Heterochromatin imparts regional, promoter-independent repression of genes and is epigenetically heritable. However, how silencing achieves regional repression is not well understood. Here, Thurtle and Rine probe the chromatin architecture at the silent mating type loci and telomeres at high resolution in budding yeast. The authors find evidence of a higher-order structure for silenced chromatin that is likely imposed by Sir proteins. This study illustrates the importance of the topology of heterochromatic regions to regional repression.
Keywords: Sir complex, ChIP-seq, heterochromatin, H4K16-acetyl, histones
Abstract
Heterochromatin imparts regional, promoter-independent repression of genes and is epigenetically heritable. Understanding how silencing achieves this regional repression is a fundamental problem in genetics and development. Current models of yeast silencing posit that Sir proteins, recruited by transcription factors bound to the silencers, spread throughout the silenced region. To test this model directly at high resolution, we probed the silenced chromatin architecture by chromatin immunoprecipitation (ChIP) followed by next-generation sequencing (ChIP-seq) of Sir proteins, histones, and a key histone modification, H4K16-acetyl. These analyses revealed that Sir proteins are strikingly concentrated at and immediately adjacent to the silencers, with lower levels of enrichment over the promoters at HML and HMR, the critical targets for transcriptional repression. The telomeres also showed discrete peaks of Sir enrichment yet a continuous domain of hypoacetylated histone H4K16. Surprisingly, ChIP-seq of cross-linked chromatin revealed a distribution of nucleosomes at silenced loci that was similar to Sir proteins, whereas native nucleosome maps showed a regular distribution throughout silenced loci, indicating that cross-linking captured a specialized chromatin organization imposed by Sir proteins. This specialized chromatin architecture observed in yeast informs the importance of a steric contribution to regional repression in other organisms.
Eukaryotic cells exhibit many different scales of genetic regulation. The most thoroughly understood examples include mechanisms of promoter-dependent activation and repression of individual genes. At the other extreme, entire chromosomes are subject to a common regulatory influence, such as mechanisms of dosage compensation of sex chromosomes (Meyer 2010; Conrad and Akhtar 2012; Jeon et al. 2012). At intermediate scales, heterochromatin facilitates regional, promoter-independent repression either constitutively, as in the pericentric regions of chromosomes in many species, or in a facultative fashion, such as repression of Hox genes by Polycomb group proteins in Drosophila (Ringrose and Paro 2004).
There are four key features of heterochromatic repression essential to its role in genetic regulation: (1) its nucleation, (2) the mechanism of apparent spreading, (3) the mechanism of transcriptional repression, and (4) the mechanism of its epigenetic inheritance. Understanding the first two features of heterochromatin, its ability to nucleate from a specific site and its ability to then spread to create larger areas of regional repression, is critical to understanding the other two features. Formally, the apparent spreading of repression could occur through two classes of mechanisms: (1) physical spreading of chromatin-associating proteins or histone marks from the nucleation site to the distal sites of repression or (2) “action at a distance,” where the nucleation site interacts with the distal sites of repression through long-range and potentially stochastic transient interactions. The function of heterochromatin in many organisms depends on an interplay between noncoding RNA, enzymes that modify nucleosomes, and proteins that recognize and bind those modifications (Beisel and Paro 2011). In Saccharomyces, the silenced chromatin at HML, HMR (the silenced mating type loci), and telomeres bear all of the hallmarks of classically defined heterochromatin yet achieve the four features of heterochromatin with no contribution from noncoding RNA (Grunstein and Gasser 2013). Hence, this example offers the opportunity to address fundamental mechanisms with one less layer of complexity.
The silent mating loci in Saccharomyces cerevisiae harbor unexpressed copies of the mating type alleles that contain the same promoters as the actively transcribed alleles of the MAT locus (Hicks et al. 1979; Rine et al. 1979). Repression at the silent mating loci is due to the nucleation and spreading of the Sir complex. The nucleation of silenced chromatin formation at HML and HMR is achieved by the E and I silencers, which are specific DNA sequences that flank both loci and contain binding sites for the widely acting transcriptional activators Abf1 and/or Rap1 as well as the origin recognition complex (ORC), the initiation factor for DNA replication common to all eukaryotes (Brand et al. 1985; Rusche et al. 2003). These nucleation sites recruit the Sir proteins, which, through modification and subsequent binding to histone tails of nucleosomes, constitute silenced chromatin. The mechanism of silencing is at least in part due to steric occlusion of the transcriptional machinery from their recognition sequences in silenced chromatin (Singh and Klar 1992; Loo and Rine 1994; Chen and Widom 2005; Gao and Gross 2008).
Sir1 is recruited to the silencers through direct binding to ORC (Triolo and Sternglanz 1996; Zhang et al. 2002; Hou 2005), facilitating the subsequent recruitment of Sir4–Sir2 dimers and Sir3, which also bind to Rap1 and Abf1 (Ghidelli et al. 2001; Hoppe et al. 2002; Chen et al. 2011). Sir2, the founding member of the Sirtuin family conserved throughout eukaryotes, is an NAD-dependent histone deacetylase that deacetylates the N-terminal tail of histone H4 on Lys16 (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). The deacetylase activity of Sir2 is necessary for silencing (Tanny et al. 1999; Imai et al. 2000). Chromatin immunoprecipitation (ChIP) studies have shown Sir2, Sir3, and Sir4 to be enriched throughout the silenced domain at HML and HMR (Hoppe et al. 2002; Rusche et al. 2002). Sir3 has a higher affinity for deacetylated tails of histone H4 than for acetylated tails (Johnson et al. 2009; Martino et al. 2009). These observations and others have led to a widely adopted model for how Sir proteins appear to spread throughout a silenced domain: Following nucleation of a Sir protein complex at the silencers, Sir2 deacetylates histone tails of adjacent nucleosomes, creating new high-affinity binding sites for additional Sir protein complexes. Iterative rounds of deacetylation followed by recruitment of additional Sir protein complexes leads to spreading of repression across the silenced locus. Boundary elements, best defined at HMR, restrict silencing to one side of the boundary, preventing aberrant Sir spreading into euchromatin (Donze and Kamakaka 2001; Oki and Kamakaka 2005).
Sir2, Sir3, and Sir4 also mediate telomere position effects in Saccharomyces, causing genes positioned near telomeres to be stochastically expressed or repressed (Gottschling et al. 1990). Telomeres in S. cerevisiae consist of tandem repeats of sequences produced by telomerase and other telomere-associated sequences, referred to as X elements or Y′ elements (Louis 1995). Studies have reported considerable variation in the silencing capacity among telomeres. At natural telomeres, silencing is more pronounced directly next to the X elements, diminishing substantially within a few kilobase pairs or less (Fourel et al. 1999; Pryde and Louis 1999; Takahashi et al. 2011). In contrast, studies of silencing at synthetic telomeres, especially with overexpression of Sir3, have indicated a more extensive domain of silencing (Gottschling et al. 1990; Hecht et al. 1996; Radman-Livaja et al. 2011).
Classic chromatin mapping studies at HML and HMR have identified well-positioned nucleosomes at the silent mating type loci, with the silencers being nucleosome-free, reflecting their occupancy by ORC, Abf1, and/or Rap1 (Weiss and Simpson 1998; Ravindra et al. 1999; Henikoff et al. 2011). In contrast to the regular distribution of nucleosomes in silenced chromatin, limited higher-resolution ChIP followed by next-generation sequencing (referred to throughout this study as ChIP-seq) studies revealed less homogeneous distributions of individual Sir proteins than suggested by prior methods and by the model for Sir protein spreading (Zill et al. 2010; Radman-Livaja et al. 2011; Li et al. 2013). To provide a definitive assessment of the silenced chromatin architecture as revealed by Sir protein distributions at HML, HMR, and telomeres and across the genome, we performed high-resolution ChIP-seq analysis of Sir2, Sir3, Sir4, and a catalytically inactive Sir2 and an evaluation of the distribution of nucleosomes and acetylated H4K16. At this high resolution, we found that the Sir proteins had a strikingly heterogeneous and highly reproducible pattern of enrichment at the HML and HMR loci, with the lowest apparent level of enrichment paradoxically over the promoter regions that are the targets of the silencing mechanism. In contrast, H4K16 hypoacetylation was homogeneous across the silenced loci. An integrated suite of chromatin analyses revealed a molecular topography of Sir proteins that suggested the existence of a chromatin superstructure at both HML and HMR, which provided insight into the steric dimension of the mechanism of silencing.
