This study investigates the biogenesis and propagation of nucleoli, sites of ribosome biogenesis and key regulators of cellular growth. Nucleoli disappear during mitosis and reform around nucleolar organizer regions (NORs). By examining the effects of Pol I transcription factor UBF depletion on endogenous NORs and synthetic neo-NORs, McStay and colleagues show that UBF-dependent mitotic bookmarking precedes function-dependent nucleolar assembly. This study not only reveals that nucleolar assembly is a staged process but also establishes the sequence requirements for nucleolar biogenesis.
Keywords: nucleolus, nucleolar organizer region (NOR), UBF, mitotic bookmarking, pseudo-NOR, neo-NOR
Abstract
Human cell nuclei are functionally organized into structurally stable yet dynamic bodies whose cell cycle inheritance is poorly understood. Here, we investigate the biogenesis and propagation of nucleoli, sites of ribosome biogenesis and key regulators of cellular growth. Nucleolar and cell cycles are intimately connected. Nucleoli disappear during mitosis, reforming around prominent uncharacterized chromosomal features, nucleolar organizer regions (NORs). By examining the effects of UBF depletion on both endogenous NORs and synthetic pseudo-NORs, we reveal its essential role in maintaining competency and establishing a bookmark on mitotic NORs. Furthermore, we demonstrate that neo-NORs, UBF-binding site arrays coupled with rDNA transcription units, direct the de novo biogenesis of functional compartmentalized neonucleoli irrespective of their site of chromosomal integration. For the first time, we establish the sequence requirements for nucleolar biogenesis and provide proof that this is a staged process where UBF-dependent mitotic bookmarking precedes function-dependent nucleolar assembly.
Nuclear bodies range between those closely associated with chromatin (i.e., nucleoli and histone locus bodies) and others nonassociated with chromatin (i.e., Cajal bodies and paraspeckles) (Dundr and Misteli 2010). The latter are present in high copy number and persist through cell division as mitotic bodies. Conversely, chromatin-associated bodies disassemble during mitosis and must rapidly reassemble in early G1 to ensure full functionality of daughter nuclei. Until now, the mechanism underpinning cell cycle inheritance of these nuclear bodies has not been determined. The nucleolus is the prototypical and largest chromatin-associated nuclear body. Although its primary role is ribosome biogenesis, it is now often described as plurifunctional with defined additional roles, including cell cycle regulation, stress sensing, viral replication, and RNP biogenesis (Boisvert et al. 2007; Pederson 2010). A detailed understanding of nucleolar biology is essential to address how misregulation of ribosome biogenesis is associated with tumorigenesis and an increasing spectrum of diseases known as ribosomopathies. To understand the underlying mechanisms organizing this essential functional domain, we must identify the DNA sequences, key factors, and mechanisms involved.
Nucleoli form around prominent chromosomal features originally described as secondary constrictions but now more commonly referred to as nucleolar organizer regions (NORs) (Heitz 1931; McClintock 1934). In humans, NORs containing 50-kb to 6-Mb ribosomal gene (rDNA) arrays (Stults et al. 2008) are positioned on the short arms (p-arms) of the five acrocentric chromosomes (Henderson et al. 1972). It has been generally assumed, but never proven, that rDNA sequences within NORs are sufficient to organize nucleolar formation in all eukaryotes. There is some preliminary evidence in support of this hypothesis. Insertion of a single rDNA into Drosophila polytene chromosomes produced pre-rRNA and recruited a 47-kDa nucleolar antigen (Karpen et al. 1988). However, only in a model lower eukaryote, the yeast Saccharomyces cerevisiae, have rDNA arrays been demonstrated to be sufficient to organize nucleoli (Oakes et al. 2006). The yeast nucleolus forms around a single NOR, contains only two compartments, and is intimately connected to the nuclear envelope, which remains intact through its “closed” mitosis (Thiry and Lafontaine 2005). Importantly, yeast nucleoli remain intact throughout mitosis, and transcription is only momentarily inhibited during anaphase to ensure rDNA condensation and nucleolar segregation (Clemente-Blanco et al. 2009). Nucleolar biology is more complex in humans, with most cells containing more than one nucleolus and most nucleoli deriving from multiple NORs. Furthermore, human nucleoli are tripartite in structure and break down during their “open” mitosis (Sirri et al. 2008). Thus, unlike yeast, post-mitotic human cells face the critical challenge of regenerating nucleoli around competent NORs. To date, the DNA requirement and molecular mechanism allowing the propagation through mitosis of human nucleoli remain to be elucidated. The positioning of rDNA arrays within heterochromatic regions is an evolutionarily conserved feature (Henderson et al. 1972; Britton-Davidian et al. 2012), hinting at a role for chromosome context in nucleolar biology. Furthermore, even though acrocentric chromosome p-arms are still missing from current human genome assemblies, our recent work has established that sequences surrounding rDNA arrays are conserved among all human acrocentric chromosomes (Floutsakou et al. 2013). Interestingly, the distal side to the rDNA array, termed the distal junction (DJ), appears to be functional and involved in nucleolar organization by anchoring the rDNA array to the perinucleolar heterochromatin (Floutsakou et al. 2013). Hence, open questions remain regarding what actually constitutes a human NOR and whether non-rDNA sequences are required.
NORs competent for nucleolar formation contain relatively undercondensed chromatin (Heliot et al. 1997), exhibit the classical mitotic secondary constriction, and can be identified by silver staining (Goodpasture and Bloom 1975). Noncompetent or silent NORs lack these hallmarks, being entirely heterochromatic and not associated with nucleoli during interphase (McStay and Grummt 2008). At mitotic entry, rDNA transcription is repressed, and human nucleoli are disassembled (Sirri et al. 2008). The rDNA transcription machinery, including the HMG-box protein UBF (Roussel et al. 1993; O'Sullivan et al. 2002), remains associated with secondary constrictions, while the preribosomal RNA (pre-rRNA) processing machinery localizes to the periphery of metaphase chromosomes (Sirri et al. 2008). As cells exit mitosis, CDK1 inhibition leads to the resumption of rDNA transcription by RNA polymerase I (Pol I) (Sirri et al. 2000), yielding pre-rRNA comprised of 18S, 5.8S, and 28S rRNAs with external transcribed spacers (ETSs) and internal transcribed spacers (ITSs) (McStay and Grummt 2008). Coordinated with transcription resumption, the processing machinery assembles into prenucleolar bodies (PNBs). Early processing factors are then recruited from PNBs to nascent transcripts, and in a subsequent G1 CDK-dependent manner, late processing factors are also recruited to maturing nucleoli (Sirri et al. 2002). Resulting nucleoli are functionally compartmentalized, with their inner core or fibrillar center (FC) containing pools of unengaged transcription factors and nontranscribed rDNA (Raska et al. 2006). FCs are surrounded by the dense fibrillar component (DFC), with Pol I transcription occurring at the interface between these compartments. Early pre-rRNA processing, mediated by the small subunit (SSU) processome, takes place in the DFC. Later processing events, carried out by the large subunit (LSU) processome and assembly of ribosome subunits, occur in the largest nucleolar compartment, the granular component (GC). It is unknown whether this functional compartmentalization of nucleoli is intrinsic to the process of ribosome biogenesis or imposed by some external cytoskeletal framework.
While cell biology has provided a detailed description of the events in the nucleolar cycle, we believe that a detailed understanding of the molecular basis of nucleolar biogenesis and propagation can be achieved by using a synthetic biology approach. In this study, we use chromosome engineering to demonstrate first a central role for UBF in maintaining NOR competency and establishing the mitotic hallmarks of competent NORs. Second, we establish the DNA sequence requirements for the formation of a fully functional ectopic nucleolus. Third, we provide the first demonstration that NORs occupy distinct territories within large nucleoli. Finally, our synthetic biology approach provides compelling evidence that nucleolar biogenesis is a staged and evolutionarily conserved process independent of chromosomal context in which UBF-dependent mitotic bookmarking underpins function-dependent assembly of nucleoli.
