Abstract
Overdiagnosis of breast cancer, i.e. the detection of slow-growing tumors that would never have caused symptoms or death, became more prevalent with the implementation of population-based screening. Only rough estimates have been made of the proportion of patients that are overdiagnosed and identification of those patients is difficult. Therefore, the aim of this study is to evaluate whether tumor biology can help identify patients with screen-detected tumors at such a low risk of recurrence that they are likely to be overdiagnosed. Furthermore, we wish to evaluate the impact of the transition from film-screen mammography (FSM) to the more sensitive full-field digital mammography (FFDM) on the biology of the tumors detected by each screening-modality. All Dutch breast cancer patients enrolled in the MINDACT trial (EORTC-10041) accrued 2007–2011, who participated in the national screening program (biennial screening ages 50–75) were included (n = 1,165). We calculated the proportions of high-, low- and among those the ultralow-risk tumors according to the 70-gene signature for patients with screen-detected (n = 775) and interval (n = 390) cancers for FSM and FFDM. Screen-detected cancers had significantly more often a low-risk tumor biology (68 %) of which 54 % even an ultralow-risk compared to interval cancers (53 % low-, of which 45 % ultralow-risk (p = 0.001) with an OR of 2.33 (p < 0.0001; 95 % CI 1.73–3.15). FFDM detected significantly more high-risk tumors (35 %) compared to FSM (27 %) (p = 0.011). Aside from favorable clinico-pathological factors, screen-detected cancers were also more likely to have a biologically low-risk or even ultralow-risk tumor. Especially for patients with screen-detected cancers the use of tools, such as the 70-gene signature, to differentiate breast cancers by risk of recurrence may minimize overtreatment. The recent transition in screening-modalities led to an increase in the detection of biologically high-risk cancers using FFDM.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-013-2830-5) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Screening, 70-Gene signature, Film-screen mammography, Full-field digital mammography
Introduction
The increasing incidence in breast cancer after implementation of population-based mammographic screening programs has been suggested to be partly due to the detection of slow-growing tumors that would never have caused symptoms or death, i.e. breast cancer overdiagnosis [1]. This lead time bias is related to the phenomenon of length time bias, as slow-growing tumors have a longer window of opportunity to be detected in screening and therefore they are overrepresented in screen-detected cancers [1]. Whether this actually results in an increase in the detection of low-risk tumors or even clinically indolent disease is still being investigated [2, 3]. The concept of overdiagnosis due to screening was first reported in 1982 by Lundgren and Helleberg [4]. Estimates of the proportion of overdiagnosis were made by different study groups and are reported between 1 and 54 %, depending on the denominators that are used [5, 6]. In The Netherlands, there is an estimated 2.8 % overdiagnosis [6].
Previous analyses, including our own, reported that screen-detection is associated with a better prognosis for overall and breast-cancer-specific survival, independent of other favorable prognostic clinico-pathological factors [7]. Screen-detected cancers are more often tumors of smaller size, lymph node-negative, low grade, and estrogen-receptor-positive than interval cancers [7]. Identification of the patients with screen-detected cancers that are likely to be overdiagnosed based on clinico-pathological factors remains difficult. Therefore, the hypothesis was generated that knowledge of the biological background of the tumor may be helpful in the identification of patients with screen-detected tumors at such a low risk of recurrence that they are likely to be overdiagnosed. Nowadays, gene-expression classifiers are used in addition to clinico-pathological factors to identify patients with a favorable prognosis based on the biology of their tumor [8]. One of these gene-expression classifiers is the 70-gene signature (MammaPrint™), developed to improve the selection of those patients who may benefit from adjuvant systemic treatment [9]. The prognostic value of the 70-gene signature has been validated in several studies, both retrospectively and prospectively [10–13]. We previously reported on the tumor biology of screen-detected cancers and suggested that screen-detection might also be associated with a higher likelihood of a biologically low-risk or even ultralow-risk tumor assessed by the 70-gene signature [2].
