Abstract
Despite the progress made in targeted anticancer therapies in recent years, challenges remain. The identification of new potential targets will ensure that the arsenal of cancer therapies continues to expand. FAM83B was recently discovered in a forward genetic screen for novel oncogenes that drive human mammary epithelial cell (HMEC) transformation. We report here that elevated FAM83B expression increases Phospholipase D (PLD) activity, and that suppression of PLD1 activity prevents FAM83B-mediated transformation. The increased PLD activity is engaged by hyperactivation of epidermal growth factor receptor (EGFR), which is regulated by an interaction involving FAM83B and EGFR. Preventing the FAM83B/EGFR interaction by site-directed mutation of lysine 230 of FAM83B suppressed PLD activity and MAPK signaling. Furthermore, ablation of FAM83B expression from breast cancer cells inhibited EGFR phosphorylation and suppressed cell proliferation. We propose that understanding the mechanism of FAM83B-mediated transformation will provide a foundation for future therapies aimed at targeting its function as an intermediary in EGFR, MAPK, and mTOR activation.
Keywords: FAM83B, EGFR, PLD1, MAPK, mTOR, HMEC transformation
Introduction
Receptor tyrosine kinase (RTK) signaling cascades have become the subject of intense research aimed at identifying pharmacological inhibitors that will suppress growth signaling and prevent cancer cell proliferation. Targeted therapies aimed at disrupting RTKs (including antibodies and small molecule inhibitors targeting EGFR, HER2 and VEGF), RAS (Farnesyltransferase Inhibitors Tipifarnib and lonafarnib), RAF (Sorafenib, RAF265 and PLX4032), MEK (PD0325901, AZD6244, ARRY-142886 and ARRY-438162), AKT (VDQ-002) and mTOR (Rapamycin, CCI-779, RAD001, and AP-23573), have been developed and are currently being evaluated in a number of clinical trials (1;2). However, the complexity of signaling interactions continues to limit the effectiveness of these therapies, since resistance is easily obtained due to the plasticity of cancer cells.
Phospholipase D (PLD) activity can be altered by many extracellular signals, one being Epidermal Growth Factor (EGF) (3). Downstream of Epidermal Growth Factor Receptor (EGFR) activation, RAS stimulates RalA, RalB, and small GTPases that facilitate intracellular signal transduction. RAL proteins are required for RAS-mediated tumorigenesis and tumor survival (4;5), due to their stimulation of PLD activity, which hydrolyzes phosphatidylcholine into phosphatidic acid (PA) and choline (6;7). PA is an important signaling lipid involved in recruiting cytosolic proteins to the membrane where they can be activated to potentiate growth signaling. Two important signaling molecules that require PA for their activation are CRAF and mTOR (8;9). In fact, the activation of PLD1 has been proven to be critical for the transforming activity of RAS and EGFR. Inhibition of PLD activity prevented the anchorage-independent growth (AIG) and tumorigenicity of both H-RAS and K-RAS-expressing cells. This effect was specific for PA, since transformation could be restored simply by providing the cells exogenous PA (10). Given that many tumors have elevated PLD activity, considerations for the role of PA in RAS-mediated signaling pathways will be important for devising future therapies (11).
Studies of CRAF activation have defined an important regulatory process mediated by the direct binding of PA to a site in the C-terminus of CRAF (9). Upon growth factor receptor activation, PLD-mediated PA production induces the translocation of CRAF to the plasma membrane. Mutation of key basic residues located in the putative PA binding region (PA-BR) of CRAF efficiently prevents CRAF translocation and activation (9;12;13). Moreover, the production of PA is also required to couple ERK activation at the cell membrane (14). In addition to regulating CRAF activation, the role of PA in activating mammalian Target of Rapamycin (mTOR) is a developing research area with important implications for cancer therapeutics. Analysis of human tumors indicates that the mTOR pathway is commonly hyperactivated (15). This provides tumor cells with a growth advantage, since mTOR promotes protein synthesis and is involved in cell growth, proliferation, and survival.
Using an innovative forward genetic screen, we recently identified FAM83B (Family with Sequence Similarity 83, member B), based on its ability to promote the transformation of human mammary epithelial cells (HMEC; (16)). Analysis of Mitogen Activated Protein Kinase (MAPK) and mTOR signaling in FAM83B-mediated transformation confirmed that substrates downstream of CRAF and mTOR, including ERK1/2, S6K and 4E-BP1 were significantly activated in FAM83B-expressing cells. Furthermore, elevated FAM83B expression also activates the PI3K/AKT signaling pathway and confers a decreased sensitivity to PI3K, AKT, and mTOR inhibitors (17). Analysis of FAM83B expression in human tumor specimens revealed its overexpression in a variety of human cancers. Ablation of FAM83B from breast cancer cells with elevated EGFR or HER2, or in HMECs transformed by activated RAS, inhibited their proliferation, AIG and tumorigenicity, supporting a role for FAM83B as an important intermediary in aberrant EGFR/RAS signaling. Despite the presence of a putative phospholipase D (PLD)-like motif in FAM83B, we found that FAM83B itself does not have conventional PLD activity. However, we show here that elevated FAM83B expression increases PLD activity by binding and hyperactivating EGFR, which drives tumorigenesis in numerous model systems. In addition, we demonstrate that chemical inhibition or shRNA-mediated suppression of PLD1 prevents FAM83B-mediated transformation, implicating PLD1 activity as a key downstream effector of FAM83B. Site-directed mutation of lysine 230 of FAM83B attenuates FAM83B/EGFR interactions and suppresses PLD activity and downstream MAPK signaling. Our studies demonstrate that FAM83B is a key EGFR signaling intermediate, that when overexpressed promotes aberrant growth signaling. We propose that understanding the mechanism of FAM83B-mediated transformation will provide a foundation for future therapies aimed at targeting its function, which is required for EGFR and MAPK activation.
