Extradenticle protein is a selective cofactor for the Drosophila homeotics: role of the homeodomain and YPWM amino acid motif in the interaction. Proc Natl Acad Sci U S A. 1995;92:739–43. doi: 10.1073/pnas.92.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Wold BJ. Stable reduction of thymidine kinase activity in cells expressing high levels of anti-sense RNA. Cell. 1985;42:129–38. doi: 10.1016/s0092-8674(85)80108-2. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Kamps MP. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol Cell Biol. 1995;15:5811–9. doi: 10.1128/mcb.15.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Kamps MP. The Pbx family of proteins is strongly upregulated by a post-transcriptional mechanism during retinoic acid-induced differentiation of P19 embryonal carcinoma cells. Mech Dev. 1997;63:5–14. doi: 10.1016/s0925-4773(97)00669-2. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Lu Q, Kamps MP. Pbx-1 Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acids Res. 1996;24:2288–94. doi: 10.1093/nar/24.12.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuves LG, Shen WF, Kwong A, Stelnicki E, Rozenfeld S, Oda Y, Blink A, Krishnan K, Lau B, Mauro T, Largman C. Changes in HOXB6 homeodomain protein structure and localization during human epidermal development and differentiation. Dev Dyn. 2000;218:636–47. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1014>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38:217–27. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- Langston AW, Gudas LJ. Retinoic acid and homeobox gene regulation. Curr Opin Genet Dev. 1994;4:550–5. doi: 10.1016/0959-437x(94)90071-a. [DOI] [PubMed] [Google Scholar]
- Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J Biol Chem. 1997;272:2167–75. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Gudas LJ. An early effect of retinoic acid: cloning of an mRNA (Era-1) exhibiting rapid and protein synthesis-independent induction during teratocarcinoma stem cell differentiation. Proc Natl Acad Sci U S A. 1988a;85:329–33. doi: 10.1073/pnas.85.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa GJ, Gudas LJ. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol Cell Biol. 1988b;8:3906–17. doi: 10.1128/mcb.8.9.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–19. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T, Moriuchi T. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114:436–41. doi: 10.1002/ijc.20706. [DOI] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, Gorry P, Lumsden A, Chambon P. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–38. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- Marshall H, Morrison A, Studer M, Popperl H, Krumlauf R. Retinoids and Hox genes. Faseb J. 1996;10:969–78. [PubMed] [Google Scholar]
- Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–98. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- Martinez-Ceballos E, Gudas LJ. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J Neurosci Res. 2008 doi: 10.1002/jnr.21729. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–33. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, Gluckman PD, Sukumar S, Emerald BS, Lobie PE. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007;26:3998–4008. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Neuteboom ST, Murre C. Pbx raises the DNA binding specificity but not the selectivity of antennapedia Hox proteins. Mol Cell Biol. 1997;17:4696–706. doi: 10.1128/mcb.17.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuteboom ST, Peltenburg LT, van Dijk MA, Murre C. The hexapeptide LFPWMR in Hoxb-8 is required for cooperative DNA binding with Pbx1 and Pbx2 proteins. Proc Natl Acad Sci U S A. 1995;92:9166–70. doi: 10.1073/pnas.92.20.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20:1636–50. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraguison RC, Higaki K, Sakamoto Y, Hashimoto O, Miyake N, Matsumoto H, Yamamoto K, Sasaki T, Kato N, Nanba E. Polyhistidine tract expansions in HOXA1 result in intranuclear aggregation and increased cell death. Biochem Biophys Res Commun. 2005;336:1033–9. doi: 10.1016/j.bbrc.2005.08.212. [DOI] [PubMed] [Google Scholar]
- Pecorino LT, Rickles RJ, Strickland S. Anti-sense inhibition of tissue plasminogen activator production in differentiated F9 teratocarcinoma cells. Dev Biol. 1988;129:408–16. doi: 10.1016/0012-1606(88)90388-0. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Featherstone MS. Distinct HOX N-terminal arm residues are responsible for specificity of DNA recognition by HOX monomers and HOX.PBX heterodimers. J Biol Chem. 1997;272:8635–43. doi: 10.1074/jbc.272.13.8635. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–97. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sadoul R, Featherstone MS. Functional differences between HOX proteins conferred by two residues in the homeodomain N-terminal arm. Mol Cell Biol. 1994;14:5066–75. doi: 10.1128/mcb.14.8.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–42. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Qin P, Haberbusch JM, Soprano KJ, Soprano DR. Retinoic acid regulates the expression of PBX1, PBX2, and PBX3 in P19 cells both transcriptionally and post-translationally. J Cell Biochem. 2004;92:147–63. doi: 10.1002/jcb.20057. [DOI] [PubMed] [Google Scholar]
- Shen J, Wu H, Gudas LJ. Molecular cloning and analysis of a group of genes differentially expressed in cells which overexpress the Hoxa-1 homeobox gene. Exp Cell Res. 2000;259:274–83. doi: 10.1006/excr.2000.4963. [DOI] [PubMed] [Google Scholar]
- Shen WF, Chang CP, Rozenfeld S, Sauvageau G, Humphries RK, Lu M, Lawrence HJ, Cleary ML, Largman C. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Detmer K, Simonitch-Eason TA, Lawrence HJ, Largman C. Alternative splicing of the HOX 2.2 homeobox gene in human hematopoietic cells and murine embryonic and adult tissues. Nucleic Acids Res. 1991;19:539–45. doi: 10.1093/nar/19.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763–6. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Nigro V, Faiella A, D’Esposito M, Stornaiuolo A, Mavilio F, Boncinelli E. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech Dev. 1991;33:215–27. doi: 10.1016/0925-4773(91)90029-6. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–36. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–4. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2005;37:1035–7. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–51. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Wright CV, Cho KW, Fritz A, Burglin TR, De Robertis EM. A Xenopus laevis gene encodes both homeobox-containing and homeobox-less transcripts. Embo J. 1987;6:4083–94. doi: 10.1002/j.1460-2075.1987.tb02754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, Gluckman PD, Lee KO, Lobie PE. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J Biol Chem. 2006;281:6471–81. doi: 10.1074/jbc.M512666200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu T, Chen Y, Mertani HC, Lee KO, Lobie PE. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J Biol Chem. 2003;278:7580–90. doi: 10.1074/jbc.M212050200. [DOI] [PubMed] [Google Scholar]