Abstract
Advanced cholangiocarcinoma continues to harbor a difficult prognosis and therapeutic options have been limited. During the course of a clinical trial of whole genomic sequencing seeking druggable targets, we examined six patients with advanced cholangiocarcinoma. Integrated genome-wide and whole transcriptome sequence analyses were performed on tumors from six patients with advanced, sporadic intrahepatic cholangiocarcinoma (SIC) to identify potential therapeutically actionable events. Among the somatic events captured in our analysis, we uncovered two novel therapeutically relevant genomic contexts that when acted upon, resulted in preliminary evidence of anti-tumor activity. Genome-wide structural analysis of sequence data revealed recurrent translocation events involving the FGFR2 locus in three of six assessed patients. These observations and supporting evidence triggered the use of FGFR inhibitors in these patients. In one example, preliminary anti-tumor activity of pazopanib (in vitro FGFR2 IC50≈350 nM) was noted in a patient with an FGFR2-TACC3 fusion. After progression on pazopanib, the same patient also had stable disease on ponatinib, a pan-FGFR inhibitor (in vitro, FGFR2 IC50≈8 nM). In an independent non-FGFR2 translocation patient, exome and transcriptome analysis revealed an allele specific somatic nonsense mutation (E384X) in ERRFI1, a direct negative regulator of EGFR activation. Rapid and robust disease regression was noted in this ERRFI1 inactivated tumor when treated with erlotinib, an EGFR kinase inhibitor. FGFR2 fusions and ERRFI mutations may represent novel targets in sporadic intrahepatic cholangiocarcinoma and trials should be characterized in larger cohorts of patients with these aberrations.
Author Summary
Cholangiocarcinoma is a cancer that affects the bile ducts. Unfortunately, many patients diagnosed with cholangiocarcinoma have disease that cannot be treated with surgery or has spread to other parts of the body, thus severely limiting treatment options. New advances in drug treatment have enabled treatment of these cancers with “targeted therapy” that exploits an error in the normal functioning of a tumor cell, compared to other cells in the body, thus allowing only tumor cells to be killed by the drug. We sought to identify changes in the genetic material of cholangiocarcinoma patient tumors in order to identify potential errors in cellular functioning by utilizing cutting edge genetic sequencing technology. We identified three patient tumors possessing an FGFR2 gene that was aberrantly fused to another gene. Two of these patients were able to receive targeted therapy for FGFR2 with resulting tumor shrinkage. A fourth tumor contained an error in a gene that controls a very important cellular mechanism in cancer, termed epidermal growth factor pathway (EGFR). This patient received therapy targeting this mechanism and also demonstrated response to treatment. Thus, we have been able to utilize cutting edge technology with targeted drug treatment to personalize medical treatment for cancer in cholangiocarcinoma patients.
Introduction
Biliary tract cancers (BTC) comprise malignant tumors of the intrahepatic and extrahepatic bile ducts. Known risk factors for BTC are the liver flukes O. viverrini and C. sinensis in high prevalence endemic regions in southeast Asia [1]–[3], as well as primary sclerosing cholangitis [4]–[7], Caroli's disease [8], hepatitis B and hepatitis C [9]–[14], obesity [13], hepatolithiasis [15], [16] and thorotrast contrast exposure [17], [18]. Surgical approaches such as resection and liver transplantation represent the only curative treatment approaches for BTC [19]. Unfortunately, most patients present with surgically unresectable and/or metastatic disease at diagnosis. Systemic therapy with gemcitabine and cisplatin has been established as the standard of care for patients with advanced disease, but is only palliative [20], emphasizing the imminent need for novel therapies.
Multiple studies have reported the presence of mutations/allelic loss of known cancer genes in BTC [21]–[39] and recently, a prevalence set of 46 patients was used to validate 15 of these genes including: TP53, KRAS, CDKN2A and SMAD4 as well as MLL3, ROBO2, RNF43, GNAS, PEG3, XIRP2, PTEN, RADIL, NCD80, LAMA2 and PCDHA13. Recent studies have also identified recurrent mutations in IDH1 (codon 132) and IDH2 (codons 140 and 172) with a prevalence of 22–23% associated with clear cell/poorly differentiated histology and intrahepatic primary [40], [41]. Fusions with oncogenic potential involving the kinase gene ROS1 have been identified in patients with BTC with a prevalence of 8.7% in a recent study [42]. Less frequently, mutations in sporadic BTC have been reported in EGFR [43], [44], BRAF [45], NRAS [40], [46], PIK3CA [40], [46], [47], APC [40], CTNNB1 [40], AKT1 [40], PTEN [40], ABCB4 [48], ABCB11 [49], [50], and CDH1 [51] as well as amplifications in ERRB2 [52].
Recently, two independent studies reported the presence of FGFR fusions in cholangiocarcinoma; a single case with FGFR2-AHCYL1 [53] as well as several cases identifying FGFR2-BICC1 fusions [53], [54]. Arai et al. evaluated the presence of FGFR2 fusions in a cohort of 102 cholangiocarcinoma patients observing that the fusions occurred exclusively in the intrahepatic cases with a prevalence of 13.6% [53]. Due to the presence of known dimerization motifs in the fusion partners, Wu et al. conducted mechanistic studies that demonstrated the in vitro interaction of FGFR2-BICC1 and other fusions that was not observed in the presence of wildtype FGFR2 [54]. Furthermore, overexpression of the FGFR2-BICC1 and other selected fusions resulted in altered cell morphology and increased cell proliferation [54]. These data led to the conclusion that the fusion partners are facilitating oligomerization, resulting in FGFR kinase activation in tumors possessing FGFR fusions. In addition, in vitro and in vivo assessment of the sensitivity of cell lines containing an FGFR2 fusion to an FGFR inhibitor demonstrated sensitivity to treatment only in the fusion containing cells [53], [54], suggesting the presence of FGFR fusions may be a useful predictor of tumor response to FGFR inhibitors.
To comprehensively explore the genetic basis of sporadic intrahepatic cholangiocarcinoma (SIC), with emphasis on elucidation of therapeutically relevant targets, we performed integrated whole genome and whole transcriptome analyses on tumors from 6 patients with advanced, sporadic intrahepatic cholangiocarcinoma (SIC). Notably, recurrent fusions involving the oncogene FGFR2 (n = 3) were identified. A patient whose tumor presented with an FGFR2-MGEA5 fusion has demonstrated preliminary evidence of anti-tumor activity manifest as stable disease accompanied by CA19-9 reduction and tumor necrosis to ponatinib, a pan-FGFR inhibitor (in vitro FGFR1 IC50≈24 nM, FGFR2 IC50≈8 nM, FGFR3 IC50≈8 nM and FGFR4 IC50≈34 nM). In another patient whose tumor possessed an FGFR2-TACC3 fusion, preliminary anti-tumor activity of pazopanib (in vitro FGFR2 IC50≈350 nM) was also noted. After progression on pazopanib, the same patient also responded to ponatinib and again demonstrated tumor shrinkage. Additionally, a non-FGFR fusion patient was found to have allele-specific preferential expression of a loss of function mutation in ERRFI1, a direct negative regulator of EGFR activation. Similarly, rapid and robust disease regression was noted in the patient with an ERRFI1 mutant tumor when treated with erlotinib, an EGFR kinase inhibitor. Results suggest that these novel targets in the EGFR and FGFR pathways may be therapeutically relevant in patients with sporadic cholangiocarcinoma.
Results
Genomic landscape
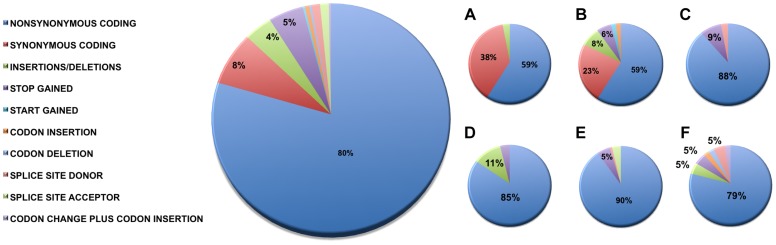
We identified 327 somatic coding mutations, with an average of 55 mutations/tumor (range 34–112), within our cohort ( Table 1 , Figure 1 ). Nonsynonymous single nucleotide variations were the predominant class in all of the patients. Patients 1 and 2 accumulated a high number of synonymous mutations in comparison to the other patients. Patient 5 carried the most stops gained likely contributing to a higher number of pseudogenes in comparison to the others and was also the only patient to carry several predicted high impact mutations affecting splice site acceptor regions ( Figure 1 , light green, percentage <5%). In addition, patient 6 also carried a codon change plus insertion variation. Sequencing statistics are provided in Table 2 . Genes with mutations in more than one case included CSPG4 (n = 2), GRIN3A (n = 2) and PLXBN3 (n = 2) (Table S1); with half of these predicted to be potentially damaging by SIFT [55], Polyphen [56], Mutation Assessor [57] and Mutation Taster [58]. While there was overlap in the somatic landscape of SIC with liver-fluke associated cholangiocarcinoma, hepatocellular cancer and pancreatic cancer, most of the aberrations detected in our study were distinct ( Table 3 ). More importantly, using previously published methods [59], we identified molecular fusions involving FGFR2 that were felt to be therapeutically relevant in 3 patients. Additionally, these fusions were validated with a break apart Fluorescent In situ Hybridization (FISH) assay ( Figure 2 ). Notably, the patients who did not harbor the FGFR2 fusions were negative using the same assay. Two of the three patients with FGFR2 fusions (Patients 4 and 6) were treated with FGFR inhibitors while the third patient (Patient 5), experienced clinical decline prior to the availability of results and as such did not receive any further therapy. Furthermore, overexpression of an SNV in ERRFI1 (E384X), a negative regulator of EGFR, was detected in a non-FGFR2 translocation patient's tumor. Taken together, our results constitute important therapeutically actionable alterations in patients with advanced SIC (Text S1).
Table 1. Summary of mutation type by patient.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
| Nonsynonymous coding | 20 | 30 | 31 | 44 | 101 | 34 |
| Synonymous coding | 13 | 12 | 0 | 0 | 0 | 0 |
| Insertions/deletions | 1 | 4 | 0 | 6 | 0 | 2 |
| Stop gained | 0 | 3 | 3 | 2 | 6 | 2 |
| Start gained | 0 | 1 | 0 | 0 | 0 | 0 |
| Codon insertion | 0 | 1 | 0 | 0 | 0 | 1 |
| Codon deletion | 0 | 0 | 0 | 0 | 0 | 1 |
| Splice site donor | 0 | 0 | 1 | 0 | 1 | 2 |
| Splice site acceptor | 0 | 0 | 0 | 0 | 4 | 0 |
| Total | 34 | 51 | 35 | 52 | 112 | 42 |
Figure 1. Sequence variation effects.
Functional effects of high confidence sequence variations for all of the patients were identified as described in the Methods. The abundance of variations in each functional category is provided as percentages relative to the total number of high confidence variations and raw counts are provided in Table 1 . For categories where the percentage was less than 5%, values are not shown. Summaries by individual patients are shown as follows: A) Patient 1, B) Patient 2, C) Patient 3, D) Patient 4, E) Patient 5, and F) Patient 6. Nonsynonymous single nucleotide variations were the predominant class in all of the patients. Two patients, Patients 1 and 2 also accumulated a high number of synonymous mutations in comparison to the other patients; Patient 5 carries the most stops gained likely contributing to a higher number of pseudogenes in comparison to the others; Patient 5 was also the only patient to carry several predicted high impact mutations that affect the splice site acceptor regions (light green, percentage <5%). In addition to the major functional classes summarized, Patient 6 also carried a codon change plus insertion variation.
Table 2. Sequencing metrics of 6 advanced, sporadic biliary tract cancer patients.
| Exome | Whole Genome | RNA Seq | ||||||||
| Patient | Tissue | Aligned Reads (Millions) | Mean Target Coverage | % Target Bases 10× | # of Functional Coding Variants | Aligned Reads (Millions) | Aligned Bases (Billions) | Physical Coverage | Aligned Reads (Millions) | Aligned Bases (Billions) |
| 1 | N | 161 | 100 | 94% | - | 266 | 22 | 37 | - | - |
| T | 156 | 112 | 94% | 21 | 228 | 18 | 35 | 100 | 8.1 | |
| 2 | N | 176 | 74 | 94% | - | 179 | 14 | 5 | - | - |
| T | 202 | 81 | 94% | 34 | 370 | 30 | 10 | 341 | 26 | |
| 3 | N | 226 | 110 | 58% | - | 296 | 24 | 50 | 163 | 13 |
| T | 195 | 92 | 58% | 52 | 321 | 26 | 50 | 101 | 8.1 | |
| 4 | N | 167 | 80 | 95% | - | 317 | 26 | 42 | - | - |
| T | 202 | 93 | 96% | 52 | 163 | 13 | 12 | 264 | 20 | |
| 5 | N | 257 | 146 | 96% | - | 335 | 27 | 51 | - | - |
| T | 133 | 78 | 93% | 250 | 349 | 28 | 39 | 401 | 31 | |
| 6 | N | 350 | 243 | 92% | - | - | - | - | - | - |
| T | 340 | 245 | 92% | 43 | - | - | - | 713 | 31 | |
| Liver Control | - | - | - | - | - | - | - | - | 118 | 9.6 |
N = Normal, T = Tumor.