Results
Sir proteins showed heterogeneous distribution at HML and HMR
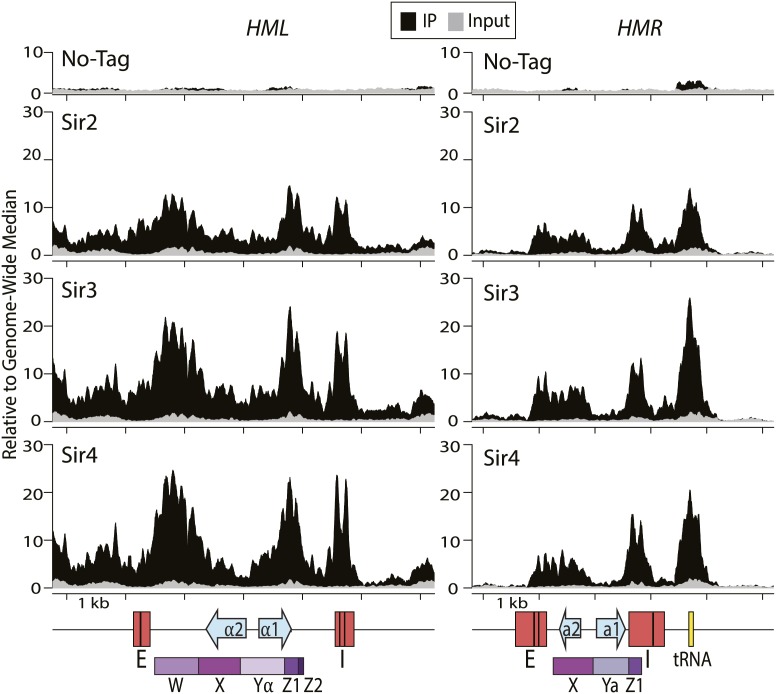
To characterize the chromatin positions occupied by the Sir proteins more precisely than previously possible, we performed ChIP-seq of myc-tagged Sir2, Sir3, and Sir4. Chromatin that was not immunoprecipitated, the input chromatin, was also sequenced, providing a baseline reference for the immunoprecipitated sample. Based on previous ChIP-qPCR and ChIP–chip results, we expected a homogenous distribution of Sir proteins across HML and HMR (Hoppe et al. 2002; Rusche et al. 2002). However the picture at both loci was more complex than expected. As anticipated, Sir proteins were enriched at the E and the I silencers well above the genome-wide median (Fig. 1). Enrichment at these positions presumably reflects Sir2, Sir3, and Sir4 association with Rap1, Abf1, ORC, and Sir1. The most striking enrichment for all three Sir proteins was immediately internal to each of the silencers, corresponding to the well-positioned nucleosomes adjacent to the silencers at HMR and HML (Weiss and Simpson 1998; Ravindra et al. 1999). However, the pattern of Sir protein enrichment appeared to be more heterogeneous than expected throughout the rest of the loci (Fig. 1). The extent of Sir protein enrichment at the promoter regions of HML and HMR and at the a1 ORF was somewhat higher than the signal from the input chromatin yet was much lower than the enrichment at the silencers and at positions immediately adjacent to the silencers. This striking quantitative difference was surprising given that the promoters of HML and HMR are the critical targets of silencing.
Figure 1.
Sir protein ChIP-seq of sonicated chromatin showed heterogeneous distribution at HML and HMR. ChIP-seq of formaldehyde cross-linked chromatin sheared by sonication was performed on a no-tag control sample and on strains with Sir2, Sir3, or Sir4 tagged with the 13xmyc epitope. The left panel shows 6 kb centered on HML (Chr. III coordinates 10,000–16,000), and the right panel shows 5 kb around HMR (Chr. III coordinates 291,793–296,793). Tick marks denote 1-kb increments. Salient features are shown below for each locus: The red rectangles denote the silencers, as defined in the Saccharomyces Genome Database; black bars within the red rectangles indicate the binding sites for ORC, Rap1, or Abf1; blue arrows denote the ORFs; and the yellow box denotes the position of the tRNA-Thr. The purple boxes show the W, X, Y, and Z regions (as defined in Nasmyth and Tatchell 1980). The distribution of sequence reads for the immunoprecipitate (IP) for each sample is shown in black, and the sequence reads from the input chromatin for that sample are in gray. The Y-axis units are sequencing reads normalized to the genome-wide median for that sample. To facilitate accurate and unambiguous mapping of the sequence reads to both HML and HMR, the MAT locus was deleted. Still, some reads could not be uniquely mapped because the X region is identical between HML and HMR. However, there were reads within the X region whose paired-end read mapped uniquely to either HML or HMR, indicating that Sir proteins were at both X regions.
In addition to the peaks associated with the silencers and the adjacent nucleosomes, we observed a third large peak of Sir protein enrichment at both HML and HMR. At HML, this third peak was in the middle of the locus, centered approximately over the Yα and Z1 border, corresponding to the HO endonuclease recognition sequence cleaved by HO to facilitate mating type switching (Strathern et al. 1982). This peak of Sir protein enrichment was consistent with genetic evidence of a protosilencer element that stabilizes Sir protein association within HML independently of the E and I silencers, possibly involving interactions between Rap1 bound to the promoter and Sir proteins (Cheng and Gartenberg 2000). At HMR, the third peak of enrichment of Sir proteins was immediately to the right of the locus and corresponded to the tRNA-Thr gene that serves as a boundary element for the silenced domain at HMR (Donze and Kamakaka 2001; Li et al. 2013). A slight increase in nonspecific binding of the tRNA gene to the anti-myc beads was evident in the immunoprecipitated sample from cells lacking any myc-tagged proteins (Fig. 1, top right panel). However, this enrichment was minor compared with the Sir protein enrichment and is thoroughly documented and discussed in a report on nonspecific ChIP-seq enrichment (Teytelman et al. 2013). The left side of HMR also exhibited a discrete boundary for enrichment of all three Sir proteins directly to the left of the ORC-, Abf1-, and Rap1-binding sites in the E silencer (Fig. 1, right panels). This discrete decrease in Sir protein enrichment was in contrast to reporter assays showing the silenced domain to extend ∼500 base pairs (bp) to the left of HMR-E (Donze et al. 1999).
Consistent with genetic studies of HML and unlike the discrete boundary for Sir protein enrichment at HMR, enrichment at HML was continuous for >15 kb from the left telomere of chromosome III to HML (Supplemental Fig. S1). Curiously, the extent of enrichment oscillated with a period of ∼1 kb. This extended domain of Sir protein enrichment corresponded to the extended domain of telomeric silencing between the telomere and HML and was also consistent with lower-resolution ChIP–chip studies of Sir proteins (Hecht et al. 1996; Lieb et al. 2001). Additionally, to the right of HML-I, Sir protein enrichment was evident for ∼1 kb, although the level of enrichment was lower than within or to the left of HML (Fig. 1). Further to the right, there was no detectable enrichment. Together, these data supported earlier suggestions of a lack of a clear boundary for silencing on the telomere-proximal side of HML (Bi et al. 1999) and the lack of a distinct boundary element on the centromere-proximal side of HML (Fig. 1; Supplemental Fig. S1) and established distinct structural differences between the silenced chromatin in and near HML and the more discrete domain at HMR.
The genome-wide reproducibility between the immunoprecipitated samples from a biological replicate of each of the Sir proteins showed high correlation between each biological replicate: 0.920, 0.989, and 0.964 for Sir2, Sir3, and Sir4, respectively. Additionally, the pattern from one Sir protein was nearly indistinguishable from that of the others, emphasizing the robustness of this surprisingly complex topography (Fig. 1). Thus, the features of chromatin responsible for this pattern were robust and highly reproducible.
