Abstract
Barnacles are major sessile components of the intertidal areas worldwide, and also one of the most dominant fouling organisms in fouling communities. Larval settlement has a crucial ecological effect not only on the distribution of the barnacle population but also intertidal community structures. However, the molecular mechanisms involved in the transition process from the larval to the juvenile stage remain largely unclear. In this study, we carried out comparative proteomic profiles of stage II nauplii, stage VI nauplii, cyprids, and juveniles of the barnacle Balanus amphitrite using label-free quantitative proteomics, followed by the measurement of the gene expression levels of candidate proteins. More than 700 proteins were identified at each stage; 80 were significantly up-regulated in cyprids and 95 in juveniles vs other stages. Specifically, proteins involved in energy and metabolism, the nervous system and signal transduction were significantly up-regulated in cyprids, whereas proteins involved in cytoskeletal remodeling, transcription and translation, cell proliferation and differentiation, and biomineralization were up-regulated in juveniles, consistent with changes associated with larval metamorphosis and tissue remodeling in juveniles. These findings provided molecular evidence for the morphological, physiological and biological changes that occur during the transition process from the larval to the juvenile stages in B. amphitrite.
Introduction
The life cycles of most sessile marine invertebrates include a microscopic and planktonic larval stage that may last for minutes to months, as well as benthic juvenile and adult stages during which individuals attach to a submerged surface [1]. The transition from pelagic to sessile stages is referred to as larval attachment and metamorphosis (collectively known as larval settlement), which is associated with morphological, physiological and biochemical changes. Larval settlement is crucial not only for recruitment but also for species distribution and community structures [2]. Although details of the signal transduction pathways and mechanisms that regulate larval settlement have been partially reported in some species [3], [4], the associated molecular mechanisms remain largely unknown in most marine invertebrate species, due to their high biological diversity.
The barnacle Balanus amphitrite is a dominant fouling organism worldwide. B. amphitrite larvae released from adults molt 6 times and transit to cyprids, the competent stage for subsequent settlement. The process of settlement can be divided into 3 phases: attainment of competency, attachment to a suitable substratum, and metamorphosis into juveniles [5]. The morphogenetic development associated with metamorphosis includes decortication of the cyprid carapace, formation of a new chitinous layer, migration of the naupliar eye, degeneration of the compound eyes and antenna, and development of the feeding cirri [6]. In addition, physiological, structural and functional changes occur, all of which are regulated by functional genes and proteins [7]. Six cyprid-specific genes were first isolated from a cyprid cDNA library [8], and responded differentially to settlement cues [9]. Recently, we conducted a comparative transcriptomic study and identified several genes with potential roles in the larval settlement process [5].
There is no predictive correlation between mRNA and protein levels. Because proteins directly mediate most biological events, evaluation of changes in their levels could provide comprehensive biological insights [10]. An earlier 2-DE-based proteomic study from our laboratory revealed approximately 400 spots and identified some proteins that were differentially expressed during barnacle larval settlement [10]. Furthermore, a significantly higher number of protein spots were obtained when implementing additional solution-phase IEF sample prefractionation and narrow-pH-range IEF [11]. However, the 2-DE method has a relatively poor reproducibility, low sensitivity, and narrow linear dynamic ranges [12]. In addition, few proteins exhibiting a relatively lower expression level could be identified using mass spectrometry in a 2-DE-based analysis. In contrast, a gel-free proteomics technique incorporating a combination of multidimensional liquid chromatography (LC) separation, MS analysis and sequence database searches could provide a robust and effective platform for direct analysis of the proteome of the bryozoan Bugula neritina [13]. In the present study, we used a label-free quantitative proteomic platform to profile the proteomes of 4 developmental stages of B. amphitrite, compared 4 proteomes to identify many differentially expressed proteins that might play key roles in the settlement of B. amphitrite, and confirmed the expression patterns of numerous proteins using quantitative real-time polymer chain reaction (qRT-PCR).
Materials and Methods
Sample preparation
The barnacle, Balanus amphitrite, that we used for this study is a common species of marine invertebrates. It is a biofouling species and not endangered or protected. Balanus amphitrite adults were collected from a dock in Pak Sha Wan, Hong Kong (22.21′45″ N, 114.15′35″ E). No specific permits were required for the described field studies. The dock does not belong to any national parks, protected areas, or privately-owned places. The filed studies did not involve any endangered or protected species.
Larvae of different stages were obtained and cultured according to the methods described by Thiyagarajan and Qian [10]. Briefly, newly released larvae were maintained in filtered seawater (FSW) for 2 h and then collected as stage II nauplii. Other larvae were cultured at 27°C and fed with Chaetoceros gracilis Schutt for 3 to 4 d until they had developed into stage VI nauplii with 2 compound eyes. After 18–24 h, a portion of the cyprids undergoing molting from stage VI nauplii was collected; the remaining cyprids attached to polystyrene Petri dishes (Falcon no. 1006) in the dark. Most of the cyprids attached to the dishes within 24 h and completed metamorphosis into juveniles within 48 h. The juveniles were then scraped off the dishes. All of the samples were stored at −80°C until use.
Protein extraction and digestion
The samples were resuspended in 1 mL protein lysis buffer containing 0.1 M Tris-HCl (pH 7.6), 2% SDS, 0.1 M dithiothreitol and protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After homogenization, the samples were sonicated 3 times using a Misonix sonicator-XL2020 (Misonix, Farmingdale, NY) and then incubated in boiling water for 5 min. Larvae and debris that were not lysed were pelleted by centrifugation at 15,000 g for 10 min at 4°C. The protein-containing supernatant was transferred into a new tube, and the protein concentration was quantified using the RC/DC protein assay kit (BioRad, Hercules, CA). Due to the presence of pigments and other contaminants in the barnacle protein extract, one dimensional gel electrophoresis was performed before trypsin digestion to remove impurities, including pigments, detergents, and buffer components. This purification step facilitated the subsequent liquid chromatographic separation. Specifically, protein samples were loaded in a 10% SDS-PAGE gel and separated for 25 min to purify the proteins from other non-proteins/small molecules. Following Coomassie blue staining, the concentrated protein bands were removed from the gel and ready for in-gel digestion as previously described [14].
Liquid chromatography/mass spectrometry (LC/MS) analysis
The digests were resuspended and fractionated by strong cation exchange chromatography [15]. Each dried fraction was reconstituted in 30 μL of 0.1% formic acid. The samples were run as 3 replicates using a Proxeon EASY-nLC unit (Thermo Scientific, San Jose, USA) with an LTQ-Orbitrap mass spectrometer (Velos, Thermo Scientific). Peptide separation was conducted in a capillary column (0.1×150 mm, with C18 AQ of 3 μm particles and 200 Å pore size, Bruker Michrom BioResources). Mobile phase A (0.1% formic acid in H2O) and mobile phase B (0.1% formic acid in ACN) were used to establish a 75-min gradient consisting of 45 min from 100 to 65% A, 10 min from 65 to 20% A, and 20 min at 20% A. The LC was operated at a constant flow rate of 0.5 μL/min. The ion source was set as a capillary voltage of 1.5 kV and a source temperature of 160°C. The LTQ-Orbitrap was set to perform data-dependent acquisition in positive ion mode with a selected MS survey mass range of 350-1600 m/z. The 10 most intense ions above a 500-count threshold and carrying a charge from 2+ to 4+, were selected for MS/MS fragmentation. Dynamic exclusion was activated using a repeat count of 2, an exclusion duration of 45 s, and a mass tolerance of ±5 ppm. The CID parameters included a normalized collision energy of 35%, an activation Q of 0.25, an isolation width of 3.0 and an activation time for 10 ms.
Database search and data analysis
The raw MS data were converted into mascot generic files using Proteome Discover (1.2) and then submitted to Mascot version 2.2 (Matrix Sciences Ltd., London, UK) for searching against an in-house protein database developed in our transcriptome study [5]. The mass tolerances were set at 10 ppm for the peptide precursors and 0.5 Da for the fragment ions. A decoy option was included. Carboxamidomethylation at cysteine residues was set as a fixed modification, and oxidation at methionine residues was set as a variable modification. Up to 1 missed trypsin cleavage was permitted.
The resulting .dat files from the Mascot search were processed using Scaffold (version 4.0, Proteome Software Inc., OR, USA) to validate the MS/MS identification. Peptide identification was accepted if the result could established at >95.0% probability by the Scaffold Local FDR, whereas protein identification was accepted if the result had a probability of >99.0% and contained at least 1 identified peptide. Peptide and protein probabilities assigned by the PeptideProphet [16] and ProteinProphet [17]. Protein XML files were then exported from Scaffold to calculate the protein abundances were calculated using APEX quantitation proteomics tools [18], [19]. A 1.5-fold change was set as the cutoff, and only proteins with >5,000 molecules per cell during at least one stage were considered for significantly up-regulated proteins and proteins with >5,000 molecules per cell during at least 3 stages were considered for significantly down-regulated proteins [20].
Phylogenetic analysis of vitellogenin
Amino acid sequences of vitellogenin from various species were downloaded from GenBank and aligned using MUSCLE [21]via the CIPRES Portal v2.2 [22]. Neighbor-joining analysis was performed with MEGA4. All of the positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons (pairwise deletion option). The topological stability was evaluated based on 1,000 bootstrapping (BS) replications.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from each stage as well as from adults was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The quantity and quality of the RNA were assessed using agarose gel electrophoresis and a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), respectively. Trace genomic DNA in the RNA solution was removed using the Turbo DNAfree Kit (Ambion Inc, Austin, TX). First-strand cDNA was synthesized from RNA using MMLV reverse transcriptase (USB, Cleveland, OH) with oligo dT(18) primer. Gene-specific primers were designed with Primer3 software [23], and the sequences of all primers used are listed in Table S1. According to the standard protocol, the qRT-PCR assays were conducted using SYBR Green Supermix (BioRad) on an ABI 7500 fast real-time PCR machine (Applied Biosystems, Foster City, CA). Cytochrome b (Cyb) was employed as an internal control for normalization [24]; the relative expression patterns were calculated based on the 2−ΔΔCt method [25], [26]. Significant differences in expression patterns were analyzed by one-way ANOVA followed by the Tukey's post-hoc test.
Results
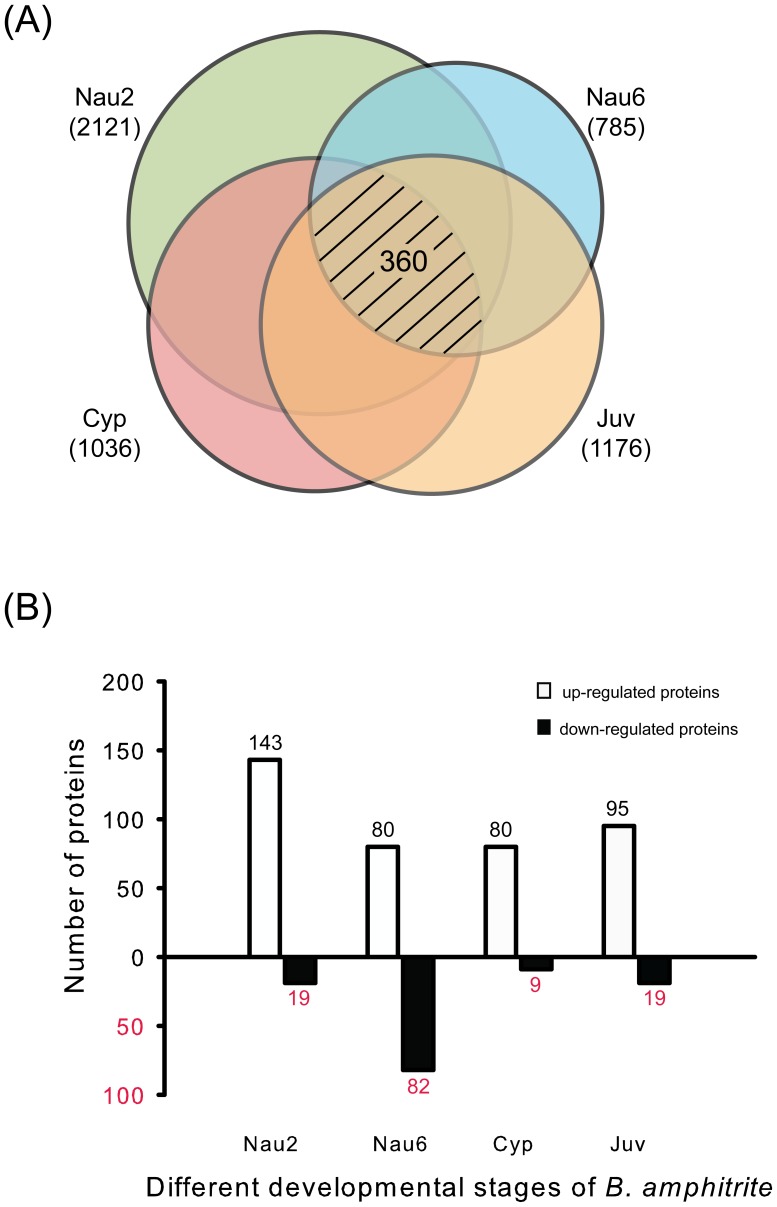
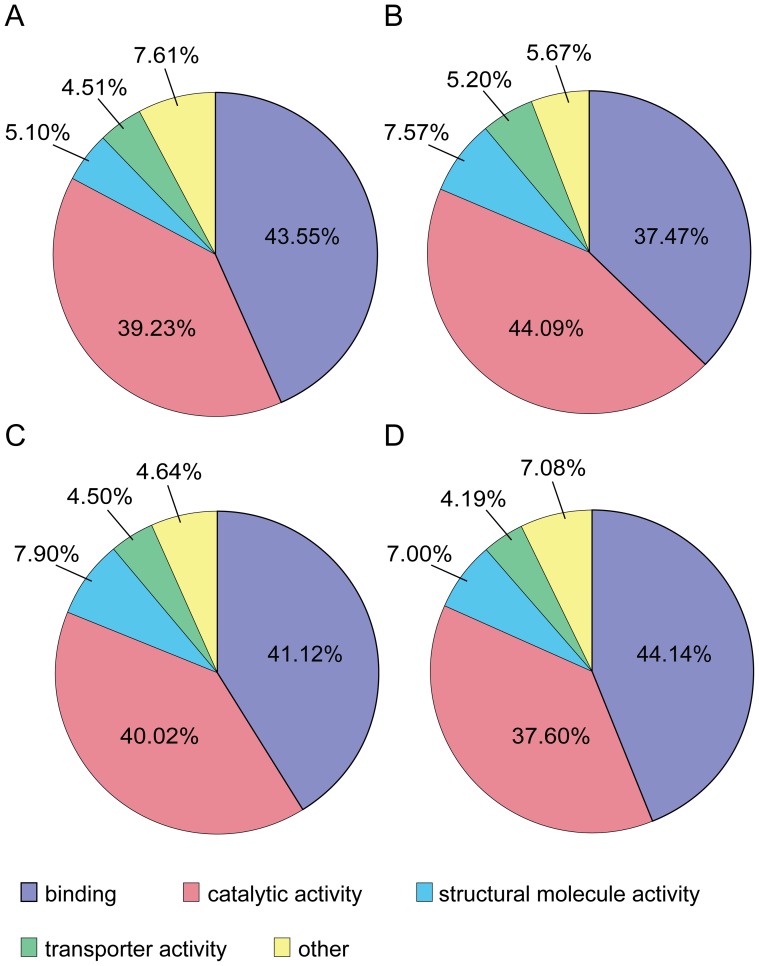
Four developmental stages of B. amphitrite were collected for our proteomic profiling. After MS profiling, 2,121 proteins were isolated from stage II nauplii, 785 from stage VI nauplii, 1,036 from cyprids, and 1176 from juveniles (Figure 1A, Table S2). Among the total 2,520 proteins, 360 proteins were common to all 4 developmental stages. Using gene ontology (GO), 1,793 of 2,520 proteins (accounting for 71.15% of the total proteins) were categorized into several functional groups, including 1,555 of 2,118 (73.42%) in stage II nauplii, 570 of 783 (72.80%) in stage VI nauplii, 790 of 1,034 (76.40%) in cyprids and 895 of 1,174 (76.24%) in juveniles (GO categories are shown in Figure 2). The majority of the proteins displayed binding and catalytic activity (37.47%–44.14%), and the remainder were associated with structural molecule activity and transporter functions (4.19%–7.90%). Unlike the other 3 developmental stages, stage VI nauplii had more proteins that displayed catalytic (44.09%) rather than with binding activity (37.47%). The remaining categories included electron carrier activity, enzyme regulator activity, receptor activity, antioxidant activity, nucleic acid binding, and transcription factor activity, among others.