Over the past decade, a transition in diagnostic imaging has occurred. Most screening facilities switched from film-screen mammography (FSM) to full-field digital mammography (FFDM). In the Netherlands, this transition started in 2008 and as of 2010, 94 % of the women participating in the Dutch screening program have been screened using FFDM [14]. Several studies have evaluated the performance of FFDM compared to FSM and showed comparable or even better results for FFDM in the detection of clinically relevant tumors [15, 16]. FFDM showed a higher sensitivity compared to FSM and detects more ductal carcinoma in situ (DCIS) and invasive cancers [17], especially in women under the age of 50 years and in pre- or peri-menopausal women with radiographically dense breasts [16, 17]. Recent studies indicate that FFDM-detected cancers are more often estrogen-receptor-negative tumors [17, 18]. A more sensitive screening-modality such as FFDM may also lead to an increase in the detection of biologically high-risk tumors as assessed by the 70-gene signature. No differences in other clinico-pathological factors, such as tumor size or grade, are described in the literature [15, 16].
The aim of this study is to determine the proportion of biologically high-, low-, and among those ultralow-risk tumors among the screen-detected and interval tumors and to evaluate the impact of the transition from FSM to the more sensitive FFDM on the biology of the tumors detected by each screening-modality.
Patients and methods
Patients and Clinico-pathological characteristics
All Dutch breast cancer patients enrolled in the MINDACT trial (EORTC-10041) [19, 20], who were invited for the Dutch screening program, were included in this study.
The MINDACT trial enrolled women aged 18–70 years with histologically proven operable invasive breast cancer, no distant metastases, and for whom a frozen tumor sample was available between 2007 and 2011 [19, 20]. Eligibility criteria included tumor stage T1, T2, or operable T3, and unilateral; DCIS or lobular carcinoma in situ (LCIS) provided invasive cancer is present; surgery options included breast-conserving surgery or mastectomy combined with either a sentinel node procedure or full axillary clearance; WHO performance status of 0 or 1 and adequate bone marrow, liver, and renal functions. Main exclusion criteria were: previous or concurrent cancer, previous chemotherapy, anticancer endocrine therapy or radiotherapy, and clinically significant impaired cardiac function. The protocol was amended in April 2008 to allow inclusion of 1–3 lymph node positive (N1) disease and genomic test in samples containing >30 % of tumor cells [19, 20]. Clinico-pathological characteristics were obtained from the EORTC-10041 trial database. In case of discordance between a patients’ clinical risk estimation (based on Adjuvant! Online) and 70-gene signature result, the patient was randomized between treatment according to their clinical risk estimation or according to the 70-gene signature result.
Screening program
The Dutch Screening Program started on April 1, 1990. First, women aged 50–69 years old and from 1998 women up to 75 years old were invited to participate in the screening program based on area code regions. Full coverage for woman aged 50–69 was achieved in 1997 [7, 21]. Women were invited for biennial mammography. Screening mammograms were performed in independent and (mostly) mobile screening units (3–8 units per region). The images are read double-blind by trained radiologists. The current attendance rate is around 80 % [7, 14]. FFDM was rolled out as from 2007 and fully implemented in 2011. From each patient in this study, data were collected on whether the most recent screening was by FSM or FFDM.
Method of detection
Data on the method of detection were retrieved from the database of the Dutch screening organization. Data of all five regions are centrally collected in the iBob database [14]. The screening data for the eligible Dutch MINDACT patients were derived from the iBob database based on demographic information. Patients were eligible if they were 49 years or older at the time of diagnoses and were invited to participate in the Dutch screening program (n = 1,475). One hospital excluded their patients (n = 4) from the linkage protocol and 62 patients could not be matched to the iBob database, due to incomplete demographic information. Of the 1,409 patients that were matched to the iBob database, 1,165 were identified as participants of the screening program.
Two types of breast cancer were identified based on the method of detection. First, the screen-detected cancers, defined as breast cancers that were mammographically detected in the first (prevalent cancers, n = 115) or a subsequent screening round (incident cancers, n = 660) (total n = 775). Second, the interval cancers, defined as symptomatic cancers that were diagnosed within 30 months of a negative screening (n = 390). Screening is biennial, giving a window of 24 months for an interval cancer to become symptomatic after a negative screening mammography. When a woman moves to another area code, her next screening could be delayed up to 6 months. Therefore, the interval of 30 months was chosen.
70-Gene signature
In this study, we used the 70-gene signature to evaluate tumor biology. For all patients included in the MINDACT trial, a 70-gene signature result was available. The 70-gene signature, MammaPrint® (Agendia Inc, Amsterdam, The Netherlands), is a gene-expression classifier, used to estimate the risk of developing distant metastasis. The result of the 70-gene signature is presented as a binary result (good or poor prognosis), which is derived from an index-score (−1 to 1) [9, 10]. An index-score greater than 0.4 is classified as good prognosis (low risk) and an index-score less than 0.4 is classified as poor prognosis (high risk). For this study, we also applied the previously set threshold to identify patients with an ultralow risk of distant recurrence (index-score >0.6) [2]. Within the low-risk group of the original 78 patients used to develop this classifier, no distant metastases were observed at 5 years in patients who had an index-score greater than 0.6 [2, 9].