Results
FAM83B expression increases PLD activity
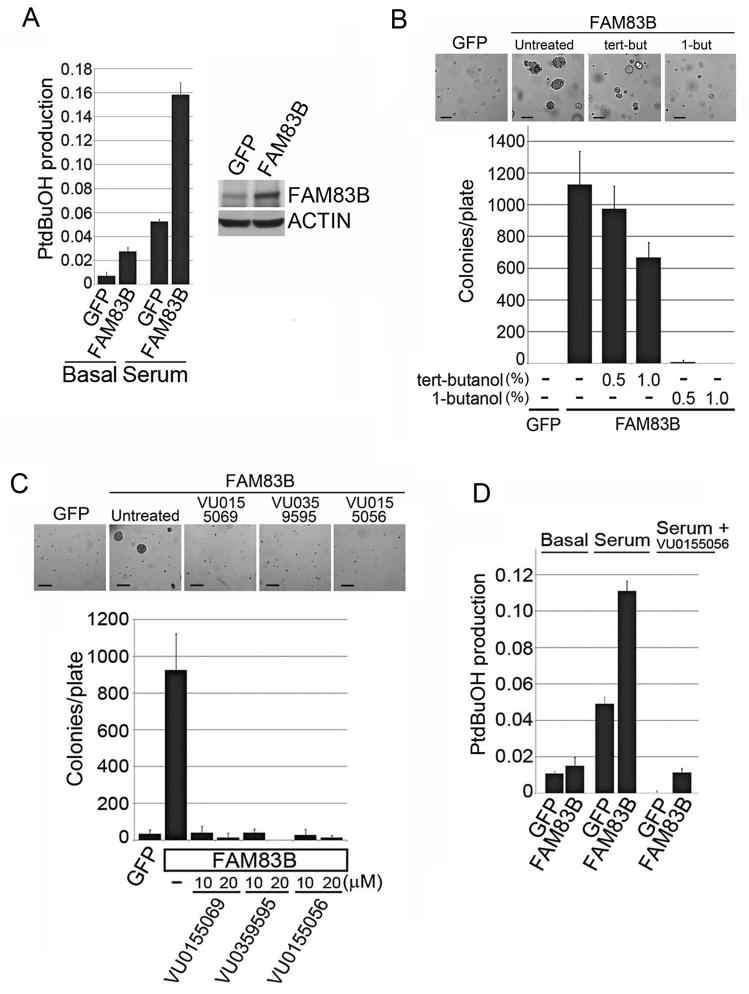
Phospholipase D (PLD) activity results in the hydrolysis of phosphatidylcholine into phosphatidic acid (PA) and choline (6;7). PA is an important signaling lipid required for the activation of both CRAF and mTOR (8), two signaling pathways commonly altered in cancer and required for FAM83B-mediated transformation (16;17). While the FAM83B protein was annotated as being a potential phospholipase, the motif in FAM83B (HxKxxxKxxxD) varies from the highly conserved consensus sequence of conventional PLDs (HxKxxxxD; (18)). To examine whether FAM83B-expressing cells have elevated PLD activity, a cell-based mass spectrometric PLD assay was performed (19). HME1 cells expressing GFP or FAM83B were left untreated or stimulated with serum in the presence of 1-butanol-d10 to trap the PLD product as phosphatidylbutanol-d9 (PtdBuOH), which was analyzed by mass spectrometry. FAM83B-expressing cells had elevated PLD activity in both basal and serum-stimulated conditions relative to control GFP-expressing cells (Fig. 1A). However, the PLD activity observed in FAM83B-expressing HME1 cells was modest (ranging from 1.5–3 fold higher than control cells) when compared to PLD1- or PLD2-expressing HME1 cells (which ranged from 5–30 fold higher than control cells; Supplementary Fig. S1). Furthermore, treatment of FAM83B-expressing cells with 1-butanol, a competitive nucleophile that prevents phosphatidic acid production, strongly inhibited FAM83B-mediated AIG. Tert-butanol, which is unable to serve as a competitive nucleophile in the transphosphatidylation reaction was used as a negative control (Fig. 1B). However, consistent with the altered PLD motif in FAM83B and the stringent requirements of PLD active sites, we determined that neither recombinant FAM83B nor FAM83B immunoprecipitated from human cells conferred hydrolysis of phosphatidylcholine (16). Therefore, we examined whether conventional PLD enzymes were responsible for the elevated PLD activity in FAM83B-expressing HME1 cells using isoform-selective phospholipase D inhibitors. A dual PLD1/PLD2 inhibitor (VU0155056), and two PLD1-specific inhibitors (VU0155069 and VU0359595) each significantly suppressed FAM83B-mediated AIG ((20); Fig. 1C). Also, the dual PLD1/PLD2 inhibitor (VU0155056) suppressed the PLD activity in FAM83B-expressing HME1 cells, as well as the PLD1- and PLD2-expressing HME1 cells (Fig. 1D; Supplementary Fig. S1). GFP-expressing HME1 cells failed to form colonies in soft agar in the absence or presence of tert-butanol, 1-butanol, or the selective PLD1/2 inhibitors. Together, these data support a model in which elevated FAM83B expression increases PLD1 activity, which is critical for FAM83B-mediated transformation.
Figure 1. FAM83B expression increases conventional PLD activity.
(A) Analysis for PLD activity. HME1 cells expressing GFP or FAM83B were left untreated or stimulated with serum in the presence of 1-butanol-d10 to trap the PLD product as phosphatidylbutanol-d9 (PtdBuOH), which was detected by mass spectrometric analysis. Data is plotted as a ratio of PtdBuOH to phosphatidylmethanol (PtdMeOH), which was used as an internal standard. Western analysis with a FAM83B monoclonal antibody (clone 7D11) confirmed the expression of FAM83B. (B) Inhibition of FAM83B-mediated transformation by 1-butanol. FAM83B-expressing HME1 cells (or GFP-expressing control cells) were plated in soft agar and left untreated (−), or treated with 0.5% or 1.0% tert-butanol or 1-butanol, a competitive nucleophile that prevents phosphatidic acid production, and AIG was assessed. Tert-butanol, which is unable to serve as a competitive nucleophile in the transphosphatidylation reaction was used as a negative control. Representative images are shown for GFP and FAM83B, either untreated or treated with tert-butanol or 1-butanol. Bar depicts 200 μm (C) PLD1 inhibition abolishes FAM83B-mediated AIG. HME1 cells expressing GFP or FAM83B were analyzed for AIG in the presence of PLD1 inhibitors VU0359595 and VU0155069, or a dual PLD1/PLD2 inhibitor VU0155056. Representative images are shown for GFP and FAM83B, either untreated or treated with the indicated PLD inhibitor. Bar depicts 200 μm. (D) A PLD1/2 dual inhibitor inhibits PLD activity in FAM83B-expressing cells. HME1 cells expressing GFP or FAM83B were incubated with media containing 1-butanol-d10, in the presence or absence of 2μM VU0155056 and 20% serum as indicated. PtdBuOH was detected by mass spectrometric analysis and data is plotted as a ratio of PtdBuOH to internal standards.