Table 3. Comparison of mutation frequency in cholangiocarcinoma, pancreatic and liver cancers.
| Gene | Non-liver fluke CCA (n = 6) | Liver fluke associated CCA [111] (n = 54) | CCA [40] (n = 62) | PDAC [112] (n = 142) | HCC [113] (n = 149) |
| AKT1 | 0% | 0% | 1.6% | 0% | 0% |
| APC | 0% | 0% | 0% | 0% | 1.3% |
| ARID2 | 0% | 0% | NA | 2.1% | 6.0% |
| BAP1 | 16.7% | 0% | NA | 0% | 0% |
| BRAF | 0% | 0% | 1.6% | 0.7% | 0% |
| CDKN2A | 0% | 5.6% | NA | 2.4% | 7.4% |
| CSPG4 | 33.3% | 0% | NA | 0% | 0.7% |
| CTNNB1 | 0% | 0% | NA | 0% | 34.9% |
| DMXL1 | 0% | 0% | NA | 0% | 0% |
| EGFR | 0% | 0% | 0% | 0% | 0% |
| ERRFI1 | 16.7% | 0% | NA | 0% | 0.7% |
| FLT3 | 0% | 0% | 0% | 0% | 0% |
| GNAS | 0% | 9.3% | NA | 0.7% | 0% |
| GRIN3A | 33.3% | 0% | NA | 0% | 0% |
| IDH1 | 0% | 0% | 13% | 0% | 0% |
| IDH2 | 16.7% | 0% | 2% | 0% | 0% |
| JAK2 | 0% | 0% | 0% | 0% | 0% |
| KIT | 0% | 0% | 0% | 0% | 0% |
| KRAS | 0% | 16.7% | NA | 66.2% | 1.3% |
| LAMA2 | 16.7% | 3.7% | NA | 0% | 0% |
| MLL3 | 16.7% | 14.8% | NA | 4.9% | 0% |
| NDC80 | 0% | 3.7% | NA | 0% | 0% |
| NLRP1 | 16.7% | 0% | NA | 0% | 0% |
| NOTCH1 | 16.7% | 0% | 0% | 0% | 0% |
| NRAS | 16.7% | 0% | 3.2% | 0% | 0% |
| PCDHA13 | 16.7% | 3.7% | NA | 0.7% | 0% |
| PAK1 | 16.7% | 0% | NA | 0% | 0% |
| PEG3 | 0% | 5.6% | NA | 1.4% | 0% |
| PIK3CA | 0% | 0% | 0% | 0% | 1.3% |
| PLXNB3 | 33.3% | 0% | NA | 0% | 0% |
| PTEN | 0% | 3.7% | 2% | 0% | 0% |
| PTK2 | 16.7% | 0% | NA | 0% | 0% |
| RADIL | 0% | 3.7% | NA | 0% | 0% |
| RNF43 | 0% | 9.3% | NA | 0% | 0% |
| ROBO2 | 0% | 9.3% | NA | 1.4% | 0% |
| SMAD4 | 0% | 16.7% | NA | 11.3% | 0% |
| TP53 | 33.3% | 44.4% | 8% | 23.2% | 19.5% |
| XIRP2 | 0% | 5.6% | NA | 3.5% | 0% |
CCA, cholangiocarcinoma; PDAC, pancreatic ductal adenocarcinoma; HCC, hepatocellular carcinoma; NA, not assessed.
Figure 2. Representative fluorescent in situ hybridization (FISH) demonstrating the presence of FGFR2 fusion.
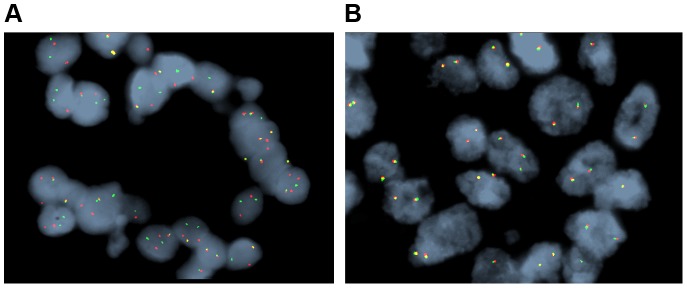
A) Cholangiocarcinoma with FGFR2 rearrangement (distinct orange and green signals are present in most of the cells). B) Cholangiocarcinoma negative for FGFR2 rearrangement (orange and green signals remain fused).
Pathway analysis
Comparative pathway analysis of genes carrying small scale nucleotide variations (SsNVs) has implicated several major pathways, possibly interacting as a network, that are predicted to underlie disease in all of our studied biliary carcinoma patients. These shared pathways include EGFR, EPHB, PDGFR-beta, Netrin-mediated and Beta1 integrin mediated signaling pathways ( Figure 3 and Tables S2 and S3). Interestingly, most of these pathways have known roles in mediating epithelial-to-mesenchymal cell transitions, which occur frequently during development as well as tumorigenesis. Cell growth and motility is inherent to the successful progression of both biological processes. Studies of the nervous system and lung development have shown that Netrins act to inhibit FGF7 and FGF10 mediated growth or cell guidance [60]. In addition, Netrin-1 has a known role in mediating cell migration during pancreatic organogenesis [60]. Furthermore, Netrin-1 acts as a ligand for α3β1 and α6β4 integrins, both of which are involved in supporting adhesion of developing pancreatic epithelial cells with Netrin-1 although it is thought that α6β4 plays the principle role during this process [60]. Interestingly, α3β1 has been hypothesized to play a role during the process of angiogenesis, when chemoattractants and chemorepellents act to guide filopodia during migration [60]. The α3β1 integrin receptor may act together with additional pathways proposed to play a role during angiogenesis such as VEGF, PDGFR-beta [61], and EphrinB [62] as well as tumorigenesis [60]. Patients 3 and 4 also shared several genes acting in cadherin signaling pathways (Tables S3, S4), which are important for maintaining cell-cell adhesion and are known to be intimately integrated with EGFR and FGFR signaling pathways [63].
Figure 3. Gene Ontology pathway analysis.
Genes carrying single nucleotide or frameshift variations, or aberrant in copy number were annotated and clustered by GO term functional classes, some of which are known to play a role in Cancer (Tables S2 and S3). Major classes for A) SNVs and B) CNVs are labeled in the figure. Proteins predicted to be integral to the membrane and involved in transport, as well as transcriptional regulators were among the most abundant class in all of the patients affected by small scale sequence variations and copy number variations. Variations specifically affecting the EGFR or FGFR gene families were prevalent in Patients 4, 5, and 6 and are highlighted in the figure with the gene name provided in parenthesis next to the pathway name.
In addition to the variations identified in genes acting in EGFR and/or FGFR signaling pathways, we also report multiple sSNVs and copy number variations (CNVs) ( Figure 4 ) in genes such as HDAC1, TP53, MDM2 and AKT1, acting in interaction networks or regulatory pathways involving the fusion partner genes in patients 5 (BICC1), and 6 (TACC3) ( Table 4 ). Known mutations in BICC1 have been shown to disrupt canonical Wnt signaling [64] and genes, such as BCL9, involved in this pathway are known to regulate a range of biological processes such as transcription and cell proliferation and carry variations in patient 5 ( Table 4 ). CSPG4, a target that is being investigated for antibody-based immunotherapy in preclinical studies of triple negative breast cancer [65], is involved in the Wnt signaling pathway, and carries variations in both patients 1 and 2, however, it is not mutated in patient 5. TACC3 is known to mediate central spindle assembly and multiple genes including CDCA8, BUB1, and TACC1, belonging to the TACC3 interaction network exhibit aberrant copy number in patient 6 ( Table 4 ). A recent study has also implicated TACC3 in EGF-mediated EMT when overexpressed [64], and we find that the PLCG1, MAP2K1, and MAPK8 genes, which act in both FGFR and EGFR regulatory pathways, exhibit CNV in patient 6. We also note that the DNAH5 gene encoding a dynein protein which is part of the microtubule-associated motor protein complex carries two G→C missense mutations in patient 6 (Table S1). Several genes carrying more than one variation in either the same patient or different patients also included genes with known roles similar to genes in FGFR/EGFR pathways including axon guidance, invasive growth, or cell differentiation (NAV3, LAMC3, PLXNB3, and PTPRK) (Table S1). In the case of patient 4, our studies suggest that the primary effect of the FGFR2-MGEA5 fusion is on FGFR2 related signaling, since changes in expression were observed in FGF8 (p<0.05) and the genome of this patient also carries a 4-bp insertion (∧GTGT) in the FGFR4 gene (Table S1).
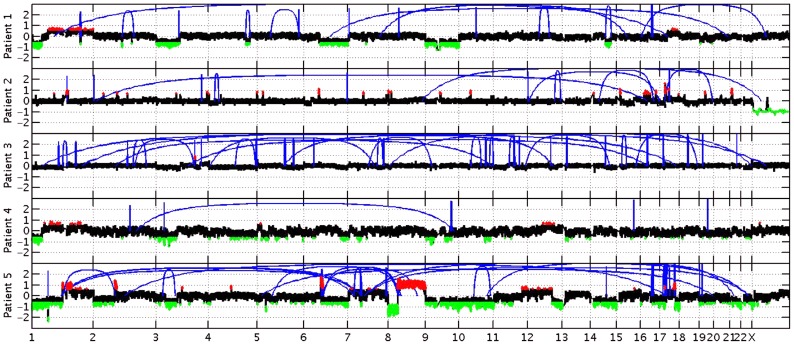
Figure 4. Copy number changes and structural rearrangements.
Whole genome data was utilized to determine copy number alterations and structural rearrangements in the genome for Patients 1–5. WGS was not conducted for patient 6. Red indicates copy number gain, green copy number loss and blue lines indicate structural rearrangements. Significant variability between samples was observed for both copy number changes and structural rearrangements. Patient 5 presented with numerous copy number changes and structural rearrangements contrasting with patient 4 who had minimal structural rearrangements and much smaller regions of copy number changes. Patient 3 is characterized by a large number of structural rearrangements with almost no copy number alterations; in contrast, Patient 1 has a moderate number of structural variations, but has large regions of copy number gain and loss. Patient 2 has a moderate number of structural rearrangements with multiple focal amplifications across the genome.
Table 4. Stable fusion partner gene pathways.