Results
UBF is required to maintain NOR competency and establish the mitotic hallmarks of competent NORs
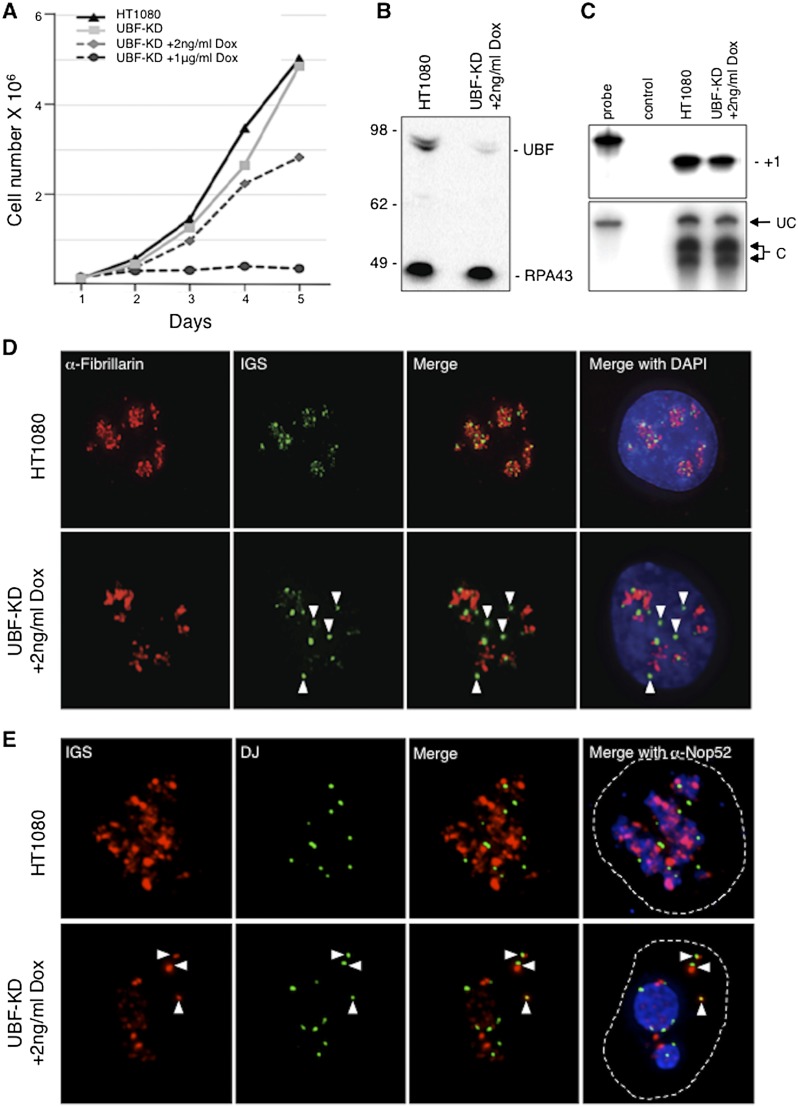
The correlation between NOR competency and extensive UBF binding across rDNA repeats throughout the cell cycle (Roussel et al. 1993; O'Sullivan et al. 2002) implies that UBF plays a central role in the nucleolar cycle. To test this, we generated UBF-KD cells, a stable derivative of human HT1080 cells containing a doxycycline (Dox)-inducible shRNA that targets UBF messenger RNA (mRNA). HT1080 cells were chosen because three-dimensional (3D) immuno-FISH revealed that all of their rDNA repeats are present within nucleoli (Fig. 1; Supplemental Fig. S1), and consequently all of their NORs are competent. Culturing the UBF-KD cell line with 1 μg/mL Dox induced an ∼50-fold depletion of UBF, resulted in complete growth inhibition (Fig. 1A), and increased cell death, as evidenced by an increase in the proportion of cells with sub-G1 DNA content (Supplemental Fig. S2). Thus, UBF is essential for cell growth and survival. However, growing UBF-KD cells with 2 ng/mL Dox induced a 6.5-fold depletion of UBF cellular level without markedly affecting cell growth or survival for up to 2 wk (Fig. 1A,B). Furthermore, this 6.5-fold depletion of UBF did not noticeably affect the level of rDNA transcription or the efficiency of A′ cleavage within pre-rRNA 5′ ETS, as assessed by S1 nuclease protection assays (Fig. 1C). 3D immuno-FISH revealed that UBF depletion clearly led to the compaction of rDNA repeats and the dissociation of these condensed repeats from the nucleolus (Fig. 1D). These nucleoplasmic rDNA foci were devoid of any of the remaining UBF (Supplemental Fig. S3). To address whether these noncompetent rDNA foci represented entire NORs, we took advantage of our recently described DJ probes (Supplemental Fig. S1), which can be used to identify and count individual NORs (Floutsakou et al. 2013). Importantly, DJ sequences linked to rDNA in competent NORs are embedded in perinucleolar heterochromatin. 3D immuno-FISH using combined DJ and rDNA probes together with Nop52 immunostaining of the GC compartments of nucleoli identified DJ signals immediately adjacent to rDNA foci that were clearly separated from nucleoli in UBF-depleted cells (Fig. 1E). These combined results demonstrate that foci of compacted rDNA induced by UBF depletion represent newly silenced NORs and point to an essential role for UBF in maintaining NOR competency.
Figure 1.
UBF depletion induces NOR silencing. (A) Growth curves of HT1080 cells and UBF-KD cells cultured in the absence and presence of 2 ng/mL or 1 μg/mL Dox. (B) Western blot of proteins extracted from HT1080 cells and UBF-KD cells cultured with 2 ng/mL Dox. Molecular weight markers in kilodaltons are indicated at the left. Anti-UBF antibodies revealed a 6.5-fold depletion of UBF1 (97 kDa) and UBF2 (94 kDa), while anti-RPA43 antibodies reveal the conserved level of Pol I subunit RPA43 (43 kDa). (C) S1 nuclease protection assays performed on equal amounts of RNA extracted from HT1080 and UBF-KD cells indicate that the rDNA transcription level is conserved in UBF-KD cells cultured with 2 ng/mL Dox. S1 assays performed on equal amounts of RNA extracted from HT1080 and UBF-KD cells cultured in 2 ng/mL Dox reveal that UBF depletion does not affect the efficiency of cleavage at the A′ site in the 5′ ETS of pre-rRNA. Escherichia coli RNA served as a negative control. Protected fragments arising from cleaved (C) and uncleaved (UC) transcripts are indicated. (D) Combined 3D immuno-FISH performed on HT1080 and UBF-KD cells cultured with 2 ng/mL Dox reveals the dissociation of rDNA repeats (IGS probe) from the nucleolus (fibrillarin immunostaining) upon UBF depletion (arrowheads). (E) Combined 3D immuno-FISH performed on HT1080 and UBF-KD cells cultured with 2 ng/mL Dox reveals that foci of rDNA staining dissociated from nucleoli represent individual silent NORs (arrowheads). NORs are identified using a DJ BAC (green) that maps to the distal side of rDNA arrays, rDNA is identified using an IGS probe (red), and nucleoli are visualized by staining with anti-Nop52 antibodies (blue). Dashed lines indicate the edges of nuclei.