Figure 1. The numbers of overlap proteins among the 4 developmental stages of B. amphitrite (A) and the numbers of differentially expressed proteins during each developmental stage (B).
In the Venn diagram, the numbers in parentheses are the number of quantified proteins expressed during each stage. Shaded numbers show the number of the common proteins; Nau2: stage II nauplii, Nau6: stage VI nauplii, Cyp: cyprids, and Juv: juveniles.
Figure 2. Percentage distribution of the molecular functions of identified proteins in 4 developmental stages, including stage II nauplii (A), stage VI nauplii (B), cyprids (C) and juveniles (D).
The top 4 categories were binding, catalytic activity, structural molecule activity and transporter activity, followed by regulators of the activity of enzymes, receptors, molecular transducers, electron carriers, nucleic acid binding transcription factors, protein binding transcription factors, antioxidants, translation regulators, metallochaperones, and receptor regulators.
Comparative proteomic analysis among the 4 developmental stages of B. amphitrite revealed that 398 proteins met the established criteria, displaying a 1.5-fold change and >5,000 molecules per cell in at least one stage (Figure 1B). Among them, 143 in stage II nauplii, 80 in stage VI nauplii, 80 in cyprids and 95 in juveniles were significantly up-regulated (listed in Tables 1, 2, S3 and S4). Interestingly, 118 proteins were uniquely expressed in stage II nauplii, accounting for 82.51% of the total 143 proteins. In contrast, 36, 22 and 27 proteins were uniquely expressed in stage VI nauplii, cyprids and juveniles, respectively. In addition, 129 proteins were significantly down-regulated in the 4 developmental stages, which is less than the up-regulated ones (Figure 1B). Among them, 19 were in stage II nauplii, 82 in stage VI nauplii, 9 in cyprids and 19 in juveniles (listed in Table S5, S6, S7 and S8). To identify the important proteins involved in the larval settlement process of barnacles, proteins that were significantly up-regulated in cyprids (listed in Table 1) were categorized into a diverse set of functional groups using GO and eukaryotic orthologous groups (KOG). Approximately 30% proteins were related to energy and metabolism; others were associated with structural molecules, nervous system-related molecules, and signaling molecules, among others. Additionally, proteins that were significantly up-regulated in juveniles (Table 2) might be crucial for tissue reorganization and development postlarval settlement. The top 3 categories detected were structural, transcription and translation, and energy and metabolism proteins. The other proteins were categorized into various functional groups such as cell differentiation-related, cell proliferation-related, shell calcification-related, and stress-induced proteins.
Table 1. Significantly Up-regulated Proteins in Cyprids a .
| Protein abundance (×1000 molecules/cell) | Protein ratios | |||||||||||||||||
| nau2 | nau6 | cyp | juv | nau2/cyp | nau6/cyp | juv/cyp | ||||||||||||
| Accession No. | Protein Description | ave | SD | ave | SD | ave | SD | ave | SD | ave | SD | p | ave | SD | p | ave | SD | p |
| Energy and Metabolism | ||||||||||||||||||
| CL5556.Contig1 | Chromosome 14 open reading frame 149 | N.D. | N.D. | 8.78 | 2.01 | 29.75 | 0.93 | N.D. | N.D. | 0.00 | 0.00 | 0.000 | 0.30 | 0.07 | 0.001 | 0.00 | 0.00 | 0.000 |
| Unigene12197 | Serine hydroxymethyl-transferase, mitochondria | 4.03 | 2.33 | N.D. | N.D. | 16.26 | 2.86 | N.D. | N.D. | 0.25 | 0.14 | 0.005 | 0.00 | 0.00 | 0.002 | 0.00 | 0.00 | 0.000 |
| CL13389.Contig1 | prolyl endopeptidase | 2.59 | 0.90 | 1.54 | 0.07 | 21.40 | 1.52 | 6.53 | 0.62 | 0.12 | 0.04 | 0.000 | 0.07 | 0.00 | 0.000 | 0.30 | 0.03 | 0.000 |
| CL6434.Contig1 | Cytosolic non-specific dipeptidase | 16.25 | 1.39 | 23.57 | 6.75 | 47.51 | 3.40 | 26.53 | 2.30 | 0.34 | 0.03 | 0.000 | 0.50 | 0.14 | 0.015 | 0.56 | 0.05 | 0.001 |
| Unigene15104 | CG3999 CG3999-PA | N.D. | N.D. | N.D. | N.D. | 23.99 | 5.36 | N.D. | N.D. | only in cyp | 0.004 | only in cyp | 0.004 | only in cyp | 0.004 | |||
| Unigene6569 | dimethylglycine dehydrogenase, mitochondrial-like | N.D. | N.D. | N.D. | N.D. | 8.65 | 2.53 | N.D. | N.D. | only in cyp | 0.007 | only in cyp | 0.007 | only in cyp | 0.007 | |||
| CL346.Contig4 | 4-aminobutyrate aminotransferase | N.D. | N.D. | N.D. | N.D. | 10.55 | 5.06 | N.D. | N.D. | only in cyp | 0.020 | only in cyp | 0.020 | only in cyp | 0.020 | |||
| CL10509.Contig1 | betaine homocysteine methyl transferase | 5.65 | 0.45 | N.D. | N.D. | 13.64 | 1.93 | N.D. | N.D. | 0.41 | 0.03 | 0.001 | 0.08 | 0.00 | 0.000 | 0.00 | 0.00 | 0.002 |
| CL8097.Contig1 | betaine homocysteine methyltransferase | N.D. | N.D. | N.D. | N.D. | 10.79 | 2.59 | N.D. | N.D. | only in cyp | 0.005 | only in cyp | 0.005 | only in cyp | 0.005 | |||
| CL4185.Contig1 | mannose-6-phosphate isomerase | 3.34 | 0.11 | N.D. | N.D. | 6.14 | 0.29 | 0.00 | 0.00 | 0.54 | 0.02 | 0.000 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| CL16245.Contig1 | fructose 1,6-bisphosphatase | 9.63 | 0.67 | 11.37 | 2.59 | 62.13 | 2.63 | 18.61 | 3.70 | 0.16 | 0.01 | 0.000 | 0.18 | 0.04 | 0.000 | 0.30 | 0.06 | 0.000 |
| CL288.Contig4 | fructose-biphosphate aldolase | 40.69 | 2.27 | 13.01 | 3.27 | 108.11 | 10.44 | 66.50 | 5.09 | 0.38 | 0.02 | 0.000 | 0.12 | 0.03 | 0.001 | 0.62 | 0.05 | 0.003 |
| CL1136.Contig1 | glucose-6-phosphate isomerase | 13.97 | 3.48 | N.D. | N.D. | 31.90 | 3.55 | 18.55 | 2.05 | 0.44 | 0.11 | 0.004 | 0.00 | 0.00 | 0.001 | 0.58 | 0.06 | 0.004 |
| CL280.Contig1 | CG11255 CG11255-PB | 10.29 | 3.39 | N.D. | N.D. | 44.27 | 2.72 | 16.98 | 7.27 | 0.23 | 0.08 | 0.001 | 0.00 | 0.00 | 0.000 | 0.38 | 0.16 | 0.007 |
| CL9352.Contig1 | glucosamine-6-phosphate isomerase 1 | 3.33 | 1.28 | 2.06 | 0.02 | 19.44 | 2.63 | 10.73 | 4.13 | 0.17 | 0.07 | 0.001 | 0.11 | 0.00 | 0.003 | 0.55 | 0.21 | 0.040 |
| CL1148.Contig1 | neutral alpha-glucosidase AB | 8.25 | 1.89 | N.D. | N.D. | 17.70 | 2.65 | 7.53 | 1.70 | 0.47 | 0.11 | 0.007 | 0.00 | 0.00 | 0.002 | 0.43 | 0.10 | 0.005 |
| CL3803.Contig1 | Acetyl-CoA acetyltransferase, mitochondrial | 10.95 | 0.56 | N.D. | N.D. | 19.65 | 3.45 | 6.09 | 2.61 | 0.56 | 0.03 | 0.008 | 0.00 | 0.00 | 0.002 | 0.31 | 0.13 | 0.007 |
| CL4830.Contig1 | Acad8 protein | N.D. | N.D. | N.D. | N.D. | 12.29 | 3.92 | N.D. | N.D. | only in cyp | 0.009 | only in cyp | 0.009 | only in cyp | 0.009 | |||
| CL10791.Contig1 | acyl-CoA oxidase | 21.55 | 1.27 | 5.72 | 0.21 | 46.67 | 0.50 | 29.83 | 3.10 | 0.46 | 0.03 | 0.000 | 0.12 | 0.00 | 0.000 | 0.64 | 0.07 | 0.013 |
| Unigene30254 | acetyl-CoA carboxylase 2 | 6.81 | 2.06 | N.D. | N.D. | 18.31 | 0.73 | N.D. | N.D. | 0.37 | 0.11 | 0.002 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| Unigene4811 | uridine 5′-monophosphate synthas | N.D. | N.D. | N.D. | N.D. | 21.71 | 11.80 | N.D. | N.D. | only in cyp | 0.026 | only in cyp | 0.026 | only in cyp | 0.026 | |||
| CL1575.Contig1 | aldehyde dehydrogenase 1 family, member L1 | 7.02 | 0.63 | N.D. | N.D. | 21.42 | 3.69 | 6.15 | 3.29 | 0.33 | 0.03 | 0.001 | 0.00 | 0.00 | 0.003 | 0.29 | 0.15 | 0.011 |
| CL1120.Contig2 | pyrroline-5-carboxylate dehydrogenase | 20.82 | 0.76 | 26.34 | 6.95 | 69.73 | 3.99 | 28.92 | 3.05 | 0.30 | 0.01 | 0.000 | 0.38 | 0.10 | 0.004 | 0.41 | 0.04 | 0.000 |
| CL6520.Contig1 | hydroxyacid-oxoacid transhydrogenase, mitochondrial | 5.39 | 2.42 | N.D. | N.D. | 20.87 | 1.39 | N.D. | N.D. | 0.26 | 0.12 | 0.002 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| CL1939.Contig1 | malate synthase, glyoxysomal | N.D. | N.D. | N.D. | N.D. | 14.20 | 3.58 | 4.67 | 1.18 | 0.00 | 0.00 | 0.005 | 0.00 | 0.00 | 0.005 | 0.33 | 0.08 | 0.007 |
| CL5668.Contig1 | CG7280 CG7280-PA | 1.62 | 0.69 | N.D. | N.D. | 17.34 | 4.89 | 4.95 | 4.05 | 0.09 | 0.04 | 0.001 | 0.07 | 0.00 | 0.006 | 0.29 | 0.23 | 0.028 |
| CL271.Contig3 | mitochondrial aldehyde dehydrogenase precursor | 27.10 | 1.68 | N.D. | N.D. | 63.61 | 5.66 | 32.13 | 5.27 | 0.43 | 0.03 | 0.000 | 0.00 | 0.00 | 0.001 | 0.51 | 0.08 | 0.003 |
| Structural Molecules | ||||||||||||||||||
| Unigene13912 | projectin | 6.34 | 0.39 | 8.77 | 0.11 | 23.62 | 1.19 | 6.36 | 0.53 | 0.27 | 0.02 | 0.000 | 0.37 | 0.00 | 0.000 | 0.27 | 0.02 | 0.000 |
| Unigene28824 | Titin | 6.83 | 0.69 | N.D. | N.D. | 14.87 | 0.34 | N.D. | N.D. | 0.46 | 0.05 | 0.000 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| CL682.Contig1 | Muscle M-line assembly protein unc-89 | 1.11 | 0.39 | N.D. | N.D. | 6.62 | 1.26 | N.D. | N.D. | 0.17 | 0.06 | 0.001 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.000 |