Chromatin structures can impact the efficiency of fragmentation by sonication, especially of silenced chromatin, as we discovered previously and as can be seen in the input chromatin distribution (Teytelman et al. 2009). To provide an independent assessment of this unexpected distribution of Sir proteins and as a further control for fragmentation bias, we employed a different method of chromatin fragmentation to characterize the distribution of Sir proteins. Specifically, Sir protein chromatin association was evaluated through ChIP-seq of Sir proteins with cross-linked chromatin that was digested to mononucleosomes with micrococcal nuclease (MNase) rather than sonicated, as above. Additionally, to ensure that the extended cross-linking time often used to capture chromatin-associating proteins that do not directly bind DNA did not influence the topology observed, we used a more limited, 15-min cross-linking time for the following MNase preparations.
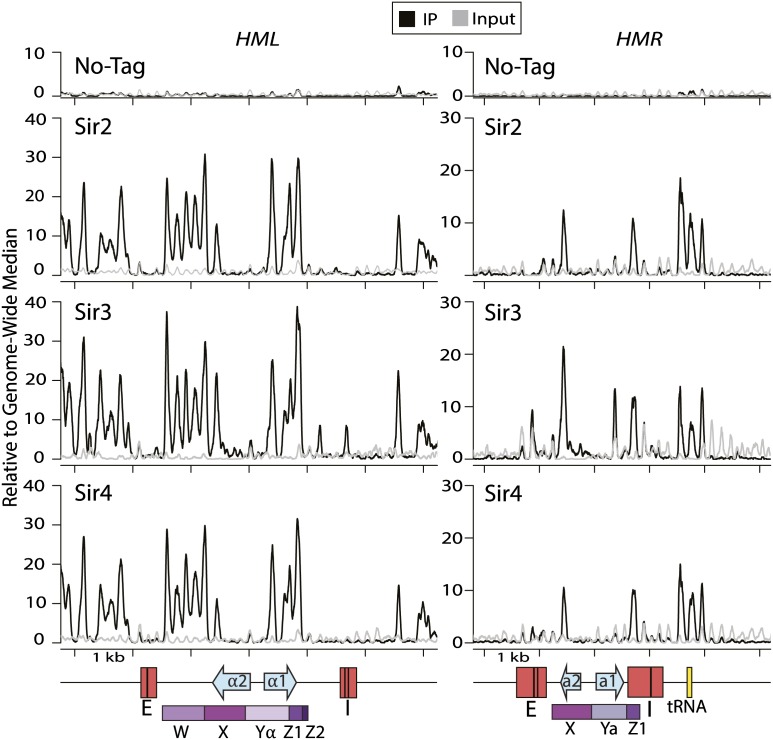
At the level of mononucleosomes, Sir proteins appeared to be primarily associated with the nucleosomes adjacent to the silencers (Fig. 2). The extent of enrichment of Sir proteins at the silencers in the MNase-treated chromatin was somewhat lower than their enrichment as assayed with sonicated chromatin. At this higher resolution, Sir3 and Sir4 at HMR-E showed more association with the silencer than did Sir2, likely reflecting the directness of the interaction between Sir3, Rap1, and Abf1 (Chen et al. 2011) and between Sir4, Sir1, and Rap1 (Triolo and Sternglanz 1996). At mononucleosome resolution, the peak of enrichment at HML internal to the silencers and adjacent nucleosomes was resolved into distinct features: a single peak and a double peak (Fig. 2, left panels). The double peak over the Yα–Z1 border is consistent with the previously described tightly packed nucleosome pair over the HO recognition site (Weiss and Simpson 1998). The single peak possibly reflected protein association with the internal protosilencer element that stabilizes Sir protein associations within HML (Cheng and Gartenberg 2000).
Figure 2.
Sir protein ChIP-seq at HML and HMR on cross-linked chromatin digested with MNase. The left panel shows 6 kb centered on HML (Chr. III coordinates 10,000–16,000), and the right panel shows 5 kb around HMR (Chr. III coordinates 291,793–296,793). Tick marks denote 1-kb increments. Salient features are as in Figure 1. The distribution of sequence reads for the immunoprecipitate (IP) for each sample is shown as a black line, and the sequence reads from the input chromatin for that sample are shown as a gray line. To better define the peaks to nucleosome resolution, trimming of the reads was performed with nucleR (Flores and Orozco 2011).
The third peak at HMR at the tRNA gene resolved into three peaks of Sir protein association. Two peaks corresponded to nucleosomes on either side of the tRNA gene flanking a third distinct peak of enrichment directly over the tRNA gene, which is nucleosome-free (Oki and Kamakaka 2005). There, the Sir proteins formed stable associations with nonnucleosomal factors that bound the tRNA gene, as discussed below. The paradoxical paucity of Sir enrichment over the promoters at both HML and HMR was even more evident than in the sonicated chromatin data (Fig. 2). (The MNase ChIP no-tag sample also showed low nonspecific binding to the myc beads, as did the sonication no-tag sample.) Thus, two independent chromatin preparation methods clearly demonstrated that the unusual pattern of enrichment of Sir proteins in silenced chromatin was a fundamental property of silenced chromatin rather than an artifact of either method of analysis. Importantly, the greater resolution of the mononucleosome analysis highlighted what appeared to be positions at which the nucleosomes lacked Sir proteins. As discussed below, this appearance resulted from a property of a chromatin superstructure in the domain of silenced chromatin.
H4K16 was hypoacetylated across silent mating type loci
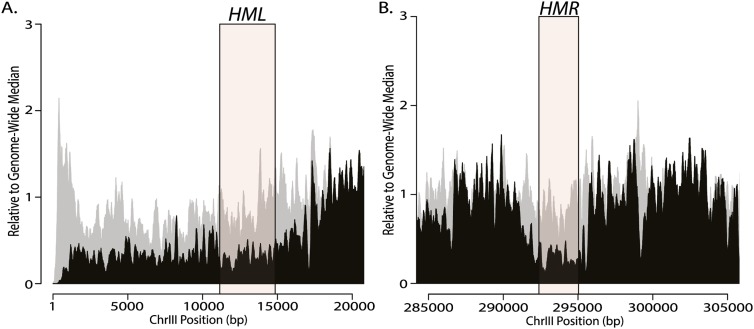
Sir2, an NAD-dependent histone deacetylase, acts on lysines on the N-terminal tails of histones H3 and H4, creating high-affinity binding sites for Sir3 and Sir4 (Grunstein and Gasser 2013). This hypoacetylation, especially of H4K16-acetyl, is a key feature of silenced chromatin in yeast (Johnson et al. 1990; Park and Szostak 1990). Thus, we tested whether the distribution of hypoacetylated H4K16 was homogeneous across silenced loci or exhibited the uneven distribution of the Sir2 deacetylase. ChIP-seq analysis of the distribution of H4K16 acetylation revealed that H4K16 was uniformly hypoacetylated across HML and HMR (Fig. 3). Additionally, H4K16 hypoacetylation was apparent for the entire left end of Chromosome III, from the telomere to HML (Fig. 3A). Thus, although Sir2 exhibited a complex pattern of enrichment at positions across HML and HMR, the consequence of its catalytic activity, H4K16 hypoacetylation, was more uniformly distributed across these loci than were the apparent distributions of the Sir proteins themselves (Fig. 3B).
Figure 3.
H4K16 was hypoacetylated throughout HML and at HMR. ChIP-seq of sonicated sheared chromatin against H4K16-acetyl from the left telomere of Chromosome III through HML (A) and 20 kb centered around HMR (B). (A) HML from the E to the I silencer (chromosomal coordinates 11,146–14,849) is shown as a shaded pink box. (B) HMR from the E to the I silencers (chromosomal coordinates 292,388–295,034) is shown as a shaded pink box. The X-axis represents chromosomal position in 5-kb increments. The distribution of sequence reads for the immunoprecipitate for each sample is shown in black, and the sequence reads from the input chromatin for that sample are in gray.