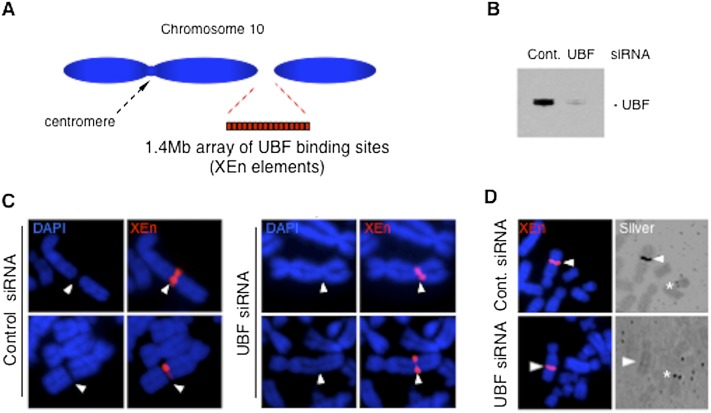
If NOR competency is associated with the presence of a secondary constriction on metaphase chromosomes, silencing of NORs following UBF depletion should be associated with the loss of this morphology. To validate this hypothesis, we exploited the cell line clone 3D that contains a prominent synthetic secondary constriction termed pseudo-NOR. This pseudo-NOR was generated by integration of a 1.4-Mb array of a heterologous UBF-binding site (XEn) into the middle of a chromosome 10 long arm (q-arm) of HT1080 cells (Fig. 2A; Mais et al. 2005). Using a pool of siRNAs that target UBF, we were able to deplete UBF levels ∼10-fold in pseudo-NOR clone 3D cells (Fig. 2B). In contrast with the level of depletion in UBF-KD cells grown with 1 μg/mL Dox, this level of UBF depletion did not markedly repress cell growth (data not shown). Moreover, FISH experiments performed on chromosome spreads revealed loss of pseudo-NOR secondary constriction upon UBF depletion (Fig. 2C). Silver staining, another mitotic hallmark of NOR competency, was also lost from pseudo-NORs upon UBF depletion (Fig. 2D). Note that silver staining of endogenous NORs is retained in the UBF-depleted clone 3D cells, consistent with their maintained growth capacity. These findings from pseudo-NORs indicated that UBF is key in the establishment of competent NOR mitotic hallmarks. To demonstrate that synthetic pseudo-NORs model the behavior of endogenous mitotic NORs, we sought to determine the effects of UBF depletion on secondary constrictions in cells of the marsupial Potorous tridactylus. The Potorous genome contains a single large NOR near the centromere of its X chromosomes, while human NORs are positioned close to the ends of acrocentric p-arms, making it difficult to unequivocally demonstrate loss of secondary constrictions. siRNA-mediated depletion of UBF in male Ptk-2 cells resulted in loss of the secondary constriction and silver staining associated with their single NOR (Supplemental Fig. S4). These results combined with those on pseudo-NORs demonstrate that UBF is essential in establishing the unique morphology of mitotic competent NORs.
Figure 2.
Formation of secondary constriction on mitotic chromosomes is UBF-dependent. (A) Schematic representation of the novel secondary constriction formed by the insertion of a 1.4-Mb XEn array into the q-arm of chromosome 10 in pseudo-NOR clone 3D. (B) Western blot of proteins extracted from pseudo-NOR clone 3D cells transfected with control or UBF siRNAs revealed a 13-fold depletion of UBF1 and UBF2. (C) Metaphase chromosome spreads from pseudo-NOR clone 3D were hybridized with an XEn probe. Arrowheads indicate the site of pseudo-NOR insertion. Transfection of UBF siRNA induces the loss of pseudo-NOR secondary constriction. (D) Combined FISH and silver staining of metaphase spreads from pseudo-NOR clone 3D. UBF depletion results in the loss of silver staining at pseudo-NORs (arrowheads). Note the silver staining of an endogenous NOR (*) in the bottom right panel.
Construction of transcriptionally active neo-NORs
Remarkably, pseudo-NORs recruit not only human UBF but also the entire Pol I transcription machinery in a UBF-dependent manner despite their lack of sequence homology with human rDNA and their lack of promoter sequences (Mais et al. 2005). In interphase, pseudo-NORs direct the formation of novel subnuclear bodies, related in protein composition to nucleolar FCs. Thus, pseudo-NORs were so called because they lack promoters, are nontranscribed, fail to recruit the majority of nucleolar proteins (including pre-rRNA processing factors), and do not form nucleoli. The silencing of endogenous NORs upon UBF depletion suggests that retention of the Pol I machinery on UBF-loaded chromatin at mitotic NORs, modeled by pseudo-NORs, is a prerequisite for nucleolar formation.
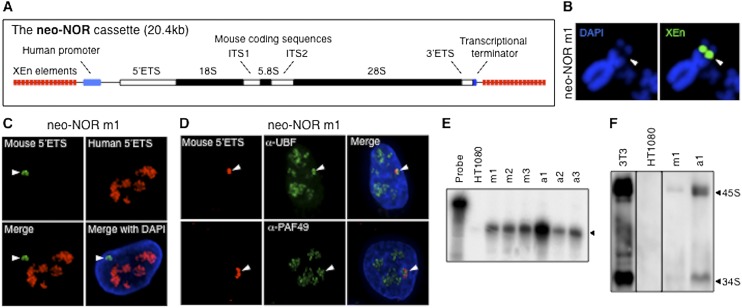
To test the above hypothesis and determine the sequence requirements for nucleolar formation, we next sought to construct synthetic nucleoli. To this end, we integrated ectopic arrays of a synthetic neo-NOR cassette into various chromosomal contexts within HT1080 cells. The neo-NOR cassette is composed of blocks of XEn elements interspersed with rDNA transcription units. As with pseudo-NORs, the repetitive nature of the XEn blocks promoted concatenation and formation of large integrated arrays. The neo-NOR rDNA transcription unit was comprised of an engineered human rDNA promoter, mouse pre-rRNA coding sequences, and mouse transcriptional terminators (Fig. 3A). Since Pol I transcription is remarkably species-specific (Heix and Grummt 1995; Sullivan et al. 2001), transcription of neo-NOR coding sequences is driven by a human rDNA promoter. However, human pre-rRNA coding sequences were not used, since it was essential that neo-NOR products could be distinguished from those of endogenous NORs. Since the sequence requirements for production of mature rRNAs from a pre-rRNA are poorly characterized in any species, we chose to use mouse rather than engineered human pre-rRNA coding sequences. Hence, the sequence divergence between mouse and human pre-rRNAs was exploited to establish the functionality of integrated neo-NOR arrays.
Figure 3.
Neo-NORs are undercondensed during mitosis and transcribed by Pol I into 45S pre-rRNA in interphase. (A) Schematic representation of the neo-NOR cassette. (B) Metaphase chromosome spreads from neo-NOR clone m1 were hybridized with an XEn probe. In clone m1, an array of neo-NOR cassettes is inserted into the p-arm of chromosome 4 (Supplemental Fig. S4). Arrowheads indicate novel secondary constrictions at the site of neo-NOR insertion. (C) RNA-FISH using mouse (green) and human (red) 5′ ETS probes identifies neo-NOR-derived transcripts (arrowheads) in clone m1. (D) Combined 3D immuno-RNA-FISH reveals that UBF and PAF49 colocalize with neo-NOR-derived transcripts in clone m1. (E) S1 nuclease protection assays on RNA extracted from HT1080 and neo-NOR cell lines (20 μg each) were performed at nucleotide resolution. These assays indicate that neo-NOR-derived transcripts initiate from the expected nucleotide +1 of neo-NOR 5′ ETS, resulting in the protection of 41 base pairs (bp) from the 60-bp probe. (F) Northern blotting of RNA extracted from 3T3 cells (1 μg), HT1080 cells (10 μg), and neo-NOR clones a1 and m1 (10 μg each) probed with mouse 5′ ETS sequences identifies neo-NOR-derived 45S pre-rRNAs and 34S pre-rRNA processing intermediates.
Following transfection of the 20.4-kb neo-NOR cassette into HT1080 cells, six neo-NOR clones, classified into two groups, were selected. In acrocentric neo-NOR clones a1–3, single integration events occurred on p-arms of acrocentric chromosomes 15, 22, and 13, respectively, while in metacentric neo-NOR clones m1–3, single integration events occurred on metacentric chromosomes (Supplemental Fig. S5A). By comparing the behavior of these two neo-NOR groups, we could assess the contribution of chromosomal context in the nucleolar cycle. Southern blotting was used to establish that integrated neo-NORs are comprised of large arrays containing between 50 and 180 intact transcription units with a significant number of rearranged units (Supplemental Fig. S5B,C). We thus estimated that the size of these arrays ranges between 1 and 4 Mb, which is comparable with endogenous NORs (Stults et al. 2008). Interestingly, rearranged rDNA repeats also appear to be a feature occurring in endogenous human NORs (Caburet et al. 2005). The analysis of metaphase chromosomes from clone m1 cells revealed that its neo-NOR, integrated into chromosome 4, induced the formation of a novel silver-staining secondary constriction identical to that of pseudo-NORs and endogenous competent NORs (Fig. 3B; Supplemental Fig. S5D). From this and the analysis of the other metacentric lines (data not shown), we were able to conclude that neo-NORs mimic the specific chromatin structure of mitotic competent NORs.