| Unigene3292 | translocase of outer membrane 34 | N.D. | N.D. | N.D. | N.D. | 5.96 | 0.34 | N.D. | N.D. | only in cyp | 0.000 | only in cyp | 0.000 | only in cyp | 0.000 | |||
| Unigene5637 | twitchin | 3.99 | 0.15 | N.D. | N.D. | 20.24 | 1.31 | N.D. | N.D. | 0.20 | 0.01 | 0.000 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| Unigene10292 | Titin | 7.42 | 2.30 | 8.04 | 0.19 | 16.36 | 2.76 | 6.73 | 2.09 | 0.45 | 0.14 | 0.012 | 0.49 | 0.01 | 0.018 | 0.41 | 0.13 | 0.009 |
| Unigene2708 | BMKETTIN | 3.60 | 3.14 | N.D. | N.D. | 21.33 | 6.94 | N.D. | N.D. | 0.17 | 0.15 | 0.029 | 0.00 | 0.00 | 0.010 | 0.00 | 0.00 | 0.010 |
| CL1129.Contig1 | Echinoderm microtubule-associated protein | 4.68 | 0.86 | N.D. | N.D. | 11.25 | 2.12 | 2.50 | 2.20 | 0.42 | 0.08 | 0.005 | 0.00 | 0.00 | 0.003 | 0.22 | 0.20 | 0.037 |
| Transcription and Translation | ||||||||||||||||||
| CL128.Contig1 | ribosomal protein L10 | 16.26 | 4.07 | 5.93 | 1.62 | 29.64 | 1.00 | 18.58 | 1.00 | 0.55 | 0.14 | 0.009 | 0.20 | 0.05 | 0.001 | 0.63 | 0.03 | 0.000 |
| CL136.Contig1 | ribosomal protein L18A family member | 5.59 | 1.02 | 11.50 | 1.33 | 20.06 | 1.63 | N.D. | N.D. | 0.28 | 0.05 | 0.000 | 0.57 | 0.07 | 0.008 | 0.00 | 0.00 | 0.001 |
| CL814.Contig1 | 40S ribosomal protein S2 | 9.17 | 1.01 | 7.44 | 1.78 | 25.55 | 2.99 | 15.14 | 1.26 | 0.36 | 0.04 | 0.001 | 0.29 | 0.07 | 0.004 | 0.59 | 0.05 | 0.004 |
| Unigene11657 | ribosomal protein L9e | 15.02 | 3.74 | 16.52 | 1.52 | 42.90 | 2.09 | 23.35 | 7.07 | 0.35 | 0.09 | 0.001 | 0.39 | 0.04 | 0.001 | 0.54 | 0.16 | 0.014 |
| CL6677.Contig1 | translational activator GCN1 | 12.85 | 1.02 | N.D. | N.D. | 21.49 | 1.69 | 11.36 | 3.60 | 0.60 | 0.05 | 0.001 | 0.00 | 0.00 | 0.001 | 0.53 | 0.17 | 0.019 |
| CL2773.Contig1 | 10-formyltetrahydrofolate dehydrogenase | 1.59 | 0.13 | N.D. | N.D. | 11.22 | 2.23 | N.D. | N.D. | 0.14 | 0.01 | 0.008 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.003 |
| Stressed-Induced Protein | ||||||||||||||||||
| CL1841.Contig1 | catalase | 10.20 | 1.92 | N.D. | N.D. | 27.14 | 2.84 | 5.48 | 1.22 | 0.38 | 0.07 | 0.001 | 0.00 | 0.00 | 0.001 | 0.20 | 0.05 | 0.000 |
| CL7722.Contig1 | glutathione S-transferase mu | 3.85 | 1.84 | N.D. | N.D. | 12.42 | 2.48 | N.D. | N.D. | 0.31 | 0.15 | 0.011 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.206 |
| CL1155.Contig1 | glutathione S-transferase Mu 3 | 3.40 | 0.95 | N.D. | N.D. | 20.55 | 3.13 | N.D. | N.D. | 0.17 | 0.05 | 0.000 | 0.00 | 0.00 | 0.002 | 0.00 | 0.00 | 0.002 |
| Unigene5253 | glutathione peroxidase | 6.94 | 1.21 | N.D. | N.D. | 10.88 | 1.95 | N.D. | N.D. | 0.64 | 0.11 | 0.036 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.003 |
| Nervous System-Related Molecules | ||||||||||||||||||
| Unigene6214 | N-acetylated alpha-linked acidic dipeptidase 2 | 5.39 | 4.28 | 11.05 | 0.74 | 22.38 | 1.63 | 6.42 | 0.38 | 0.24 | 0.19 | 0.008 | 0.49 | 0.03 | 0.002 | 0.29 | 0.02 | 0.000 |
| CL1083.Contig2 | neuroglian | 3.30 | 0.63 | N.D. | N.D. | 20.66 | 1.85 | 8.32 | 0.41 | 0.16 | 0.03 | 0.000 | 0.00 | 0.00 | 0.001 | 0.40 | 0.02 | 0.000 |
| Unigene14124 | Cysteine string protein | N.D. | N.D. | N.D. | N.D. | 6.50 | 0.63 | N.D. | N.D. | only in cyp | 0.001 | only in cyp | 0.001 | only in cyp | 0.001 | |||
| CL745.Contig1 | Fasciclin-2 | 4.75 | 0.73 | N.D. | N.D. | 17.03 | 3.42 | 3.67 | 0.12 | 0.28 | 0.04 | 0.002 | 0.00 | 0.00 | 0.004 | 0.22 | 0.01 | 0.001 |
| CL716.Contig1 | Fasciclin-2 | 7.65 | 1.22 | 1.40 | 0.01 | 28.50 | 5.46 | 6.16 | 2.20 | 0.27 | 0.04 | 0.001 | 0.05 | 0.00 | 0.005 | 0.22 | 0.08 | 0.002 |
| CL6416.Contig1 | Acetylcholine receptor subunit alpha-type acr-16 | N.D. | N.D. | 4.82 | 0.36 | 7.27 | 0.53 | N.D. | N.D. | 0.00 | 0.00 | 0.000 | 0.66 | 0.04 | 0.008 | 0.00 | 0.00 | 0.000 |
| Unigene11480 | ELAV-like protein 2 | N.D. | N.D. | N.D. | N.D. | 9.65 | 3.46 | N.D. | N.D. | only in cyp | 0.011 | only in cyp | 0.011 | only in cyp | 0.011 | |||
| Protein Modification | ||||||||||||||||||
| CL78.Contig2 | cyclophilin type peptidyl-prolyl cis-trans isomeras | 43.10 | 6.70 | 19.85 | 3.57 | 86.99 | 9.51 | 26.99 | 4.82 | 0.50 | 0.08 | 0.003 | 0.23 | 0.04 | 0.002 | 0.31 | 0.06 | 0.001 |
| CL3896.Contig1 | malectin-A | 5.27 | 1.15 | N.D. | N.D. | 13.15 | 3.71 | N.D. | N.D. | 0.40 | 0.09 | 0.015 | 0.00 | 0.00 | 0.006 | 0.00 | 0.00 | 0.006 |
| Unigene7031 | Putative protein disulfide-isomerase A4 | 12.76 | 1.02 | N.D. | N.D. | 43.82 | 15.45 | N.D. | N.D. | 0.29 | 0.02 | 0.043 | 0.00 | 0.00 | 0.010 | 0.00 | 0.00 | 0.001 |
| CL18.Contig2 | Peptidyl-prolyl cis-trans isomerase | 12.15 | 0.56 | 6.79 | 0.22 | 53.30 | 5.77 | 15.90 | 1.71 | 0.07 | 0.02 | 0.000 | 0.13 | 0.00 | 0.000 | 0.30 | 0.03 | 0.000 |
| CL7127.Contig1 | cyclophilin B | 3.47 | 1.41 | N.D. | N.D. | 6.50 | 0.47 | N.D. | N.D. | 0.53 | 0.22 | 0.031 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| CL14028.Contig1 | receptor accessory protein 5 | N.D. | N.D. | N.D. | N.D. | 6.15 | 0.49 | N.D. | N.D. | only in cyp | 0.001 | only in cyp | 0.001 | only in cyp | 0.001 | |||
| Replication, Recombination and Repair | ||||||||||||||||||
| CL4480.Contig1 | Replication protein A 32 kDa subunit | N.D. | N.D. | N.D. | N.D. | 12.43 | 4.10 | N.D. | N.D. | only in cyp | 0.010 | only in cyp | 0.010 | only in cyp | 0.010 | |||
| Signaling Molecules | ||||||||||||||||||
| CL4060.Contig1 | Probable G-protein coupled receptor 158 | 5.55 | 0.80 | N.D. | N.D. | 11.39 | 0.58 | 3.51 | 1.65 | 0.49 | 0.07 | 0.001 | 0.00 | 0.00 | 0.000 | 0.31 | 0.14 | 0.004 |
| CL188.Contig2 | Protein phosphatase 2 | 2.42 | 1.15 | N.D. | N.D. | 17.05 | 4.88 | 7.71 | 0.67 | 0.14 | 0.07 | 0.003 | 0.00 | 0.00 | 0.007 | 0.45 | 0.04 | 0.018 |
| CL7.Contig12 | CUB-serine protease | N.D. | N.D. | N.D. | N.D. | 7.69 | 2.34 | N.D. | N.D. | only in cyp | 0.009 | only in cyp | 0.009 | only in cyp | 0.009 | |||
| CL8242.Contig1 | Dipeptidyl peptidase 4 | N.D. | N.D. | N.D. | N.D. | 5.77 | 1.80 | N.D. | N.D. | only in cyp | 0.009 | only in cyp | 0.009 | only in cyp | 0.009 | |||
| Unclassified and Function Unkown | ||||||||||||||||||
| CL62.Contig1 | intracellular fatty acid-binding protein | 10.22 | 2.47 | 5.56 | 0.21 | 34.45 | 5.10 | N.D. | N.D. | 0.30 | 0.07 | 0.001 | 0.16 | 0.01 | 0.002 | 0.00 | 0.00 | 0.000 |
| CL3340.Contig1 | chorion peroxidase | N.D. | N.D. | N.D. | N.D. | 7.71 | 0.54 | 1.70 | 0.23 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 | 0.22 | 0.03 | 0.000 |
| Unigene8333 | Sterile alpha and TIR motif-containing protein | N.D. | N.D. | N.D. | N.D. | 11.14 | 0.41 | N.D. | N.D. | only in cyp | 0.000 | only in cyp | 0.000 | only in cyp | 0.000 | |||
| CL1705.Contig1 | serine-threonine kinase receptor-associated protein | 12.77 | 0.33 | N.D. | N.D. | 27.71 | 3.93 | 3.79 | 0.57 | 0.46 | 0.01 | 0.016 | 0.00 | 0.00 | 0.002 | 0.14 | 0.02 | 0.000 |
| Unigene25709 | mannosidase alpha class 2a | N.D. | N.D. | N.D. | N.D. | 13.00 | 0.65 | N.D. | N.D. | only in cyp | 0.000 | only in cyp | 0.000 | only in cyp | 0.000 | |||
| CL6638.Contig1 | BCS-2 | 2.61 | 0.18 | N.D. | N.D. | 48.27 | 4.50 | 10.27 | 2.65 | 0.05 | 0.00 | 0.000 | 0.00 | 0.00 | 0.001 | 0.21 | 0.05 | 0.000 |
| Unigene6188 | CG11190 CG11190-PA | N.D. | N.D. | N.D. | N.D. | 17.22 | 1.06 | N.D. | N.D. | only in cyp | 0.000 | only in cyp | 0.000 | only in cyp | 0.000 | |||
| CL3280.Contig1 | proprotein convertase subtilisin/kexin type 9 preproprotein | N.D. | N.D. | N.D. | N.D. | 9.09 | 0.68 | N.D. | N.D. | only in cyp | 0.000 | only in cyp | 0.000 | only in cyp | 0.000 | |||
| CL6163.Contig1 | AGAP003142-PA | N.D. | N.D. | N.D. | N.D. | 4.94 | 1.30 | N.D. | N.D. | only in cyp | 0.006 | only in cyp | 0.006 | only in cyp | 0.006 | |||
| CL16604.Contig1 | opsin 5 | 8.39 | 2.28 | N.D. | N.D. | 20.66 | 6.92 | N.D. | N.D. | 0.41 | 0.11 | 0.035 | 0.00 | 0.00 | 0.010 | 0.00 | 0.00 | 0.010 |
| CL1826.Contig1 | nodal modulator | N.D. | N.D. | N.D. | N.D. | 3.66 | 1.40 | N.D. | N.D. | only in cyp | 0.011 | only in cyp | 0.011 | only in cyp | 0.011 | |||
| Unigene8029 | RE59172p | N.D. | N.D. | N.D. | N.D. | 7.87 | 3.89 | N.D. | N.D. | only in cyp | 0.019 | only in cyp | 0.019 | only in cyp | 0.019 | |||
| CL4128.Contig1 | MGC84319 protein | N.D. | N.D. | N.D. | N.D. | 3.77 | 1.92 | N.D. | N.D. | only in cyp | 0.020 | only in cyp | 0.020 | only in cyp | 0.020 | |||
nau2, the stage II nauplii; nau6, the stage VI nauplii; cyp, the cyprids; juv, the juveniles; Ave, average; SD, standard deviation; N.D., not detected, p value was obtained by Student t-test.