Knockdown of PLD1 causes the growth suppression of cells dependent on FAM83B expression
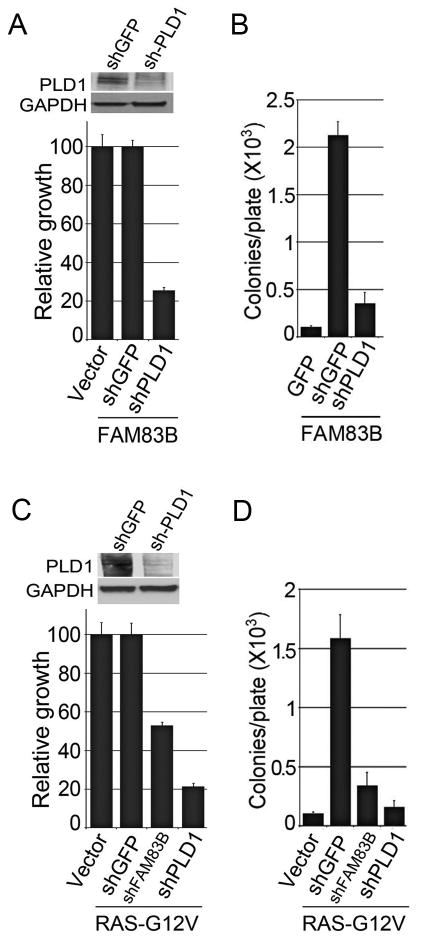
Small molecule PLD1 inhibitors can also partially inhibit PLD2 at the doses used in our experiments. Therefore, to further confirm a role for PLD1 in FAM83B-mediated transformation, shRNAs targeting PLD1 or GFP were delivered to FAM83B-expressing HME1 cells by lentiviral infection. The efficiency of PLD1 knock-down in FAM83B-expressing HME1 cells was examined by Western analysis and the effects on proliferation and AIG were assessed. Ablation of PLD1 decreased both the proliferation and AIG of FAM83B-expressing HME1 cells, again implicating elevated PLD1 activity as a critical signal necessary for FAM83B-mediated transformation (Fig. 2A and 2B). In addition, we recently demonstrated that shRNA-mediated ablation of FAM83B from RAS-G12V transformed HME1 cells suppressed their transformed phenotype. To determine whether ablation of PLD1 would recapitulate the growth inhibition observed following ablation of FAM83B, RAS-expressing HME1 cells were infected with shRNAs targeting GFP, PLD1, or FAM83B. The efficiency of PLD1 knock-down in RAS-expressing HME1 cells was examined by Western analysis (Fig. 2C), and the effects on proliferation and AIG were assessed. Importantly, ablation of either PLD1 or FAM83B suppressed the growth and AIG of RAS-G12V-transformed HME1 cells (Fig. 2C and 2D). Together, these data demonstrate that both FAM83B- and RAS-mediated transformation requires PLD1 activity.
Figure 2. Knockdown of PLD1 causes growth suppression of cells dependent on FAM83B expression.
(A) HME1 cells expressing FAM83B were infected with lentiviruses encoding shRNAs targeting GFP or PLD1. The cells were plated and growth was assessed after 5 days. Western analysis of FAM83B-expressing HME1 cells expressing shPLD1. (B) HME1 cells expressing FAM83B were infected with lentiviruses encoding shRNAs targeting GFP or PLD1 and assessed for AIG. (C) HME1 cells expressing RAS were infected with lentiviruses encoding shRNAs targeting GFP, FAM83B, or PLD1. The cells were plated for 5 days, cell number was assessed, and Western analysis confirmed the knockdown of PLD1. (D) HME1 cells expressing RAS were infected with lentiviruses encoding shRNAs targeting GFP, FAM83B (B2), or PLD1 and assessed for AIG.
Breast cancer cells dependent on FAM83B expression are sensitive to PLD inhibitors
MCF7 and MDA468 breast cancer cell lines express elevated FAM83B protein and require sustained FAM83B expression for growth, AIG, and tumorgenicity (16). We next examined whether knockdown of PLD1 in MDA468 and MCF7 cells would result in growth inhibition, similar to the inhibition observed by FAM83B ablation. MDA468 and MCF7 cells were infected with shRNAs targeting GFP, PLD1, or FAM83B, the efficiency of PLD1 and FAM83B knock-down was examined by Western analysis, and the effects on proliferation and AIG were assessed (Fig. 3A). Similar to the results obtained with RAS-transformed HME1 cells, ablation of either PLD1 or FAM83B resulted in the suppression of growth and AIG in MDA468 and MCF7 cells (Fig. 3A and 3B). In addition, we examined whether MDA468 and MCF7 cells would also be susceptible to growth inhibition by PLD inhibitors. MDA468 and MCF7 cells were plated and grown in the presence of VU0155056 (0.2, 2, 10, and 20 μM) for 5 days and cell number was determined (Fig. 3C and Fig. 3D). Similar to our results with FAM83B-expressing HME1 cells, the growth of cancer cell lines sensitive to FAM83B ablation (MCF7 and MDA468) were also suppressed by PLD inhibitors. Taken together, these data demonstrate that tumor-derived cells and RAS-transformed HMEC that have a dependency for sustained FAM83B expression also require PLD activity to maintain their growth. Our findings are consistent with previous observations in which elevated PLD activity induced by RAS, CRAF, and SRC is required for their transforming activities (21).
Figure 3. Breast cancer cells dependent on FAM83B expression are sensitive to PLD inhibition.
(A) MCF7 and MDA468 cells were infected with lentiviruses encoding shRNAs targeting GFP, PLD1, or FAM83B. Western analysis confirmed knockdown of PLD1 and FAM83B in both MCF7 and MDA468 cells (+ control lane includes extract from 293T cells transfected with a plasmid encoding FAM83B). The cells were plated, grown for 5 days, and cell number determined. (B) MDA468 cells were infected with lentiviruses encoding shRNAs targeting GFP, FAM83B (B2), or PLD1 and assessed for AIG. (C and D) MDA468 and MCF7 cells were plated and treated with a PLD1/PLD2 dual inhibitor VU0155056 (0.2, 2, 10, and 20 μM) and cell number was assessed 5 days later.
FAM83B expression increases EGFR activity
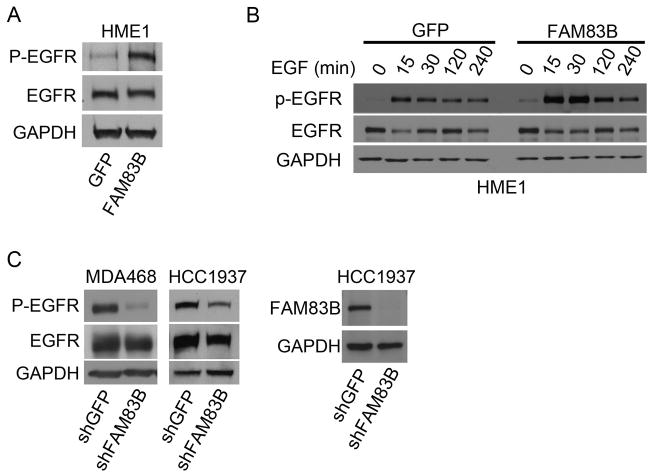
EGFR stimulation is a known activator of PLD1 activity. Since we previously demonstrated that FAM83B is a key intermediate in EGFR signaling, we hypothesized that the increase in PLD activity resulting from elevated FAM83B expression was due to EGFR hyperactivation. To test this hypothesis, we compared EGFR phosphorylation on a number of tyrosine phosphorylation sites, including Y992, Y1045, and Y1086 in GFP- and FAM83B-expressing HME1 cells. Using whole cell lysates from GFP- or FAM83B-expressing HME1 cells grown in laminin-rich basement membrane (lrBM) for 10 days, we noted that EGFR-Y1086 phosphorylation was basally elevated in the FAM83B-expressing cells (Fig. 4A). EGFR phosphorylated Y1068 recruits the Grb2 SH2 domain, activating downstream RAL proteins, which can stimulate PLD activity. Next, GFP- or FAM83B-expressing HME1 cells deprived of growth factors were stimulated with EGF and analyzed at various times after stimulation. Elevated FAM83B expression resulted in increased EGF-stimulated EGFR-Y1068 phosphorylation at the 15, 30 and 120 minute time points, further supporting a role for FAM83B in EGFR activation (Fig. 4B). Finally, we examined breast cancer cell lines to determine whether ablation of FAM83B from cancer cells suppresses EGFR activation. MDA468 and HCC1937 breast cancer cells overexpress EGFR and require sustained FAM83B expression for growth (Figure 3A and 3B; (16)). Both cell lines were infected with lentiviruses encoding shRNA targeting FAM83B or GFP and EGFR-Y1068 phosphorylation was assessed. Importantly, suppression of FAM83B expression led to diminished EGFR phosphorylation in both cell lines compared to control cells (Fig. 4C). Our data suggest that, in tumor cells harboring elevated FAM83B expression, FAM83B-mediated EGFR activation may play a key role in activating downstream PLD activity to drive epithelial cell transformation and maintain breast cancer cell growth.