To establish neo-NOR transcriptional status, we performed RNA-FISH using mouse- and human-specific 5′ ETS probes that distinguish between neo-NOR-derived and endogenous transcripts. In acrocentric clones a1–3, neo-NOR-derived transcripts were observed close to endogenous pre-rRNAs (Supplemental Fig. S6A). Conversely, in most metacentric neo-NOR cells (∼60%), neo-NOR-derived transcripts were clearly separate from endogenous nucleoli (Fig. 3C; Supplemental Fig. S6A). These RNA-FISH experiments also revealed that 94% of clone m1 cells contained a transcriptionally active neo-NOR. Further RNA-FISH experiments using mouse-specific ITS2 and 3′ ETS probes revealed that neo-NOR transcription extends across their entire coding sequences (Supplemental Fig. S6B,C). Additionally, 3D immuno-RNA-FISH indicated that FC components UBF, t-UTP10, Treacle, and Pol I subunit PAF49 colocalize with neo-NOR-derived transcripts in clone m1 (Fig. 3D; Supplemental Fig. S7). In line with this, neo-NOR-derived transcripts initiate from the expected +1 site used by Pol I (Fig. 3E) and are the same size as mouse 45S primary pre-rRNA (Fig. 3F). Noticeably, quantitation reveals that transcription levels are lower than expected, with only ∼2% of pre-rRNAs originating from neo-NORs. Possible explanations for this observation are discussed below. Nevertheless, these results establish that neo-NORs recruit FC components enabling their transcription by Pol I, yielding full-length mouse pre-rRNA. This suggests that neo-NORs organize formation of synthetic nucleoli (neonucleoli).
Neonucleoli produce mature rRNAs
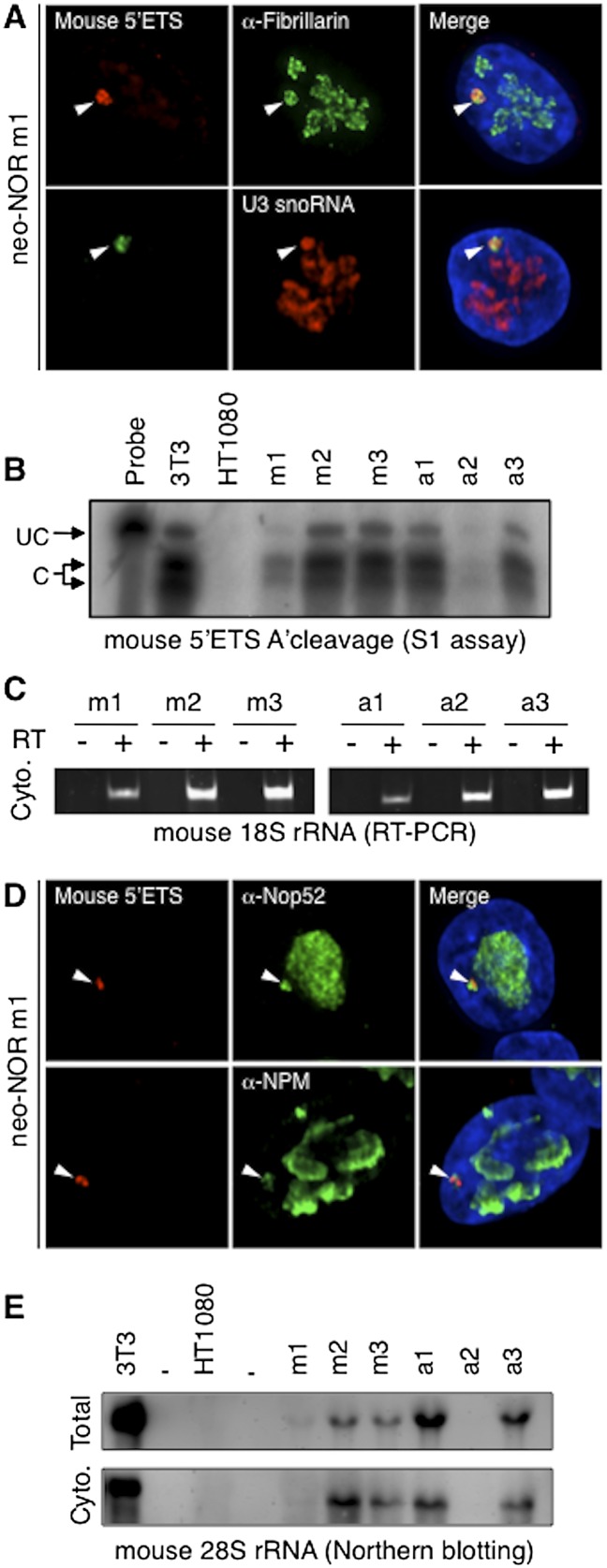
The next aspect of neonucleolar function that we investigated was pre-rRNA processing into cytoplasmic mature rRNAs. To this end, we first concentrated on early processing events leading to the production of 18S rRNA by the SSU processome. 3D immuno-RNA-FISH clearly established that DFC components of U3 (U3 small nucleolar RNA [snoRNA] and hU3-55k), box C/D (fibrillarin), and box H/ACA (Nap57) small nucleolar ribonucleoproteins (snoRNPs) are recruited to isolated neo-NOR-derived transcripts in clone m1 (Fig. 4A; Supplemental Fig. S6). In mouse and human cells, the initial SSU processome-dependent cleavage event occurs at the A′ site and comprises alternative cleavages 6 nucleotides (nt) apart (Financsek et al. 1982; Craig et al. 1987). S1 assays with a mouse-specific probe reveal that this A′ cleavage event is carried out on neo-NOR-derived transcripts with the same pattern and efficiency as observed in mouse 3T3 cells (Fig. 4B; Supplemental Fig. S8A). In both humans and mice, subsequent cleavages resulting in mature 18S rRNA can occur by alternative pathways. The main processing pathway involves complete removal of the 5′ ETS prior to cleavage in ITS1, while the alternative pathway generates a 34S processing intermediate by cleaving initially within ITS1 (Mullineux and Lafontaine 2012). A RT–PCR assay (Supplemental Fig. S8A,B) was used to demonstrate the presence of neo-NOR-derived mouse 18S rRNA within the cytoplasm of neo-NOR lines (Fig. 4C). This, combined with the presence in neo-NOR lines of mouse 34S pre-rRNA (Fig. 3F), provides compelling evidence that the SSU processome is fully functional in neonucleoli.
Figure 4.
Neonucleoli recruit DFC and GC nucleolar components and produce mature neo-NOR-derived rRNAs. (A) Combined 3D immuno-RNA-FISH reveals that early pre-rRNA processing factors fibrillarin and U3 snoRNA colocalize with neo-NOR-derived transcripts in clone m1. (B) S1 assays performed on RNA extracted from 3T3 (0.2 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines indicate that neo-NOR-derived transcripts are accurately and efficiently cleaved at the A′ site of their 5′ ETS. Protected fragments arising from cleaved (C) and uncleaved (UC) transcripts are indicated. (C) RT–PCR reveals the presence of mouse 18S rRNAs in the cytoplasm of neo-NOR cell lines. Reactions in which reverse transcriptase (RT) was omitted served as negative controls. (D) Combined 3D immuno-RNA-FISH reveals that late pre-rRNA processing factors Nop52 and NPM colocalize with neo-NOR-derived transcripts in clone m1. (E) Northern blots of cellular (Total) and cytoplasmic (Cyto.) RNA extracted from 3T3 (0.5 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines probed with a mouse-specific oligonucleotide reveal the presence of cytoplasmic mouse 28S rRNAs in neo-NOR lines.
We then investigated the later processing events that result in production of 5.8S and 28S rRNAs by the LSU processome. Initially, 3D immuno-RNA-FISH revealed that GC components Nop52, nucleophosmin (NPM), and nucleolin are also recruited to isolated neo-NOR-derived transcripts in clone m1 (Fig. 4D; Supplemental Fig. S7). We then used a mouse-specific 28S probe (Supplemental Fig. S9B) to assess the ability of neonucleoli to produce mature 28S rRNA. Northern blots of total RNA revealed the production of neo-NOR-derived 28S rRNAs, indicating full functionality of the LSU processome within neonucleoli (Fig. 4E). Further Northern blots of cytoplasmic RNA showed that neo-NOR-derived 28S rRNAs are successfully exported from the nucleus (Fig. 4E). Taken together, these results demonstrate that mouse pre-rRNAs are accurately and efficiently processed by human SSU and LSU processomes, resulting in the recruitment of DFC and GC nucleolar components and ultimate formation of ectopic neonucleoli. Hence, even though the initiation of rDNA transcription by Pol I is highly species-specific, neo-NORs have revealed unexpected levels of evolutionary conservation in post-transcriptional stages of ribosome biogenesis.