Statistical analysis
Baseline characteristics for screen-detected and interval cancers were compared and the proportions of 70-gene signature high-, low-, and among the latter the ultralow risk were calculated. We performed separate analyses for FSM and FFDM. Prognostic factors, such as age, tumor size, histological type, estrogen receptor (ER), progesterone receptor (PR), and HER2/neu-oncoprotein (ERBB2), were evaluated in a logistic regression model. Hereafter, tumor biology-related factors are referred to as “prognostic factors.” Only factors that resulted in <10 % change in the coefficient of association of the 70-gene signature with the method of detection were included in the multivariate analyses. Calculations were done using SPSS (version 19.0). A two-sided p value of less than 0.05 was considered statistically significant.
Results
Patient characteristics
The clinico-pathological characteristics of the 1,165 included patients are described in Table 1, stratified by method of detection, and in supplementary Table 1 also stratified by 70-gene signature result. Screen-detected cancers were more often of smaller size (<2 cm), ER and PR positive, HER2 negative, grade I, without nodal involvement compared to interval cancers.
Table 1.
Breast cancer patients eligible to participate in the Dutch screening program
| Screen-detected cancers (SD) | Interval cancers (IC) | p valuea SD vs. IC | |
|---|---|---|---|
| n = 775 | n = 390 | ||
| 70-Gene signature | |||
| High risk | 244 (32 %) | 185 (47 %) | <0.0001 |
| Low risk | 242 (31 %) | 111 (29 %) | |
| Ultralow risk | 289 (37 %) | 94 (24 %) | |
| Age (years) | |||
| 49–54 | 208 (27 %) | 103 (26 %) | 0.896 |
| 55–59 | 193 (25 %) | 104 (27 %) | |
| 60–64 | 200 (26 %) | 94 (24 %) | |
| 65–69 | 170 (22 %) | 88 (23 %) | |
| Size | |||
| T1 (<20 mm) | 613 (79 %) | 247 (63 %) | <0.0001 |
| T2 (20–50 mm) | 160 (21 %) | 139 (36 %) | |
| T3 (>50 mm) | 2 (0.3 %) | 4 (1 %) | |
| Lymph node status | |||
| Negative | 680 (88 %) | 315 (81 %) | 0.001 |
| 1–3 positive nodes | 95 (12 %) | 75 (19 %) | |
| Histological type | |||
| Ductal | 643 (83 %) | 316 (81 %) | 0.390 |
| Lobular | 76 (10 %) | 49 (13 %) | |
| Mixed | 28 (4 %) | 9 (2 %) | |
| Other | 28 (4 %) | 16 (4 %) | |
| Grade | |||
| Grade I | 244 (32 %) | 59 (15 %) | <0.0001 |
| Grade II | 356 (46 %) | 170 (44 %) | |
| Grade III | 174 (23 %) | 160 (41 %) | |
| Undefined | 1 | 1 | |
| ER status | |||
| Negative | 77 (10 %) | 80 (21 %) | <0.0001 |
| Positive | 698 (90 %) | 310 (80 %) | |
| PR status | |||
| Negative | 188 (24 %) | 138 (35 %) | <0.0001 |
| Positive | 573 (74 %) | 242 (62 %) | |
| Unknown | 14 | 10 | |
| HER2 status | |||
| Negative | 680 (88 %) | 325 (83 %) | 0.043 |
| Positive | 94 (12 %) | 64 (16 %) | |
| Unknown | 1 | 1 | |
Patient and tumor characteristics stratified by method of detection
ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor 2
aChi square test
70-Gene signature for screen-detected and interval cancers
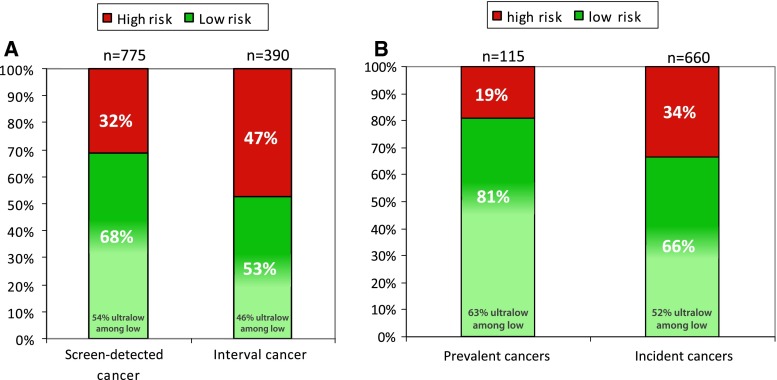
Among the screen-detected cancers, 32 % had a 70-gene signature high-risk and 68 % a low-risk tumor, of which 54 % had a ultralow-risk tumor (37 % of total) (Fig. 1a; Table 1). Among the interval cancers, 47 % had a high-risk and 53 % a low-risk tumor, of which 46 % could be defined as ultralow-risk tumor (24 % of total). A significant difference was seen between screen-detected and interval cancers (p X 2 test = 0.001) in 70-gene signature high-, low-, and ultralow-risk groups.