| Patients | Gene in Interaction or Regulatory Network | Small-scale Variation (sSNV)/Copy Number Variation (CNV) | Associated Network | Associated Pathway |
| 4 | FGFR4 | ssNV | FGFR | glucose homeostasis |
| 5 | RAF1 | CNV | EGFR/FGFR | axon guidance |
| 5 | RPS6KA5 | CNV | FGFR | innate immune response |
| 5 | HGF | CNV | FGFR | mitosis |
| 5 | FRS2 | CNV | FGFR | ventricular septum development |
| 5 | FGFR2 | CNV | FGFR | apoptotic process |
| 5 | FGFR4 | CNV | FGFR | glucose homeostasis |
| 5 | FGFR1OP2 | CNV | FGFR | response to wounding |
| 5 | FGFR1 | CNV | FGFR | transcription, DNA-dependent |
| 5 | ANTXR1 | CNV | BICC1 | actin cytoskeleton reorganization |
| 5 | ARL3 | CNV | BICC1 | cell cycle |
| 5 | NKX3-1 | CNV | BICC1 | multicellular organismal development |
| 5 | WIF1 | CNV | BICC1 | multicellular organismal development |
| 5 | AXIN2 | CNV | BICC1 | negative regulation of cell proliferation |
| 5 | SFRP1 | CNV | BICC1 | negative regulation of cell proliferation |
| 5 | HDAC1 | CNV | BICC1 | negative regulation of transcription from RNA polymerase II promoter |
| 5 | HNF1A | CNV | BICC1 | positive regulation of transcription, DNA-dependent |
| 5 | NR5A2 | CNV | BICC1 | positive regulation of transcription, DNA-dependent |
| 5 | IPO13 | CNV | BICC1 | protein import into nucleus |
| 5 | MAP3K7 | CNV | BICC1 | transcription, DNA-dependent |
| 5 | SLC6A20 | CNV | BICC1 | transmembrane transport |
| 5 | BTRC | CNV | BICC1 | ubiquitin-dependent protein catabolic process |
| 5 | BCL9 | CNV | BICC1 | Wnt receptor signaling pathway |
| 5 | TP53 | ssNV | BICC1 | transcription, DNA-dependent |
| 6 | PLCG1 | CNV | EGFR/FGFR | axon guidance |
| 6 | MAP2K1 | CNV | EGFR/FGFR | innate immune response |
| 6 | MAPK8 | CNV | EGFR/FGFR | peptidyl-threonine phosphorylation |
| 6 | GAB1 | CNV | FGFR | heart development |
| 6 | ATF2 | CNV | FGFR | innate immune response |
| 6 | MAPKAPK2 | CNV | FGFR | innate immune response |
| 6 | RPS6KA5 | CNV | FGFR | innate immune response |
| 6 | HGF | CNV | FGFR | mitosis |
| 6 | FRS2 | CNV | FGFR | ventricular septum development |
| 6 | FGF2 | CNV | FGFR | apoptotic process |
| 6 | FGFR2 | CNV | FGFR | apoptotic process |
| 6 | FGFR4 | CNV | FGFR | glucose homeostasis |
| 6 | FGF17 | CNV | FGFR | positive regulation of cell proliferation |
| 6 | FGF18 | CNV | FGFR | positive regulation of cell proliferation |
| 6 | FGF20 | CNV | FGFR | positive regulation of cell proliferation |
| 6 | FGFR1OP | CNV | FGFR | positive regulation of cell proliferation |
| 6 | FGFR1 | CNV | FGFR | transcription, DNA-dependent |
| 6 | MDM2 | CNV | TACC3 | protein ubiquitination |
| 6 | E2F2 | CNV | TACC3 | apoptotic process |
| 6 | GADD45A | CNV | TACC3 | apoptotic process |
| 6 | HMGB2 | CNV | TACC3 | apoptotic process |
| 6 | RHOA | CNV | TACC3 | axon guidance |
| 6 | PEBP1 | CNV | TACC3 | brain development |
| 6 | EVI5 | CNV | TACC3 | cell cycle |
| 6 | CDCA8 | CNV | TACC3 | cell division |
| 6 | CKAP5 | CNV | TACC3 | cell division |
| 6 | PPP1CC | CNV | TACC3 | cell division |
| 6 | BUB1 | CNV | TACC3 | cell proliferation |
| 6 | GTSE1 | CNV | TACC3 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest |
| 6 | TACC1 | CNV | TACC3 | microtubule cytoskeleton organization |
| 6 | KIF20A | CNV | TACC3 | microtubule-based movement |
| 6 | KIF2C | CNV | TACC3 | microtubule-based movement |
| 6 | NCAPH | CNV | TACC3 | mitosis |
| 6 | NSUN2 | CNV | TACC3 | mitosis |
| 6 | AKAP9 | CNV | TACC3 | mitotic cell cycle |
| 6 | KIF23 | CNV | TACC3 | mitotic cell cycle |
| 6 | MCM5 | CNV | TACC3 | mitotic cell cycle |
| 6 | NPM1 | CNV | TACC3 | negative regulation of cell proliferation |
| 6 | CBX5 | CNV | TACC3 | negative regulation of transcription, DNA-dependent |
| 6 | MKI67 | CNV | TACC3 | organ regeneration |
| 6 | AURKAIP1 | CNV | TACC3 | positive regulation of proteolysis |
| 6 | AKT1 | CNV | TACC3 | protein ubiquitination |
| 6 | BRCA1 | CNV | TACC3 | protein ubiquitination |
| 6 | KLHL13 | CNV | TACC3 | protein ubiquitination |
| 6 | KLHL9 | CNV | TACC3 | protein ubiquitination |
| 6 | TTF2 | CNV | TACC3 | regulation of transcription, DNA-dependent |
| 6 | RACGAP1 | CNV | TACC3 | signal transduction |
| 6 | TDRD7 | CNV | TACC3 | spermatogenesis |
| 6 | PRKACA | CNV | TACC3 | transmembrane transport |
FGFR2-MGEA5 as a putative therapeutic target
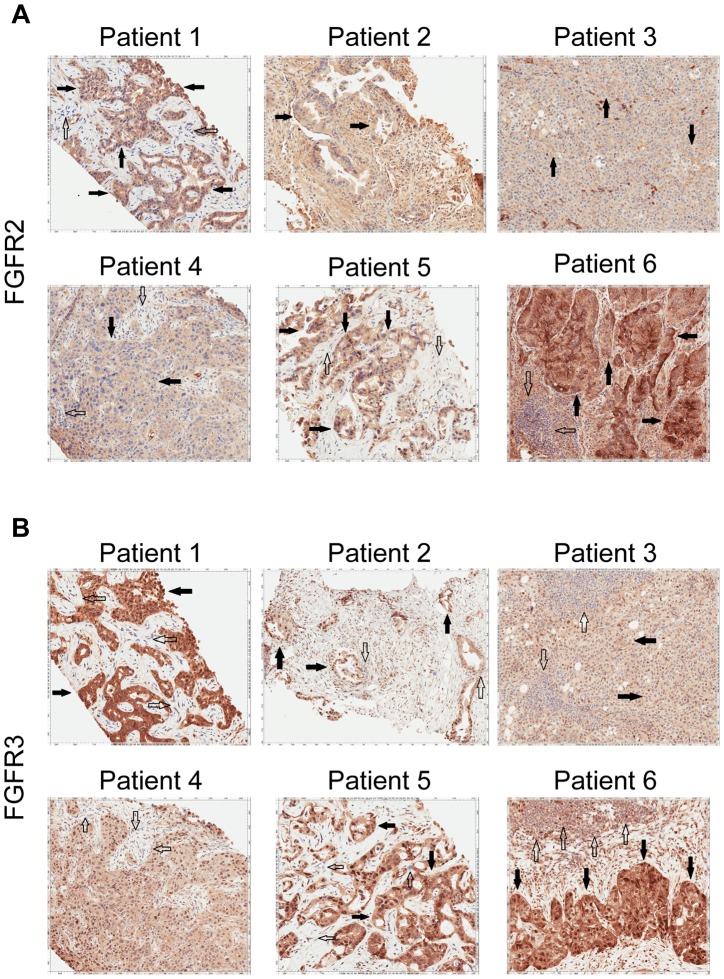
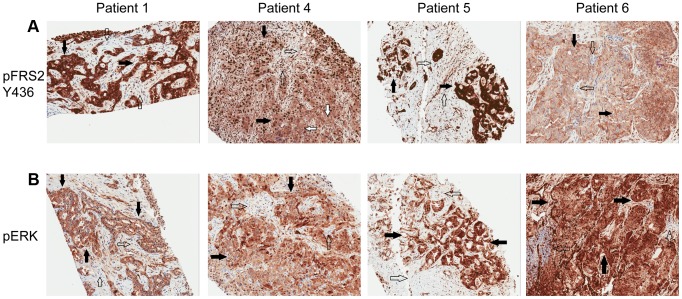
Patient 4 is a 62 year-old white female found to have a left-sided intrahepatic mass with satellite lesions, with metastasis to regional lymph nodes ( Table 5 ). A biopsy of the liver mass revealed the presence of a poorly differentiated adenocarcinoma that was consistent with intrahepatic cholangiocarcinoma (CK7+, CEA+, CK20+, Hep-par 1−, TTF-1−) ( Table 6 ). She received gemcitabine and cisplatin and obtained clinical benefit in the form of stable disease for 6 months, followed by disease progression. She was re-treated with gemcitabine and capecitabine systemic therapy and attained stable disease for 6 months, followed by disease progression. A clinical trial of pegylated hyaluronidase (PEGPH20) produced only stable disease for 4 months, followed again by disease progression. At this juncture, she underwent a liver biopsy to obtain tissue for whole genome characterization of her tumor. She was found to have an FGFR2-MGEA5 fusion ( Table 7 , Figure 2 ) and ponatinib monotherapy was pursued as salvage treatment. Evaluation of pre-treatment immunohistochemistry demonstrated increased expression of FGFR2 and FGFR3 ( Figure 5 ) and Clinical Laboratory Improvement Amendments (CLIA) validation by quantitative PCR revealed increased expression of FGFR3 (Table S5). In order to further validate the activation of the receptor, we conducted immunohistochemistry (IHC) of pFRS2 Y436 and pERK(MAPK) that revealed strong expression of pFRS2 Y436 and pERK ( Figure 6 ), thus confirming activation of the receptor.
Table 5. Clinical characteristics of 6 advanced, sporadic biliary tract cancer patients.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
| Age (years) | 64 | 66 | 50 | 62 | 50 | 43 |
| Gender | F | M | M | F | F | F |
| Location of Primary Tumor | Intrahepatic | Intrahepatic/Gallbladder | Intrahepatic | Intrahepatic | Intrahepatic | Intrahepatic |
| Stage | III | IV | IV | IV | IV | IV |
| CA19-9 (Units/ml) | WNL | 1008 | WNL | WNL* | N/A | 56 |
| Sites of Metastasis | N/A | Abdominal Lymph Nodes | Cervical,Thoracic, Abdominal, Pelvic Lymph Nodes | Abdominal, Pelvic Lymph Nodes, Liver | Liver, Lungs, Peritoneum | Lungs |
| Underlying Etiology | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown |
| Liver fluke | No | No | No | No | No | No |
| Hepatitis B | Unknown | Unknown | Negative | Unknown | Unknown | Unknown |
| Hepatitis C | Unknown | Unknown | Negative | Unknown | Unknown | Unknown |
| Prior Surgical Resection | No | Yes | Yes | No | Yes | No |
| Prior Radiation Therapy | No | No | No | No | No | No |
| Systemic Chemotherapy | Gem/Cis | Gem/Cis, Capecitabine | Gem/Cis | Gem/Cis, Gem/Cape, PEGPH20 | Gem/Cis, 5-FU/Carbo | Gem/Cis, FOLFOX, Pazopanib |
| Survival Status | Alive | Dead | Dead | Alive | Dead | Alive |
| Survival Duration from biopsy (months) | 14.5+ | 8.8 | 9.0 | 9.3+ | 4.1 | 5.5+ |
F = female; M = male; WNL = Within Normal Limits; Gem/Cis = Gemcitabine and Cisplatin; Gem/Cape = Gemcitabine and Capecitabine; PEGPH20 = pegylated hyaluronidase; 5-FU/Carbo = 5-Fluorouracil and Carboplatin; FOLFOX – 5-FU, Leucovorin and Oxaliplatin,
= WNL at baseline but 1408 U/ml prior to therapy and N/A = Not Available.
Table 6. Pathological characteristics of 6 advanced, sporadic biliary tract cancer patients.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
| Grade/differentiation | Moderate | Moderate | Undifferentiated** | Poor | Moderate | Poor |
| Biopsy Procedure | U/S Guided Liver Biopsy | U/S Guided Liver Biopsy | Excisional Biopsy Lymph Node | U/S Guided Liver Biopsy | U/S Guided Liver Biopsy | Excisional Lung Biopsy |
| %Necrosis (aliquots) | 5 (1) | 0 (2) | 0–35 (7) | 0 (3) | 0–5 (3) | 0 |
| %Tumor | 50 | 10–20 | 25–75 | 0–20 | 40–50 | 30 |
| %Stroma and normal elements | 50 | 80–90 | 25–75 | 80–100 | 50–60 | 70 |
| Histological Type | NST* | NST | NST | NST | NST | NST |
| Clear Cell Histology (Yes/No) | No | No | No | No | No | No |
U/S = Ultrasound.
NST: No special type.
Rare gland formation with expression of cytokeratin, polyclonal CEA, and MOC-31.
All were adenocarcinomas of no special types and high grades as defined by the World Health Organization Classification of Tumors of the Digestive System (Lyon 2000). Degree of differentiation is based on the percentage of glands (defined as having visible lumens by visual estimate) as follow: 95% or more glands-well differentiated, 40–94% glands-moderately differentiated, 5–39% glands-poorly differentiated, <5% glands-undifferentiated.
Table 7. Fusion events.
| Gene1 | Gene2 | Gene1 break location | Gene2 break location | Predicted Reciprocal Translocation | Patient | |
| Fusions | FGFR2 | MGEA5 | chr10:123243211 | chr10:103552699 | No | 4 |
| FGFR2 | BICC1 | chr10:123237843 | chr10:60380614 | Yes | 5 | |
| BICC1 | FGFR2 | chr10:60272900 | chr10:123237848 | Yes | 5 | |
| FGFR2 | TACC3 | chr10:123243211 | chr4:1741428 | No | 6 |
Figure 5. Immunohistochemistry demonstrating FGFR2 and FGFR3 expression.