Sir proteins showed discrete enrichment at natural telomeres
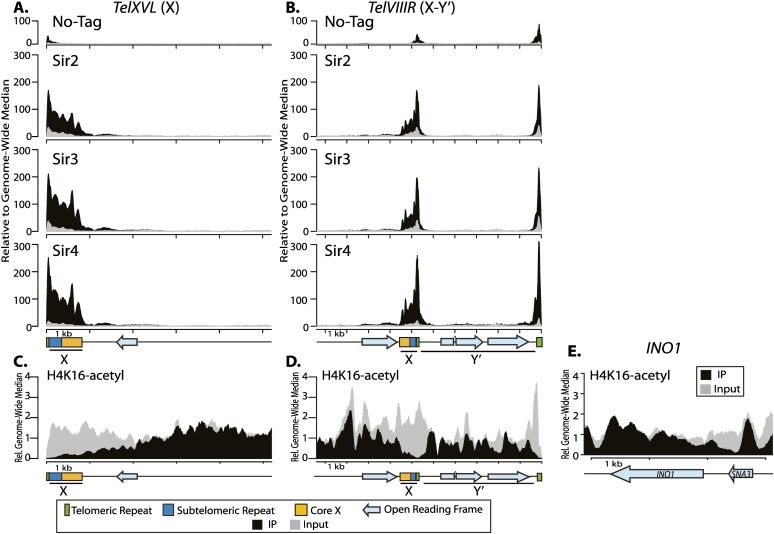
Given the surprising discrepancy between the heterogeneous Sir protein distribution and the larger, more homogeneous H4K16 hypoacetylation, we determined whether this difference applied to the other major class of Sir silenced loci: the chromatin of telomeres. S. cerevisiae has telomeres of two structural classes: those containing just an X element (X), and those with both an X element and a Y′ element (X–Y′). The X element contains the core X, where ORC and Abf1 bind, as well as the subtelomeric repeat sequence. Y′ elements contain an ORF that encodes a protein with sequence similarity with helicases. At telomeres that contain only an X element, the X element is adjacent to the telomerase-generated telomeric repeats, where Rap1 binds. At telomeres that contain both a Y′ element and X element, the X element is immediately centromere-proximal to the Y′ element, which is adjacent to the telomeric repeats (Fig. 4; Louis 1995). We examined enrichment of all three Sir proteins at an X-element telomere (TELXVL) and an X–Y′ telomere (TELVIIIR). Sir protein enrichment was in discrete peaks, similar to the large peaks at the silencers at HML and HMR, rather than a continuous gradient of Sir protein association expected from the studies of telomere position effects with the synthetic telomeres (Gottschling et al. 1990). At the X-element telomere, enrichment of all three Sir proteins overlapped and was coincident with both the Rap1-binding site array in the telomeric repeats and the ORC- and Abf1-binding sites within the core X (Fig. 4A). These binding patterns were consistent with the reported silencing capacity of natural telomeres, which is primarily evident immediately adjacent to the X element (Fourel et al. 1999; Pryde and Louis 1999). Similar Sir protein distributions were also seen at the Y′ telomere, where enrichment was concentrated at two discrete regions: at the sites of the telomeric repeats and at the X element. No Sir protein enrichment was observed within the Y′ region (Fig. 4A). These enrichment patterns were consistent with the URA3 reporter gene not being silenced when inserted within the Y′ element (Pryde and Louis 1999) and with previous studies that found little Sir protein enrichment within the Y′ element (Zhu and Gustafsson 2009; Zill et al. 2010).
Figure 4.
Sir protein enrichment at telomeres was discrete. (A) Enrichment for each of the Sir proteins in sonicated ChIP-seq samples at an X-element telomere (5-kb TELXVL) (A) and a X–Y′ telomere (10 kb of TELVIIIR) (B). C and D show ChIP-seq of cross-linked and sonicated chromatin of H4K16-acetyl for TELXVL and TELVIIIR, respectively. Salient features are shown below the X-axis, with telomeric repeats in green, subtelomeric repeats in dark blue, and the core X in orange. ORFs are blue arrows, and the X and Y′ elements are underlined. (E) H4K16-acetyl ChIP-seq data for INO1, where the 3′ end of the ORF has previously been shown to be acetylated (Suka et al. 2002). The X-axis tick marks represent 1 kb on all graphs. The immunoprecipitate (IP) samples are in black, and the input samples are gray. The Y-axis is reported at reads relative to the genome-wide median.
Next, we compared the distribution of H4K16-acetyl, a mark imparted by the histone acetyltransferase Sas2, with the restricted Sir protein enrichment at the natural telomeres (Suka et al. 2001; Kimura et al. 2002). Interestingly, similar to the theme at the silenced mating type loci, the region of H4K16 hypoacetylation was much larger than the region of Sir protein enrichment (Fig. 4C,D). At the X-element telomere, H4K16 was hypoacetylated for ∼3 kb inward from the end of the chromosome, but Sir protein enrichment was limited to 1 kb from the end (Fig. 4B). This extended domain of H4K16 hypoacetylation was even more apparent at X–Y′ telomeres with hypoacetylation extending throughout the Y′ element, although without Sir protein enrichment in this region (Fig. 4D). The nucleosome distribution at telomeres is needed to interpret the H4K16-acetyl signal. For example, telomeric repeats and the core X showed extremely low coverage in the H4K16-acetyl immunoprecipitate not because the nucleosomes there were hypoacetylated but rather because these areas are nucleosome-free (Loney et al. 2009; Zhu and Gustafsson 2009). However, the amount of H4K16-acetyl immunoprecipitated signal across the Y′ element was still below the genome-wide average for this sample and less than the input signal for this region, indicating that it was hypoacetylated. Additionally, a well-studied region previously reported to be acetylated at H4K16, the 3′ end of the INO1 gene (Suka et al. 2002), showed clear H4K16-acetyl enrichment in the immunoprecipitated chromatin (Fig. 4E), as expected. The region of H4K16 hypoacetylation was much larger than the region of Sir protein enrichment. Therefore, models invoking an antagonism of Sir2 function, such as a positioned H4K16 acetylase near telomeres, could not account for the discrete enrichment of Sir proteins at telomeres.
Restricted Sir3 and Sir4 enrichment in cells lacking Sir2 catalytic activity
In all previous models of Sir-based silencing, sequential deacetylation of histone tails was envisioned to create new binding sites for additional Sir protein complexes (Grunstein and Gasser 2013). Such models predict that in the absence of Sir2's catalytic activity, the distribution of Sir proteins should be restricted to the sites of initial recruitment. Previous lower-resolution studies established that in the sir2N345A mutant, Sir proteins nucleate at the silencers but do not spread within the locus (Rusche et al. 2002). To distinguish recruitment sites from sites of secondary occupation, we performed ChIP-seq on myc-tagged Sir3 and Sir4 in a sir2N345A mutant, which has an amino acid substitution in the active site of Sir2, rendering it catalytically inactive but structurally intact (Imai et al. 2000).
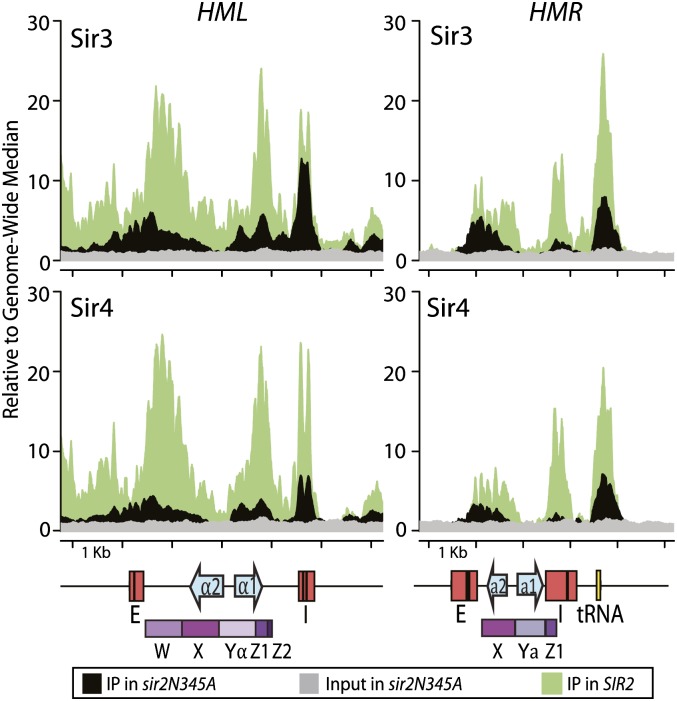
Enrichment of both Sir3 and Sir4 at HML and HMR in cells lacking Sir2 catalytic activity was overall reduced. However, enrichment was evident at three of the four silencers, confirming that these three silencers could be considered nucleation sites for the silenced domain (Fig. 5). Sir3 and Sir4 enrichment was almost completely absent from HMR-I in the absence of Sir2 function. The absence of a signal at HMR-I was consistent with genetic observations that HMR-I is neither necessary nor sufficient for silencing (Rivier et al. 1999) presumably because, as shown here, HMR-I is unable to nucleate Sir proteins. The widespread reduction in Sir3 and Sir4 enrichment at internal positions and apparent absence from others implied that Sir2's catalytic activity was necessary for either interactions between Sir3 and Sir4 with nucleosomes at some positions within these loci or stabilizing these interactions. Interestingly, Sir2's catalytic activity was not necessary for establishing the Sir3 and Sir4 association at the internal region near the HO recognition sequence and protosilencer at HML or at the tRNA gene flanking HMR (Fig. 5). There was also clear evidence, albeit at slightly lower levels in the sir2N345A mutant, for some Sir3 and Sir4 occupancy of nucleosomes internal to the silencers at HML and HMR. These data suggested that deacetylated H4K16 was not an absolute requirement for Sir protein enrichment on nucleosomes at these positions.