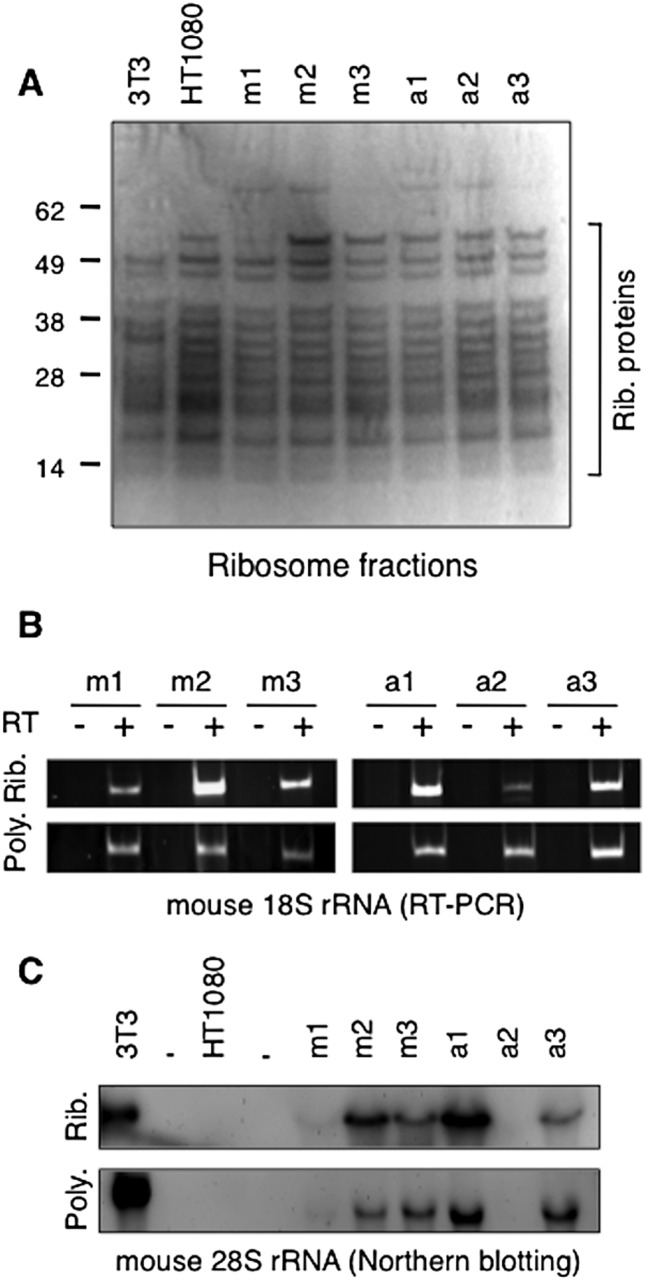
Neonucleoli produce polysome-associated ribosomes
To further address the functionality of neonucleoli, we investigated whether neo-NOR-derived mature rRNAs assemble with ribosomal protein into polysome-associated ribosomes. To assess this, we used a well-established ribosome purification protocol (Belin et al. 2010). In this protocol, cells are lysed, after which nuclei and mitochondria are removed. The salt concentration of the resulting post-mitochondrial fraction (PMT) is then raised to disrupt polysomes and dissociate ribosomes into small 40S and large 60S subunits. Resulting ribosome subunits are further purified by ultracentrifugation through a sucrose cushion. Purity is assessed by the lack of histones and high-molecular-weight proteins and by enrichment of ribosomal proteins ranging in size from 11.5 to 47.3 kDa (Supplemental Fig. S10A). Using this protocol, cytoplasmic ribosomes were purified from HT1080, 3T3, and neo-NOR cell lines (Fig. 5A). RT–PCR and Northern blotting revealed that cytoplasmic ribosomes purified from mouse and neo-NOR lines contain mouse 18S and 28S rRNAs, thus demonstrating that human ribosomal proteins assembled with mouse rRNAs into intact ribosome subunits (Fig. 5B,C). This provides further confirmation of SSU and LSU processome functionality within neonucleoli and indicates that the ribosomal subunit export pathway from neonucleoli to the cytoplasm is intact. To assess whether these neo-NOR-derived ribosome subunits can be loaded onto mRNA, we determined whether they appeared in polysomes. For this purpose, polysome fractions were generated from HT1080, 3T3, and neo-NOR cell lines by ultracentrifugation of cytoplasmic extracts through a sucrose gradient (Supplemental Fig. S10B). We observed mouse 18S and 28S rRNAs in polysomes prepared from each neo-NOR line (Fig. 5B,C). Quantitation reveals a consistent ratio of mouse to human 28S rRNA in total, cytoplasmic, and polysomal RNA (Supplemental Fig. S8B). Thus, we conclude that although transcription of neo-NORs is inefficient, processing and ribosome assembly around neo-NOR-derived transcripts are highly efficient. The appearance of neo-NOR-derived ribosomes in polysomes provides compelling evidence that neo-NORs organize the formation of fully functional neonucleoli.
Figure 5.
Neo-NOR-derived rRNAs are assembled into polysome-associated ribosomes. (A) Coomassie-stained SDS protein gel of ribosome fractions purified from 3T3, HT1080, and neo-NOR cell lines. Ribosomal proteins are indicated. (B) RT–PCR detects neo-NOR-derived 18S rRNA in purified ribosome (Rib.) and polysome (Poly.) fractions. (C) Northern blots of RNA extracted from purified ribosomes (Rib.) and polysomes (Poly.) of 3T3 (0.5 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines probed with a mouse-specific oligonucleotide indicate that neo-NOR-derived 28S rRNAs are incorporated into polysome-associated ribosomes within neo-NOR lines.
Spatial organization of neonucleoli
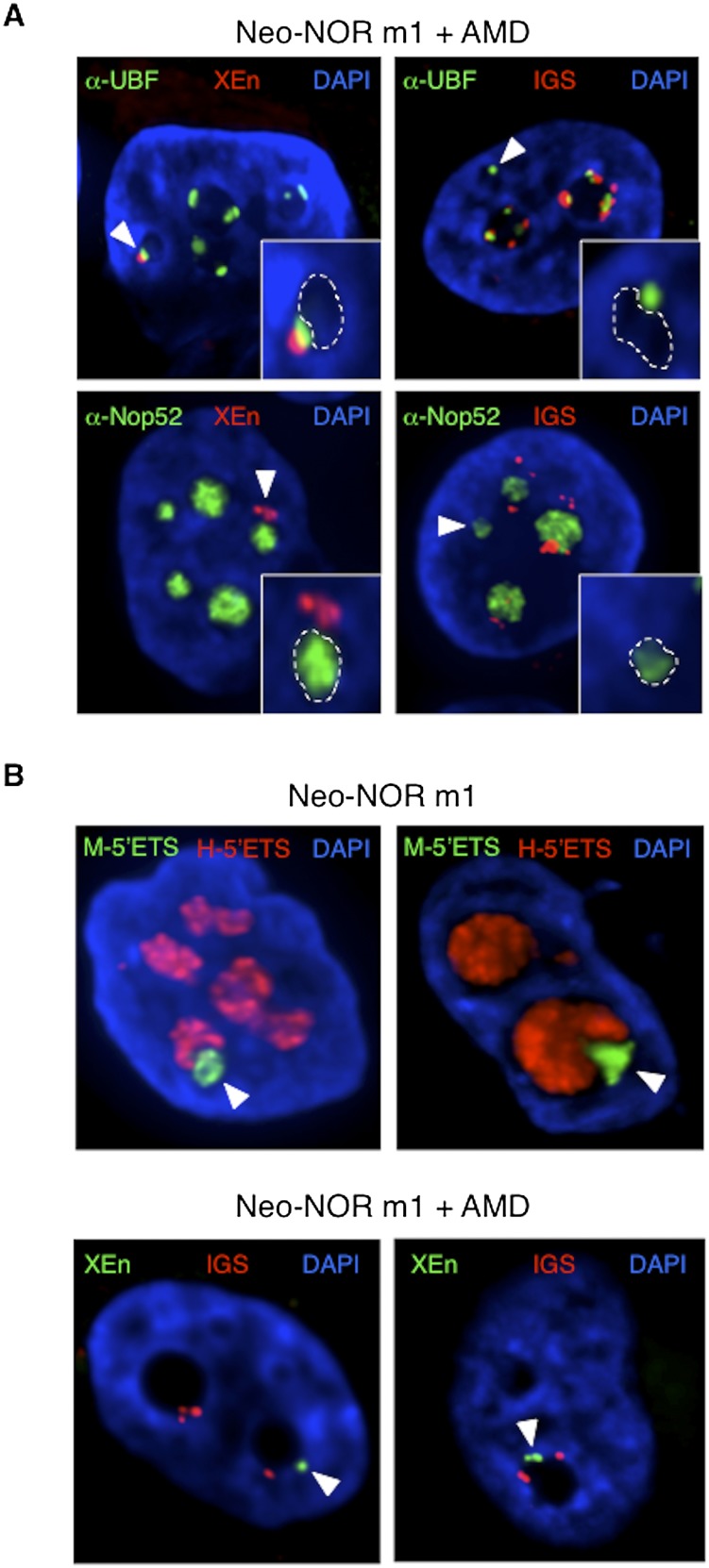
One striking aspect of mature nucleoli is their compartmentalization, yet the mechanism establishing such complex functional organization is unknown. Thus, we investigated whether neo-NORs directed the formation of compartmentalized neonucleoli. Internal FC/DFC regions can be identified by the punctate staining observed with UBF antibodies, while diffuse nucleolar staining observed with antibodies to Nop52 identifies the surrounding GC. However, nucleolar compartmentalization is more dramatically revealed upon inhibition of Pol I transcription with Actinomycin D (AMD) (Hadjiolov 1985). AMD induces the reorganization of nucleolar compartments, with formation of nucleolar caps containing the FC/DFC at the periphery of a low-DAPI-staining nucleolar interior derived from the GC (Supplemental Fig. S11). 3D immuno-FISH of AMD-treated cells from clone m1 identified small nucleoli containing a single UBF-positive, neo-NOR-derived cap (Fig. 6A). The appearance of a low-DAPI-staining and Nop52-positive region associated with these neo-NOR-derived caps (Fig. 6A) demonstrates that neo-NORs direct the de novo biogenesis of appropriately compartmentalized neonucleoli. Hence, formation of compartmentalized nucleoli depends on rDNA activities.