Table 2. Significantly Up-regulated Proteins in Juveniles b .
| Protein abundance (×1000 molecules/cell) | Protein ratios | |||||||||||||||||
| nau2 | nau6 | cyp | juv | nau2/juv | nau6/juv | cyp/juv | ||||||||||||
| Accession No. | Protein Description | ave | SD | ave | SD | ave | SD | ave | SD | ave | SD | p | ave | SD | p | ave | SD | p |
| Structural Molecules | ||||||||||||||||||
| CL8091.Contig1 | calponin/transgelin | 3.90 | 1.58 | N.D. | N.D. | N.D. | N.D. | 16.86 | 3.24 | 0.23 | 0.09 | 0.003 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.000 |
| Unigene7914 | AGAP007532-PA isoform 2 | 6.38 | 2.53 | N.D. | N.D. | 5.18 | 2.31 | 16.55 | 2.37 | 0.39 | 0.15 | 0.010 | 0.00 | 0.00 | 0.002 | 0.31 | 0.14 | 0.006 |
| CL3067.Contig1 | TPA: TPA_inf: troponin H isoform 2 | 6.16 | 1.36 | N.D. | N.D. | 2.64 | 1.10 | 12.09 | 1.37 | 0.51 | 0.11 | 0.007 | 0.00 | 0.00 | 0.001 | 0.22 | 0.09 | 0.001 |
| CL400.Contig3 | beta chain spectrin isoform 2 | 10.33 | 1.56 | 2.55 | 1.32 | 3.39 | 1.03 | 20.09 | 3.64 | 0.51 | 0.08 | 0.010 | 0.13 | 0.07 | 0.004 | 0.17 | 0.05 | 0.001 |
| CL3753.Contig1 | myosin heavy chain, non-muscle | 11.78 | 1.10 | N.D. | N.D. | 1.74 | 0.52 | 25.31 | 2.03 | 0.47 | 0.04 | 0.000 | 0.00 | 0.00 | 0.001 | 0.07 | 0.02 | 0.000 |
| CL3442.Contig1 | laminin beta chain | 10.59 | 1.84 | 1.53 | 0.73 | 5.66 | 1.45 | 17.23 | 2.33 | 0.61 | 0.11 | 0.017 | 0.09 | 0.04 | 0.001 | 0.33 | 0.08 | 0.002 |
| Unigene11825 | TPA: TPA_inf: troponin T isoform 1 | 6.52 | 2.55 | N.D. | N.D. | 5.07 | 1.08 | 29.72 | 1.83 | 0.22 | 0.09 | 0.001 | 0.00 | 0.00 | 0.000 | 0.17 | 0.04 | 0.000 |
| CL352.Contig1 | Talin-1 | 9.02 | 0.24 | N.D. | N.D. | 3.55 | 0.19 | 15.24 | 0.91 | 0.59 | 0.02 | 0.000 | 0.00 | 0.00 | 0.000 | 0.23 | 0.01 | 0.000 |
| CL8008.Contig1 | myosin regulatory light chain | 13.53 | 3.34 | N.D. | N.D. | N.D. | N.D. | 30.63 | 3.10 | 0.44 | 0.11 | 0.004 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 |
| Unigene2229 | vinculin | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 21.09 | 2.19 | only in juv | 0.000 | only in juv | 0.001 | only in juv | 0.001 | |||
| CL1498.Contig1 | Beta-parvin | 0.68 | 0.09 | N.D. | N.D. | N.D. | N.D. | 7.96 | 2.66 | 0.09 | 0.01 | 0.002 | 0.00 | 0.00 | 0.010 | 0.00 | 0.00 | 0.010 |
| CL14241.Contig1 | I-connectin | 4.68 | 2.60 | N.D. | N.D. | N.D. | N.D. | 10.58 | 1.68 | 0.44 | 0.25 | 0.041 | 0.00 | 0.00 | 0.002 | 0.00 | 0.00 | 0.002 |
| Unigene27904 | titin | 4.22 | 0.58 | N.D. | N.D. | N.D. | N.D. | 7.30 | 0.59 | 0.58 | 0.08 | 0.003 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 |
| CL8128.Contig1 | thymosin beta | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 5.23 | 2.60 | only in juv | 0.019 | only in juv | 0.019 | only in juv | 0.019 | |||
| Unigene17377 | Titin | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 10.82 | 0.12 | only in juv | 0.000 | only in juv | 0.000 | only in juv | 0.000 | |||
| Unigene27606 | Basic proline-rich protein | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 23.82 | 6.25 | only in juv | 0.006 | only in juv | 0.006 | only in juv | 0.006 | |||
| Unigene28460 | LOW QUALITY PROTEIN: twitchin | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 9.90 | 3.91 | only in juv | 0.012 | only in juv | 0.012 | only in juv | 0.012 | |||
| Transcription and Translation | ||||||||||||||||||
| CL129.Contig2 | Filamin-like | 2.98 | 0.32 | N.D. | N.D. | 2.55 | 1.68 | 16.60 | 0.89 | 0.18 | 0.02 | 0.000 | 0.00 | 0.00 | 0.000 | 0.15 | 0.10 | 0.001 |
| CL7751.Contig1 | vacuolar protein sorting 35 | 16.07 | 3.96 | N.D. | N.D. | 9.60 | 5.63 | 30.55 | 7.95 | 0.53 | 0.13 | 0.038 | 0.00 | 0.00 | 0.005 | 0.31 | 0.18 | 0.024 |
| CL6818.Contig1 | heterogeneous nuclear ribonucleoprotein F | 10.35 | 1.77 | N.D. | N.D. | 5.72 | 3.01 | 17.83 | 0.99 | 0.58 | 0.10 | 0.004 | 0.00 | 0.00 | 0.000 | 0.32 | 0.17 | 0.008 |
| CL6551.Contig1 | heterogeneous nuclear ribonucleoprotein Q | 14.95 | 1.68 | N.D. | N.D. | 12.18 | 1.13 | 26.43 | 2.50 | 0.57 | 0.06 | 0.003 | 0.00 | 0.00 | 0.001 | 0.46 | 0.04 | 0.001 |
| CL887.Contig1 | ribosomal protein S6 | 7.82 | 0.16 | N.D. | N.D. | 8.17 | 1.61 | 24.43 | 3.27 | 0.32 | 0.01 | 0.008 | 0.00 | 0.00 | 0.002 | 0.33 | 0.07 | 0.001 |
| CL268.Contig1 | ribosomal protein S3 | 27.26 | 3.26 | 21.75 | 0.44 | 19.26 | 3.23 | 42.32 | 5.08 | 0.64 | 0.08 | 0.011 | 0.51 | 0.01 | 0.008 | 0.46 | 0.08 | 0.002 |
| CL11.Contig4 | Polyadenylate-binding protein 1 | 7.49 | 1.11 | 1.94 | 0.88 | 8.85 | 1.29 | 14.32 | 2.57 | 0.52 | 0.08 | 0.010 | 0.14 | 0.06 | 0.004 | 0.62 | 0.09 | 0.027 |
| CL1783.Contig1 | histone cluster 1, H2ad | 24.18 | 2.70 | 7.38 | 3.48 | 20.69 | 0.71 | 39.18 | 2.48 | 0.62 | 0.07 | 0.002 | 0.19 | 0.09 | 0.002 | 0.53 | 0.02 | 0.000 |
| CL6555.Contig1 | Histone H1 | N.D. | N.D. | N.D. | N.D. | 27.55 | 4.67 | 44.74 | 5.73 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 | 0.61 | 0.10 | 0.016 |
| CL6298.Contig1 | Elongation factor 2 | 27.72 | 0.80 | 3.13 | 0.56 | 29.21 | 0.85 | 51.52 | 4.68 | 0.54 | 0.02 | 0.001 | 0.06 | 0.01 | 0.000 | 0.57 | 0.02 | 0.001 |
| Unigene18926 | DNA topoisomerase 2 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 6.58 | 2.50 | only in juv | 0.001 | only in juv | 0.014 | only in juv | 0.014 | |||
| CL1214.Contig3 | DNA-directed RNA polymerase II largest subunit isoform 12 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 15.44 | 7.46 | only in juv | 0.018 | only in juv | 0.018 | only in juv | 0.018 | |||
| Unigene23156 | mannose-binding protein | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 5.42 | 0.24 | only in juv | 0.000 | only in juv | 0.000 | only in juv | 0.000 | |||
| CL7413.Contig1 | trans-sialidase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 6.41 | 1.90 | only in juv | 0.008 | only in juv | 0.008 | only in juv | 0.008 | |||
| Energy and Metabolism | ||||||||||||||||||
| CL2238.Contig1 | phosphate carrier protein, mitochondrial-like isoform 1 | 24.12 | 4.27 | 33.15 | 2.76 | 27.08 | 2.17 | 56.52 | 4.46 | 0.43 | 0.08 | 0.001 | 0.59 | 0.05 | 0.006 | 0.48 | 0.04 | 0.000 |
| CL436.Contig4 | anion exchange protein 3-like isoform 1 | 1.66 | 0.07 | N.D. | N.D. | N.D. | N.D. | 12.77 | 3.17 | 0.13 | 0.01 | 0.012 | 0.00 | 0.00 | 0.005 | 0.00 | 0.00 | 0.005 |
| CL3616.Contig1 | 6-phosphogluconate dehydrogenase, decarboxylating | 16.24 | 5.81 | 9.02 | 2.76 | 9.25 | 3.00 | 30.72 | 6.34 | 0.53 | 0.19 | 0.047 | 0.29 | 0.09 | 0.015 | 0.30 | 0.10 | 0.005 |
| CL6215.Contig1 | phosphoacetylglucosamine mutase | 5.90 | 0.45 | N.D. | N.D. | N.D. | N.D. | 10.72 | 3.36 | 0.55 | 0.04 | 0.048 | 0.00 | 0.00 | 0.008 | 0.00 | 0.00 | 0.008 |
| CL1940.Contig2 | galactokinase 2 | 2.45 | 0.15 | N.D. | N.D. | N.D. | N.D. | 10.98 | 0.82 | 0.22 | 0.01 | 0.000 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.000 |
| CL4661.Contig3 | MOXD1-like protein 2 | 4.95 | 0.70 | N.D. | N.D. | N.D. | N.D. | 8.12 | 0.68 | 0.61 | 0.09 | 0.006 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 |
| Unigene32056 | Putative glycogen [starch] synthase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 16.80 | 1.84 | only in juv | 0.001 | only in juv | 0.001 | only in juv | 0.001 | |||
| CL10045.Contig1 | dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial-lik | 3.62 | 1.36 | N.D. | N.D. | N.D. | N.D. | 10.61 | 4.21 | 0.34 | 0.13 | 0.037 | 0.00 | 0.00 | 0.014 | 0.00 | 0.00 | 0.014 |
| CL339.Contig5 | ATP citrate lyase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 17.56 | 10.48 | only in juv | 0.040 | only in juv | 0.040 | only in juv | 0.040 | |||
| CL1080.Contig3 | Acetyl-coenzyme A synthetase, cytoplasmic | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 7.68 | 3.65 | only in juv | 0.003 | only in juv | 0.022 | only in juv | 0.022 | |||
| Unigene26389 | probable 4-coumarate--CoA ligase 1 | 4.27 | 2.19 | N.D. | N.D. | N.D. | N.D. | 11.56 | 3.12 | 0.37 | 0.19 | 0.034 | 0.00 | 0.00 | 0.006 | 0.00 | 0.00 | 0.006 |
| CL1.Contig245 | arginine kinase | 19.32 | 3.92 | 26.89 | 8.83 | 4.00 | 4.00 | 46.72 | 6.30 | 0.41 | 0.08 | 0.002 | 0.58 | 0.02 | 0.011 | 0.09 | 0.09 | 0.001 |
| Unigene11220 | Estradiol 17-beta-dehydrogenase 12 | N.D. | N.D. | N.D. | N.D. | 6.14 | 2.87 | 12.61 | 1.17 | 0.00 | 0.00 | 0.000 | 0.00 | 0.00 | 0.001 | 0.49 | 0.23 | 0.039 |
| Extracellular Matrix and Adhesion Molecules | ||||||||||||||||||
| CL14344.Contig1 | integrin beta-PS | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 15.10 | 6.78 | only in juv | 0.020 | only in juv | 0.020 | only in juv | 0.002 | |||
| CL475.Contig2 | neuroblast differentiation-associated protein AHNAK | 13.63 | 2.17 | N.D. | N.D. | 3.18 | 1.48 | 35.33 | 0.44 | 0.39 | 0.06 | 0.005 | 0.00 | 0.00 | 0.000 | 0.09 | 0.04 | 0.003 |
| Unigene6163 | Basement membrane-specific heparan sulfate proteoglycan core protein | 8.58 | 3.68 | N.D. | N.D. | 15.42 | 0.96 | 40.67 | 7.04 | 0.21 | 0.09 | 0.002 | 0.00 | 0.00 | 0.003 | 0.38 | 0.02 | 0.002 |
| Unigene32829 | chorion peroxidas | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 8.17 | 3.05 | only in juv | 0.011 | only in juv | 0.011 | only in juv | 0.011 | |||
| Unigene9044 | probable chitinase 3 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 7.82 | 3.57 | only in juv | 0.016 | only in juv | 0.016 | only in juv | 0.016 | |||
| Cell Signaling | ||||||||||||||||||
| CL260.Contig1 | 14-3-3 zeta | 41.16 | 3.94 | 18.00 | 5.25 | 34.99 | 3.48 | 72.75 | 2.10 | 0.57 | 0.05 | 0.000 | 0.25 | 0.07 | 0.001 | 0.48 | 0.05 | 0.000 |