Figure 5.
Sir3 and Sir4 enrichment in a strain lacking Sir2 catalytic activity revealed nucleation sites. 13xmyc-tagged Sir3 (top) and Sir4 (bottom) enrichment in a sir2N345A mutant shown at HML (left) and HMR (right). Salient features in the genome are shown below the X-axis, as above. Values are reported as sequence reads relative to the genome-wide median for the immunoprecipitate (IP; black) and the input (gray). For a reference, the green track shows the distribution of the Sir3 or Sir4 immunoprecipitate in chromatin from cells with wild-type Sir2, as in Figure 1.
Formaldehyde cross-linking captured a unique feature of silenced chromatin architecture
Because Sir proteins associate with nucleosomes, it was possible that their binding to nucleosomes disturbed the underlying nucleosome architecture, explaining the heterogeneous distribution of Sir proteins. However, nucleosome position has been interrogated extensively in yeast both globally and specifically at the silent mating type loci, with no indication of unusual distributions (Weiss and Simpson 1998; Ravindra et al. 1999; Henikoff et al. 2011). To assess independently whether the previously reported distribution of nucleosomes applied to our strains, we used the standard anti-H3 antibody to map nucleosomes on native chromatin digested with MNase by ChIP-seq, as others have done. The critical difference between this analysis and the analysis of Sir protein ChIP-seq above was that no formaldehyde was needed due to the tight associations of nucleosomes and DNA. We found a regular distribution of nucleosomes very similar to those previously published for both silenced loci (Fig. 6A). Thus, the heterogeneous distribution of the Sir proteins could not be explained by a peculiar distribution of nucleosomes in our strains.
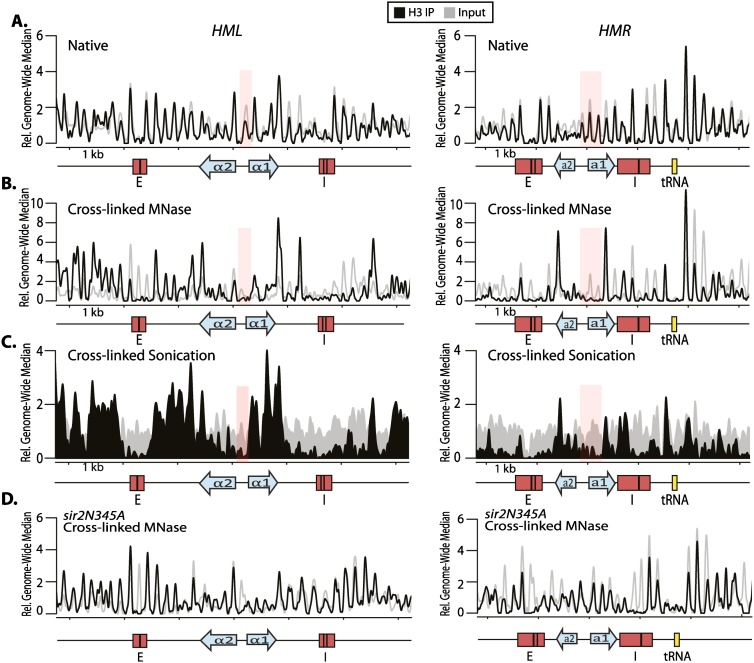
Figure 6.
H3 was differentially recovered in immunoprecipitates from native versus cross-linked chromatin. (A) H3 ChIP-seq of native chromatin treated with MNase. (B) H3 ChIP-seq from chromatin cross-linked for 15 min at room temperature and treated with MNase. (C) H3 ChIP-seq of sonicated chromatin cross-linked for 15 min at room temperature. The pink-shaded region in each graph highlights an area in which nucleosomes are detected in the H3 immunoprecipitate in the native preparation (A) but not in either of the cross-linked chromatin preparations (B,C). (D) H3 ChIP-seq in cells expressing only the catalytically inactive version of Sir2 (sir2N345A; JRY9498). Chromatin was cross-linked for 15 min and treated with MNase. The black track shows the reads for the H3 immunoprecipitate (IP), and input is shown in gray. Nucleosomes appeared less well positioned in the X region because reads could not be uniquely mapped between HML and HMR. Tick marks denote 1-kb intervals, and salient features are shown below the X-axis, as above. All reads have been trimmed and mapped with nucleR to facilitate nucleosome mapping (Flores and Orozco 2011).
One clear, possible explanation for the discrepancy between the nucleosome positioning data and Sir protein enrichment data could be that the use of formaldehyde cross-linking allowed the Sir protein ChIP-seq procedure to capture a unique chromatin architecture that was not preserved on non-cross-linked, native chromatin preparations. To investigate this possibility, we performed ChIP-seq against H3 of cross-linked chromatin fragmented by sonication or MNase digestion. This approach allowed us to interrogate the potential contribution of formaldehyde cross-linking to nucleosome position independently of the fragmentation method. There were two possible distributions of H3 from such an experiment that would be illuminating: (1) If the cross-linked H3 distribution indicated regularly spaced nucleosomes, as in their native distribution, then the Sir proteins must occupy only some of the nucleosomes in the silenced domain. (2) If the cross-linked H3 distribution were heterogeneous, similar to the heterogeneous Sir protein distribution, then cross-linking would cause the specific underrecovery of certain nucleosome and the proteins bound to them. Unlike the distribution of H3 in native chromatin, the distribution of H3 in cross-linked chromatin either sheared by sonication or cleaved by MNase was heterogeneous, qualitatively similar to each other, and similar to the Sir protein distribution (Fig. 6B,C). Most notably, at both HML and HMR, there were fewer nucleosomes captured in the H3 immunoprecipitate at the promoter region and across α1 and a1, respectively, in cross-linked chromatin than in the native chromatin (Fig. 6, highlighted by pink shading), which was especially evident when nucleosome occupancy was determined by peak calling (Supplemental Fig. S2). This paucity in nucleosome enrichment in the cross-linked chromatin coincided precisely with the positions of the lowest Sir protein enrichment. The lack of H3 at positions within HML and HMR in the cross-linked chromatin could not be explained by underrecovery of this chromatin per se because these regions were well represented in the data from the input chromatin (Fig. 6, gray lines). The input chromatin trace represented data from a MNase-seq experiment, which allowed the determination of nucleosome position by patterns of sequences protected from MNase, further emphasizing the presence of nucleosomes in this region. Rather, the “missing nucleosomes” resulted from underrecovery due to position-dependent epitope masking of certain positions, specifically in the immunoprecipitated chromatin from cross-linked samples. H3 enrichment was also evaluated using the 1-h cross-linking time originally used for Sir protein preparations (Supplemental Fig. S3). These samples showed a distribution similar to the 15 min cross-linking preparations, indicating that varying cross-linking time did not change the H3 ChIP profile. As discussed below, it was the inability to detect both H3, which was present in the native H3 distribution, and Sir proteins at key positions in cross-linked chromatin that provided strong evidence that both were there but in an inaccessible chromatin superstructure that resulted in epitope masking of both H3 and Sir proteins within the core promoter region of the silenced loci but not in flanking regions.