Figure 4. Elevated FAM83B expression increases EGFR phosphorylation.
(A) HME1 cells expressing GFP or FAM83B were grown in lrBM for 10 days and western analysis was performed for phosho-Y1068 on EGFR, total EGFR, and GAPDH. (B) HME1 cells expressing GFP or FAM83B were grown in the absence of mammary epithelial growth supplement (MEGS) for 24 hours and then treated with 10 ng/mL epidermal growth factor (EGF) for 15, 30, 120, and 240 minutes. Immunoblot analysis of phosho-Y1068 on EGFR, total EGFR, and GAPDH was performed. (C) MDA468 and HCC1937 cells expressing shRNAs targeting GFP or FAM83B were grown in lrBM for 10 days and western analysis was performed for phosho-Y1068 on EGFR, total EGFR, and GAPDH. Western analysis of FAM83B confirmed the shRNA efficiency.
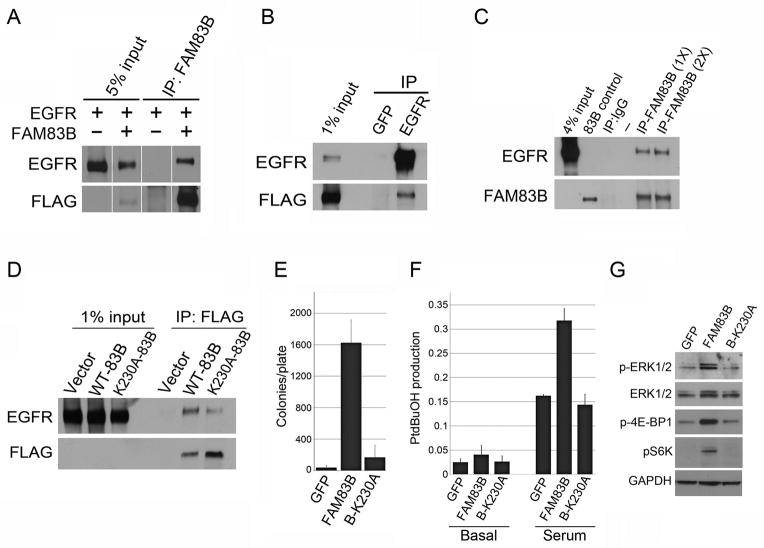
Next, we examined the mechanism by which elevated FAM83B expression increases EGFR phosphorylation. Analysis of global mass spectrometry data available on the protein modification resource PhophoSitePlus indicates that FAM83B is a target of EGFR tyrosine kinase activity. Therefore, we hypothesized that FAM83B interacts with EGFR and this interaction increases EGFR activity. To examine whether FAM83B and EGFR could be co-precipitated, plasmids encoding FLAG-FAM83B and EGFR were co-expressed, cell lysates were subjected to immunoprecipitation using a control (GFP), FLAG, or EGFR antibody, and Western analysis was performed. Consistent with our hypothesis, EGFR co-precipitated with FAM83B following either FAM83B or EGFR immunoprecipitation (Fig. 5A and 5B). In addition, we confirmed that endogenous EGFR could be co-precipitated with FAM83B from MDA468 cancer cells, which require sustained FAM83B expression for the maintenance of phosphorylated EGFR (Fig. 5C and Fig. 4C).
Figure 5. Elevated FAM83B expression hyperactivates EGFR leading to increased PLD activity.
(A and B) 293T cells were transfected with expression constructs encoding EGFR and FLAG-FAM83B as indicated. Immunoprecipitation was performed using a FLAG, EGFR, or GFP (control) antibody, and precipitated proteins analyzed by Western analysis to determine the amount of EGFR bound to FAM83B. Discontiguous lanes are from the same membrane and the same exposure. (C)) Immunoprecipitation was performed by incubating lysates from MDA468 breast cancer cells with either 500 μL (1X) or 1000 μL (2X) of FAM83B hybridoma supernatant or IgG (control) antibody. Precipitated proteins were analyzed by Western analysis for EGFR and FAM83B. The lane labeled 83B control contains lysate from 293T cells transfected with a plasmid encoding FAM83B. (D) 293T cells were transfected with expression constructs encoding GFP (Vector), EGFR, FLAG-FAM83B, and FLAG-FAM83B-K230A as indicated. Immunoprecipitation was performed using a FLAG antibody, and precipitated proteins analyzed by Western analysis to determine the amount of EGFR bound to FAM83B. (E) HME1 cells expressing full-length (FL/WT) FAM83B or FAM83B with a site-directed mutation at K230A were assessed for AIG. (F) HME1 cells expressing GFP, FAM83B, or FAM83B-K230A were left untreated or stimulated with serum in the presence of 1-butanol-d10 to trap the PLD product as phosphatidylbutanol-d9 (PtdBuOH), which was detected by mass spectrometric analysis. Data is plotted as a ratio of PtdBuOH to phosphatidylmethanol (PtdMeOH), which was added as an internal standard (G) Western analysis of HME1 cells expressing GFP, FAM83B, or FAM83B-K230A for phospho-ERK1/2, ERK1/2, phospho-4E-BP1, phospho-S6K, and GAPDH.