A) Tumor stained with FGFR2 antibody. Patient 1 demonstrates moderate cytoplasmic positivity (solid arrows); background fibro-inflammatory tissue is negative (empty arrows). Patient 2 demonstrates moderate cytoplasmic expression for FGFR2; tumor nuclei are negative. Patient 3 demonstrates tumor cells with negative nuclear and weak cytoplasmic expression of FGFR2 (solid arrows) with cells demonstrating moderate basolateral or complete membranous staining as well. Patient 4 demonstrates weak/moderate cytoplasmic positivity with multi-focal weak/moderate membranous expression (solid arrows); background fibro-inflammatory tissue demonstrates negative/weak staining (empty arrows). Patient 5 demonstrates weak/moderate cytoplasmic positivity with multi-focal moderate/strong membranous expression (solid arrows); background fibro-inflammatory tissue is negative (empty arrows). Patient 6 demonstrates moderate/strong cytoplasmic positivity (solid arrows); background lymphocytes are negative (empty arrows). B) Tumor stained with FGFR3 antibody. Patient 1 demonstrates strong cytoplasmic positivity, variable nuclear expression and occasional moderate/strong membranous expression (solid arrows); background fibrous tissue is negative (empty arrows). Patient 2 demonstrates negatively staining background neutrophils (focally intraepithelial-far right) (empty arrows) and tumor cells with strong nuclear expression and moderate cytoplasmic positivity (solid arrows). Patient 3 demonstrates negatively staining background inflammation (empty arrows) and tumor cells with weak nuclear expression and moderate cytoplasmic positivity (solid arrows). Patient 4 demonstrates weak/moderate cytoplasmic positivity and variable nuclear expression; background fibro-inflammatory tissue demonstrates negative/weak positivity (empty arrows). Patient 5 demonstrates moderate cytoplasmic positivity, variable nuclear expression and strong multi-focal membranous expression (solid arrows); background fibrous tissue is negative. Patient 6 demonstrates diffuse/moderate/strong cytoplasmic and membranous positivity and variable nuclear expression (solid arrows); background lymphocytes are negative (empty arrows).
Figure 6. Immunohistochemistry demonstrating pFRS2 Y436, and pERK expression in Patients 1, 4, 5 and 6.
A) Tumor stained with pFRS2 Y436 antibody. Patient 1 tumor cells demonstrating both strong cytoplasmic and nuclear expression of pFRS2 (solid arrows); background fibrous stroma is negative (empty arrows). Patient 4 tumor cells show strong nuclear expression and moderate to strong cytoplasmic positivity (solid arrows); occasional background fibrous stromal cells are negative for pFRS2 (empty arrows) and scattered tumor cells show basolateral/membranous staining as well (white arrows). Patient 5 tumor cells show intensely strong expression in both nuclei and cytoplasm (solid arrows); scattered background fibrous stromal cells are negative (empty arrows). Patient 6 tumor cells show negative nuclear expression of pFRS2, moderate cytoplasmic expression and basolateral or membranous expression of varying intensity (solid arrows); background fibrous stromal cells are negative (empty arrows). B) Tumor stained with pERK(MAPK) antibody. Patient 1 demonstrates negative/weak fibrous stroma (empty arrows) and tumor cells with negative nuclei and moderate to strong cytoplasmic expression (solid arrows). Patient 4 demonstrates negative inflammatory background (empty arrows) tumor cells with variable negative to strong nuclear expression and moderate to strong cytoplasmic positivity (solid arrows). Patient 5 demonstrates negative/weak fibrous stroma (empty arrows) and tumor cells with strong nuclear and cytoplasmic expression (solid arrows). Patient 6 demonstrates negative background lymphocytes/mononuclear inflammatory cells (empty arrows) and tumor cells with strong nuclear and cytoplasmic expression (solid arrows).
Ponatinib was initiated at 45 mg given orally on a daily schedule. Approximately 6 weeks after initiation of therapy she was noted to have anti-tumor activity that was characterized by central necrosis of a caudate liver lobe mass, shrinkage of metastatic lymph nodes involving the right cardiophrenic angle, central necrosis and shrinkage of a metastatic supraceliac axis lymph node ( Figure 7 ) and reduction in CA 19-9 from 1408 U/ml to 142 U/ml. Per RECIST criteria, she exhibited stable disease with a 14% decrease in the sum of largest diameters but with tumor necrosis and reduction in the CA19-9 tumor marker (89.8%). While the evidence is preliminary in nature, it was felt that the combination of tumor shrinkage not meeting the RECIST criteria definition of partial response, tumor necrosis and reduction in CA19-9 constituted preliminary evidence of anti-tumor activity. She has experienced no treatment related toxicities thus far and remains on therapy of approximately 3.5 months duration thus far.
Figure 7. Anti-tumor activity in Patient 4 harboring an FGFR2-MGEA5 fusion, to FGFR inhibitors.
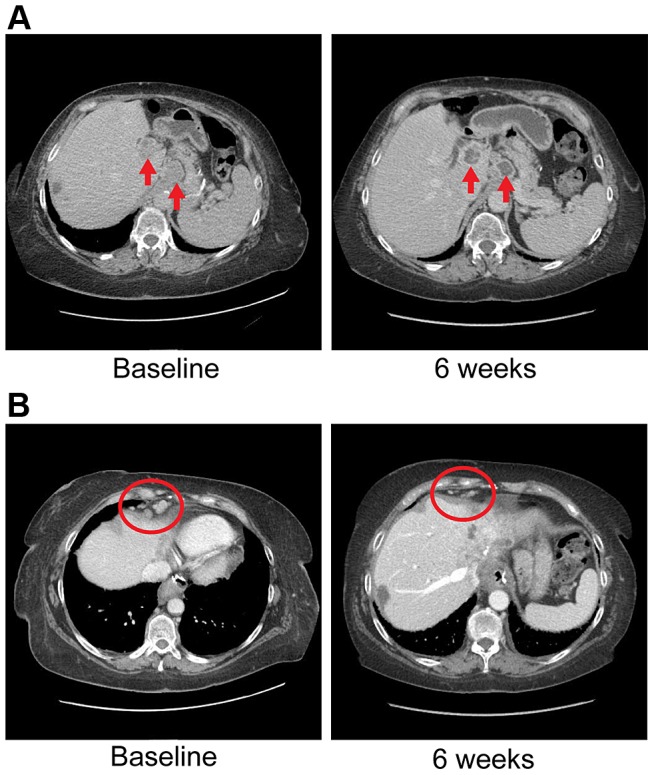
A) CT images of patient 4, whose tumor possessed an FGFR2-MGEA5 fusion, at baseline and 6 weeks demonstrate central necrosis of a caudate liver lobe mass (left arrow), 2.6 cm at baseline and 6 weeks, and shrinkage of a metastatic supraceliac axis lymph node (right arrow), 3.1 cm and 2.9 cm at baseline and 6 weeks respectively. B) CT images of patient 4 showing shrinkage of metastatic lymph nodes involving the right cardiophrenic angle (red circles), 1.3 cm and 0.5 cm at baseline and 6 weeks respectively.
The FGFR2 fusion partner observed in this patient, MGEA5, is an enzyme responsible for the removal of O-GlcNAc from proteins [66]. Interestingly, soft tissue tumors myxoinflammatory fibroblastic sarcoma (MIFS) and hemosiderotic fibrolipomatous tumor (HFLT) both share a translocation event resulting in rearrangements in TGFBR3 and MGEA5 [67], [68]. Associated with this translocation event is the upregulation of NPM3 and FGF8 [68], of which both genes are upregulated in this patient (fold change: NPM3 = 6.17865, FGF8 = 1.79769e+308). In breast cancer, grade III tumors had significantly lower MGEA5 expression than grade I tumors with a trend of decreasing expression observed with increasing tumor grade [66]. In summary, MGEA5 may play an important role in carcinogenesis as an FGFR fusion partner.
FGFR2-TACC3 as a putative therapeutic target
Patient 6 is a 43 year-old white female who underwent a right salpingo-oophorectomy and endometrial ablation in the context of a ruptured ovarian cyst ( Table 5 ). Postoperatively she developed dyspnea and was found to have pulmonary nodules as well as a 5 cm left sided liver mass. Pathological evaluation of the liver mass was consistent with a moderately differentiated intrahepatic cholangiocarcinoma (CK7+, CK20−, TTF-1−) in the absence of any known risk factors ( Table 6 ). She was treated systemically with gemcitabine and cisplatin and had stable disease for approximately 6 months, but was subsequently found to have disease progression. She was treated with FOLFOX for 7 months and again attained stable disease as best response to therapy but eventually experienced disease progression. Upon disease progression, she was enrolled on a clinical study with the multi-kinase inhibitor pazopanib that is FDA-approved for the treatment of advanced renal cancer or sarcoma – and fortuitously has nanomolar activity against FGFR2 (in vitro IC50 to FGFR2≈350 nM) [69]. Transcriptome analysis revealed the presence of an FGFR2-TACC3 fusion ( Table 7 , Figure 2 ). Evaluation of post-pazopanib tissue by immunohistochemistry revealed increased expression of FGFR2 and FGFR3 ( Figure 5 ) Further evaluation of phosphorylation of downstream targets FRS2 Y436, and ERK(MAPK) revealed strong expression of pERK and moderate expression of pFRS2 Y436 ( Figure 6 ), confirming activation of the receptor. She had been treated with pazopanib 800 mg orally daily for 4 months and demonstrated tumor shrinkage, which in retrospect, was postulated to be secondary to the FGFR2 inhibitory activity of pazopanib ( Figure 8A ). By RECIST criteria v1.1, the patient had a partial response to therapy as evidenced by a 71% decrease in the sum of diameters. Subsequently, the same patient was treated with a dedicated pan-FGFR inhibitor, ponatinib, (45 mg daily orally; in vitro IC50 : FGFR1≈24 nM, FGFR2≈8 nM, FGFR3≈8 nM and FGFR4≈34 nM). She again attained minor tumor shrinkage (stable disease by RECIST criteria v1.1, decrease of 4% in sum of largest diameters) in multiple lesions after 2 months of therapy, despite undergoing a 50% dose reduction for abdominal pain felt to be related to drug ( Figure 8B ). She remains on therapy approximately 4 months since the initiation of ponatinib. As such, anti-tumor activity was obtained with two distinct FGFR inhibitors in the same patient.
Figure 8. Anti-tumor activity in Patient 6, harboring an FGFR2-TACC3 fusion, to FGFR inhibitors.
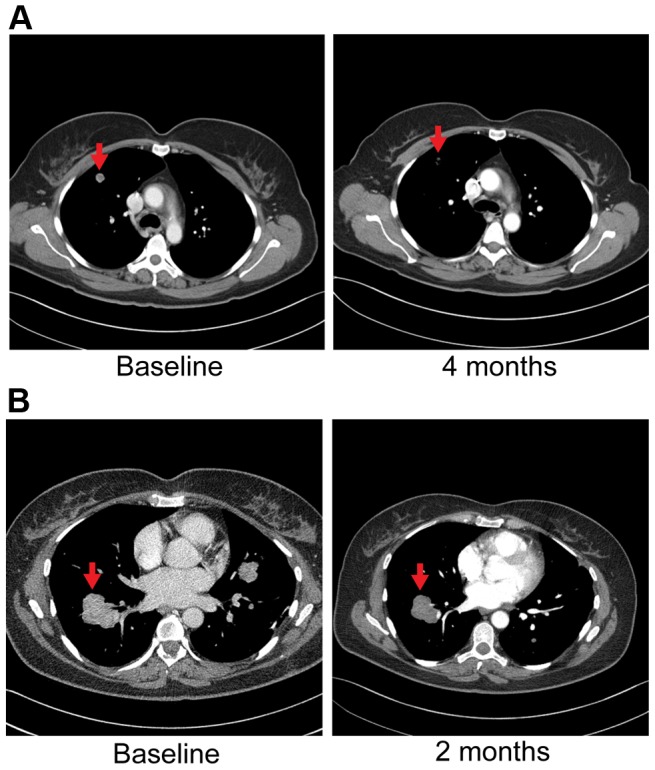
A) CT images of patient 6, whose tumor possessed an FGFR2-TACC3 fusion, at baseline and after four months of pazopanib demonstrate significant tumor shrinkage (red arrows), 10.8 mm and 3.1 mm respectively. B) CT images of patient 6 at baseline and two months demonstrate significant tumor shrinkage (red arrows), 41.1 mm and 39.4 mm respectively after subsequent ponatinib treatment, 45 mg/daily, was begun.
The FGFR2 fusion partner observed in this patient's tumor, TACC3, is overexpressed in many tumor types with enhanced cell proliferation, migration, and transformation observed in cells overexpressing TACC3 [70]. Furthermore regulation of ERK and PI3K/AKT by TACC3 may contribute in part to epithelial-mesenchymal transition (EMT) in cancer [70], a significant contributor to carcinogenesis. Interestingly, TACC3 has been identified as a fusion partner to FGFR3 in bladder cancer, squamous cell lung cancer, oral cancer, head and neck cancer and glioblastoma multiforme [54].