To test whether the inability of the antibody to recover H3 across the promoter region in cross-linked treated chromatin was specific to silenced cells, we performed MNase-seq followed by an H3 immunoprecipitation of cross-linked chromatin in cells expressing only the catalytically inactive version of Sir2 (sir2N345A). As with the native chromatin, in this strain, all nucleosomes were recovered in the immunoprecipitate, indicating that silencing was necessary for the inaccessible chromatin structure, resulting in position-dependent epitope masking of H3 (Fig. 6D).
The chromatin superstructure appeared specific to silenced but not repressed genes
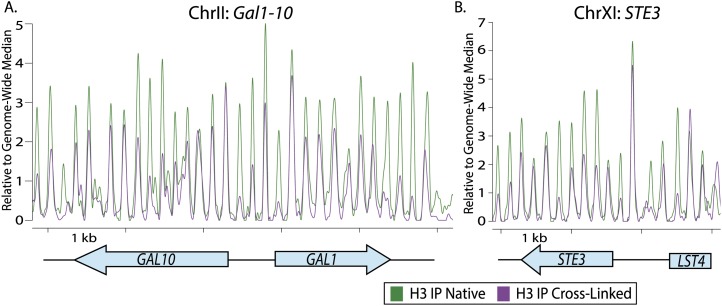
We tested whether the chromatin structure implied by the difference in the H3 distribution in cross-linked versus native H3 mapping was a general property of the chromatin at repressed genes or was potentially particular to silenced chromatin. To answer this question, we compared nucleosome positions in native and cross-linked chromatin digested with MNase at the repressed GAL1–GAL10 gene pair (Fig. 7A) and at the α-specific gene STE3 (Fig. 7B), which are repressed in these strains. Unlike at HML and HMR, the nucleosomes mapped on native chromatin occupied positions essentially identical to those mapped on cross-linked chromatin (Fig. 7; Supplemental Fig. S4). Thus, in contrast to the transcriptionally repressed locations tested, the silenced chromatin exhibited a unique architecture captured by formaldehyde cross-linking and recognized by its underrecovery in the immunoprecipitation of specific and highly reproducible positions within the silenced domain.
Figure 7.
H3 was recovered in cross-linked samples at loci of site-specific transcriptional repression. Comparison of ChIP-seq of MNase-treated H3 immunoprecipitate (IP) from native chromatin (green) and chromatin cross-linked for 15 min (purple) at the GAL1–10 gene pair (A) and STE3 (B). Tick marks denote 1-kb intervals, and ORFs are shown below the X-axis. Reads were trimmed with nucleR to facilitate nucleosome mapping (Flores and Orozco 2011).
Discussion
At higher resolution than previously possible, we characterized the structure of the silenced chromatin of Saccharomyces formed by the association of Sir2, Sir3, and Sir4 at the two canonical silenced regions of the yeast genome: the silent mating type loci, which harbor the unexpressed copies of mating type regulatory genes, and representatives of the two classes of telomeres. This characterization revealed a much more complex topology of Sir protein distribution at HML and HMR than anticipated, provided a high-resolution test of the nucleation and spreading model for the formation of silenced chromatin, and uncovered several unanticipated dimensions that informed the mechanism of Sir protein silencing.
Cross-linking captured a higher-order chromatin superstructure
The surprising patterns of Sir protein enrichment described here and for Sir2 in Li et al. (2013), along with evidence that sonication is influenced by specific chromatin structures, motivated a thorough investigation of the structure of the underlying chromatin of silenced regions (Teytelman et al. 2009). Through interrogation of the chromatin by two different fragmentation methods (MNase and sonication) and two different chromatin preparations (not cross-linked [native] or cross-linked by formaldehyde), we found that in the cross-linked samples, the nucleosome distribution was similar to the pattern of Sir protein enrichment, with some positions appearing devoid of nucleosomes as assayed by recognition by the H3 antibody even though these same positions were fully occupied by nucleosomes in the native chromatin.
We considered multiple explanations for how the distribution of nucleosomes and presumably the distribution of Sir proteins could be so dramatically influenced by cross-linking. Underrecovery of certain regions of the genome in the chromatin preparation from cross-linked samples could not explain the paucity of Sir proteins or nucleosomes at particular positions because there was abundant signal at those positions in the input samples (Fig. 6, gray lines). Similarly, the cross-linking procedure per se did not harm the H3 epitope recognized by the antibody, as shown by the robust mapping of nucleosomes in cross-linked chromatin over the rest of the genome. Thus, at HML and HMR, cross-linking appeared to capture a chromatin superstructure resulting in position-dependent masking of the H3 and anti-myc epitopes, leading to the appearance of an uneven distributions of nucleosomes, which the native samples and nonsilenced chromatin showed were evenly distributed.
The H3 antibody used in these and most nucleosome mapping studies recognizes the last 20 residues of the C terminus of H3 (ab1791). Thus, the cross-linking may capture a local chromatin architecture in which the nucleosomes at the promoter region of HMR and HML, for example, are positioned such that cross-linking occludes the antibody recognition site for H3. Because the regions of underrecovery of the H3 immunoprecipitate correlated with the regions of low Sir protein occupancy shown above, the chromatin architecture captured by the cross-linking likely contributed to the heterogeneous distribution of Sir proteins observed at the same positions, indicating that a chromatin superstructure resulted in masking of both the H3 and anti-myc epitopes at the promoter region of silenced chromatin. It was not possible to test this inference directly because the association of Sir proteins with nucleosomes was insufficiently tight to allow ChIP-seq analysis in the absence of cross-linking (DM Thurtle and J Rine, unpubl.). However, we hypothesize that Sir proteins are associated throughout the locus but, like the H3 distribution, are underrecovered in the immunoprecipitate when cross-linked.
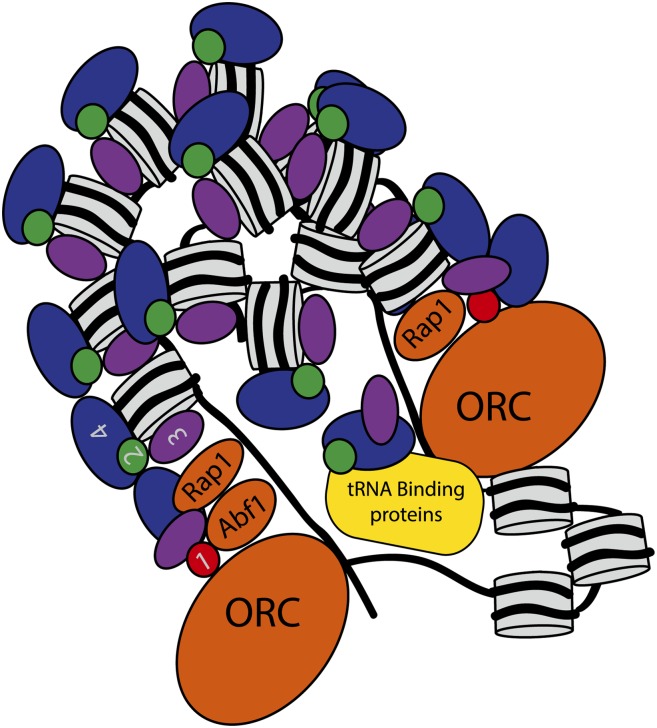
Previous studies have also implicated a higher-order structure for silenced chromatin, although none have addressed the structure with this resolution (Sperling and Grunstein 2009; Yu et al. 2011). Chromatin conformation capture (3C) studies, which exploit formaldehyde cross-linking to capture long-range interactions between sequences, suggest that the E and I silencers of HMR physically interact to form a looped structure in a Sir-dependent manner (Dekker et al. 2002; Valenzuela et al. 2008). Our data implied that the loop's structure was sufficiently complex to facilitate cross-links that cause differential recovery of relatively precise regions in immunoprecipitations, perhaps burying the promoter region and associated Sir proteins in the silenced superstructure. One possible model consistent with the data presented and also compatible with current views of the possible higher-order chromatin structures would be an additional, internal looping or coiling beyond the loop detected by 3C between the two silencers (Fig. 8). This structure would allow nucleosomes and associated Sir proteins immediately adjacent to the silencers to be accessible to antibody recognition by being on the outer part of the loop, and the promoter region buried within the silenced structure would have less accessibility to antibodies upon cross-linking, resulting in epitope masking of the proteins buried within the structure.
Figure 8.