| CL16749.Contig1 | CAMP-dependent protein kinase | 5.69 | 1.37 | N.D. | N.D. | N.D. | N.D. | 11.69 | 3.06 | 0.49 | 0.12 | 0.032 | 0.00 | 0.00 | 0.006 | 0.00 | 0.00 | 0.006 |
| Shell Calcification | ||||||||||||||||||
| CL13807.Contig1 | Carbonic anhydrase 2 | 13.78 | 1.84 | 5.09 | 2.44 | N.D. | N.D. | 24.43 | 1.76 | 0.56 | 0.08 | 0.002 | 0.21 | 0.10 | 0.003 | 0.00 | 0.00 | 0.000 |
| CL15286.Contig1 | alpha-carbonic anhydrase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 31.20 | 3.76 | only in juv | 0.001 | only in juv | 0.001 | only in juv | 0.001 | |||
| Protein Degradation | ||||||||||||||||||
| CL7237.Contig1 | Ubiquitin-like modifier-activating enzyme 1 | 14.57 | 4.98 | N.D. | N.D. | N.D. | N.D. | 30.21 | 4.63 | 0.48 | 0.16 | 0.021 | 0.00 | 0.00 | 0.002 | 0.00 | 0.00 | 0.000 |
| CL6617.Contig2 | 26S proteasome non-ATPase regulatory subunit 2 | 11.37 | 2.49 | N.D. | N.D. | 9.92 | 0.80 | 18.94 | 2.15 | 0.60 | 0.13 | 0.019 | 0.00 | 0.00 | 0.001 | 0.52 | 0.04 | 0.002 |
| Protein Modification | ||||||||||||||||||
| Unigene11234 | AGAP009694-PA | 14.51 | 0.20 | 5.10 | 1.23 | 6.33 | 2.24 | 22.59 | 1.70 | 0.64 | 0.01 | 0.011 | 0.23 | 0.05 | 0.001 | 0.28 | 0.10 | 0.001 |
| CL210.Contig1 | AGAP007393-PB | 21.04 | 0.59 | 5.71 | 3.41 | 12.50 | 2.86 | 37.85 | 3.43 | 0.56 | 0.02 | 0.001 | 0.15 | 0.09 | 0.003 | 0.33 | 0.08 | 0.001 |
| Cell Differentiation | ||||||||||||||||||
| CL9909.Contig1 | n-myc downstream regulated | 29.52 | 5.94 | N.D. | N.D. | 27.01 | 4.12 | 59.86 | 1.69 | 0.49 | 0.10 | 0.002 | 0.00 | 0.00 | 0.000 | 0.45 | 0.07 | 0.000 |
| CL439.Contig1 | kakapo | 4.36 | 1.36 | 0.33 | 0.15 | 1.18 | 0.18 | 18.20 | 0.40 | 0.24 | 0.07 | 0.007 | 0.02 | 0.01 | 0.000 | 0.07 | 0.01 | 0.000 |
| CL4061.Contig1 | kakapo | 6.09 | 0.48 | N.D. | N.D. | 1.07 | 0.07 | 12.33 | 0.36 | 0.49 | 0.04 | 0.000 | 0.00 | 0.00 | 0.000 | 0.09 | 0.01 | 0.000 |
| Unigene14025 | kakapo | 1.46 | 0.16 | N.D. | N.D. | N.D. | N.D. | 10.26 | 2.18 | 0.14 | 0.02 | 0.001 | 0.00 | 0.00 | 0.004 | 0.00 | 0.00 | 0.004 |
| CL3810.Contig1 | kakapo | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 6.52 | 2.23 | only in juv | 0.010 | only in juv | 0.010 | only in juv | 0.010 | |||
| Cell Proliferation | ||||||||||||||||||
| CL3176.Contig1 | annexin 1 | N.D. | N.D. | N.D. | N.D. | 2.21 | 1.90 | 10.63 | 1.41 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 | 0.21 | 0.18 | 0.030 |
| Unigene14024 | ovulatory protein-2 precursor | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 23.76 | 7.63 | only in juv | 0.009 | only in juv | 0.009 | only in juv | 0.009 | |||
| Stress-induced Protein | ||||||||||||||||||
| CL364.Contig1 | Glycoprotein 93 CG5520-PA | 8.11 | 1.68 | 12.89 | 0.41 | 6.12 | 0.64 | 31.49 | 2.17 | 0.26 | 0.05 | 0.000 | 0.41 | 0.01 | 0.001 | 0.19 | 0.02 | 0.000 |
| CL405.Contig3 | 70kDa heat shock protein | 20.10 | 2.71 | 21.29 | 0.08 | 19.17 | 2.38 | 40.46 | 4.34 | 0.50 | 0.07 | ????? | 0.53 | 0.00 | 0.012 | 0.47 | 0.06 | 0.001 |
| CL695.Contig1 | Cct5-prov protein | 6.48 | 0.94 | 2.52 | 0.10 | 3.05 | 0.20 | 13.78 | 1.22 | 0.47 | 0.07 | 0.001 | 0.18 | 0.01 | 0.000 | 0.22 | 0.01 | 0.000 |
| Inorganic Ion Transportaion | ||||||||||||||||||
| CL3156.Contig2 | Na+/K+ ATPase alpha subunit | 35.37 | 1.81 | 10.17 | 0.91 | 22.21 | 0.32 | 61.04 | 2.01 | 0.58 | 0.03 | 0.000 | 0.17 | 0.01 | 0.000 | 0.36 | 0.01 | 0.000 |
| CL12979.Contig1 | Na+/K+ ATPase beta subunit | 22.21 | 1.72 | 11.59 | 2.01 | N.D. | N.D. | 35.44 | 3.73 | 0.63 | 0.05 | 0.004 | 0.33 | 0.06 | 0.003 | 0.00 | 0.00 | 0.001 |
| Unigene13663 | nitric oxide synthase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 7.09 | 2.68 | only in juv | 0.001 | only in juv | 0.014 | only in juv | 0.014 | |||
| CL1831.Contig1 | ribophorin I | 9.08 | 0.45 | N.D. | N.D. | N.D. | N.D. | 32.37 | 6.47 | 0.28 | 0.01 | 0.013 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.003 |
| Cell Wall/Membrane/Envelope Biogenesis | ||||||||||||||||||
| CL7195.Contig1 | Vigilin | 10.00 | 0.71 | N.D. | N.D. | 4.06 | 1.08 | 18.07 | 3.72 | 0.55 | 0.04 | 0.016 | 0.00 | 0.00 | 0.004 | 0.22 | 0.06 | 0.002 |
| Cell Cycle Control | ||||||||||||||||||
| CL184.Contig1 | uncharacterized protein C6orf168 | 16.46 | 1.09 | 8.23 | 0.01 | 10.49 | 2.86 | 32.76 | 2.27 | 0.50 | 0.03 | 0.000 | 0.25 | 0.00 | 0.002 | 0.32 | 0.09 | 0.001 |
| Intracellular Trafficking, Secretion, and Vesicular Transport | ||||||||||||||||||
| CL1315.Contig1 | clathrin heavy chain-like isoform 1 | 18.28 | 0.30 | 3.21 | 1.39 | 13.58 | 0.22 | 33.17 | 1.29 | 0.55 | 0.01 | 0.000 | 0.10 | 0.04 | 0.000 | 0.41 | 0.01 | 0.000 |
| CL1268.Contig2 | signal recognition particle receptor subunit alpha homolog isoform | 1.70 | 0.51 | N.D. | N.D. | N.D. | N.D. | 5.16 | 1.36 | 0.33 | 0.10 | 0.009 | 0.00 | 0.00 | 0.005 | 0.00 | 0.00 | 0.005 |
| Unclassified and Function Unkown | ||||||||||||||||||
| Unigene24129 | endothelin-converting enzyme 1 | 3.99 | 0.31 | N.D. | N.D. | N.D. | N.D. | 20.63 | 5.45 | 0.19 | 0.02 | 0.017 | 0.00 | 0.00 | 0.006 | 0.00 | 0.00 | 0.000 |
| Unigene29044 | Nuclear receptor coactivator 5 | 4.10 | 1.55 | N.D. | N.D. | N.D. | N.D. | 13.59 | 1.78 | 0.30 | 0.11 | 0.003 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.000 |
| CL3782.Contig1 | sporozoite surface protein | 3.16 | 0.65 | 1.15 | 0.01 | N.D. | N.D. | 7.27 | 2.70 | 0.43 | 0.09 | 0.045 | 0.16 | 0.00 | 0.032 | 0.00 | 0.00 | 0.012 |
| CL6558.Contig1 | anion exchange protein 2-like isoform 2 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 7.42 | 0.42 | only in juv | 0.000 | only in juv | 0.000 | only in juv | 0.000 | |||
| CL9869.Contig1 | dendritic cell protein | 9.57 | 1.13 | 2.68 | 0.10 | 4.73 | 1.77 | 16.63 | 3.49 | 0.58 | 0.07 | 0.023 | 0.16 | 0.01 | 0.005 | 0.28 | 0.11 | 0.005 |
| Unigene26106 | CD109 antigen | N.D. | N.D. | N.D. | N.D. | 2.14 | 0.26 | 5.40 | 1.06 | 0.00 | 0.00 | 0.003 | 0.00 | 0.00 | 0.003 | 0.40 | 0.05 | 0.003 |
| CL7633.Contig1 | Aldose reductase | 15.90 | 2.74 | 3.86 | 1.69 | 16.73 | 1.15 | 30.80 | 3.24 | 0.52 | 0.09 | 0.004 | 0.13 | 0.05 | 0.001 | 0.54 | 0.04 | 0.002 |
| CL987.Contig1 | CG5149 | 6.16 | 1.60 | 2.24 | 0.56 | N.D. | N.D. | 12.88 | 4.05 | 0.48 | 0.12 | 0.040 | 0.17 | 0.04 | 0.016 | 0.00 | 0.00 | 0.008 |
| CL6595.Contig1 | settlement inducing protein complex | N.D. | N.D. | 0.87 | 0.03 | N.D. | N.D. | 5.29 | 1.34 | 0.00 | 0.00 | 0.005 | 0.17 | 0.01 | 0.008 | 0.00 | 0.00 | 0.005 |
| CL217.Contig3 | cross-beta structure silk protein 1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 8.58 | 0.80 | only in juv | 0.000 | only in juv | 0.001 | only in juv | 0.001 | |||
| CL7617.Contig1 | sodium-driven chloride bicarbonate exchanger-like isoform 3 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 5.97 | 1.04 | only in juv | 0.000 | only in juv | 0.003 | only in juv | 0.003 | |||
| CL608.Contig2 | trans-sialidase | 2.21 | 1.24 | N.D. | N.D. | N.D. | N.D. | 4.70 | 0.40 | 0.47 | 0.26 | 0.047 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 |
| CL4385.Contig1 | NSFL1 cofactor p47 | 2.83 | 1.08 | N.D. | N.D. | N.D. | N.D. | 10.94 | 0.96 | 0.26 | 0.10 | 0.002 | 0.00 | 0.00 | 0.001 | 0.00 | 0.00 | 0.001 |
| CL5942.Contig1 | lipoprotein receptor | 8.46 | 4.91 | 15.12 | 7.57 | 3.13 | 2.73 | 33.71 | 2.88 | 0.25 | 0.15 | 0.004 | 0.45 | 0.22 | 0.038 | 0.09 | 0.08 | 0.004 |
| CL1729.Contig1 | C-type lectin 4 | 8.03 | 2.13 | N.D. | N.D. | N.D. | N.D. | 13.94 | 2.22 | 0.58 | 0.15 | 0.032 | 0.00 | 0.00 | 0.002 | 0.00 | 0.00 | 0.002 |
| Unigene4943 | cystathionine-beta-synthase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 5.63 | 0.55 | only in juv | 0.001 | only in juv | 0.001 | only in juv | 0.001 | |||
| Unigene26120 | cysteine-rich-protein | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 36.29 | 3.56 | only in juv | 0.001 | only in juv | 0.001 | only in juv | 0.001 | |||
| CL2143.Contig2 | cement protein-100k | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 9.77 | 3.13 | only in juv | 0.009 | only in juv | 0.009 | only in juv | 0.009 | |||
| CL1152.Contig1 | BCS-4 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 35.37 | 4.58 | only in juv | 0.001 | only in juv | 0.001 | only in juv | 0.001 | |||
| Unigene7077 | transglutaminase | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 8.11 | 4.26 | only in juv | 0.021 | only in juv | 0.021 | only in juv | 0.021 | |||
nau2, the stage II nauplii; nau6, the stage VI nauplii; cyp, the cyprids; juv, the juveniles; Ave, average; SD, standard deviation; N.D., not detected, p value was obtained by Student t-test.
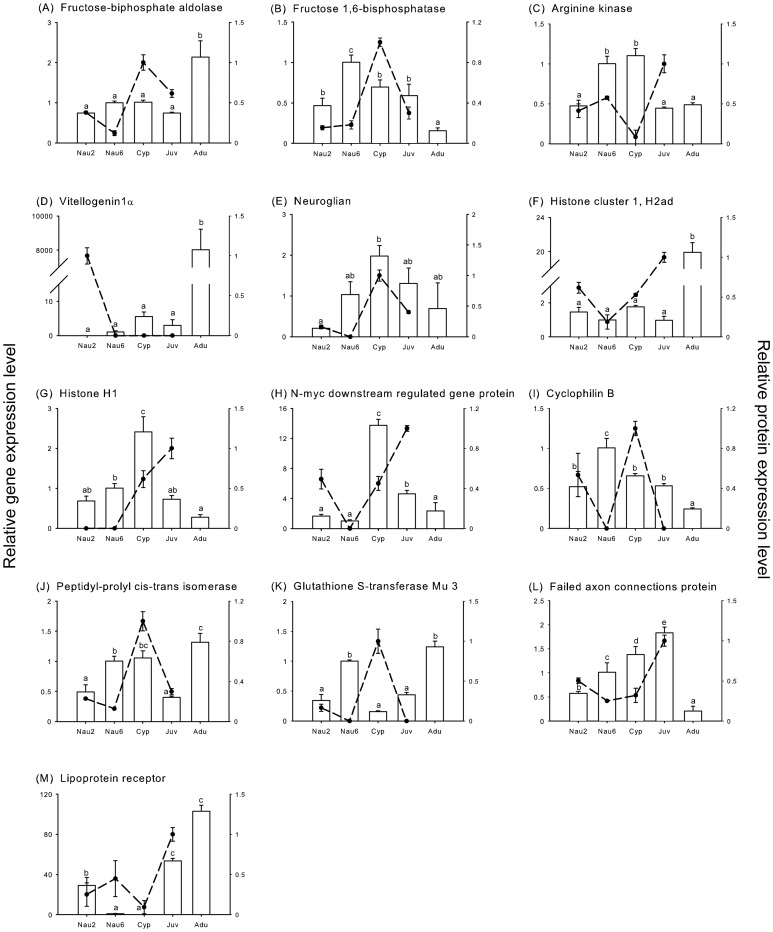
To analyze the gene expression of the selected proteins, qRT-PCR was conducted. As shown in Figure 3, a positive correlation between transcription and translation expression patterns was detected in several proteins, such as neuroglian, failed axon connection protein and lipoprotein receptor. In contrast, the expression trends observed for some proteins, including fructose-bisphosphate aldolase, histone cluster 1 H2ad and glutathione S-transferase Mu 3, did not display any similarity with their gene expression profiles. Interestingly, some transcripts accumulated prior to the peak expression of the protein products, as observed for fructose 1,6-bisphosphatase, arginine kinase, histone H1, n-myc downstream gene protein, cyclophilin B and peptidyl-prolyl cis-trans isomerase.