ERRFI1 as a putative therapeutic target
Patient 3 was a 50 year-old white male who presented with fevers and night sweats ( Table 5 ). He was found to have a 4 cm tumor in his liver determined to be a poorly differentiated intrahepatic cholangiocarcinoma (CK7+, CK20−, TTF1−, CD56−, synatophysin−, Hep-par 1−) with sclerotic features ( Table 6 ). No overt risk factors for cholangiocarcinoma were identified. A left hepatectomy was undertaken three months later. In addition to the primary tumor in segment 4, limited resections of segments 6 and 8 were undertaken to remove two tumor nodules. He was soon noted to have increased hypermetabolism in the left lower cervical, upper mediastinal and abdomino-retroperitoneal lymph nodes related to metastatic disease from his cholangiocarcinoma. He was treated with gemcitabine and cisplatin for 9 months and obtained stable disease as his best response, followed by eventual progression. He received treatment with pegylated hyaluronidase (PEGPH20) in the setting of an investigational study for one month and had no response to therapy. A biopsy of a left supraclavicular lymph node was obtained two months prior to the initiation of PEGPH20 in the context of a clinical study employing whole genome analysis for putative therapeutic target selection.
Since our study goal was to identify potential therapeutically relevant events, the novel loss of function mutation in ERRFI1 (E384X) detected in Patient 3's metastatic, recurrent/refractory SIC (Table S1) warranted additional examination. Specifically, the allelic fraction of the DNA mutation constituted only 11% of the sequencing reads, is consistent with tissue heterogeneity, and constituted 78% of the sequencing reads within the RNASeq data. Such allele specific expression of the mutated allele from the same tissue specimen suggests nearly complete loss of function of ERRFI1 in this patient's tumor. Notably, the patient's tumor did not harbor any mutations or amplifications in other EGFR signaling members such as EGFR and BRAF.
Upon availability of CLIA validated sequencing data (Table S5), the patient was treated with erlotinib 150 mg orally/daily. After 3 months, RECIST v1.1 partial response evidenced by a decrease of 58% in the sum of largest diameters was observed ( Figure 9 ). Evaluation of pretreatment tumor tissue by immunohistochemistry revealed increased expression of EGFR pathway members ( Figure 10 ).
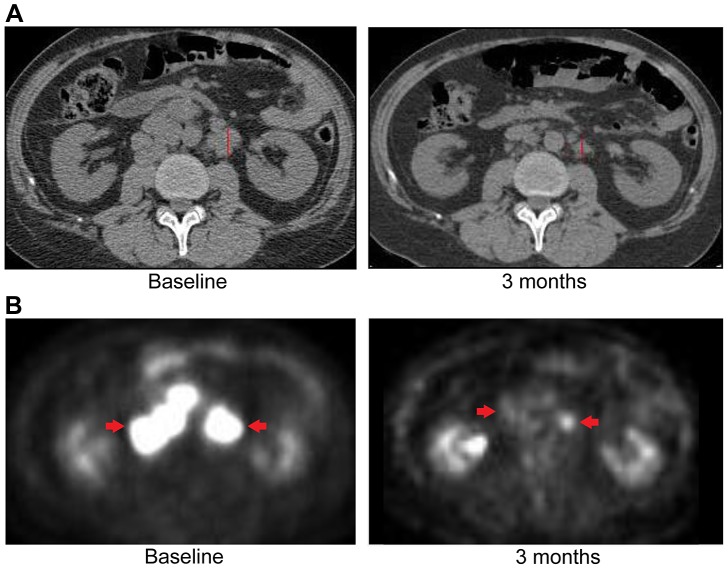
Figure 9. Anti-tumor activity of Patient 3, harboring an ERRFI1 mutation, to erlotinib, an EGFR inhibitor.
A) CT images of patient 3 at baseline and three months demonstrate significant tumor shrinkage (red marks). CT demonstrates right retroperitoneal lymph nodes decreasing from 7.6 cm to 2.9 cm and left retroperitoneal lymph nodes decreasing from 3.3 cm to 1.7 cm. B) PET images of patient 3 at baseline and three months demonstrate significant tumor shrinkage (red arrows). Hypermetabolic areas corresponding to right retroperitoneal lymph nodes demonstrate decrease from 8 cm longest diameter to imperceptible and left retroperitoneal lymph nodes decreasing from 4.2 cm to 1.4 cm. Both regions demonstrated significant reduction in metabolic activity.
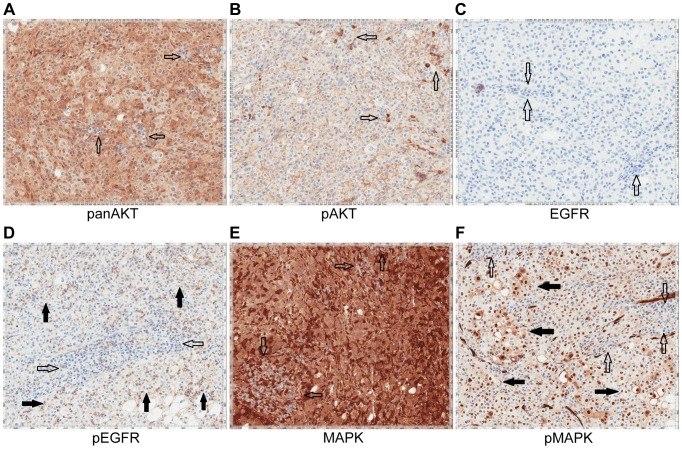
Figure 10. Immunohistochemistry of Patient 3's tumor demonstrating activation of the EGFR pathway.
A) Tumor stained with panAKT demonstrating diffuse cytoplasmic positivity with negative background lymphocytes (empty arrows). B) Tumor stained with pAKT demonstrating diffuse membranous staining and negative cytoplasmic expression; scattered background inflammatory cells showing strong cytoplasmic staining (empty arrows). C) Tumor stained with EGFR. Tumor cells are EGFR negative with background lymphocytes also negative (empty arrows). D) Tumor stained with pEGFR showing membranous positivity (solid arrows) with negative background lymphocytes (empty arrows). E) Tumor stained with MAPK/ERK1/2 demonstrating moderate to strong cytoplasmic staining of total MAPK with negative background lymphocytes (empty arrows). F) Tumor stained with pMAPK/pERK demonstrating increased expression compared to the negative background lymphocytes (empty arrows).
Discussion
Integrated analysis of sporadic intrahepatic cholangiocarcinoma (SIC) genomic and transcriptomic data led to the discovery of FGFR2 fusion products in three of six assessed patients ( Table 7 , Figures 4 and 11 ). Members of the FGFR family (FGFR1-4) have been associated with mutations, amplifications and translocation events with oncogenic potential [71]. FGFR fusions with oncogenic activity have been previously identified in bladder cancer (FGFR3) [72], lymphoma (FGFR1 and FGFR3) [73], [74], acute myeloid leukemia (FGFR1) [75], multiple myeloma [76], myeloproliferative neoplasms [77], and most recently glioblastoma multiforme (FGFR1 and FGFR3) [78]. FGFR2, FGFR3 and FGFR4 have been found to be overexpressed in IDH1/IDH2 mutant biliary cancers [79], a context seen within Patient 1 in our study (Tables S1 and S6, Figure 5 ); although, no fusion events were depicted in these studies or in Patient 1.
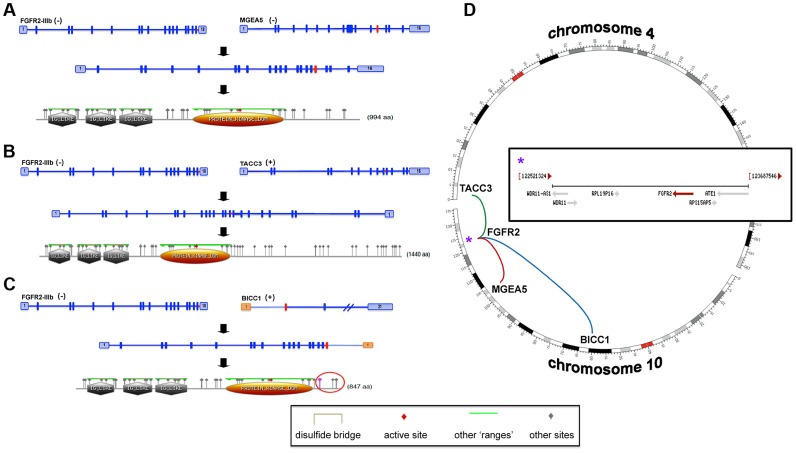
Figure 11. FGFR2-IIIb fusion events.
Transcripts and hypothetical protein products are modeled to illustrate the potential functional impact of fusion events involving FGFR2 (A–C). The identified fusion events involving MGEA5 (patient 4) (A) and BICC1 (patient 5, reciprocal event) (C) are chromosome 10 intrachromosomal (D). In addition, patient 6 carried an interchromosomal fusion event (D) involving FGFR2 and TACC3 (B). The FGFR2 gene encodes for several isoforms with eleven representative transcripts and patients 4, 5, and 6 carry fusions involving the epithelial cell specific transcript isoform (FGFR2-IIIb). All identified fusion breakpoints are close in proximity and are predicted to occur within the last intron of the transcript and terminal to a known protein tyrosine kinase domain (A–C, gold domain). Predicted “Other” sites for all of the fusion protein models are the same and include the following: Casein kinase II phosphorylation sites, N-glycosylation sites, Protein kinase C phosphorylation sites, N-myristoylation sites, Tyrosine kinase phosphorylation sites, and cAMP-/cGMP-dependent protein kinase phosphorylation sites (A–C, grey triangle annotations). In all cases, fusions result in a predicted expansion of Casein kinase II phosphorylation and Protein kinase C phosphorylation sites. A protein product model is shown only for one of the reciprocal events involving the FGFR2 and BICC1 genes (FGFR2→BICC1, C). The fusion breakpoints of the reciprocal events effect Exons 1 and 2 of the BICC1 gene, which translates to a difference of a predicted phosphoserine site within the Casein kinase II phosphorylation region (C, purple triangle within red circle). The FGFR2 gene is located within a fragile site region (FRA10F) and is flanked by two ribosomal protein pseudogenes, RPS15AP5 and RPL19P16 (see D inset (*)), whose repetitive sequence content may also contribute to genomic instability at the FGFR2 initiation site.
Although the gene partner fused to FGFR2 was different for each patient (MGEA5, BICC1 and TACC3), the breakpoints in FGFR2 all occurred within the last intron distal to the last coding exon and terminal protein tyrosine kinase domain ( Figure 11 ). All three fusions were validated at the DNA and/or RNA level ( Table 8 ). Amongst these fusions, the FGFR2-BICC1 fusion has recently been independently identified in SIC [53], [54]. For this particular fusion product we observed, and validated, the presence of two fusion isoforms (FGFR2-BICC1 and BICC1-FGFR2). Interestingly, BICC1 is a negative regulator of Wnt signaling [80] and when comparing expression of tumor and normal tissue we observed differentially expressed Wnt signaling genes, APC (fold change -4.75027), GSK3B (fold change -3.35309), and CTNNB1 (fold change -1.73148), yet when the expression was compared to other cholangiocarincomas, no difference was observed.
Table 8. DNA and RNA validation of FGFR2 fusions in 3 patients with advanced sporadic biliary tract cancer.
| Fusion | Annealing site | PCR input | Direction | Primer sequence |
| FGFR2-MGEA5 | FGFR2 | gDNA | F | 5′-CTGACTATAACCACGTACCC-3′ |
| MGEA5 | gDNA | R | 5′-AGGGAGAAATTAAAGAACTTGG-3′ | |
| FGFR2 | cDNA | F | 5′-TGATGATGAGGGACTGTTG-3′ | |
| MGEA5 | cDNA | R | 5′-GAGTTCCTTGTCACCATTTG-3′ | |
| FGFR2-BICC1 | FGFR2 | gDNA | F | 5′-GGCAGAAGAAGAAAGTTGG-3′ |
| BICC1 | gDNA | R | 5′-ACTACTGCAGTTTGTTCAAT-3′ | |
| FGFR2 | cDNA | F | 5′-TGATGATGAGGGACTGTTG-3′ | |
| BICC1 | cDNA | R | 5′-TGTGTGCTCACAGGAATAG-3′ | |
| BICC1-FGFR2 | BICC1 | cDNA | F | 5′ CGTGGACAGGAAGAAACT-3′ |
| FGFR2 | cDNA | R | 5′-GTGTGGATACTGAGGAAG-3′ | |
| FGFR2-TACC3 | FGFR2 | gDNA | F | 5′-TGACCCCCTAATCTAGTTGC-3′ |
| TACC3 | gDNA | R | 5′-AACCTGTCCATGATCTTCCT-3′ |
F - forward, R - reverse.
The FGFR genes encode multiple structural variants through alternative splicing. Notably, RNASeq data revealed that the FGFR2-IIIb isoform was present in all fusions detected in our study and has been shown to have selectivity for epithelial cells as opposed to the FGFR2-IIIc isoform, which is found selectively in mesenchymal cells [81]. Paradoxically, wildtype FGFR2-IIIb has been described as a tumor suppressor in pre-clinical systems of bladder cancer and prostate cancer [82], [83]. As such, FGFR signaling appears context-dependent and exhibits variability in disparate tumor types.