We previously demonstrated that the N-terminal DUF1669 of FAM83B was necessary and sufficient to drive HME1 transformation (16). Therefore, we created a site-directed mutation within the DUF1669. We choose Lysine 230 because it is highly conserved between the 8 FAM83 proteins, including FAM83A which also drives HMEC transformation similar to FAM83B (16;22). Importantly, the K230A mutation in FAM83B inhibited FAM83B-EGFR complex formation and suppressed FAM83B-mediated AIG (Fig. 5D and 5E). Previously, we showed that elevated FAM83B expression resulted in increased ERK1/2 phosphorylation and mTOR activation, as measured by S6K and 4E-BP1 phosphorylation. Expression of the FAM83B-K230A protein failed to increase basal or serum-stimulated PLD activity or the basal phosphorylation of ERK or mTOR targets S6K and 4E-BP1 when compared to wild-type FAM83B (Fig. 5F and 5G). Cumulatively, these results suggest that elevated FAM83B expression increases PLD activity by binding to EGFR, and increasing EGFR-mediated PLD activity. A number of experiments were performed to examine the regulation of FAM83B-EGFR complexes. First, we examined whether FAM83B could co-precipitate phosphorylated EGFR and whether EGF stimulation enhanced FAM83B-EGFR complexes. In our experiments, we could not detect phosphorylated EGFR in complex with FAM83B (Supplementary Fig. S2). Moreover, upon EGF stimulation, EGFR-FAM83B complex formation was disrupted rather than increased (Supplementary Fig. S3). In agreement, inhibition of EGFR phosphorylation by Erlotinib treatment failed to disrupt FAM83B-EGFR complexes (Supplementary Fig. S4). Finally, we examined whether inhibition of PLD activity would alter the FAM83B-EGFR complexes. Addition of the PLD1-specific inhibitor VU0155069 did not alter FAM83B-EGFR complex formation. Taken together, our findings suggest that in cancer cells harboring elevated FAM83B expression, unphosphorylated EGFR and FAM83B complexes result in the increased steady-state autophosphorylation of EGFR on Tyrosine 1068. Upon EGFR autophosphorylation, FAM83B would be released from the complex to activate downstream MAPK and PI3K/AKT/mTOR signaling as previously described (16;17)
Elevated PLD activity fails to recapitulate FAM83B phenotypes or rescue growth suppression following FAM83B ablation
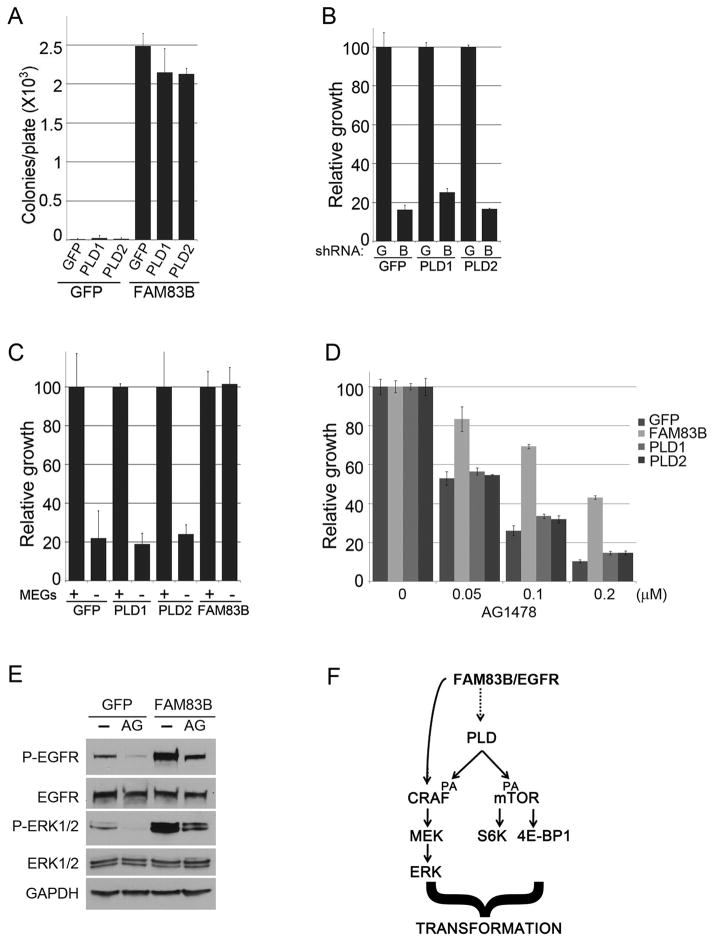
Given that PLD activity was required for the AIG of FAM83B-expressing HME1 cells, we next analyzed whether PLD1 or PLD2 overexpression alone is sufficient to drive AIG, or whether additional activities imparted by FAM83B are required. The basal activity of PLD1 is low, and is activated by EGFR, PKC, ADP-ribosylation factor (ARF) Rac, Rho, and Cdc42. In contrast, PLD2 exhibits high basal activity and is not affected by PLD1 activators (23). HME1 cells expressing GFP, PLD1, a lipase-inactive mutant of PLD1 (PLD1-K830R), or PLD2 or FAM83B were plated into soft agar to assess AIG. Despite a clear elevation of PLD activity, neither PLD1 or PLD2 expression promoted AIG. These results demonstrate that simply elevating PLD activity, while necessary for FAM83B-mediated transformation, is not sufficient to drive transformation alone (Supplementary Fig. S1 and S6).
We next examined whether further enhancement of PLD activity in FAM83B-expressing cells could synergize with FAM83B to enhance FAM83B-mediated transformation. GFP or FAM83B-expressing HME1 cells were infected with retroviruses encoding GFP, PLD1, or PLD2 and plated into soft agar. Again, PLD1 or PLD2 expression alone was unable to promote significant AIG (Fig. 6A). Furthermore, PLD1 or PLD2 expression in combination with FAM83B failed to enhance the AIG conferred by FAM83B expression alone (Fig. 6A). Finally, since RAS-expressing HME1 cells require sustained expression of FAM83B for growth and AIG (Fig. 2C and 2D), we examined whether elevated PLD activity conferred by PLD1 or PLD2 expression could compensate for the loss of FAM83B in RAS-mediated transformation. To test this, exogenous PLD1, PLD2 or GFP (as a control) were expressed in RAS-HME1 cells, and each derivative was subsequently infected with lentivirues encoding shRNAs targeting GFP (G) or FAM83B (B). The resulting cells were plated, grown for 7 days, and cell number was determined (Fig. 6B). Exogenous expression of either PLD1 or PLD2 failed to rescue RAS-expressing HME1 cells from the growth suppression engaged by FAM83B ablation, further arguing that elevated PLD activity is insufficient to functionally replace FAM83B. Taken together, our data suggest that elevated FAM83B expression activates sufficient PLD activity to drive HMEC transformation, yet additional FAM83B-mediated signals, independent of elevating PLD1 activity, are also required for HME1 transformation.
Figure 6. Elevated PLD activity fails to recapitulate FAM83B phenotypes or rescue growth suppression following FAM83B ablation.