Figure 3. Quantitative real-time PCR (qRT-PCR) results of the levels of 13 genes with protein products that were significantly up-regulated in either cyprids or juveniles (white bars).
The corresponding protein expression levels are presented as line charts. The detected developmental stages included stage II nauplii (Nau2), stage VI nauplii (Nau6), cyprids (Cyp), young juveniles (Juv) and adults (Adu). Values are expressed as the mean ± SD of 3 different experimental replicates. In the figures, a, b, c, d and e in the figures showed significantly different expression patterns among the samples detected, as determined by one-way ANOVA followed by Tukey's post-hoc test (P<0.05).
Discussion
Barnacle larval settlement includes the attainment of competency by swimming cyprids, attachment to the substratum, and the metamorphosis of attached cyprids into juveniles, which involve energy-related, neurotransmission-related, and cell proliferation and differentiation-related molecules are involved [5]. With a special focus on larval settlement and subsequent juvenile development, we identified 80 and 95 proteins that were significantly up-regulated in the cyprid and the juvenile proteome, respectively.
Proteins that were significantly up-regulated in cyprids
Energy and metabolism proteins
Twenty-four energy-related proteins were significantly up-regulated in cyprids, accounting for approximately 30% of the up-regulated proteins in the larval stage (shown in Table 1). Fructose-bisphosphate aldolase, fructose 1,6-bisphosphatase, glucose-6-phosphate isomerase, glucosamine-6-phosphate isomerase 1 and mannose-6-phosphate isomerase are involved in the metabolism of fructose 6-phosphate, which is one of the key products in glycolysis and its reverse process, gluconeogenesis, for converting a non-carbohydrate to glucose [27]. Because cyprids do not feed, they rely on stored lipids and proteins as primary energy sources to swim, search for a suitable surface for attachment and metamorphosis [28]. The up-regulation of fructose 1,6-bisphosphatase in cyprids suggested that lipids and proteins were converted to glucose via active gluconeogenesis to support cyprid energy consumption. Interestingly, the transcript displayed a higher expression level in stage VI nauplii than in other stages (Figure 3B). Furthermore, because two of the enzymes catalyzed reactions in both glycolysis and gluconeogenesis, fructose-bisphosphate aldolase and glucose-6-phosphate isomerase might play important roles in maintaining the balance between these two metabolic pathways in cyprids.
In the present study, acetyl-CoA acetyltransferase was significantly up-regulated during the cyprid stage, which is consistent to the results of an early study demonstrating that the acetyl-CoA acetyltransferase gene was significantly up-regulated during this stage [29]. Acetyl-CoA carboxylase 2 and acyl-CoA oxidase participate in the catabolism of lipids through fatty acid oxidation [30], [31]. The product of acetyl-CoA enters citric acid cycle to produce energy. Acetyl-CoA acetyltransferase is involved in several metabolic pathways, including ketone body synthesis and degradation, fatty acid metabolism and pyruvate metabolism. Cyprids become less active and their settlement rate decreases in response to treatment with butenolide [32], as acetyl-CoA acetyltransferase is one of the binding targets of butenolide in barnacle cyprids and is involved in the inhibition of larval settlement triggered by butenolide [29]. Taken together, these results indicated that acetyl-CoA acetyltransferase is a crucial enzyme for maintaining barnacle cyprid activity and their subsequent successful settlement.
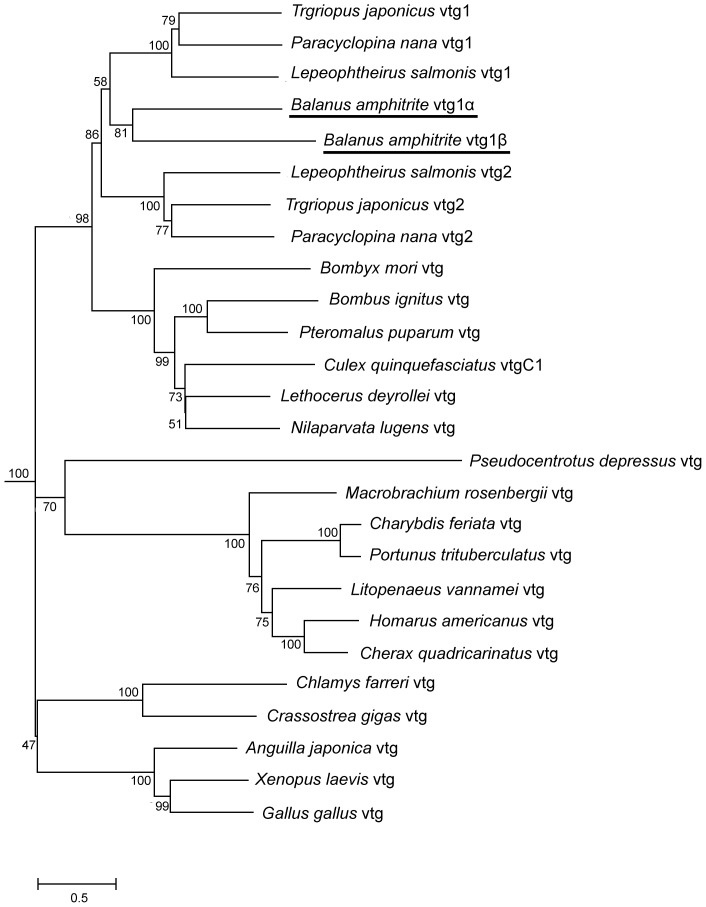
In the barnacle proteome, two vitellogenins were identified. Vitellogenin 1α was only detected in the stage II nauplii (Table S2); however, its gene expression level was 1,000 times higher in adults (Figure 3D), suggesting that this maternal vitellogenin 1α might function as a storage protein for embryonic and early larval development, similar to the general vitellogenin in other species. Interestingly, unlike vitellogenin 1α, vitellogenin 1β was highly expressed in stage VI nauplii and cyprids (Table S2), and its expression pattern was similar to that of the cyprid major protein. Cyprid major protein is utilized as an energy resource during cyprid settlment and metamorphosis [33]. It was identified as a vitellogenin 1β-like protein based on the results of SDS-PAGE and LC-MS analysis [34]. To better explore the relationship between the two vitellogenins detected in B. amphitrite, we conducted phylogenetic reconstruction (Figure 4). Our results revealed the monophyly of copepod Vitellogenin 1 (BS = 100), copepod Vitellogenin 2 (BS = 100) and the sister-relationship between Vitellogenin 1α and 1β (BS = 81). Furthermore, the last two genes also formed the sister taxon of copepod Vitellogenin 1 (BS = 58). These relationships suggest that barnacle Vitellogenins have functions similar to other vitellogenins; however, Vitellogenin 1β has evolved a novel and related function after gene duplication such that it is highly expressed in stage VI nauplii and cyprids rather than embryos or adults.
Figure 4. Neighbor-joining tree based on vitellogenin protein sequences.
Branch lengths represent substitutions per site, and numbers at each node represent bootstrap values. The sequences used were as follows: Tigriopus japonicas vtg1 (ABZ91537), Tigriopus japonicas vtg2 (ACJ12892), Paracyclopina nana vtg1 (ADD73551), Paracyclopina nana vtg2 (ADD73552), Lepeophtheirus salmonis vtg1 (ABU41134), Lepeophtheirus salmonis vtg2 (ABU41135), Bombyx mori vtg (BAA06397), Bombus ignites vtg (ACM46019), Pteromalus puparum vtg (ABO70318), Culex quinquefasciatus vtg (AAV31930), Lethocerus deyrollei vtg (BAG12118), Nilaparvata lugens vtg (AEL22916), Pseudocentrotus depressus vtg (AAK57983), Macrobrachium rosenbergii vtg (BAB69831), Charybdis feriata vtg (AAU93694), Portunus trituberculatus vtg (AAX94762), Litopenaeus vannamei vtg (AAP76571), Homarus americanus vtg (ABO09863), Cherax quadricarinatus vtg (AAG17936), Chlamys farreri vtg (ADE05540), Crassostrea gigas vtg (BAC22716), Anguilla japonica vtg (AAV48826), Gallus gallus vtg (AAA49139.1), and Xenopus laevis vtg (AAA49982).
Nervous system-related molecules
Cyprids possess a more complicated nervous system compared with nauplii and adults [35]. Extensive studies have been conducted to investigate neurotransmitters such as serotonin and prostanoid [36], [37]. In the current study, we detected one neurotransmitter receptor, ie, acetylcholine receptor, with a high expression level in cyprids. This receptor binds to the neurotransmitter acetylcholine, which is involved in barnacle cyprid muscular contraction and cement gland exocytosis [38]. Elevated levels of acetylcholine lead to a higher settlement rate of cyprids [38]. The up-regulation of this receptor in cyprids herein confirmed that acetylcholine has a key role in barnacle larval settlement.
N-acetylated alpha-linked acidic dipeptidase is responsible for the cleavage of the neuropeptide N-acetyl-L-aspartate-L-glutamate (NAAG), which is abundant in the central nervous system and functions as a neurotransmitter [39]. A previous study revealed the effects of NAAG on the reduction of cAMP levels and the release of GABA [40], [41]. In B. amphitrite, the intracellular cAMP level affects the patterns of cyprid settlement [42]. As a neurotransmitter, GABA controls thoracic muscle contraction and modulates eye vision and antennule activity [43]. Thus, in cyprids, N-acetylated alpha-linked acidic dipeptidase may be involved in regulating of cAMP and GABA levels by catabolizing NAAG.
In addition, we identified neuroglian, which belongs to the Ig superfamily and is essential for the development of neuroglia and the formation of synapses [44]. Neuroglian gene expression increased during larval development and peaked in cyprids, but no significant difference was observed after settlement (Figure 3E). The expression of its protein product displayed similar trends during all larval stages but decreased after settlement (Table 1). Barnacle neuroglian was found to contain 6 Ig superfamily domains and possess region homologies to cell adhesion molecules or a neuroglian-like isoform in insects. It has been reported that neuroglian regulates glial morphogenesis and antennal lobe development in Drosophila larvae undergoing metamorphosis [45].
Signaling molecules
In the present study, the CUB-serine protease was found to be uniquely expressed in cyprids, the antennules of which contain an olfactory receptor neuron-like structure. The CUB-serine protease was first located in the olfactory organ and eyestalk of the spiny lobster Panulirus argus, which suggested that it functions in the olfactory system [46]. When barnacle cyprids were treated with the anti-settlement compound meleagrin, the protein expression of the CUB-serine protease was modulated [34]. Up-regulation of the CUB-serine protease in barnacle cyprids suggested that this protein might be involved in the cyprid olfactory chemoreception system during the search for an ideal settlement spot.
Furthermore, in the present study, we identified a putative G protein-coupled receptor in the barnacle proteome that was significantly up-regulated during the cyprid stage. In the red abalone Haliotis refescens, a G protein-coupled receptor and downstream PKA-dependent cyclic AMP (cAMP) pathway are crucial participants in the pathway controlling larval settlement and metamorphosis [47]. In B. amphitrite, it has been suggested that a G protein-coupled receptor binds to the exogenous metamorphic cue while the settlement signal is conducted via a PKA independent pathway [48], [49]. Therefore, the G protein-coupled receptor identified in the current study may play a crucial role in recognizing of the settlement cue. Interestingly, we did not find any other cAMP-pathway-related proteins that were differentially expressed in cyprids, yet 1 cAMP-dependent protein kinase (PKA) was up-regulated in juveniles. Signal transduction regulated by cAMP is required for the development of a metazoan [50]. Down-regulation of PKA in cyprids and up-regulation in juveniles indicates that this PKA may be important for juvenile development rather than for larval settlement.
Structural proteins
7 structural proteins were differentially expressed during the cyprid stage. Among them, 5 belonged to the connectin/titin family, including projectin, twitchin and 3 titins (Table 1). Titins are giant proteins that contribute to muscle assembly and contraction, especially in striated muscle [51]. In barnacle cyprids, striated muscle is distributed in antennules, thoracopods and their related muscles [52]. Antennules and thoracopods function differently in cyprids: antennules are the prime locomotor and sensory apparatus, which helps these organisms contact with the substratum during the attachment process. In contrast, thoracopods enable cyprids to move or stay in the water column [53]. The up-regulation of the proteins involved in the assembly and contraction of striated muscle supported the active searching and swimming behavior displayed during barnacle larval settlement.
Functionally ungrouped proteins
In addition to the functional groups mentioned above, we also identified many proteins that belonged to some other functional groups or which unknown functions. Opsin 5 showed the most similarity to Limulus opsin 5, the protein component of visual pigment that is sensitive to visible light (400–700 nm) [54]. Barnacle cyprids, which have a pair of stage-specific compound eyes and 1 naupliar eye, display a phototactic response to light flux. The photoreception in cyprids has been suggested to be associated with a visual sensory system needed for larval settlement and distribution [55].
A previous study described the cloning of 6 barnacle cyprids larva-specific genes (bcs) by screening cDNA libraries [8]. As an ‘early’ gene, bcs-2 was highly expressed in cyprids, and then decreased dramatically when larval settlement occurred [8]. In the present study, the protein product of this gene displayed the same abundance in cyprids but could not be detected in juveniles, which suggested a specific role for Bcs-2 in barnacle cyprids.
Proteins that were significantly up-regulated in juveniles
Energy and metabolism proteins
In juveniles, we identified 12 proteins that were related to energy and metabolism. Among these 12 proteins, arginine kinase was significantly up-regulated during the juvenile stage, whereas its gene expression was 2-fold higher in stage VI nauplii and cyprids than in the other 3 stages (Figure 3C). These results indicated that the arginine kinase transcript might be accumulated prior to that of fructose 1,6-bisphosphatase. Arginine kinase often serves as a temporal energy buffer system, increasing the efficiency of the reversible phosphorylation of arginine by ATP and of fluctuating energy requirements in invertebrates [56], [57]. In addition, both pyruvate dehydrogenase and ATP citrate lyase were up-regulated in juveniles. This up-regulation might ensure the production of acetyl-CoA for subsequent energy production and fatty acid synthesis, as the pyruvate dehydrogenase complex catalyzes the transformation from pyruvate to acetyl-CoA via pyruvate decarboxylation. Alternately, this phenomenon might stimulate the generation of acetyl-CoA from acetate and CoA via the catalysis of acetyl-CoA synthetase [58]. In contrast, ATP citrate lyase is involved in the generation of acetyl CoA together with the ATP hydrolysis [59].