Importantly, one critical study has shown that FGFR2 carboxy-terminal deletion mutants induce ligand-independent transformation and clonogenic growth [84]. This is important because all of the fusion events within our study would lead to loss of the carboxy-terminus of FGFR2. Furthermore, a very recent study that described FGFR fusions in solid tumors illustrated that FGFR fusion partners in SIC resulted in dimerization domains, and suggested that activation occurred through ligand independent dimerization and oligomerization [54]. It is likely that both loss of the carboxy terminus and the addition of dimerization domains leads to oncogenic FGFR2 activity in these tumors.
Comparative pathway analysis of genes carrying mutations/aberrant in copy number identified additional potential therapeutic targets belonging to, or intimately integrated with, the EGFR and FGFR signaling pathways ( Figure 3 , Tables S2, S3, S4). Interestingly, most of these pathways also have known roles in mediating epithelial-to-mesenchymal cell transitions, which occur frequently during development as well as during tumorigenesis [60]. Patients 3 and 4 harbored aberrations in several genes acting in cadherin signaling pathways (Tables S3, S4), which are important for maintaining cell-cell adhesion [63].
The preliminary anti-tumor activity noted in a patient with FGFR2-MGEA5 (Patient 4) and FGFR2-TACC3 fusion (Patient 6) represent the first reports of application of FGFR inhibitors to the treatment of patients with cholangiocarcinoma harboring these alterations. These results suggest that oncogenic activation of FGFR2 represent a potential therapeutically actionable event. The FGFR tyrosine kinase inhibitors (TKI) dovitinib [85] and NVP-BGJ398 [86] are currently in clinical development and the FGFR TKI ponatinib [75], [87] was recently approved by the FDA for use in treating T315I mutant chronic myelogenous leukemia. FGF7 (keratinocyte growth factor) has been previously linked to poor prognosis in patients with biliary tract cancer and a small molecule FGFR kinase inhibitor, Ki23057, has demonstrated efficacy in preclinical models [88]. It should be recognized that small molecule tyrosine inhibitors are almost universally promiscuous with regards to specificity and typically significant off-target effects are resultant. Off target efficacy resulting from inhibition of angiogenic kinases in addition to FGFR2 inhibition could explain the anti-tumor activity exhibited in patient 6, as pazopanib has been shown to have nanomolar range potency towards VEGFR1-3, PDGFRA/B and CKIT as well [89].
Larger trials, preferably of a randomized nature with a control arm, need to be conducted to truly define the role of FGFR inhibitors in the treatment of patients with cholangiocarcinoma, particularly those harboring FGFR2 fusions. While our results provide impetus and enthusiasm towards this end, at this stage they should be considered preliminary in nature.
The preliminary anti-tumor activity observed in patient 6 with both pazopanib, and subsequently ponatinib, is particularly intriguing, but also raises important questions. There was an initial response to pazopanib, followed by disease progression. This is a phenomenon observed with the clinical application of most targeted therapeutic approaches. Potential explanations include tumor heterogeneity resulting from clonal selection, transcriptional up-regulation of escape pathways, epigenetic mechanisms and other yet undefined mechanisms of resistance to therapy. The patient did not have additional known alterations in key oncogenic pathways in genes such as BRAF, KRAS, EGFR and PIK3CA, which if present, could provide a putative basis for eventual escape from FGFR pathway inhibition. It is unclear why patient 6 initially responded to pazopanib followed by resistance and subsequently responded to ponatinib, another FGFR inhibitor. Putative explanations include the higher potency of ponatinib observed in vitro to FGFR2 (IC50≈8 nM for ponatinib vs. 350 nM for pazopanib) and resistance being defined as >20% increase in sum of largest diameters per RECIST v1.1 standard criteria that triggered a discontinuation from pazopanib and recapturing of anti-tumor activity by subsequent inhibition of the FGFR pathway which still maintained therapeutic relevance in that patient at a later time point.
ERRFI1 has a role as a negative regulator of EGFR dependent skin morphogenesis [90], [91], uterine steroid hormone responsiveness [92] and as a tumor suppressor gene [90], [93], [94]. ERRFI1 is an endogenous inhibitor of EGFR, ERRB2, ERRB3 and ERRB4 through direct interaction with the kinase domains of these proteins [95], [96] and endocytosis/lysosomal degradation of ERBB receptors [97]. ERRFI1 deletions have been found in glioblastoma multiforme and breast cancer [98]–[100]. Other mechanisms of ERRFI1 loss include methylation, acetylation and loss of function mutations [98], [101], [102]. Consistent with a driver role of this mutation, previously germline homozygous disruption of ERRFI1 in mice induces hyperplasia and adenoma formation in the epithelium and development of spontaneous adenocarcinomas of the lung, gallbladder and biliary tract [103]. The tyrosine kinase inhibitor gefitinib has demonstrated anti-tumor activity in mice in spontaneous tumors driven by ERRFI1 germline loss [91].
Our results suggest immediate and actionable implications for SIC patients with tumors harboring ERFFI1 loss of function mutations or FGFR fusions, given the clinical availability of FDA-approved EGFR and FGFR tyrosine kinase inhibitors. Antibodies specific to FGFR2-IIIb have also shown preclinical efficacy and may serve as an additional platform for therapeutic development in this context [104]. Additional studies to characterize the prevalence of these aberrations in both sporadic and liver fluke associated BTC will need to be conducted. Nevertheless, our results suggest that prospective clinical studies designed to treat patient's tumors harboring these novel genomic aberrations utilizing targeted agents on an individualized basis should be pursued more fully through larger clinical studies in order to explore the precise extent of clinical benefit that this tailored approach may have in patients with primary or advanced BTC.
Additionally, post-treatment biopsies to assess pathway down-regulation in patients 4 and 6 (treated with FGFR inhibitors) and patient 3 (treated with EGFR inhibitor) are not available, as the treatment was not conducted in the setting of a protocol that would allow for the collection of additional research biopsies. Incorporation of post-treatment biopsies in carefully designed prospective studies will be critical towards defining the association between the use of FGFR and EGFR inhibitors in appropriately selected patients with relevant genomic aberrations.
Materials and Methods
Ethics statement and sample collection
Clinical information was assimilated from patient records from the Mayo Clinic. Informed consent was obtained for each patient on two ongoing research protocols approved by the Mayo Clinic Institutional Review Board (10-006180 and 10-002879). Clinicopathological features collected included age, gender, stage, histological grade, sites of metastasis, tumor sample assessment for overall cellularity/necrosis and percent tumor cellularity, prior therapies and risk factors (hepatitis B and C, Caroli's disease, obesity, hepatolithiasis and cholelithiasis, primary sclerosing cholangitis, thorotrast exposure and H. pylori, H. bilis, S. typhi and S. paratyphi infections). All patients were known to not have had prior exposure to liver flukes that have been implicated in biliary carcinogenesis (O. viverrini and C. sinensis). Tissue specimens were collected fresh frozen and maintained below −80°C until nucleic acid extraction. A board certified pathologist who is experienced in biospecimen studies, evaluated a portion of each specimen to confirm the presence of tumor, the degree of necrosis and the percent cellularity.
Whole genome sequencing
Patients 1, 3, 4, and 5
1.1 µg genomic DNA was used to generate separate long insert whole genome libraries for each sample using Illumina's (San Diego, CA) TruSeq DNA Sample Prep Kit (catalog# FC-121-2001). In summary, genomic DNAs are fragmented to a target size of 900–1000 bp on the Covaris E210. 100 ng of the sample was run on a 1% TAE gel to verify fragmentation. Samples were end repaired and purified with Ampure XP beads using a 1∶1 bead volume to sample volume ratio, and ligated with indexed adapters. Samples are size selected at approximately 1000 bp by running samples on a 1.5% TAE gel and purified using Bio-Rad Freeze ‘n Squeeze columns and Ampure XP beads. Size selected products are then amplified using PCR and products were cleaned using Ampure XP beads.
Patient 2
300 ng genomic tumor and normal DNA was used to create whole genome libraries. Samples were fragmented on the Covaris E210 to a target size of 200–300 bp and 50 ng of the fragmented product was run on a 2% TAE gel to verify fragmentation. Whole genome libraries were prepared using Illumina's TruSeq DNA Sample Prep Kit.
Exome sequencing
Patients 1 and 3
1.1 µg genomic DNA for each sample was fragmented to a target size of 150–200 bp on the Covaris E210. 100 ng of fragmented product was run on TAE gel to verify fragmentation. The remaining 1 µg of fragmented DNA was prepared using Agilent's SureSelectXT and SureSelectXT Human All Exon 50 Mb kit (catalog# G7544C).
Patient 2
50 ng genomic tumor and normal DNA was used to create exome libraries using Illumina's Nextera Exome Enrichment kit (catalog# FC-121-1204) following the manufacturer's protocol.
Patients 4 and 5
1 µg of each tumor and germline DNA sample was used to generate separate exome libraries. Libraries were prepared using Illumina's TruSeq DNA Sample Prep Kit and Exome Enrichment Kit (catalog# FC-121-1008) following the manufacturer's protocols.
Patient 6
3 µg of genomic tumor and normal DNA was fragmented on the Covaris E210 to a target size of 150–200 bp. Exome libraries were prepared with Agilent's (Santa Clara, CA) SureSelectXT Human All Exon V4 library preparation kit (catalog# 5190-4632) and SureSelectXT Human All Exon V4+UTRs (catalog# 5190-4637) following the manufacturer's protocols.
RNA sequencing
Patients 1, 2 and 3
50 ng total RNA was used to generate whole transcriptome libraries for RNA sequencing. Using the Nugen Ovation RNA-Seq System v2 (catalog# 7102), total RNA was used to generate double stranded cDNA, which was subsequently amplified using Nugen's SPIA linear amplification process. Amplified products were cleaned using Qiagen's QIAquick PCR Purification Kit and quantitated using Invitrogen's Quant-iT Picogreen. 1 µg of amplified cDNA was fragmented on the Covaris E210 to a target size of 300 bp. Illumina's TruSeq DNA Sample Preparation Kit was used to prepare libraries from 1 µg amplified cDNA.
Patients 4, 5 and 6
1 µg of total RNA for each sample was used to generate RNA sequencing libraries using Illumina's TruSeq RNA Sample Prep Kit V2 (catalog# RS-122-2001) following the manufacturer's protocol.
Paired end sequencing
Libraries with a 1% phiX spike-in were used to generate clusters on HiSeq Paired End v3 flowcells on the Illumina cBot using Illumina's TruSeq PE Cluster Kit v3 (catalog# PE-401-3001). Clustered flowcells were sequenced by synthesis on the Illumina HiSeq 2000 using paired-end technology and Illumina's TruSeq SBS Kit.
Alignment and variant calling
Whole genome and whole exome
For whole genome and exome sequencing fastq files were aligned with BWA 0.6.2 to GRCh37.62 and the SAM output were converted to a sorted BAM file using SAMtools 0.1.18. BAM files were then processed through indel realignment, mark duplicates, and recalibration steps in this order with GATK 1.5 where dpsnp135 was used for known SNPs and 1000 Genomes' ALL.wgs.low_coverage_vqsr.20101123 was used for known indels. Lane level sample BAMs were then merged with Picard 1.65 if they were sequenced across multiple lanes. Comparative variant calling for exome data was conducted with Seurat [105].
Previously described copy number and translocation detection were applied to the whole genome long insert sequencing data [59] and these are made available through https://github.com/davcraig75/tgen_somaticSV. Copy number detection was based on a log2 comparison of normalized physical coverage (or clonal coverage) across tumor and normal whole genome long-insert sequencing data, where physical coverage was calculated by considering the entire region a paired-end fragment spans on the genome, then the coverage at 100 bp intervals was kept. Normal and tumor physical coverage was then normalized, smoothed and filtered for highly repetitive regions prior to calculating the log2 comparison. Translocation detection was based on discordant read evidence in the tumor whole genome sequencing data compared to its corresponding normal data. In order for the structural variant to be called there needs to be greater than 7 read pairs mapping to both sides of the breakpoint. The unique feature of the long-insert whole-genome sequencing was the long overall fragment size (∼1 kb), where by two 100 bp reads flank a region of ∼800 bp. The separation of forward and reverse reads increases the overall probability that the read pairs do not cross the breakpoint and confound mapping.
RNA
For RNA sequencing, lane level fastq files were appended together if they were across multiple lanes. These fastq files were then aligned with TopHat 2.0.6 to GRCh37.62 using ensembl.63.genes.gtf as GTF file. Changes in transcript expression were calculated with Cuffdiff 2.0.2. For novel fusion discovery reads were aligned with TopHat-Fusion 2.0.6 [106] (patients 2, 3, 4 and 6). In addition, Chimerascan 0.4.5 [107] was used to detect fusions in patient 1, deFuse 5.0 [108] used in patients 2, 3 and 5 and SnowShoes [109] for patients 2 and 5.
Somatic mutation validation
Mutations of potential clinical relevance were confirmed in a Clinical Laboratory Improvement Amendments (CLIA) laboratory with Sanger sequencing or quantitative PCR.