(A) HME1 cells expressing GFP or FAM83B were infected with retroviruses encoding cDNAs of GFP, PLD1, or PLD2 and AIG was assessed. (B) HME1 cells expressing RAS were infected with retroviruses encoding cDNAs of GFP, PLD1, or PLD2. Subsequently, the resulting cell lines were infected with lentiviruses encoding shRNAs targeting GFP (G) or FAM83B (B) and cell growth was assessed 5 days later. (C) HME1 cells expressing GFP, PLD1, PLD2, or FAM83B were plated in the presence and absence of mammary epithelial growth supplement (MEGS) and cell number quantified 5 days later. (D) FAM83B confers decreased sensitivity to EGFR inhibition. HME1 cells expressing GFP, PLD1, PLD2, or FAM83B were treated with EGFR inhibitor AG1478 at 50, 100, and 200nM and cell number quantified 7 days later. (E) HME1 cells expressing GFP or FAM83B were grown as 3-dimensional (3D) cultures in lrBM in presence or absence of AG1478 (100 nM) for 10 days. Immunoblot analysis of phospho-Y1068-EGFR, total EGFR, phospho-ERK1/2, total ERK1/2, and GAPDH was performed. (F) Model of PLD involvement in FAM83B transformation and the pathways required.
Elevated FAM83B expression allows HME1 cells to grow robustly in the absence of growth factors (minus Mammary Epithelial Growth Supplement; MEGS), while the proliferation of control HME1 cells is significantly inhibited (Fig. 6C). Given the importance of PA in regulating CRAF and mTOR signaling, we examined whether elevated PLD activity was responsible for the growth observed in the absence of growth factors. GFP-, PLD1-, PLD2-, and FAM83B-expressing HME1 cells were plated in the presence and absence of MEGS, grown for 7 days, and cell number was determined. Again, elevated PLD activity was clearly not sufficient to rescue HME1 cells from the growth suppression engaged by growth factor withdrawal (Fig. 6C). Finally, given the importance of FAM83B in EGFR signaling, we examined whether elevated expression of FAM83B could confer resistance to EGFR tyrosine kinase inhibitors (TKIs) and whether PLD activity was implicated. GFP-, PLD1-, PLD2-, and FAM83B-expressing HME1 cells were plated in the presence and absence of various doses of AG1478, an EGFR-TKI, and cell number was assessed after 7 days. Elevated PLD activity from ectopic expression of PLD1 or PLD2 failed to prevent the AG1478-mediated growth suppression. In contrast, FAM83B expression permitted significant growth in the presence of AG1478 (Fig 6D). EGFR-Y1068 phosphorylation was again increased in FAM83B-expressing cells, and while there was a decrease in EGFR-Y1068 phosphorylation in both the GFP and FAM83B-expressing cells following the addition of AG1478, the levels of active EGFR remained higher in the FAM83B-expressing cells than even the control, untreated cells (Fig 6E). When taken together, we conclude that FAM83B binds to EGFR, thereby increasing EGFR autophosphorylation and elevating PLD activity, which is required for FAM83B-mediated transformation, growth factor independence, and decreased EGFR-TKI sensitivity.
Discussion
The complexity of signaling interactions that emanate from receptors continue to limit the effectiveness of many therapies, since resistance is often easily obtained due to the plasticity of cancer cells. It is therefore important that novel oncoproteins, suitable for therapeutic targeting, continue to be identified. Our recent discovery of FAM83B as a key intermediary in EGFR/RAS signaling may lead to novel FAM83B targeted therapies that can suppress aberrant EGFR/RAS-mediated tumor growth. We have previously shown that elevated FAM83B expression results in the elevation of basal ERK1/2 and AKT phoshorylation, and mTOR activation (as measured by S6K and 4E-BP1 phosphorylation; (16;17)). We hypothesize based on our continuing research, that FAM83B is involved in a large signaling complex likely to include growth factor receptors (such as EGFR), as well as downstream effectors (RAS, CRAF, PI3K, AKT, mTOR, etc.; Fig. 6F). There are many points of crosstalk between the MAPK and mTOR signaling pathways that need to be considered when therapeutically targeting these pathways individually. Recently, two studies identified that when mTORC1 is inhibited by the Rapamycin derivative RAD001, the MAPK pathway becomes activated through a feedback loop that is dependent on S6K, PI3K, and RAS. By combining inhibitors that target MAPK and mTOR, prostate tumor growth could be significantly inhibited (24;25). Importantly, this observation demonstrates clinically the efficacy of targeting both MAPK and mTOR simultaneously.
Since FAM83B expression leads to the activation of MAPK and PI3K/AKT/mTOR signaling, as well as the EGFR-PLD-1 axis, therapeutically targeting FAM83B may represent a suitable strategy for inactivating each of these pathways in tumors. Although the full details of how FAM83B activates each will need to be further defined, our studies represent an important first step. Microarray studies have identified an association between elevated FAM83B expression and estrogen receptor negative status, increased tumor grade and increased rate of recurrence. The lack of ER and PR expression correlates with the basal or triple negative phenotype. While not necessarily synonymous, these classifications describe tumors that are not responsive to hormonal therapy and tend to be less responsive to standard systemic chemotherapies (26). In short, individuals with these tumors would benefit most from new therapies aimed at novel protein targets such as FAM83B.
The increased PLD activity observed in FAM83B-expressing cells, together with the activation of MAPK and PI3K/AKT/mTOR signaling, further implicates FAM83B as an important protein in EGFR/RAS effector activation. Additional significance for this data is provided by the observation that RAS-mediated transformation and EGFR-dependent breast cancer cell growth can be inhibited following FAM83B or PLD1 ablation or inhibition. This has important implications, since recent studies have demonstrated that breast cancers have significant elevation of overall PLD activity relative to normal adjacent tissue (27). PLD activity correlates with the loss of ER-alpha expression and increased protease secretion, both hallmarks of more aggressive tumor cell proliferation and invasion, which also correlate with FAM83B expression (3;27–31). Understanding the role of FAM83B in elevating PLD activity and increasing signaling through the MAPK and mTOR pathways are important for determining its potential as a therapeutic target.
The role of PA in the regulation of mTOR is a developing research area with important implications for cancer therapeutics (8;32). It is now clear that PA is required for the formation of active mTOR complexes (33). Clinical studies using mTOR inhibitors derived from rapamycin (including CCI-779, RAD001 and AP23573) in breast cancer have demonstrated some efficacy thus far (34). Interestingly, PA and rapamycin analogs compete for the same binding domain on mTOR, explaining why the overall PLD activity within a cancer cell influences its sensitivity to rapamycin. Recent studies have shown that by reducing the overall PLD activity in cancer cells, the sensitivity of mTOR complexes to inhibition by rapamycin could be significantly increased (35). In agreement, we recently observed that the elevated PLD activity conferred by FAM83B expression decreases the sensitivity of transformed cells to rapamycin derivatives (17).