A model for a silenced superstructure at HMR. The E and the I silencers interact to form a loop, creating distinct boundaries for Sir protein association. Nucleosomes are tightly packed throughout the loci, with a secondary loop burying the promoter in the chromatin superstructure, excluding access by RNA polymerase in vivo and antibodies in cross-linked chromatin in vitro. Sir proteins associate with the nucleosomes throughout the locus and are also buried within the conformation at the promoter region. The tRNA gene associates with the silenced superstructure and interacts with the Sir complex mediated by Sir2 cohesin binding.
The contribution of the chromatin superstructure to silencing
This architecture of silenced chromatin was distinct from two canonical examples of site-specific repression (GAL1–10 repression in cells grown on glucose medium and repression of STE3 in MATa cells), and hence the special architecture appeared specific to Sir-based gene silencing (Fig. 7). Additionally, in nonsilenced cells, nucleosomes were accessible across HML and HMR, indicating that the differential accessibility, as probed by recovery with the H3 antibody, was dependent on silencing (Fig. 6D). The paradox in the data from Figures 1 and 2 was why the very promoters that must be silenced by Sir proteins exhibited the lowest enrichment of Sir proteins. Since it now appears that the structure that exists at those promoters has the capacity, upon cross-linking, to mask recognition by the anti-H3 and anti-myc antibodies, that same structure might occlude RNA polymerase from the resident promoters, thus suggesting a structural component to the steric occlusion mechanism of regional, promoter-independent repression.
Other striking features of the enrichment data of Sir proteins at HMR were the clear boundaries, one on the left side of HMR-E and immediately to the right of the tRNA-Thr gene. That tRNA gene creates a boundary, limiting Sir-based silencing on one side from affecting the expression of a reporter gene on the other (Donze and Kamakaka 2001; Oki and Kamakaka 2005). The tRNA gene and associated cohesin might function as a boundary by providing a structural scaffold for constraining the silenced domain at HMR through interaction with HMR-E. This may be akin to cohesin's chromatin organizational role in other organisms, such as the dosage compensation complex in Caenorhabditis elegans and CTCF/cohesin interactions in mammals (Meyer 2010; Phillips-Cremins et al. 2013). If so, perhaps the selective pressure for a functional I silencer at HMR would be reduced. HML, however, showed a less discrete domain of enrichment than HMR, possibly reflecting its relative proximity to the Chromosome III left telomere.
Nucleation and Sir protein association at silenced chromatin
The working model for the assembly of silenced chromatin, drawn from low-resolution ChIP-qPCR studies, has been the nucleation of a Sir protein complex at the silencers through association of individual Sir proteins with ORC, Abf1, and Rap1. In the model, nucleation is followed by spreading of Sir complexes across the region by iterative cycles of deacetylation of histones by Sir2, leading to recruitment of additional Sir protein complexes, ultimately occupying a region of chromatin and repressing transcription within it (Rusche et al. 2002). Our data have substantially refined this view. Sir3 and Sir4 enrichment in cells lacking Sir2 catalytic activity was substantially reduced or missing from internal regions of HML and HMR. However, we still observed some clear, although reduced, Sir3 and Sir4 occupancy at nucleosomes adjacent to the silencers. According to the spreading model and with support from the structure of Sir3 bound to a nucleosome, deacetylation of H4K16 is required to produce the highest-affinity interactions between Sir3 and nucleosomes (Armache et al. 2011; Wang et al. 2013). However, since Sir2 must bind to H4K16-acetyl before the deacetylation reaction, it stands to reason that the Sir2–3–4 complex would still bind H4K16-acetyl, although with less affinity and perhaps less stability than the deacetylated histone tail. Indeed, in vitro studies have demonstrated a slightly higher affinity of the Sir2–4 complex with acetylated H4K16 than with the deacetylated H4K16, supporting our in vivo studies of Sir protein association independent of Sir2 catalytic activity (Oppikofer et al. 2011).
This distribution of Sir3 and Sir4 independent of Sir2 catalytic activity also revealed additional nucleation sites at both HMR and HML. At HML, there was apparent nucleation at a region overlapping the HO recognition sequence. This internal peak may correspond to a genetic entity, called the protosilencer, internal to HML. This peak supports the hypothesis that the Rap1-binding site in the promoter region of HML might actually be occupied by Rap1 in silenced chromatin, perhaps providing stabilizing interactions with Sir3 and Sir4 just as it does at the silencer (Cheng and Gartenberg 2000). Additionally, Sir3 and Sir4 associated with the tRNA gene adjacent to HMR, largely independently of Sir2's catalytic activity. That tRNA gene recruits Scc1 cohesin, which in turn interacts with a noncatalytic domain of Sir2, presumably mediating the peak of Sir3 and Sir4 enrichment precisely over the tRNA gene (Wu et al. 2011). As a note of caution, we recently identified tRNA genes and other highly expressed loci to be susceptible to nonspecific enrichment in ChIP protocols (Teytelman et al. 2013). However, the magnitude of that nonspecific enrichment could not account for the majority of the Sir protein enrichment at this tRNA gene.
Several observations have been difficult to reconcile with the notion that spreading occurs in iterative cycles of histone deacetylation and Sir complex recruitment across a linear silenced region. For example, studies on the kinetics of silencing have yet to detect time-revolved distinctions of where in a silenced domain Sir proteins first bind (Lynch and Rusche 2008; Radman-Livaja et al. 2011). The initial nucleation of Sir proteins to the silencers, as evidenced by the Sir3 and Sir4 enrichment in the absence of Sir2 catalytic activity, and the resulting looped structure may facilitate efficient, nonlinear assembly of silent chromatin by allowing Sir2 to access all of the nucleosomes from one or more positions at the locus. Therefore, there may be no obligate order of Sir proteins associating with nuclesomes across the silenced domain.
The domains of hypoacetylated H4K16
At HML, HMR, and the telomeres, the regions of H4K16 hypoacetylation were larger than the regions of Sir protein enrichment. There were at least three possible explanations for how the hypoacetylated domain could be larger than the domain occupied by Sir proteins: (1) Sir proteins bound to one nucleosome could potentially deacetylate H4K16-acetyl on nearby nucleosomes. (2) There could be differences in the stability of Sir protein association with some hypoacetylated H4K16 nucleosomes relative to others. (3) The extended H4K16 hypoacetylation could be the result of a different histone deactylase, such as Rpd3 (Ehrentraut et al. 2010). At telomeres, these larger regions of H4K16 hypoacetylation were consistent with the apparent ability of Sir3, especially upon overexpression, to spread inward from telomeres (Hecht et al. 1996; Radman-Livaja et al. 2011), exerting a telomere position effect, due to Sir3's affinity for nucleosomes hypoacetylated at H4K16 (Armache et al. 2011; Wang et al. 2013). Formally, because we were unable to efficiently immunoprecipitate H3 from the internal silencer regions, we cannot completely discount the possibility that the chromatin is acetylated at some positions within HML and HMR yet was unable to be immunoprecipitated by the H4K16-acetyl antibody. This explanation seems less likely though, as many studies have shown that acetylated H4K16 is not conducive to a tightly packed heterochromatin structure (Johnson et al. 2009; Martino et al. 2009). Additionally, genetic evidence shows that an H4K16-acetyl mimic is detrimental to silencing (Park and Szostak 1990).
Chromatin architecture facilitating regional repression
Increasingly, a three-dimensional architecture of chromatin has become a critical missing dimension in our understanding of chromatin regulation. Looping of silencer elements has been shown to be important for heterochromatin regulation in other organisms, such as Polycomb-bound regions and promoters in Drosophila and mammals (Bantignies et al. 2011). The high-resolution picture of silenced chromatin described here will be an important benchmark in relating the structure of domains of gene silencing to mechanisms of repression.
Materials and methods
Yeast strains used
All yeast strains were derived from the W303 background, and genotypes are indicated in Supplemental Table S1. Deletions and epitope tags were constructed through one-step integration of knockout cassettes (Longtine et al. 1998). C-terminal tagging of Sir gene ORFs was also conducted using one-step gene replacement with the 13xMyc tag. H4K16-acetyl and H3 ChIP-seq experiments were performed on JRY9316.
Chromatin isolation by sonication
Seventy OD units (A600) of logarithmically growing cells was cross-linked with 1% formaldehyde for either 15 min or 1 h at room temperature. Chromatin was prepared as previously described (Aparicio et al. 2005) with modifications as described in the Supplemental Material.