Immunohistochemistry
The immunohistochemistry was performed following the procedures described previously [110]. Briefly, slides were dewaxed, rehydrated and antigen retrieved on-line on the BondMax autostainer (Leica Microsystems, INC Bannockburn, IL). Slides were then subjected to heat-induced epitope retrieval using a proprietary EDTA-based retrieval solution. Endogenous peroxidase was then blocked and slides were incubated with the following antibodies: FGFR2 (BEK, Santa Cruz, catalog# sc-20735), FGFR3 (C-15, Santa Cruz, catalog# sc-123), panAKT (Cell Signaling Technology, catalog# 4685, pAKT (Cell Signaling Technology, catalog# 4060), EGFR (Cell Signaling Technology, catalog# 4267, pEGFR (Cell Signaling Technology, catalog#2234), MAPK/ERK1/2 (Cell Signaling Technology, catalog# 4695), pMAPK/pERK (Cell Signaling Technology, catalog# 4376) and pFRS2 Y436 (Abcam, catalog# ab78195). Sections were visualized using the Polymer Refine Detection kit (Leica) using diaminobenzidine chromogen as substrate.
Fluorescent in-situ hybridization (FISH)
FISH was performed on formalin-fixed paraffin-embedded (FFPE) specimens using standard protocols and dual-color break-apart rearrangement probes specific to the FGFR2 gene (Abbott Molecular, Inc. Des Plaines, IL) located at 10q26. The 5′ FGFR2 signal was labeled with Spectrum Orange (orange) and the 3′ FGFR2 signal was labeled with Spectrum Green (green).
Supporting Information
Somatic point mutations, insertions and deletions identified in all samples.
(DOCX)
Gene Ontology (GO) functional classification of genes carrying Smallscale Nucleotide Variations (SsNVs) or in regions exhibiting Copy Number Variation (CNV).
(XLS)
Many genes carrying SsNVs act in key Cancer-associated pathways, and are differentially expressed in tumors from 6 patients with advanced sporadic biliary tract cancer.
(XLS)
Many genes identified in genomic regions exhibiting Copy Number Variation (CNV) functionally rank by significance to known Cancer-associated pathways.
(XLS)
CLIA validation of somatic mutations with therapeutic relevance in 6 patients with advanced, sporadic biliary tract cancer.
(DOCX)
Differential gene expression of fibroblast growth factor receptor pathway family members in 6 patients with advanced sporadic biliary tract cancer.
(DOCX)
Supplementary discussion.
(DOCX)
Funding Statement
This study was supported by the NIH Grant Number K12 CA90628 and the Center for Individualized Medicine. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Shin HR, Lee CU, Park HJ, Seol SY, Chung JM, et al. (1996) Hepatitis B and C virus, Clonorchis sinensis for the risk of liver cancer: a case-control study in Pusan, Korea. International journal of epidemiology 25: 933–940. [DOI] [PubMed] [Google Scholar]
- 2. Watanapa P (1996) Cholangiocarcinoma in patients with opisthorchiasis. The British journal of surgery 83: 1062–1064. [DOI] [PubMed] [Google Scholar]
- 3. Watanapa P, Watanapa WB (2002) Liver fluke-associated cholangiocarcinoma. The British journal of surgery 89: 962–970. [DOI] [PubMed] [Google Scholar]
- 4. Bergquist A, Ekbom A, Olsson R, Kornfeldt D, Loof L, et al. (2002) Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. Journal of hepatology 36: 321–327. [DOI] [PubMed] [Google Scholar]
- 5. Bergquist A, Glaumann H, Persson B, Broome U (1998) Risk factors and clinical presentation of hepatobiliary carcinoma in patients with primary sclerosing cholangitis: a case-control study. Hepatology 27: 311–316. [DOI] [PubMed] [Google Scholar]
- 6. Burak K, Angulo P, Pasha TM, Egan K, Petz J, et al. (2004) Incidence and risk factors for cholangiocarcinoma in primary sclerosing cholangitis. The American journal of gastroenterology 99: 523–526. [DOI] [PubMed] [Google Scholar]
- 7. Claessen MM, Vleggaar FP, Tytgat KM, Siersema PD, van Buuren HR (2009) High lifetime risk of cancer in primary sclerosing cholangitis. Journal of hepatology 50: 158–164. [DOI] [PubMed] [Google Scholar]
- 8. Visser BC, Suh I, Way LW, Kang SM (2004) Congenital choledochal cysts in adults. Archives of surgery 139: 855–860 discussion 860–852. [DOI] [PubMed] [Google Scholar]
- 9. Hsing AW, Zhang M, Rashid A, McGlynn KA, Wang BS, et al. (2008) Hepatitis B and C virus infection and the risk of biliary tract cancer: a population-based study in China. International journal of cancer Journal international du cancer 122: 1849–1853. [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi M, Ikeda K, Saitoh S, Suzuki F, Tsubota A, et al. (2000) Incidence of primary cholangiocellular carcinoma of the liver in japanese patients with hepatitis C virus-related cirrhosis. Cancer 88: 2471–2477. [DOI] [PubMed] [Google Scholar]
- 11. Liu XF, Zou SQ, Qiu FZ (2003) Pathogenesis of cholangiocarcinoma in the porta hepatis and infection of hepatitis virus. Hepatobiliary & pancreatic diseases international : HBPD INT 2: 285–289. [PubMed] [Google Scholar]
- 12. Shaib YH, El-Serag HB, Davila JA, Morgan R, McGlynn KA (2005) Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology 128: 620–626. [DOI] [PubMed] [Google Scholar]
- 13. Welzel TM, Graubard BI, El-Serag HB, Shaib YH, Hsing AW, et al. (2007) Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: a population-based case-control study. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 5: 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto S, Kubo S, Hai S, Uenishi T, Yamamoto T, et al. (2004) Hepatitis C virus infection as a likely etiology of intrahepatic cholangiocarcinoma. Cancer science 95: 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donato F, Gelatti U, Tagger A, Favret M, Ribero ML, et al. (2001) Intrahepatic cholangiocarcinoma and hepatitis C and B virus infection, alcohol intake, and hepatolithiasis: a case-control study in Italy. Cancer causes & control : CCC 12: 959–964. [DOI] [PubMed] [Google Scholar]
- 16. Lee CC, Wu CY, Chen GH (2002) What is the impact of coexistence of hepatolithiasis on cholangiocarcinoma? Journal of gastroenterology and hepatology 17: 1015–1020. [DOI] [PubMed] [Google Scholar]
- 17. Becker N, Liebermann D, Wesch H, Van Kaick G (2008) Mortality among Thorotrast-exposed patients and an unexposed comparison group in the German Thorotrast study. European journal of cancer 44: 1259–1268. [DOI] [PubMed] [Google Scholar]
- 18. Travis LB, Hauptmann M, Gaul LK, Storm HH, Goldman MB, et al. (2003) Site-specific cancer incidence and mortality after cerebral angiography with radioactive thorotrast. Radiation research 160: 691–706. [DOI] [PubMed] [Google Scholar]
- 19. Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD (2005) Cholangiocarcinoma. Lancet 366: 1303–1314. [DOI] [PubMed] [Google Scholar]
- 20. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, et al. (2010) Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. The New England journal of medicine 362: 1273–1281. [DOI] [PubMed] [Google Scholar]
- 21. Kiba T, Tsuda H, Pairojkul C, Inoue S, Sugimura T, et al. (1993) Mutations of the p53 tumor suppressor gene and the ras gene family in intrahepatic cholangiocellular carcinomas in Japan and Thailand. Molecular carcinogenesis 8: 312–318. [DOI] [PubMed] [Google Scholar]
- 22. Ohashi K, Tsutsumi M, Nakajima Y, Noguchi O, Okita S, et al. (1994) High rates of Ki-ras point mutation in both intra- and extra-hepatic cholangiocarcinomas. Japanese journal of clinical oncology 24: 305–310. [PubMed] [Google Scholar]
- 23. Petmitr S, Pinlaor S, Thousungnoen A, Karalak A, Migasena P (1998) K-ras oncogene and p53 gene mutations in cholangiocarcinoma from Thai patients. The Southeast Asian journal of tropical medicine and public health 29: 71–75. [PubMed] [Google Scholar]
- 24. Roa JC, Roa I, de Aretxabala X, Melo A, Faria G, et al. (2004) [K-ras gene mutation in gallbladder carcinoma]. Revista medica de Chile 132: 955–960. [PubMed] [Google Scholar]
- 25. Sturm PD, Baas IO, Clement MJ, Nakeeb A, Johan G, et al. (1998) Alterations of the p53 tumor-suppressor gene and K-ras oncogene in perihilar cholangiocarcinomas from a high-incidence area. International journal of cancer Journal international du cancer 78: 695–698. [DOI] [PubMed] [Google Scholar]
- 26. Tada M, Omata M, Ohto M (1990) Analysis of ras gene mutations in human hepatic malignant tumors by polymerase chain reaction and direct sequencing. Cancer research 50: 1121–1124. [PubMed] [Google Scholar]
- 27. Tada M, Omata M, Ohto M (1990) [Detection of ras gene mutations of primary hepatic malignant tumors by polymerase chain reaction and direct sequencing method]. Nihon Shokakibyo Gakkai zasshi = The Japanese journal of gastro-enterology 87: 811–815. [PubMed] [Google Scholar]
- 28. Tada M, Omata M, Ohto M (1992) High incidence of ras gene mutation in intrahepatic cholangiocarcinoma. Cancer 69: 1115–1118. [DOI] [PubMed] [Google Scholar]
- 29. Wattanasirichaigoon S, Tasanakhajorn U, Jesadapatarakul S (1998) The incidence of K-ras codon 12 mutations in cholangiocarcinoma detected by polymerase chain reaction technique. Journal of the Medical Association of Thailand = Chotmaihet thangphaet 81: 316–323. [PubMed] [Google Scholar]
- 30. Boberg KM, Schrumpf E, Bergquist A, Broome U, Pares A, et al. (2000) Cholangiocarcinoma in primary sclerosing cholangitis: K-ras mutations and Tp53 dysfunction are implicated in the neoplastic development. Journal of hepatology 32: 374–380. [DOI] [PubMed] [Google Scholar]
- 31. Tannapfel A, Weinans L, Geissler F, Schutz A, Katalinic A, et al. (2000) Mutations of p53 tumor suppressor gene, apoptosis, and proliferation in intrahepatic cholangiocellular carcinoma of the liver. Digestive diseases and sciences 45: 317–324. [DOI] [PubMed] [Google Scholar]
- 32. Tullo A, D'Erchia AM, Honda K, Kelly MD, Habib NA, et al. (2000) New p53 mutations in hilar cholangiocarcinoma. European journal of clinical investigation 30: 798–803. [DOI] [PubMed] [Google Scholar]
- 33. Hahn SA, Bartsch D, Schroers A, Galehdari H, Becker M, et al. (1998) Mutations of the DPC4/Smad4 gene in biliary tract carcinoma. Cancer research 58: 1124–1126. [PubMed] [Google Scholar]
- 34. Iacobuzio-Donahue CA, Song J, Parmiagiani G, Yeo CJ, Hruban RH, et al. (2004) Missense mutations of MADH4: characterization of the mutational hot spot and functional consequences in human tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 10: 1597–1604. [DOI] [PubMed] [Google Scholar]
- 35. Ahrendt SA, Eisenberger CF, Yip L, Rashid A, Chow JT, et al. (1999) Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. The Journal of surgical research 84: 88–93. [DOI] [PubMed] [Google Scholar]
- 36. Tadokoro H, Shigihara T, Ikeda T, Takase M, Suyama M (2007) Two distinct pathways of p16 gene inactivation in gallbladder cancer. World journal of gastroenterology : WJG 13: 6396–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taniai M, Higuchi H, Burgart LJ, Gores GJ (2002) p16INK4a promoter mutations are frequent in primary sclerosing cholangitis (PSC) and PSC-associated cholangiocarcinoma. Gastroenterology 123: 1090–1098. [DOI] [PubMed] [Google Scholar]
- 38. Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Kockerling F, et al. (2000) Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut 47: 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoshida S, Todoroki T, Ichikawa Y, Hanai S, Suzuki H, et al. (1995) Mutations of p16Ink4/CDKN2 and p15Ink4B/MTS2 genes in biliary tract cancers. Cancer research 55: 2756–2760. [PubMed] [Google Scholar]
- 40. Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, et al. (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. The oncologist 17: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, et al. (2012) Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Human pathology 43: 1552–1558. [DOI] [PubMed] [Google Scholar]
- 42. Gu TL, Deng X, Huang F, Tucker M, Crosby K, et al. (2011) Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PloS one 6: e15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gwak GY, Yoon JH, Shin CM, Ahn YJ, Chung JK, et al. (2005) Detection of response-predicting mutations in the kinase domain of the epidermal growth factor receptor gene in cholangiocarcinomas. Journal of cancer research and clinical oncology 131: 649–652. [DOI] [PubMed] [Google Scholar]
- 44. Leone F, Cavalloni G, Pignochino Y, Sarotto I, Ferraris R, et al. (2006) Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 12: 1680–1685. [DOI] [PubMed] [Google Scholar]
- 45. Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, et al. (2003) Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 52: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deshpande V, Nduaguba A, Zimmerman SM, Kehoe SM, Macconaill LE, et al. (2011) Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC cancer 11: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Riener MO, Bawohl M, Clavien PA, Jochum W (2008) Rare PIK3CA hotspot mutations in carcinomas of the biliary tract. Genes, chromosomes & cancer 47: 363–367. [DOI] [PubMed] [Google Scholar]
- 48. Wendum D, Barbu V, Rosmorduc O, Arrive L, Flejou JF, et al. (2012) Aspects of liver pathology in adult patients with MDR3/ABCB4 gene mutations. Virchows Archiv : an international journal of pathology 460: 291–298. [DOI] [PubMed] [Google Scholar]