The elevation of PLD activity by FAM83B-EGFR is required for the efficient activation of CRAF and mTOR, since a mutation of FAM83B that blocks FAM83B-EGFR complex formation prevents PLD-1 activation, as well as downstream MAPK and mTOR signaling (Figure 7E). However, while elevated PLD activity was required for FAM83B-mediated transformation, it was not by itself sufficient to drive transformation or protect cells from growth suppression following ablation of FAM83B, withdrawal of growth factors, or EGFR-TKI treatment. These data argue that FAM83B expression confers additional functions independent of PLD activity, which are required for each of these phenotypes. Similar to our findings with FAM83B, previous studies of EGFR-, RAS-, CRAF-, and SRC-mediated transformation have identified a requirement for PLD activity, yet each oncogene also has independent functions which cannot be recapitulated simply by elevating PLD activity. The yet undefined functions of FAM83B are the focus of future studies into the mechanism of FAM83B-mediated transformation.
Materials and Methods
Retroviral Constructs
pcDNA5/TO-myc-PLD1 and pcDNA5/TO-myc-PLD2 (20) was subcloned into pLNCX2 (Clontech). phCMV2-HA-PLD1 and phCMV2-HA-PLD1-K830R were kindly provided by Julian Gomez-Cambronero (Wright State University) and subsequently subcloned into pLNCX2. LXSN-EGFR was provided by Lucia Pirisi (University of South Carolina School of Public Health). The LPCX-FLAG-FAM83B was described previously (16). The LPCX-FLAG-FAM83B-K230A point mutant was made by amplifying the LPCX-FLAG-FAM83B plasmid with primers (5′ GCATTTTTGTTAGTTGACTGCCAG 3′ and 5′ CTG TTC CAT TTT TCC ATG GAA 3′) which was subsequently ligated with T4 DNA ligase and sequence verified.
Virus Production and Infection
Retroviruses were produced as described (36;37). Briefly, retroviral vectors were transfected into Phoenix-Ampho cells. Plasmids encoding shRNAs targeting FAM83B (B2), PLD1, or GFP in pLKO.1 were acquired from Open Biosystems and Sigma-Aldrich. Viruses were packaged in 293T cells using the second-generation packaging constructs pCMV-dR8.74 and pMD2G, were kind gifts from Didier Trono (University of Geneva, Switzerland). Supernatants containing virus, were collected at 24 and 48 hours and supplemented with 4 μg/ml of polybrene before being frozen in aliquots or used to infect cells for 6–24 hours.
Cell Lines and Culture Conditions
hTERT-HME1 and HME1-RAS G12V cells were described previously (16). MCF7, HCC1937, and MDA468 breast cancer cell lines were grown in a humidified atmosphere containing 5% CO2 in DMEM (with glucose and L-glutamine; Life Technologies) with 5% fetal bovine serum and 50 units/mL of penicillin and 50 μg/mL of streptomycin sulfate (U.S. Biochemical Corp.).
Immunoprecipitation and Western Analysis
Western analysis was performed as described (38). Antibodies to HA (F-7), MYC (9E10), and PC-PLD1 (F-12) were from Santa Cruz Biotechnology, antibodies to β-actin (pan Ab-5) were from Neomarkers, antibodies to GAPDH were from Calbiochem, antibodies to FLAG (M2) were from Sigma-Aldrich, and antibodies for ERK1/2, P-ERK1/2 (Thr202/Tyr204), P-EGFR (Tyr1068), EGFR were from Cell Signaling. Polyclonal FAM83B antibodies were generated against recombinant FAM83B protein (Covance) and monoclonal antibodies were created at the Case Comprehensive Cancer Center Hybridoma Core Facility. Primary antibodies were detected with goat anti-mouse or goat anti-rabbit conjugated to horseradish peroxidase (Hoffman-La Roche), using enhanced chemiluminescence (Perkin-Elmer). For immunoprecipitation studies, 293T cells transfected with LXSN-EGFR and LPCX-FLAG-FAM83B or LPCX-FLAG-FAM83B-K230A using lipofectamine 2000 (Invitrogen). Transfected cells were fixed in 4% formaldehyde-PBS for 10 minutes at room temperature, harvested in ice-cold PBS and lysed in NP-40 lysis buffer with protease inhibitor cocktail (Sigma Aldrich) and phosphatase inhibitor cocktail (Thermo-Fisher) by sonication. One milligram of each lysate was pre-cleared and then incubated with the indicated primary antibodies (Sigma) and Protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoprecipitates were separated by SDS-PAGE and processed for Western analysis.
Relative Growth and Soft Agar Assays
For growth assays, hTERT-HME1, MCF7, and MDA468 cells were plated in triplicate at 20,000 cells/well in triplicate and cell number was determined on a Beckman Coulter counter. The PLD1/2 inhibitor VU0155056 (0.2, 2, 10, and 20 μM), was added to indicated experiments. Soft agar assays were performed as described (39) using 1 × 105 HME1 cells or 2 × 105 cells MCF7 or MDA468 cells. Tert-Butanol (0.5% & 1.0%) and 1-Butanol (0.5% & 1.0%) were obtained from Fisher Scientific and the specific pharmacological inhibitors of PLD1 and PLD2, compounds VU0155056 (10 and 20μM), VU0359595 (10 and 20μM), and VU0155069 (10 and 20 μM) were added to the medium during feedings for indicated experiments (20). To quantify colonies, each plate was scanned using an automated multi-panel scanning microscope, and the digital images were analyzed using MetaMorph image quantification software.
Cell-Based Mass Spectrometric PLD Assay
Cell-based PLD activity was determined using a modified in vivo deuterated 1-butanol PLD assay (19). HME1 cells were seeded at 3.5 × 105 cells/well in 6-well tissue culture plates 48 hours prior to the assay in complete growth medium. At the time of the assay cells were treated with either Medium 171 alone, Medium 171 + 0.3% (v/v) 1-butanol-d10 or Medium 171 + 0.3% (v/v) 1-butanol-d10 + 20 % fetal bovine serum for 60 min at 37 °C. The PLD inhibitor VU0155056 (2 uM) was added for indicated experiments (20). After treatment, cellular lipids were extracted as previously described (19) using 100 ng of 32:0 phosphatidyl methanol as an internal standard. After phase separation and drying the resulting lipid film was resuspended in 80 μL of 9:1 methanol: chloroform (v:v) for MS analysis. Mass spectral analysis was performed on a Finnigan TSQ Quantum triple quadrupole mass spectrometer (Thermo Finnigan, San Jose, CA) equipped with a Harvard Apparatus syringe pump and an electrospray source. Samples were analyzed at an infusion rate of 10 μL/min in negative ionization mode with the scan range from m/z 400–900. Data were collected with the Xcalibur software package (Thermo Finnigan) and analyzed with software developed in our laboratory. Data are represented as a ratio of major phosphatidylbutanol-d9 to internal standard. Background signal was subtracted using cells not treated with 1-butanol-d10 as a negative control. Samples were generated in triplicate in multiple independent experiments.