Structural proteins
In the juvenile proteome, 17 structural proteins were significantly up-regulated. Among them, several proteins were found to be involved in muscle assembly and contraction, including 4 connectin/titin family proteins, 2 troponin isoforms, myosin light chain and transgelin (Table 2). In addition, several actin-binding proteins were detected, such as vinculin, laminin, talin, β-parvin, and thymosin β. Spectrin is another actin scaffold protein that helps maintain the shape of the cell and plays a crucial role in the survival and development of Drosophila larval [60]. The up-regulation of these cytoskeletal proteins was not unexpected, as barnacles must undergo drastic tissue degeneration and organ remodeling during metamorphosis, such as the reduction of larval muscles, the formation of juvenile muscles, rotation of the thorax, the development of cirri, and raising of the body, among others [61].
Proteins related to transcription and translation
Thirteen proteins related to transcription and translation were significantly up-regulated during the juvenile stage. By comparison, only 6 proteins were up-regulated during the cyprid stage. We identified two heterogeneous nuclear ribonucleoproteins (F and Q) and 1 polyadenylate-binding protein, all of which were found to be involved in gene transcription and post-transcriptional modification by regulating mRNA metabolism [62], [63]. A similar expression pattern was observed for histone H2ad and histone H1, which are coupled to DNA replication, and elongation factor 2, which facilitates translation elongation. Interestingly, the gene expression of histone H2ad did not show much fluctuation during the larval stage or in juveniles but increased dramatically in adults. However, the histone H1 gene was expressed at least 2.4-times higher in cyprids than during any other stage (Figures 3F & 3G). These results suggested that histone H1 might be important for juvenile development only and that histone H2ad might function in both juveniles and adults. In short, the up-regulation of these proteins indicated that active transcription and translation might occur in barnacle juveniles.
Proteins related to cell proliferation and differentiation
Proteins involved in cell proliferation and differentiation were significantly up-regulated in juveniles only. In total, we identified 4 kakapos, one of which was detected only in juveniles. Kakapo, a cytoskeletal-associated protein expressed in Drosophila tendon cells, may be important for muscle-dependent tendon cell differentiation [64]. In barnacles, tendon cells, which are specially organized epithelial cells, function as a link between muscles and the overlying chitinous exoskeleton [65]. Up-regulation of these 4 kakapo proteins indicated that they have crucial functions in cell differentiation, mainly in the barnacle integument. In addition, N-myc downstream regulated gene (NDRG) protein was highly abundant in the juvenile proteome. This protein was down-regulated during the larval stage and increased in juveniles (Table 2); however, its gene expression peaked in cyprids (Figure 3H). The NDRG protein family contributes to cell differentiation and proliferation in diverse tissues of various animals [66]. Our results suggested that NDRG functions in cell differentiation processes associated with tissue reorganization in barnacle juveniles.
In the current study, we identified one annexin that was differentially expressed in juveniles. The expression of this protein was first detected in cyprids and then peaked in juveniles (4.8-fold higher than in cyprids) (Table 2). Annexins comprise a group of proteins that bind to phospholipids in a calcium-dependent manner. Previous studies have shown that the expression patterns of annexins change significantly when cells undergo proliferation or differentiation [67]. During silkworm metamorphosis of Bombyx mori, 20-hydroxyecdysone (20-HE) triggers programmed cell death to remove larval-specific tissues. Annexin, identified as a 20-HE inducible gene, was shown to be involved in this process [68]. The 20-HE gene has also been detected in barnacle cyprids, and its regulation of barnacle larval metamorphosis has been verified [69]. Although the detailed mechanism by which 20-HE affects barnacle larval metamorphosis remains unclear, the expression levels of annexin might be modulated by the tissue degeneration reorganization that occurs during barnacle larval development and metamorphosis, which in turn might be regulated by 20-HE.
Shell calcification proteins
Two carbonic anhydrases related to shell calcification were significantly up-regulated in juveniles and might be involved in biomineralization in B. amphitrite, especially α CA, which was uniquely expressed in juveniles. Carbonic anhydrase (CA) participates in a variety of metabolic pathways and is widely distributed in the tissues that are responsible for the formation of calcium carbonate [70]. It is also important for the molting cycle in crustaceans [71]. Inhibition of CA activity prevents shell development and growth of the barnacle Balanus improvisus, resulting in a failure in the initiation of normal development [70].
Functionally ungrouped proteins
In the present study, we discovered that the protein product of the bcs-4 gene was uniquely expressed in juveniles. Compared with bcs-2, bcs-4 is a ‘late’ gene showing weak expression levels in young cyprids that have recently molted from nauplii, increasing gradually as cyprids age, and beginning to decline in settled larvae [8]. This time lag between the expression of gene and protein supports the possibility that accumulation of the gene is required prior to protein function in juveniles.
In addition to Bsc-4, one cement protein-100k was found to be uniquely expressed during the juvenile stage. Its gene transcript was detected in the adult but not in the larval transcriptome [5]. In fact, several proteins have been identified, such as two 20 kDa-cement proteins characterized in the cyprids of B. amphitrite and large cement proteins fractionated from the adult cement of Megabalanus rosa (100 kDa, 68 kDa & 52 kDa) [72], [73].
Three proteins belonging to the heat shock protein (HSP) family were significantly up-regulated in juveniles. HSPs were initially known as stress-responsive proteins and later found to interface with various developmental pathways, as molecular chaperones associated with protein folding, assembly and transport [74], [75]. HSP70 is developmentally regulated in a diverse range of organisms, and HSP90 functions as a regulator during metamorphosis and the molt cycle of invertebrates [76]–[78]. Additionally, the expression levels of HSP70 and HSP90 increase in response to cell differentiation and tissue morphogenesis occur in the vetigastropod Haliotis asinina [79].
Comparison of proteome profiles with other marine invertebrates
In addition to the present study, proteomic approaches have been applied to study larval attachment and metamorphosis in several marine invertebrates. Although the species were from several evolutionarily distant phyla, common changes in the proteome were observed. First, during attachment and metamorphosis, a larva goes through serial body reconstructions, including the degeneration of larval structures and the subsequent emergence of juvenile tissues. The larvae of the bryozoan Bugula neritina and the polychaete Hydroides elegans, swim via beating cilia, and all of these ciliated tissues degenerate during metamorphosis, resulting in drastic changes in structural proteins, such as actin and tubulin, during larval settlement [20], [80]. Barnacle larvae swim by beating thoracopods driven by striated muscles. Actin and tubulin were not differentially expressed in the barnacle proteomes, but several proteins related to muscle structures and muscle contraction were up-regulated. In addition to structural proteins, in the present study, proteins involved in transcription and translation, such as ribosomal proteins, histones and elongation factors, were highly expressed during larval settlement in the bryozoan B. neritina [20], the polychaete Pseudopolydara vexillosa [81] and so as in B. amphitrite. These findings indicated that transcription and translation level were active when larvae underwent metamorphosis.
Larval settlement is an extremely energy-consuming process, especially for the whole larval settlement process. Proteins involved in the citric acid cycle, glycolysis, and fatty acid metabolism have been found to be up-regulated during the competent larval stage of B. neritina, P. vexillosa and the polychaete Capitella sp. I [20], [80], [81]. Similarly, these proteins were found to be up-regulated in barnacle cyprids herein. In addition, some proteins related to energy and metabolism were also up-regulated in barnacle juveniles, as described for P. vexillosa [81].These findings indicated that these organisms might require different energy metabolism pathways for larval competency and metamorphosis. Interestingly, vitellogenins have been described in competent larvae of B. neritina, B. anphitrite and Capitalla [20], [82], indicating that the non-feeding larvae of different phyla all consume vitellogenins as an energy source.
Carbonic anhydrase was detected in juveniles of both B. neritina and B. anphitrite, which require a calcified body wall or a calcareous shell [83]. This result indicated that carbonic anhydrase is important for the calcification of marine invertebrates.
Conclusions
This study investigated the regulation of protein expression patterns in response to larval development and settlement of the barnacle B. amphitrite. The utilization of label-free quantitative proteomics allowed us to conduct a comparative proteomics analysis among different developmental stages and to identify protein candidates that might be involved in barnacle larval settlement. Functional analysis of 4 proteins revealed their significant up-regulation of proteins involved in energy and metabolism, the nervous system, and signaling transduction in the cyprid stage. In addition, proteins related to cytoskeletal remodeling, translation and transcription, cell proliferation and differentiation, and biomineralization were up-regulated in the juvenile proteome. Notably, the expression patterns of some proteins, such as neuroglian, failed axon connection protein, and lipoprotein receptor, displayed the same trends as their transcripts; In contrast, the expression of fructose-1, 6-bisphosphatase, cyclophilin B, NDRG, and histone H1 transcripts occurred 1 developmental stage earlier than their protein products. The expression levels of some other proteins, such as fructose-biphosphate aldolase, were not associated with their mRNA expression. These results provide information about the molecular activities related to the changes in morphology, physiology, structure, and function that occur during barnacle larval settlement and juvenile development. Additional functional assays and characterization of protein candidates, for instance, nervous system-related proteins, signaling molecules, shell calcification proteins and heat shock proteins, could help identify more detailed molecular mechanisms underlying barnacle larval settlement.
Supporting Information
Primers for genes under real-time PCR assay.
(XLSX)
Total proteins identified and quantified from each LC-MS experiments.
(XLSX)
Significantly up-regulated proteins in stage II nauplii.
(XLSX)
Significantly up-regulated proteins in stage VI nauplii.
(XLSX)
Significantly down-regulated proteins in stage II nauplii.
(XLSX)
Significantly down-regulated proteins in stage VI nauplii.
(XLSX)
Significantly down-regulated proteins in cyprids.
(XLSX)
Significantly down-regulated proteins in juveniles.
(XLSX)
Acknowledgments
We thank Gen Zhang for his help with the sample collection and larval cultures; Dr. Kai He, Dr. Jin Sun and Ms. Xing-Cheng Yan for their technical advice and constructive discussions.
Funding Statement
This study was supported by grants from the Research Grants Council of the Hong Kong Special Administrative Region (GRF662413 and AoE/P-04/04-II) and an award from the King Abdullah University of Science and Technology (SA-C0040/UK-C0016) to P.-Y. Qian. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Qian PY (1999) Larval settlement of polychaetes. Hydrobiologia 402: 239–253. [Google Scholar]
- 2. Underwood AJ, Fairweather PG (1989) Supply-side ecology and benthic marine assemblages. Trends in Ecology & Evolution 4: 16–20. [DOI] [PubMed] [Google Scholar]
- 3. Pechenik JA, Cochrane DE, Li W, West ET, Pires A, et al. (2007) Nitric oxide inhibits metamorphosis in larvae of Crepidula fornicata, the slippershell snail. Biol Bull 213: 160–171. [DOI] [PubMed] [Google Scholar]
- 4. Bishop CD, Bates WR, Brandhorst BP (2001) Regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. J Exp Zool 289: 374–384. [DOI] [PubMed] [Google Scholar]
- 5. Chen Z-F, Matsumura K, Wang H, Arellano SM, Yan X, et al. (2011) Toward an understanding of the molecular mechanisms of barnacle larval settlement: a comparative transcriptomic approach. PLoS One 6: e22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernard FJ, Lane CE (1962) Early settlement and metamorphosis of the barnacle Balanus amphitrite niveus . J Morphol 110: 19–39. [Google Scholar]
- 7. Heyland A, Moroz LL (2006) Signaling mechanisms underlying metamorphic transitions in animals. Integr Comp Biol 46: 743–759. [DOI] [PubMed] [Google Scholar]
- 8. Okazaki Y, Shizuri Y (2000) Structures of six cDNAs expressed specifically at cypris larvae of barnacles, Balanus amphitrite . Gene 250: 127–135. [DOI] [PubMed] [Google Scholar]
- 9. Li H, Thiyagarajan V, Qian P-Y (2010) Response of cyprid specific genes to natural settlement cues in the barnacle Balanus ( = Amphibalanus) amphitrite . J Exp Mar Biol Ecol 389: 45–52. [Google Scholar]
- 10. Thiyagarajan V, Qian PY (2008) Proteomic analysis of larvae during development, attachment, and metamorphosis in the fouling barnacle, Balanus amphitrite . Proteomics 8: 3164–3172. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Xu Y, Arellano SM, Xiao K, Qian P-Y (2010) Comparative proteome and phosphoproteome analyses during cyprid development of the barnacle Balanus ( = Amphibalanus) amphitrite . J Proteome Res 9: 3146–3157. [DOI] [PubMed] [Google Scholar]
- 12. Zhang H, Lin Q, Ponnusamy S, Kothandaraman N, Lim TK, et al. (2007) Differential recovery of membrane proteins after extraction by aqueous methanol and trifluoroethanol. Proteomics 7: 1654–1663. [DOI] [PubMed] [Google Scholar]
- 13. Wang H, Zhang H, Wong YH, Voolstra C, Ravasi T, et al. (2010) Rapid transcriptome and proteome profiling of a non-model marine invertebrate, Bugula neritina . Proteomics 10: 2972–2981. [DOI] [PubMed] [Google Scholar]
- 14. Meng W, Zhang H, Guo T, Pandey C, Zhu Y, et al. (2008) One-step procedure for peptide extraction from in-gel digestion sample for mass spectrometric analysis. Anal Chem 80: 9797–9805. [DOI] [PubMed] [Google Scholar]
- 15. Zhang H, Zhao C, Li X, Zhu Y, Gan CS, et al. (2010) Study of monocyte membrane proteome perturbation during lipopolysaccharide-induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics 10: 2780–2789. [DOI] [PubMed] [Google Scholar]
- 16. Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392. [DOI] [PubMed] [Google Scholar]
- 17. Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- 18. Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124. [DOI] [PubMed] [Google Scholar]
- 19. Braisted JC, Kuntumalla S, Vogel C, Marcotte EM, Rodrigues AR, et al. (2008) The APEX quantitative proteomics tool: generating protein quantitation estimates from LC-MS/MS proteomics results. Bmc Bioinformatics 9: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H, Wong YH, Wang H, Chen Z, Arellano SM, et al. (2010) Quantitative proteomics identify molecular targets that are crucial in larval settlement and metamorphosis of Bugula neritina . J Proteome Res 10: 349–360. [DOI] [PubMed] [Google Scholar]
- 21. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans LA 14 Nov. 2010: pp. 1–8.