- 49. Scheimann AO, Strautnieks SS, Knisely AS, Byrne JA, Thompson RJ, et al. (2007) Mutations in bile salt export pump (ABCB11) in two children with progressive familial intrahepatic cholestasis and cholangiocarcinoma. The Journal of pediatrics 150: 556–559. [DOI] [PubMed] [Google Scholar]
- 50. Strautnieks SS, Byrne JA, Pawlikowska L, Cebecauerova D, Rayner A, et al. (2008) Severe bile salt export pump deficiency: 82 different ABCB11 mutations in 109 families. Gastroenterology 134: 1203–1214. [DOI] [PubMed] [Google Scholar]
- 51. Endo K, Ashida K, Miyake N, Terada T (2001) E-cadherin gene mutations in human intrahepatic cholangiocarcinoma. The Journal of pathology 193: 310–317. [DOI] [PubMed] [Google Scholar]
- 52. Ukita Y, Kato M, Terada T (2002) Gene amplification and mRNA and protein overexpression of c-erbB-2 (HER-2/neu) in human intrahepatic cholangiocarcinoma as detected by fluorescence in situ hybridization, in situ hybridization, and immunohistochemistry. Journal of hepatology 36: 780–785. [DOI] [PubMed] [Google Scholar]
- 53. Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, et al. (2013) FGFR2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2013 Oct 9 doi:[]10.1002/hep.26890. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 54. Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, et al. (2013) Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reva B, Antipin Y, Sander C (2007) Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 8: R232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwarz JM, Rodelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7: 575–576. [DOI] [PubMed] [Google Scholar]
- 59. Craig DW, O'Shaughnessy JA, Kiefer JA, Aldrich J, Sinari S, et al. (2013) Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther 12: 104–116. [DOI] [PubMed] [Google Scholar]
- 60. Cirulli V, Yebra M (2007) Netrins: beyond the brain. Nat Rev Mol Cell Biol 8: 296–306. [DOI] [PubMed] [Google Scholar]
- 61. Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, et al. (2003) A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 9: 407–415. [DOI] [PubMed] [Google Scholar]
- 62. Salvucci O, Tosato G (2012) Essential roles of EphB receptors and EphrinB ligands in endothelial cell function and angiogenesis. Adv Cancer Res 114: 21–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cavallaro U, Christofori G (2004) Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer 4: 118–132. [DOI] [PubMed] [Google Scholar]
- 64. Kraus MR, Clauin S, Pfister Y, Di Maio M, Ulinski T, et al. (2012) Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum Mutat 33: 86–90. [DOI] [PubMed] [Google Scholar]
- 65. Wang X, Osada T, Wang Y, Yu L, Sakakura K, et al. (2010) CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J Natl Cancer Inst 102: 1496–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krzeslak A, Forma E, Bernaciak M, Romanowicz H, Brys M (2012) Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med 12: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Antonescu CR, Zhang L, Nielsen GP, Rosenberg AE, Dal Cin P, et al. (2011) Consistent t(1;10) with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. Genes Chromosomes Cancer 50: 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hallor KH, Sciot R, Staaf J, Heidenblad M, Rydholm A, et al. (2009) Two genetic pathways, t(1;10) and amplification of 3p11-12, in myxoinflammatory fibroblastic sarcoma, haemosiderotic fibrolipomatous tumour, and morphologically similar lesions. J Pathol 217: 716–727. [DOI] [PubMed] [Google Scholar]
- 69. Kumar R, Crouthamel MC, Rominger DH, Gontarek RR, Tummino PJ, et al. (2009) Myelosuppression and kinase selectivity of multikinase angiogenesis inhibitors. Br J Cancer 101: 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ha GH, Park JS, Breuer EK (2013) TACC3 promotes epithelial-mesenchymal transition (EMT) through the activation of PI3K/Akt and ERK signaling pathways. Cancer Lett 332: 63–73. [DOI] [PubMed] [Google Scholar]
- 71. Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nature reviews Cancer 10: 116–129. [DOI] [PubMed] [Google Scholar]
- 72. Williams SV, Hurst CD, Knowles MA (2012) Oncogenic FGFR3 gene fusions in bladder cancer. Human molecular genetics. Hum Mol Genet 22: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maeda T, Yagasaki F, Ishikawa M, Takahashi N, Bessho M (2005) Transforming property of TEL-FGFR3 mediated through PI3-K in a T-cell lymphoma that subsequently progressed to AML. Blood 105: 2115–2123. [DOI] [PubMed] [Google Scholar]
- 74. Ren M, Qin H, Ren R, Tidwell J, Cowell JK (2011) Src activation plays an important key role in lymphomagenesis induced by FGFR1 fusion kinases. Cancer research 71: 7312–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren M, Qin H, Ren R, Cowell JK (2012) Ponatinib suppresses the development of myeloid and lymphoid malignancies associated with FGFR1 abnormalities. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, et al. (1997) A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood 90: 4062–4070. [PubMed] [Google Scholar]
- 77. Cross NC, Reiter A (2008) Fibroblast growth factor receptor and platelet-derived growth factor receptor abnormalities in eosinophilic myeloproliferative disorders. Acta haematologica 119: 199–206. [DOI] [PubMed] [Google Scholar]
- 78. Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, et al. (2012) Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, et al. (2012) Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32: 3091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, et al. (2009) Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136: 3019–3030. [DOI] [PubMed] [Google Scholar]
- 81. Petiot A, Conti FJ, Grose R, Revest JM, Hodivala-Dilke KM, et al. (2003) A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development 130: 5493–5501. [DOI] [PubMed] [Google Scholar]
- 82. Feng S, Wang F, Matsubara A, Kan M, McKeehan WL (1997) Fibroblast growth factor receptor 2 limits and receptor 1 accelerates tumorigenicity of prostate epithelial cells. Cancer Res 57: 5369–5378. [PubMed] [Google Scholar]
- 83. Ricol D, Cappellen D, El Marjou A, Gil-Diez-de-Medina S, Girault JM, et al. (1999) Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene 18: 7234–7243. [DOI] [PubMed] [Google Scholar]
- 84. Lorenzi MV, Castagnino P, Chen Q, Chedid M, Miki T (1997) Ligand-independent activation of fibroblast growth factor receptor-2 by carboxyl terminal alterations. Oncogene 15: 817–826. [DOI] [PubMed] [Google Scholar]
- 85. Chase A, Grand FH, Cross NC (2007) Activity of TKI258 against primary cells and cell lines with FGFR1 fusion genes associated with the 8p11 myeloproliferative syndrome. Blood 110: 3729–3734. [DOI] [PubMed] [Google Scholar]
- 86. Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, et al. (2012) FGFR Genetic Alterations Predict for Sensitivity to NVP-BGJ398, a Selective Pan-FGFR Inhibitor. Cancer discovery 2: 1118–1133. [DOI] [PubMed] [Google Scholar]
- 87. Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, et al. (2012) Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Molecular cancer therapeutics 11: 690–699. [DOI] [PubMed] [Google Scholar]
- 88. Amano R, Yamada N, Doi Y, Yashiro M, Ohira M, et al. (2010) Significance of keratinocyte growth factor receptor in the proliferation of biliary tract cancer. Anticancer Res 30: 4115–4121. [PubMed] [Google Scholar]
- 89. Kumar R, Knick VB, Rudolph SK, Johnson JH, Crosby RM, et al. (2007) Pharmacokinetic-pharmacodynamic correlation from mouse to human with pazopanib, a multikinase angiogenesis inhibitor with potent antitumor and antiangiogenic activity. Mol Cancer Ther 6: 2012–2021. [DOI] [PubMed] [Google Scholar]
- 90. Ballaro C, Ceccarelli S, Tiveron C, Tatangelo L, Salvatore AM, et al. (2005) Targeted expression of RALT in mouse skin inhibits epidermal growth factor receptor signalling and generates a Waved-like phenotype. EMBO reports 6: 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, et al. (2006) Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nature medicine 12: 568–573. [DOI] [PubMed] [Google Scholar]
- 92. Jeong JW, Lee HS, Lee KY, White LD, Broaddus RR, et al. (2009) Mig-6 modulates uterine steroid hormone responsiveness and exhibits altered expression in endometrial disease. Proceedings of the National Academy of Sciences of the United States of America 106: 8677–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lin CI, Du J, Shen WT, Whang EE, Donner DB, et al. (2011) Mitogen-inducible gene-6 is a multifunctional adaptor protein with tumor suppressor-like activity in papillary thyroid cancer. The Journal of clinical endocrinology and metabolism 96: E554–565. [DOI] [PubMed] [Google Scholar]
- 94. Reschke M, Ferby I, Stepniak E, Seitzer N, Horst D, et al. (2010) Mitogen-inducible gene-6 is a negative regulator of epidermal growth factor receptor signaling in hepatocytes and human hepatocellular carcinoma. Hepatology 51: 1383–1390. [DOI] [PubMed] [Google Scholar]
- 95. Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, et al. (2003) Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene 22: 4221–4234. [DOI] [PubMed] [Google Scholar]
- 96. Zhang X, Pickin KA, Bose R, Jura N, Cole PA, et al. (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450: 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Frosi Y, Anastasi S, Ballaro C, Varsano G, Castellani L, et al. (2010) A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. The Journal of cell biology 189: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Anastasi S, Sala G, Huiping C, Caprini E, Russo G, et al. (2005) Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene 24: 4540–4548. [DOI] [PubMed] [Google Scholar]
- 99. Duncan CG, Killela PJ, Payne CA, Lampson B, Chen WC, et al. (2010) Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget 1: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, et al. (2010) Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America 107: 6912–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li Z, Dong Q, Wang Y, Qu L, Qiu X, et al. (2012) Downregulation of Mig-6 in nonsmall-cell lung cancer is associated with EGFR signaling. Molecular carcinogenesis 51: 522–534. [DOI] [PubMed] [Google Scholar]
- 102. Zhang YW, Staal B, Dykema KJ, Furge KA, Vande Woude GF (2012) Cancer-type regulation of MIG-6 expression by inhibitors of methylation and histone deacetylation. PloS one 7: e38955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang YW, Staal B, Su Y, Swiatek P, Zhao P, et al. (2007) Evidence that MIG-6 is a tumor-suppressor gene. Oncogene 26: 269–276. [DOI] [PubMed] [Google Scholar]
- 104. Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, et al. (2010) GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res 70: 7630–7639. [DOI] [PubMed] [Google Scholar]
- 105. Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, et al. (2013) Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC Genomics 14: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Iyer MK, Chinnaiyan AM, Maher CA (2011) ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 27: 2903–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, et al. (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7: e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, et al. (2011) A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res 39: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Diep CH, Zucker KM, Hostetter G, Watanabe A, Hu C, et al. (2012) Down-regulation of Yes Associated Protein 1 expression reduces cell proliferation and clonogenicity of pancreatic cancer cells. PLoS One 7: e32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, et al. (2012) Exome sequencing of liver fluke-associated cholangiocarcinoma. Nature genetics 44: 690–693. [DOI] [PubMed] [Google Scholar]
- 112. Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, et al. (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, et al. (2012) Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature genetics 44: 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Somatic point mutations, insertions and deletions identified in all samples.
(DOCX)
Gene Ontology (GO) functional classification of genes carrying Smallscale Nucleotide Variations (SsNVs) or in regions exhibiting Copy Number Variation (CNV).
(XLS)
Many genes carrying SsNVs act in key Cancer-associated pathways, and are differentially expressed in tumors from 6 patients with advanced sporadic biliary tract cancer.
(XLS)
Many genes identified in genomic regions exhibiting Copy Number Variation (CNV) functionally rank by significance to known Cancer-associated pathways.
(XLS)
CLIA validation of somatic mutations with therapeutic relevance in 6 patients with advanced, sporadic biliary tract cancer.
(DOCX)
Differential gene expression of fibroblast growth factor receptor pathway family members in 6 patients with advanced sporadic biliary tract cancer.
(DOCX)
Supplementary discussion.
(DOCX)