Supplementary Material
Acknowledgments
We are grateful to Chase Foy for technical help and Damian Junk for helpful discussions. The core facilities provided by the Case Comprehensive Cancer Center (P30 CA43703; Athymic Animal and Xenograft Core Facility; Gene Expression and Genotyping Core Facility; Cytometry & Imaging Microscopy Core Facility; Radiation Resources Core Facility). This work was supported by the US National Institutes of Health (R01CA138421 to M.W.J; T32CA059366 to R.C and C.B.), the Department of Defense Breast Cancer Research Program (BC095847 and BC074072 to M.W.J), the American Cancer Society (RSG-10-072-01-TBG to M.W.J) and the McDonnell Foundation for brain cancer research (H.A.B) and the NIH MLPCN U54 MH084659 (C.W.L) Vanderbilt Specialized Chemistry Center for Accelerated Probe Development.
Footnotes
Conflict of interest.
Dr Jackson’s work has been funded by the NIH, the American Cancer Society, and the Department of Defense. Dr. H. Alex Brown’s work has been funded by the McDonnell Foundation. Dr. Cipriano’s and Ms. Bartel’s work has been funded by the NIH. All other authors’ declare no conflict of interest.
Reference List
- 1.Grant S. Cotargeting survival signaling pathways in cancer. J Clin Invest. 2008 Sep;118(9):3003–6. doi: 10.1172/JCI36898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007 May 14;26(22):3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 3.Joseph T, Wooden R, Bryant A, Zhong M, Lu Z, Foster DA. Transformation of cells overexpressing a tyrosine kinase by phospholipase D1 and D2. Biochem Biophys Res Commun. 2001 Dec 21;289(5):1019–24. doi: 10.1006/bbrc.2001.6118. [DOI] [PubMed] [Google Scholar]
- 4.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006 Oct 6;127(1):157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 5.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004 Aug;6(2):171–83. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003 Sep;1(11):789–800. [PubMed] [Google Scholar]
- 7.Shi M, Zheng Y, Garcia A, Xu L, Foster DA. Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 2007 Dec 18;258(2):268–75. doi: 10.1016/j.canlet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster DA. Phosphatidic acid signaling to mTOR: Signals for the survival of human cancer cells. Biochim Biophys Acta. 2009 Mar 2; doi: 10.1016/j.bbalip.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Bell RM. Regulation of Raf-1 kinase by interaction with the lipid second messenger, phosphatidic acid. Biochem Soc Trans. 1997 May;25(2):561–5. doi: 10.1042/bst0250561. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan FG, McReynolds M, Couvillon A, Kam Y, Holla VR, Dubois RN, et al. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc Natl Acad Sci U S A. 2005 Feb 1;102(5):1638–42. doi: 10.1073/pnas.0406698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster D. Targeting Phospholipase D-mediated Survival Signals in Cancer. Current Signal Transduction Therapy. 2006 [Google Scholar]
- 12.Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, et al. Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J Biol Chem. 1999 Jan 8;274(2):1131–9. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- 13.Rizzo MA, Shome K, Watkins SC, Romero G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem. 2000 Aug 4;275(31):23911–8. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- 14.Kraft CA, Garrido JL, Fluharty E, Leiva-Vega L, Romero G. Role of phosphatidic acid in the coupling of the ERK cascade. J Biol Chem. 2008 Dec 26;283(52):36636–45. doi: 10.1074/jbc.M804633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007 Jul;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA, et al. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012 Sep 4;122(9):3197–210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013 May 11; doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Ho WT, Exton JH. Association of N- and C-terminal domains of phospholipase D is required for catalytic activity. J Biol Chem. 1998 Dec 25;273(52):34679–82. doi: 10.1074/jbc.273.52.34679. [DOI] [PubMed] [Google Scholar]
- 19.Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Biochemical analysis of phospholipase D. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- 20.Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009 Feb;5(2):108–17. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladeda V, Frankel P, Feig LA, Foster DA, Bal de Kier JE, guirre-Ghiso JA. RalA mediates v-Src, v-Ras, and v-Raf regulation of CD44 and fibronectin expression in NIH3T3 fibroblasts. Biochem Biophys Res Commun. 2001 May 18;283(4):854–61. doi: 10.1006/bbrc.2001.4845. [DOI] [PubMed] [Google Scholar]
- 22.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012 Sep 4;122(9):3211–20. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Phospholipase D: enzymology, functionality, and chemical modulation. Chem Rev. 2011 Oct 12;111(10):6064–119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008 Sep;118(9):3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H, et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008 Sep;118(9):3051–64. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassam F, Enright K, Dent R, Dranitsaris G, Myers J, Flynn C, et al. Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin Breast Cancer. 2009 Feb;9(1):29–33. doi: 10.3816/CBC.2009.n.005. [DOI] [PubMed] [Google Scholar]
- 27.Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. J Cancer Res Clin Oncol. 1997;123(5):280–5. doi: 10.1007/BF01208639. [DOI] [PubMed] [Google Scholar]
- 28.Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000 Dec 20;161(2):207–14. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 29.Reich R, Blumenthal M, Liscovitch M. Role of phospholipase D in laminin-induced production of gelatinase A (MMP-2) in metastatic cells. Clin Exp Metastasis. 1995 Mar;13(2):134–40. doi: 10.1007/BF00133618. [DOI] [PubMed] [Google Scholar]
- 30.Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999 Jan;19(1B):671–5. [PubMed] [Google Scholar]
- 31.Williger BT, Ho WT, Exton JH. Phospholipase D mediates matrix metalloproteinase-9 secretion in phorbol ester-stimulated human fibrosarcoma cells. J Biol Chem. 1999 Jan 8;274(2):735–8. doi: 10.1074/jbc.274.2.735. [DOI] [PubMed] [Google Scholar]
- 32.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007 Jan 1;67(1):1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 33.Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009 Mar;29(6):1411–20. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carraway H, Hidalgo M. New targets for therapy in breast cancer: mammalian target of rapamycin (mTOR) antagonists. Breast Cancer Res. 2004;6(5):219–24. doi: 10.1186/bcr927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene. 2003 Jun 19;22(25):3937–42. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- 36.Cipriano R, Patton JT, Mayo LD, Jackson MW. Inactivation of p53 signaling by p73 or PTEN ablation results in a transformed phenotype that remains susceptible to Nutlin-3 mediated apoptosis. Cell Cycle. 2010 Apr 1;9(7):1373–9. doi: 10.4161/cc.9.7.11193. [DOI] [PubMed] [Google Scholar]
- 37.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006 Mar 15;66(6):3169–76. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 38.Jackson MW, Agarwal MK, Agarwal ML, Agarwal A, Stanhope-Baker P, Williams BR, et al. Limited role of N-terminal phosphoserine residues in the activation of transcription by p53. Oncogene. 2004 May 27;23(25):4477–87. doi: 10.1038/sj.onc.1207575. [DOI] [PubMed] [Google Scholar]
- 39.Kan CE, Patton JT, Stark GR, Jackson MW. p53-mediated growth suppression in response to Nutlin-3 in cyclin D1 transformed cells occurs independently of p21. Cancer Res. 2007 Oct 15;67(20):9862–8. doi: 10.1158/0008-5472.CAN-07-0259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.