- 23. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 24. De Gregoris TB, Borra M, Biffali E, Bekel T, Burgess JG, et al. (2009) Construction of an adult barnacle (Balanus amphitrite) cDNA library and selection of reference genes for quantitative RT-PCR studies. BMC Mol Biol 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 26. Wang H, Yin G, Yu C, Wang Y, Sun Z (2013) Inhibitory effect of sanguinarine on PKC-CPI-17 pathway mediating by muscarinic receptors in dispersed intestinal smooth muscle cells. Res Vet Sci 95: 1125–1133. [DOI] [PubMed] [Google Scholar]
- 27.Berg JM, Tymoczko JL, Stryer L (2002) Chapter 16, Glycolysis and Gluconeogenesis. Biochemistry. 5th edition. New York: W H Freeman.
- 28. Lucas MI, Walker G, Holland DL, Crisp DJ (1979) An energy budget for the free-swimming and metamorphosing larvae of Balanus balanoides (Crustacea: Cirripedia). Mar Biol 55: 221–229. [Google Scholar]
- 29. Zhang Y-F, Zhang H, He L, Liu C, Xu Y, et al. (2013) Butenolide inhibits marine fouling by altering the primary metabolism of three target organisms. ACS Chem Biol 7: 1049–1058. [DOI] [PubMed] [Google Scholar]
- 30. Bianchi A, Evans JL, Iverson AJ, Nordlund A-C, Watts TD, et al. (1990) Identification of an isozymic form of acetyl-CoA carboxylase. J Biol Chem 265: 1502–1509. [PubMed] [Google Scholar]
- 31. Footitt S, Cornah JE, Pracharoenwattana I, Bryce JH, Smith SM (2007) The Arabidopsis 3-ketoacyl-CoA thiolase-2 (kat2-1) mutant exhibits increased flowering but reduced reproductive success. J Exp Bot 58: 2959–2968. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y-F, Wang G-C, Ying X, Sougrat R, Qian P-Y (2011) The effect of butenolide on behavioral and morphological changes in two marine fouling species, the barnacle Balanus amphitrite and the bryozoan Bugula neritina . Biofouling 27: 467–475. [DOI] [PubMed] [Google Scholar]
- 33. Shimizu K, Satuito CG, Saikawa W, Fusetani N (1996) Larval storage protein of the barnacle, Balanus amphitrite: biochemical and immunological similarities to vitellin. J Exp Zool 276: 87–94. [DOI] [PubMed] [Google Scholar]
- 34. Han Z, Sun J, Zhang Y, He F, Xu Y, et al. (2013) iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J Proteome Res 12: 2090–2100. [DOI] [PubMed] [Google Scholar]
- 35. Harrison DCS (1999) Morphology of the nervous system of the barnacle cypris larva (Balanus amphitrite Darwin) revealed by light and electron microscopy. Biol Bull 197: 144. [DOI] [PubMed] [Google Scholar]
- 36. Yamamoto H, Tachibana A, Kawaii S, Matsumura K, Fusetani N (1996) Serotonin involvement in larval settlement fo the barnacle, Balanus amphitrite . J Exp Zool 275: 339–345. [Google Scholar]
- 37. Knight J, Rowley AF, Yamazaki M, Clare AS (2000) Eicosanoids are modulators of larval settlement in the barnacle, Balanus amphitrite . J Mar Biol Asso UK 80: 113–117. [Google Scholar]
- 38. Faimali M, Falugi C, Gallus L, Piazza V, Tagliafierro G (2003) Involvement of acetyl choline in settlement of Balanus amphitrite . Biofouling 19: 213–220. [DOI] [PubMed] [Google Scholar]
- 39. Neale JH, Bzdega T (2002) Wroblewska (2002) N-acetylaspartylglutamate. J Neurochem 75: 443–452. [DOI] [PubMed] [Google Scholar]
- 40. Wroblewska B, Wroblewski JT, Saab OH, Neale JH (1993) N-acetylaspartylglutamate inhibits forskolin-stimulated cyclic amp levels via a metabotropic glutamate receptor in cultured cerebellar granule cells. J Neurochem 61: 943–948. [DOI] [PubMed] [Google Scholar]
- 41. Zhao J, Ramadan E, Cappiello M, Wroblewska B, Bzdega T, et al. (2001) NAAG inhibits KCl-induced [3H]-GABA release via mGluR3, cAMP, PKA and L-type calcium conductance. Eur J Neurosci 13: 340–346. [PubMed] [Google Scholar]
- 42. Clare A, Thomas R, Rittschof D (1995) Evidence for the involvement of cyclic AMP in the pheromonal modulation of barnacle settlement. J Exp Biol 198: 655–664. [DOI] [PubMed] [Google Scholar]
- 43. Gallus L, Ferrando S, Gambardella C, Diaspro A, Bianchini P, et al. (2009) The GABAergic-like system in the cyprid of Balanus amphitrite ( = Amphibalanus amphitrite) (Cirripedia, Crustacea). Biofouling 26: 155–165. [DOI] [PubMed] [Google Scholar]
- 44. Godenschwege TA, Kristiansen LV, Uthaman SB, Hortsch M, Murphey RK (2006) A conserved role for Drosophila neuroglian and human L1-Cam in central-synapse formation. Curr Biol 16: 12–23. [DOI] [PubMed] [Google Scholar]
- 45. Chen W, Hing H (2008) The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Dev Neurobiol 68: 1029–1045 doi:10.1002/dneu.20644 [DOI] [PubMed] [Google Scholar]
- 46. Levine MZ, Harrison PJH, Walthall WW, Tai PC, Derby CD (2001) A CUB-serine protease in the olfactory organ of the spiny lobster Panulirus argus . J Neurobiol 49: 277–302. [DOI] [PubMed] [Google Scholar]
- 47. Baxter GT, Morse DE (1992) Cilia from abalone larvae contain a receptor-dependent G-protein transduction system similar to that in mammals. Biol Bull 183: 147–154. [DOI] [PubMed] [Google Scholar]
- 48. Clare AS (1996) Signal transduction in barnacle settlement: calcium re-visited. Biofouling 10: 141–159. [DOI] [PubMed] [Google Scholar]
- 49. Yamamoto H, Tachibana A, Matsumura K, Fusetani N (1995) Protein kinase c (PKC) signal transduction system involved in larval metamorphosis of the barnacle, Balanus amphitrite . Zool Sci 12: 391–396. [Google Scholar]
- 50. Lane ME, Kalderon D (1993) Genetic investigation of cAMP-dependent protein kinase function in Drosophila development. Genes Dev 7: 1229–1243. [DOI] [PubMed] [Google Scholar]
- 51. Labeit S, Kolmerer B (1995) Titins: Giant Proteins in Charge of Muscle Ultrastructure and Elasticity. Science 270: 293–296. [DOI] [PubMed] [Google Scholar]
- 52. Lagersson NC (2002) The ultrastructure of two types of muscle fibre cells in the cyprid of Balanus amphitrite (Crustacea: Cirripedia). J Mar Biol Ass 82: 573–578. [Google Scholar]
- 53. Lagersson NC, Høeg JT (2002) Settlement behavior and antennulary biomechanics in cypris larvae of Balanus amphitrite (Crustacea: Thecostraca: Cirripedia). Mar Biol 141: 513–526. [Google Scholar]
- 54. Katti C, Kempler K, Porter ML, Legg A, Gonzalez R, et al. (2010) Opsin co-expression in Limulus photoreceptors: differential regulation by light and a circadian clock. J Exp Biol 213: 2589–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anil AC, Khandeparker L, Desai DV, Baragi LV, Gaonkar CA (2010) Larval development, sensory mechanisms and physiological adaptations in acorn barnacles with special reference to Balanus amphitrite . J Exp Mar Biol Ecol 392: 89–98. [Google Scholar]
- 56. Schneider A, Wiesner RJ, Grieshaber MK (1989) On the role of arginine kinase in insect flight muscle. Insect Biochemistry 19: 471–480. [Google Scholar]
- 57. Chamberlin M (1997) Mitochondrial arginine kinase in the midgut of the tobacco hornworm (Manduca sexta). J Exp Biol 200: 2789–2796. [DOI] [PubMed] [Google Scholar]
- 58. Chou TC, Lipmann F (1952) Separation of acetyl transfer enzymes in pigeon liver extract. J Biol Chem 196: 89–103. [PubMed] [Google Scholar]
- 59. Srere PA, Lipmann F (1953) An enzymatic reaction between citrate, adenosine triphosphate and coenzyme A1. J Am Chem Soc 75: 4874–4874. [Google Scholar]
- 60. Lee JK, Coyne RS, Dubreuil RR, Goldstein LS, Branton D (1993) Cell shape and interaction defects in alpha-spectrin mutants of Drosophila melanogaster . J Cell Biol 123: 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maruzzo D, Aldred N, Clare AS, Høeg JT (2012) Metamorphosis in the cirripede crustacean Balanus amphitrite . PLoS One 5: e37408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dreyfuss G, Swanson MS, Piñol-Roma S (1988) Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci 13: 86–91. [DOI] [PubMed] [Google Scholar]
- 63. Goss DJ, Kleiman FE (2012) Poly(A) binding proteins: are they all created equal? WIREs RNA 4: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strumpf D, Volk T (1998) Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, Vein, at the muscle–tendon junction site. J Cell Biol 143: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koulish S (1973) Microtubules and muscle attachment in the integument of the balanidae. J Morphol 140: 1–13. [DOI] [PubMed] [Google Scholar]
- 66. Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruïne AP, et al. (2010) The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. The FASEB Journal 24: 4153–4166. [DOI] [PubMed] [Google Scholar]
- 67. Schlaepfer DD, Haigler HT (1990) Expression of annexins as a function of cellular growth state. J Cell Biol 111: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaneko Y, Takaki K, Iwami M, Sakurai S (2006) Developmental profile of annexin IX and its possible role in programmed cell death of the Bombyx mori anterior silk gland. Zool Sci 23: 533–542. [DOI] [PubMed] [Google Scholar]
- 69. Yamamoto H, Kawaii S, Yoshimura E, Tachibana A, Fusetani N (1997) 20-Hydroxyecdysone regulates larval metamorphosis of the barnacle, Balanus amphitrite . Zool Sci 14: 887–892. [Google Scholar]
- 70. Costlow JD (1959) Effect of carbonic anhydrase inhibitors on shell development and growth of Balanus improvisus Darwin. Physiol Zool 3: 177–184. [Google Scholar]
- 71. Giraud M-M (1981) Carbonic anhydrase activity in the integument of the crab Carcinus maenas during the intermolt cycle. Comp Biochem Physiol A Mol Integr Physiol 69: 381–387. [Google Scholar]
- 72. He L-S, Zhang G, Qian P-Y (2013) Characterization of two 20kDa-cement protein (cp20k) homologues in Amphibalanus amphitrite . PLoS One 8: e64130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kamino K, Inoue K, Maruyama T, Takamatsu N, Harayama S, et al. (2000) Barnacle cement proteins. Importance of disulfide bonds in their insolubility. J Biol Chem 275: 27360–27365. [DOI] [PubMed] [Google Scholar]
- 74. Ritossa F (1996) Discovery of the heat shock response. Cell Stress Chapersones 1: 97–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heikkila JJ, Ohan N, Tam Y, Ali A (1997) Heat shock protein gene expression during Xenopus development. Cell Mol Life Sci 53: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mahroof R, Yan Zhu K, Neven L, Subramanyam B, Bai J (2005) Expression patterns of three heat shock protein 70 genes among developmental stages of the red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae). Comp Biochem Physiol A Mol Integr Physiol 141: 247–256. [DOI] [PubMed] [Google Scholar]
- 77. Bishop CD, Brandhorst BP (2001) No/Cgmp signaling and Hsp90 activity represses metamorphosis in the sea urchin Lytechinus Pictus . Biol Bull 201: 394–404. [DOI] [PubMed] [Google Scholar]
- 78. Arbeitman MN, Hogness DS (2000) Molecular chaperones activate the Drosophila ecdysone receptor, an RXR heterodimer. Cell 101: 67–77. [DOI] [PubMed] [Google Scholar]
- 79. Gunter H, Degnan B (2007) Developmental expression of Hsp90, Hsp70 and HSF during morphogenesis in the vetigastropod Haliotis asinina . Dev Genes Evol 217: 603–612. [DOI] [PubMed] [Google Scholar]
- 80. Zhang Y, Sun J, Xiao K, Arellano SM, Thiyagarajan V, et al. (2010) 2D gel-based multiplexed proteomic analysis during larval development and metamorphosis of the biofouling polychaete tubeworm Hydroides elegans . J Proteome Res 9: 4851–4860. [DOI] [PubMed] [Google Scholar]
- 81. Chandramouli KH, Sun J, Mok FS, Liu L, Qiu J-W, et al. (2013) Transcriptome and quantitative proteome analysis reveals molecular processes associated with larval metamorphosis in the polychaete Pseudopolydora vexillosa . J Proteome Res 12: 1344–1358. [DOI] [PubMed] [Google Scholar]
- 82. Chandramouli KH, Soo L, Qian P-Y (2011) Differential expression of proteins and phosphoproteins during larval metamorphosis of the polychaete Capitella sp. . I. Proteome Sci 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Woollacott RM, Zimmer RL (1971) Attachment and metamorphosis of the cheilo-ctenostome bryozoan Bugula neritina (Linné). J Morphol 134: 351–382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for genes under real-time PCR assay.
(XLSX)
Total proteins identified and quantified from each LC-MS experiments.
(XLSX)
Significantly up-regulated proteins in stage II nauplii.
(XLSX)
Significantly up-regulated proteins in stage VI nauplii.
(XLSX)
Significantly down-regulated proteins in stage II nauplii.
(XLSX)
Significantly down-regulated proteins in stage VI nauplii.
(XLSX)
Significantly down-regulated proteins in cyprids.
(XLSX)
Significantly down-regulated proteins in juveniles.
(XLSX)