Figure 6.
Compartmentalized neonucleoli occupy distinct territories when integrated into endogenous nucleoli. (A) Combined 3D immuno-FISH performed on AMD-treated clone m1 reveals the presence of appropriately compartmentalized neonucleoli derived entirely from the neo-NOR. The top panels show UBF immunostained FC/DFC caps containing neo-NOR DNA (XEn probe; left panel) and lacking endogenous rDNA (IGS probe; right panel). The bottom panels show neonucleoli with a Nop52 immunostained GC associated with neo-NOR DNA (XEn probe; left panel) but lacking endogenous rDNA (IGS probe; right panel). Neonucleoli are indicated by arrowheads. In the enlarged insets, GCs, identified by low DAPI staining, are highlighted with a dashed line. (B, top panel) RNA-FISH performed on metacentric clone m1 identifies a distinct subnucleolar neo-NOR territory. Neo-NOR and endogenous transcripts are detected with mouse and human 5′ ETS probes, respectively. (Bottom panel) FISH performed on neo-NOR clone m1 indicates that neo-NORs and endogenous NORs are resolved into distinct nucleolar caps associated with the same GC upon AMD treatment. Neo-NORs and endogenous NORs were identified using XEn and IGS probes, respectively. GCs are identified by low DAPI staining. Images presented in the bottom panel correspond to single optical planes selected from deconvolved Z stacks.
In humans, competent NORs coalesce to form between one and three nucleoli (Savino et al. 2001). As these NORs are indistinguishable in sequence from each other, their organization within such nucleoli could not be addressed until now. In metacentric lines, ∼40% of neo-NORs associated with endogenous NORs within large nucleoli, where their transcripts occupied a distinct subnucleolar territory (Fig. 6B; Supplemental Movie S1). As further support of subnucleolar territories, we observed that neo-NOR sequences regress to form their own nucleolar caps, devoid of endogenous rDNA, following AMD treatment of clone m1 cells. Note that ∼40% of neo-NORs remain associated with nucleolar remnants following AMD treatment. These findings are the first evidence that chromosomal territories extend into nucleolar interior-generating NOR territories.
Neo-NORs mimic endogenous NORs upon UBF depletion
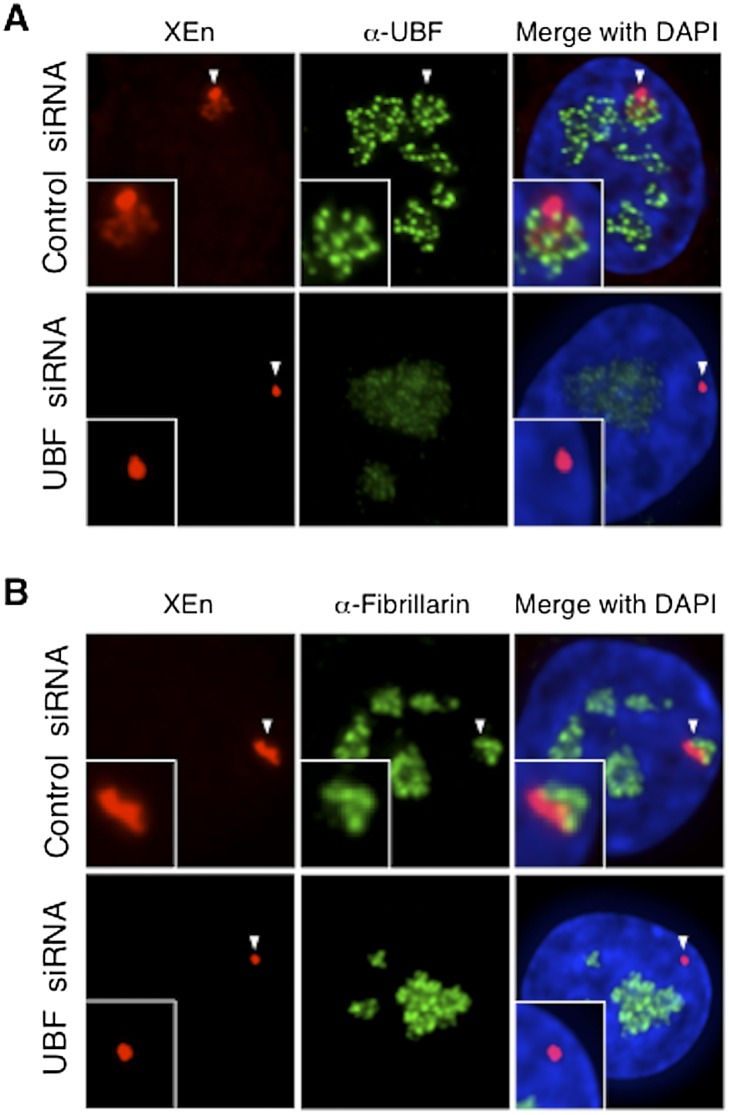
To further establish that neo-NORs mimic endogenous NOR behavior and confirm that UBF loading is a prerequisite for nucleolar formation, we transfected neo-NOR clone m1 with control or UBF siRNAs. In control cells, 3D immuno-FISH indicated that neo-NOR DNA sequences are at least partially diffuse in appearance and associated with both UBF and fibrillarin (Fig. 7; Supplemental Fig. S12). However, upon UBF depletion, ∼98% of neo-NOR DNA sequences appear as single condensed foci, devoid of associated UBF and fibrillarin and clearly separated from nucleoli. In line with expectations, RNA-FISH indicated that the number of clone m1 cells with transcribing neo-NORs drops from 94% to 4% upon UBF depletion (Supplemental Fig. S13). Note that upon UBF depletion, neo-NORs, like pseudo-NORs (Fig. 2), appear to be preferentially silenced over endogenous NORs, presumably reflecting their weaker affinity for UBF. These findings demonstrate that neo-NORs behave in a manner comparable with their endogenous counterpart, with compaction and withdrawal from nucleoli upon UBF depletion (Fig. 1). Thus, neo-NORs have defined the DNA sequence requirements to establish and maintain NOR competency and demonstrated that nucleolar formation in human cells is UBF-dependent.