MNase preparation
Three-hundred-fifty OD units (A600) of logarithmically growing cells was cross-linked with 1% formaldehyde for 15 min at room temperature. For native chromatin preparation, the cross-linking step was omitted. Nuclei were prepared as in Furuyama and Biggins (2007) with modifications as described in Supplemental Material.
Immunoprecipitations
Sir ChIP-seq samples were immunoprecipitated with 120 μL of anti-c-Myc Agarose (Sigma, catalog no. 7470). Five microliters of anti-H3 (Abcam, ab1791) or 15 μL of anti-H4K16-acetyl (Millipore, catalog no. 07-595) was used for H3 and H4K16-acetyl immunoprecipitations, respectively, each with 120 μL of protein A Sepharose bead slurry (GE Healthcare, 17-5280-01). Washes, elutions, and isolation of DNA were performed as previously described (Aparicio et al. 2005). After reversal of cross-links, RNase A was added to MNase samples. Details about immunoprecipitations can be found in the Supplemental Material.
Library preparation
The first biological replicate of Sir sonication libraries, the second biological replicate of Sir3 and Sir4 libraries, and the first replicate of the no-tag libraries were prepared with modifications to the Illumina paired-end library protocol as in Zill et al. (2010). Libraries were loaded into one lane each and sequenced using the Illumina Genome Analyzer II as 45-bp or 36-bp paired-end reads.
All MNase libraries, Sir3 and Sir4 libraries from the sir2N345A mutant, the Sir2 biological replicate, and H4K16-acetyl library were constructed using the Illumina Tru-Seq library preparation kit with the following modifications as described in the Supplemental Material. All sequences have been deposited in the NCBI Short Read Archive under accession number SRP034921.
Data analysis
Reads were mapped using BWA (Li and Durbin 2009) to a modified Sac Cer 2 genome in which the MAT locus was replaced with the Hyg-MX cassette. Duplicate reads were removed using Picard (http://picard.sourceforge.net), and per-base read counts were determined using SAMtools (Li et al. 2009). The number of reads used per sample after mapping and duplicate processing are documented in Supplemental Table S2.
For the MNase samples, mapped reads were then trimmed to 40 bp, and only reads with an insert length <250 bp were used to ensure mononucleosome resolution. The reads were further processed, including coverage calculations, signal smoothing, and peak detection, using nucleR (Flores and Orozco 2011). A threshold value of 50% was used for peak detection, and peaks were filtered for those with a score >0.4. All graphs were created using R (http://www.R-project.org). Median coverage was calculated for 100-bp windows, sliding along each chromosome in 50-bp steps. The genome-wide median was calculated by determining the median of the sliding windows for each data set (Supplemental Table S2). The genome-wide correlations were determined by pairwise comparison of the same 100-bp sliding windows between the appropriate data sets and then determining the Pearson correlation in R (http://www.R-project.org).
Acknowledgments
We thank Oliver Zill for his keen observations and assistance with initial sample preparations. We thank Meru Sadhu and members of the Rine laboratory for insightful discussions, and especially Kripa Asrani for artistic consultation. We also thank Sarah Bissonnette, Laura Lombardi, and David Steakley for critical reading of the manuscript, and reviewers for helpful comments. We thank Minyong Chung and the Vincent J. Coates Genomics Sequencing Laboratory at the University of California at Berkeley for all high-throughput sequencing. Funding was provided by the National Science Foundation predoctoral fellowship to D.M.T., and the University of California at Berkeley's Cellular, Biochemical, and Molecular Training Grant from the National Institutes of Health (NIH) and grant GM31105 from the NIH to J.R.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.230532.113.
References
- Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol 23: 17.7.1–17.7.23 [DOI] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226 [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R 2011. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Bi X, Braunstein M, Shei G-J, Broach JR 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc Natl Acad Sci 96: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K 1985. Characterization of a ‘silencer’ in yeast: A DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41: 41–48 [DOI] [PubMed] [Google Scholar]
- Chen L, Widom J 2005. Mechanism of transcriptional silencing in yeast. Cell 120: 37–48 [DOI] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou Z-R, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T-H, Gartenberg MR 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev 14: 452–463 [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A 2012. Dosage compensation in Drosophila melanogaster: Epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13: 123–134 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J 20: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S, Weber JM, Dybowski JN, Hoffmann D, Ehrenhofer-Murray AE 2010. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc Natl Acad Sci 107: 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Orozco M 2011. nucleR: A package for non-parametric nucleosome positioning. Bioinformatics 27: 2149–2150 [DOI] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci 104: 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gross DS 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol Cell Biol 28: 3979–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli S, Donze D, Dhillon N, Kamakaka RT 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J 20: 4522–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Grunstein M, Gasser SM 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5: a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Strathern JN, Klar AJS 1979. Transposable mating type genes in Saccharomyces cerevisiae. Nature 282: 478–483 [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z 2005. Structural basis of the Sir1–origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci 102: 8489–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Jeon Y, Sarma K, Lee JT 2012. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev 22: 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci 87: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D 2009. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell 35: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci 97: 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Valsakumar V, Poorey K, Bekiranov S, Smith JS 2013. Genome-wide analysis of functional sirtuin chromatin targets in yeast. Genome Biol 14: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 303–304 [DOI] [PubMed] [Google Scholar]
- Loney ER, Inglis PW, Sharp S, Pryde FE, Kent NA, Mellor J, Louis EJ 2009. Repressive and non-repressive chromatin at native telomeres in Saccharomyces cerevisiae. Epigenetics Chromatin 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J 1994. Silencers and domains of generalized repression. Science 264: 1768–1771 [DOI] [PubMed] [Google Scholar]
- Louis EJ 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- Lynch PJ, Rusche LN 2008. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol Cell Biol 29: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM 2009. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Meyer BJ 2010. Targeting X chromosomes for repression. Curr Opin Genet Dev 20: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA, Tatchell K 1980. The structure of transposable yeast mating type loci. Cell 19: 753–764 [DOI] [PubMed] [Google Scholar]
- Oki M, Kamakaka RT 2005. Barrier function at HMR. Mol Cell 19: 707–716 [DOI] [PubMed] [Google Scholar]
- Oppikofer M, Kueng S, Martino F, Soeroes S, Hancock SM, Chin JW, Fischle W, Gasser SM 2011. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J 30: 2610–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Szostak JW 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol 10: 4932–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ 1999. Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Ruben G, Weiner A, Friedman N, Kamakaka R, Rando OJ 2011. Dynamics of Sir3 spreading in budding yeast: Secondary recruitment sites and euchromatic localization. EMBO J 30: 1012–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra A, Weiss K, Simpson RT 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol Cell Biol 19: 7944–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Strathern JN, Hicks JB, Herskowitz I 1979. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: Evidence for and identification of cryptic mating-type loci. Genetics 93: 877–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R 2004. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Rivier DH, Ekena JL, Rine J 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell 13: 2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Singh J, Klar AJ 1992. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: A novel in vivo probe for chromatin structure of yeast. Genes Dev 6: 186–196 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Grunstein M 2009. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci 106: 13153–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31: 183–192 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Takahashi Y-H, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745 [DOI] [PubMed] [Google Scholar]
- Teytelman L, Özaydın B, Zill O, Lefrançois P, Snyder M, Rine J, Eisen MB 2009. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE 4: e6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci 110: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251–253 [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT 2008. Long-range communication between the silencers of HMR. Mol Cell Biol 28: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Li G, Altaf M, Lu C, Currie MA, Johnson A, Moazed D 2013. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc Natl Acad Sci 110: 8495–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol Cell Biol 18: 5392–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-S, Chen Y-F, Gartenberg MR 2011. Targeted sister chromatid cohesion by Sir2. PLoS Genet 7: e1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Olsen L, Zhang X, Boeke JD, Bi X 2011. Differential contributions of histone H3 and H4 residues to heterochromatin structure. Genetics 188: 291–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu R-M 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21: 4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gustafsson CM 2009. Distinct differences in chromatin structure at subtelomeric X and Y′ elements in budding yeast. PLoS ONE 4: e6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill OA, Scannell D, Teytelman L, Rine J 2010. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol 8: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]