Figure 7.
Formation of neonucleoli is UBF-dependent. (A) Combined 3D immuno-FISH performed on neo-NOR clone m1 transfected with control or UBF siRNA. Note that the loss of UBF is associated with the compaction of neo-NOR sequences (XEn probe). (B) Combined 3D immuno-FISH performed on neo-NOR clone m1 transfected with control or UBF siRNA reveals that neo-NOR sequences (XEn probe) devoid of associated UBF fail to recruit fibrillarin. Further images from these experiments are shown in Supplemental Figure S11.
Discussion
The relationship between secondary constrictions, NORs, and nucleoli was established in the 1930s (Heitz 1931; McClintock 1934). However, 80 years later, essential questions still remain concerning the DNA sequence requirement and the molecular mechanism involved in the biogenesis and maintenance of human nucleoli. Our synthetic biology experiments now provide for the first time definitive evidence that rDNA sequences—specifically, high-affinity UBF-binding sites coupled with rDNA transcription units—are sufficient to form a NOR competent for nucleolar biogenesis in human cells. In addition, neo-NORs have provided insights into the internal organization of mature nucleoli. Finally, construction of synthetic pseudo-NORs, neo-NORs, and neonucleoli has allowed us to provide the first experimental evidence that nucleolar biogenesis and propagation through an “open” mitosis is a staged UBF-dependent process.
The fact that metacentric neo-NORs can direct the formation of neonucleoli suggests that the DNA sequences surrounding the rDNA arrays on acrocentric p-arms and the perinucleolar heterochromatin shell they contribute to are not required for nucleolar formation. However, perinucleolar heterochromatin and chromosome context are likely to play a critical role in other aspects of nucleolar biology not assessed here. Interestingly, perinucleolar heterochromatin appears to be implicated in maintaining the stability of rDNA arrays in Drosophila (Peng and Karpen 2007) and excluding interfering activities like Pol II transcription in human cells (Gagnon-Kugler et al. 2009). In line with this, we observed significant neo-NOR rearrangements and lower neo-NOR transcription levels than expected. Furthermore, the complex and transcriptionally active chromatin landscape associated with DJ sequences (Floutsakou et al. 2013) suggests that these sequences also play some role in nucleolar biology; for example, regulating the activity status of the linked rDNA array.
The recent identification and characterization of DJ sequences have provided a valuable tool for examining the nuclear location of all NORs in human cells (Floutsakou et al. 2013). Thus, application of the DJ probe has provided the first definitive evidence that large mature nucleoli of human cells contain multiple NORs. Examination of nucleoli containing both neo-NORs and endogenous NORs establishes the existence of NOR territories reminiscent of chromosome territories. These NOR territories could provide another means to ensure rDNA array integrity by protecting NORs against interchromosomal rearrangements. UBF depletion experiments in HT1080 and neo-NOR m1 cell lines (Figs. 1, 7) have clearly established a requirement for UBF loading in nucleolar fusion. The role of transcription is more difficult to assess due to its UBF dependence. However, we do note that neo-NORs exhibit a higher level of association with endogenous nucleoli than pseudo-NORs (Mais et al. 2005).
Extensive UBF binding across endogenous rDNA repeats throughout the cell cycle (O'Sullivan et al. 2002; Mais et al. 2005) implies a key role for UBF in the nucleolar cycle. Pseudo-NORs provided the first supporting evidence for this view (Mais et al. 2005). Here we now show that depletion of UBF leads to the loss of NOR mitotic hallmarks, competency and nucleolar association, thus demonstrating a clear role for UBF in mitotic bookmarking of competent NORs. The realization that UBF is not restricted to vertebrates but present across animal phyla (Grob et al. 2011) suggests that NOR bookmarking by UBF is an evolutionarily ancient phenomenon. However, UBF is not present in plants, where secondary constrictions were first described. We suggest that a related HMG-box protein may substitute for UBF in plants and other non-UBF-containing species to influence the epigenetic state and nuclear position of NORs (Pontvianne et al. 2013). In this regard, it is interesting to point out that the HMG-box protein Hmo1 organizes rDNA chromatin in the yeast S. cerevisiae (Wittner et al. 2011). However, unlike Hmo1 in yeast, UBF is essential in mammals, as indicated by the death of UBF-KD cells cultured with 1 μg/mL Dox (Supplemental Fig. S2) and the early embryonic lethality observed in UBF knockout mice (T Moss, pers. commun.). While Hmo1 and UBF have overlapping roles in rDNA transcription, UBF has additional roles that cannot be complemented by Hmo1 (Albert et al. 2013). This may reflect the fact that in yeast and other lower eukaryotes, bookmarking of rDNA repeats may not be required, as they have a “closed” mitosis and their nucleolus remains intact throughout their cell cycle. Thus, the ability of UBF to bookmark rDNA appears to be a more recently acquired function to ensure nucleolar propagation through “open” mitosis.
UBF depletion from endogenous NORs and neo-NORs has revealed that UBF bookmarking is necessary for nucleolar formation, while pseudo-NORs reveal that it is not sufficient for nucleolar formation. Thus, we can now conclude that propagation of nucleoli through cell division in higher eukaryotes with an “open” mitosis is a staged process. Hence, NORs that were active in the previous interphase are bookmarked by UBF during mitosis, resulting in their characteristic appearance as undercondensed silver-stainable secondary constrictions. This UBF-dependent bookmarking step ensures rapid reactivation of transcription in post-mitotic cells, with pre-rRNA seeding recruitment of processing and assembly factors resulting in nucleolar compartmentalization. While neo-NORs model this entire process, we can now interpret pseudo-NORs as having uncoupled these stages. Pseudo-NORs, comprised entirely of UBF-binding site arrays, recapitulate only the bookmarking stage.
Staging is clearly an efficient strategy for rapid biogenesis of nucleoli in early G1, since it ensures nucleolar propagation through cell division and establishes a temporal order to the reformation of nucleolar compartments. Furthermore, it appears that this strategy has been conserved through evolution with the presence of UBF across animal phyla (Grob et al. 2011) and our observation that neonucleoli form in human cells around mouse pre-rRNAs. Finally, as evidence for mitotic bookmarking of other genes becomes more prevalent (Zaidi et al. 2010), this mechanism will have broader relevance for the reshaping of nuclei following cell division and their functional organization.
Materials and methods
Plasmids
The plasmid pTER+UBFshRNA contains UBF shRNA coding sequence (5′-CCGGAGATCATGAGAGACTATATTTCAAGAGAATATAGTCTCTCATGATCTCCTTTTTGGAAA-3′) cloned into pTER+. The plasmid pNeo-NOR contains a 20.4-kb neo-NOR cassette HpaI restriction fragment. To construct this plasmid, a human rDNA promoter (nucleotides −253/+37) fused to mouse pre-rRNA coding sequences and terminator (nucleotides +120/+13,698, accession no. BK000964) were inserted within XEn blocks present in pXEn8 (Mais et al. 2005).
Cell culture
HT1080 and 3T3 cells were cultured in DMEM with 10% fetal bovine serum (FBS) and antibiotics. HT1080 cells were transfected with the tetracycline repressor-expressing plasmid conferring blasticidin resistance (pcDNA6/TR plasmid, Invitrogen) to produce the HT1080 TetR cell line. The UBF-KD cell line was generated by transfection of HT1080 TetR cells with pTER+UBFshRNA. UBF-KD cells were selected and cultured in medium containing tetracycline-free FBS, 5 μg/mL blasticidin, and 200 μg/mL zeocin. The viability of UBF-KD cells was assessed by seeding 105 cells and counting surviving cells following 1–5 d of culture in the above medium with 2 ng/mL or 1 μg/mL Dox. Pseudo-NOR clone 3D cells (Mais et al. 2005) were grown in medium containing 5 μg/mL blasticidin. Ptk-2 cells were cultured in minimum essential medium (MEM) Eagle (+Earles salts, reduced NaHCO3, without L-glutamine) supplemented with 10% FBS, 200 mM 2% L-glutamine, and 1% MEM nonessential amino acid. Neo-NOR clones were obtained by cotransfection of HT1080 cells with the insert from pNeo-NOR and a plasmid conferring blasticidin resistance in a 200:1 molar ratio. Clones were maintained in medium containing 5 μg/mL blasticidin. AMD was used at 0.1 μg/mL for 2 h.
siRNA-mediated mRNA knockdown
The control siRNA corresponds to a scrambled sequence of the human nucleolar protein RNA helicase II/Guα described in Henning et al. (2003). UBF siRNA was purchased as a Smartpool of duplexes from Dharmacon. siRNA duplexes were transfected at 25 nM using DharmaFECT 1 reagent (Thermo Scientific) according to the manufacturer's instructions 24 h and 72 h following cell seeding. Following this 72-h siRNA treatment, cells were processed for chromosome or cell staining, and the efficiency of UBF depletion was monitored by Western as previously described (Prieto and McStay 2007).
Antibodies
UBF, RPA43, and t-UTP10 antibodies were previously described (Sullivan et al. 2001; Mais et al. 2005; Prieto and McStay 2007). PAF49, nucleolin, and Treacle antibodies were raised in sheep immunized with recombinant proteins. Nap57 and hU3-55k antibodies were provided by T. Meier and N. Watkins. Commercially supplied fibrillarin (mAb 72B9), NPM (mAb 32-5200), and Nop52 (NBP1-85338) antibodies were also used.
Cell and chromosome staining
FISH, 3D immuno-FISH, and antibody staining of fixed cells and chromosomes were performed as previously described (Mais et al. 2005). In RNA and 3D --immuno-RNA-FISH experiments, denatured probe was applied to nondepurinated and nondenatured slides. The probes used for cell and chromosome staining are described in Supplemental Figure S1 and Supplemental Table S1. Imaging was performed using a Photometric Coolsnap HQ camera and a 63× Plan Apochromat Zeiss objective mounted on a Zeiss Axioplan2 imaging microscope. Z-stacks were deconvolved by iterative restoration using Velocity software (PerkinElmer). Extended focus images are shown unless otherwise stated.
Ribosome purification
Cytoplasmic ribosomes were performed as described (Belin et al. 2010). For polysome purifications, cells were scraped in PBS containing 100 μg/mL cycloheximide and 1 mM ribonucleoside vanadyl complexes. Cells were then pelleted by centrifugation at 300g for 5 min at 4°C, resuspended in lysis buffer (0.5% NP-40, 100 μg/mL cycloheximide, 5 mM DTT, 300 mM KCl, 5 mM MgCl2, 10 mM HEPES, 10 mM ribonucleoside vanadyl complexes, complete), incubated for 10 min on ice, and passaged through a 25-gauge needle. Lysed cells were centrifuged at 12,000g for 10 min at 4°C. The resulting cytoplasmic supernatants were recovered and loaded onto rate zonal 15%–45% sucrose gradients. Gradient sedimentation was performed by ultracentrifugation at 43,500 rpm for 50 min at 4°C using a Beckman SW50/55 rotor. Fractionation was carried out using a 19-gauge needle piercing the bottom of the gradient.
RNA analysis
RNA was prepared using the NucleoSpin RNA II kit (Macherey-Nagel). S1 nuclease protection assays were performed as previously described (Prieto and McStay 2007). For Northerns, RNA samples were eletrophoresed in agarose–formaldehyde gels and then transferred onto Hybond-N+ membrane (GE Healthcare) and UV cross-linked. Probing with Dig-labeled probes and detection of hybridization signals were performed as per the manufacturer's instructions (Roche). The oligonucleotide probes used for S1 protection assays and Northern blotting are described in Supplemental Table S1.
Acknowledgments
We are grateful to M. van Sluis, N. Watkins, and T. Meier for providing antibodies; M. Cremer and T. Cremer for chromosome paints; D. Hernandez-Verdun for Ptk-2 cells; M. Chenut for characterization of Ptk UBF; and C. Duffy and T. Lechertier for critical reading of the manuscript. B.M. acknowledges Science Foundation Ireland (PI grant 07/IN.1/B924) for funding work in his laboratory. A.G. was the recipient of an Empower post-doctoral fellowship from Irish Research Council for Science, Engineering, and Technology (IRCSET).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.234591.113.
Freely available online through the Genes & Development Open Access option.
References
- Albert B, Colleran C, Leger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O 2013. Structure–function analysis of Hmo1 unveils an ancestral organization of HMG-box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res 41: 10135–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin S, Hacot S, Daudignon L, Therizols G, Pourpe S, Mertani HC, Rosa-Calatrava M, Diaz JJ 2010. Purification of ribosomes from human cell lines. Curr Protoc Cell Biol 49: 3.40.1–3.40.11 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Cazaux B, Catalan J 2012. Chromosomal dynamics of nucleolar organizer regions (NORs) in the house mouse: Micro-evolutionary insights. Heredity 108: 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A 2005. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res 15: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, Aragon L 2009. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458: 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N, Kass S, Sollner WB 1987. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc Natl Acad Sci 84: 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T 2010. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Financsek I, Mizumoto K, Mishima Y, Muramatsu M 1982. Human ribosomal RNA gene: Nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci 79: 3092–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B 2013. The shared genomic architecture of human nucleolar organizer regions. Genome Res 23: 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T 2009. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol Cell 35: 414–425 [DOI] [PubMed] [Google Scholar]
- Goodpasture C, Bloom SE 1975. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma 53: 37–50 [DOI] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B 2011. UBF an essential player in maintenance of active NORs and nucleolar formation. In The nucleolus (ed. Olson MOJ), pp. 83–103. Springer, New York [Google Scholar]
- Hadjiolov AA. 1985. The nucleolus and ribosome biogenesis. Springer, New York. [Google Scholar]
- Heitz E 1931. Die ursache der gesetzmassigen zahl, lage, form und grosse pflanzlicher nukleolen. Planta 12: 775–844 [Google Scholar]
- Heix J, Grummt I 1995. Species specificity of transcription by RNA polymerase I. Curr Opin Genet Dev 5: 652–656 [DOI] [PubMed] [Google Scholar]
- Heliot L, Kaplan H, Lucas L, Klein C, Beorchia A, Doco-Fenzy M, Menager M, Thiry M, O'Donohue MF, Ploton D 1997. Electron tomography of metaphase nucleolar organizer regions: Evidence for a twisted-loop organization. Mol Biol Cell 8: 2199–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC 1972. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci 69: 3394–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning D, So RB, Jin R, Lau LF, Valdez BC 2003. Silencing of RNA helicase II/Guα inhibits mammalian ribosomal RNA production. J Biol Chem 278: 52307–52314 [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD 1988. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev 2: 1745–1763 [DOI] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B 2005. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1934. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Zeit ZellforschMikAnat 21: 294–328 [Google Scholar]
- McStay B, Grummt I 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Mullineux ST, Lafontaine DL 2012. Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie 94: 1521–1532 [DOI] [PubMed] [Google Scholar]
- Oakes ML, Johzuka K, Vu L, Eliason K, Nomura M 2006. Expression of rRNA genes and nucleolus formation at ectopic chromosomal sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol 26: 6223–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 2010. The nucleolus. Cold Spring Harb Perspect Biol 3: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OM, Tucker S, Mokros P, Muchova V, Fajkus J, et al. 2013. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 27: 1545–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D 2006. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334 [DOI] [PubMed] [Google Scholar]
- Roussel P, Andre C, Masson C, Geraud G, Hernandez VD 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci 104: 327–337 [DOI] [PubMed] [Google Scholar]
- Savino TM, Gebrane-Younes J, De Mey J, Sibarita JB, Hernandez-Verdun D 2001. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol 153: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez-Verdun D 2000. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J Cell Biol 148: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Hernandez-Verdun D, Roussel P 2002. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D 2008. Nucleolus: The fascinating nuclear body. Histochem Cell Biol 129: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ 2008. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B 2001. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J 20: 2867–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Lafontaine DL 2005. Birth of a nucleolus: The evolution of nucleolar compartments. Trends Cell Biol 15: 194–199 [DOI] [PubMed] [Google Scholar]
- Wittner M, Hamperl S, Stockl U, Seufert W, Tschochner H, Milkereit P, Griesenbeck J 2011. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell 145: 543–554 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS 2010. Mitotic bookmarking of genes: A novel dimension to epigenetic control. Nat Rev Genet 11: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]