Of the prevalent tumors, detected in the first screening round, 19 % had a 70-gene signature high-risk and 81 % a low-risk tumor. Among the low-risk prevalent tumors about 63 % even had an ultralow-risk tumor (51 % of total) (Fig. 1b). Of the incident tumors, detected in subsequent screening rounds, 34 % had a 70-gene signature high-risk and 66 % a low-risk tumor. Among the low-risk incident tumors, 52 % could be defined as ultralow-risk (35 % of total) (p X 2 test prevalent vs. incident <0.0001) (Fig. 1b). When excluding the prevalent cancers from these analyses, the significant difference between screen-detected and interval cancers remained (Supplementary Table 2).
Fig. 1.
a Proportions of 70-gene signature result among screen-detected and interval cancers. b Screen-detected cancers detected in first versus subsequent screening rounds
In a univariate analyses, patients with screen-detected cancers were two-times more likely to have an ultralow-risk tumor compared to patients with an interval cancer (OR high vs. ultralow: 2.33 (95 %CI 1.73–3.15; p < 0.0001) (Table 2). When adjusting for intermediate factors such as ER status and tumor size, this significant association remained (Table 2). However, when adjusting for grade the 70-gene signature was no longer a significant factor; likely due to a substantial correlation between the 70-gene signature and grade (ρ = 0.393). The analyses mentioned above lead to similar conclusions in ER positive patients only (data not shown).
Table 2.
Unadjusted and adjusted Odd’s ratios of the tumor biology among screen-detected versus interval cancers
| Unadj. OR (95 % CI) | p valuea | Adj. ORb (95 % CI) | p valuea | |
|---|---|---|---|---|
| 70-Gene signature | ||||
| Ultralow vs. low | 1.41 (1.02–1.95) | 0.037 | 1.18 (0.84–1.65) | 0.339 |
| Ultralow vs. high | 2.33 (1.73–3.15) | <0.0001 | 1.26 (0.86–1.84) | 0.230 |
| 70-Gene signature + ER status | ||||
| 70-Gene signature ultralow vs. low | 1.40 (1.01–1.93) | 0.044 | ||
| 70-Gene signature ultralow vs. high | 1.95 (1.40–2.71) | <0.0001 | ||
| ER status positive vs. negative | 1.68 (1.14–2.47) | 0.008 | ||
| 70-Gene signature + grade | ||||
| 70-Gene signature ultralow vs. low | 1.19 (0.86–1.67) | 0.299 | ||
| 70-Gene signature ultralow vs. high | 1.37 (0.95–1.97) | 0.090 | ||
| Grade I vs. II | 1.84 (1.30–2.61) | 0.001 | ||
| Grade I vs. III | 3.15 (2.07–4.80) | <0.0001 | ||
| 70-Gene signature + tumor size | ||||
| 70-Gene signature ultralow vs. low | 1.38 (0.99–1.91) | 0.057 | ||
| 70-Gene signature ultralow vs. high | 2.15 (1.58–2.91) | <0.0001 | ||
| T1 vs. T2 | 1.97 (1.49–2.59) | <0.0001 | ||
| T1 vs. T3 | 5.4 (0.96–30.33) | 0.056 | ||
aLogistic regression model
bAdjusted for grade, estrogen receptor status, and tumor size
Film-screen versus full-field digital mammography
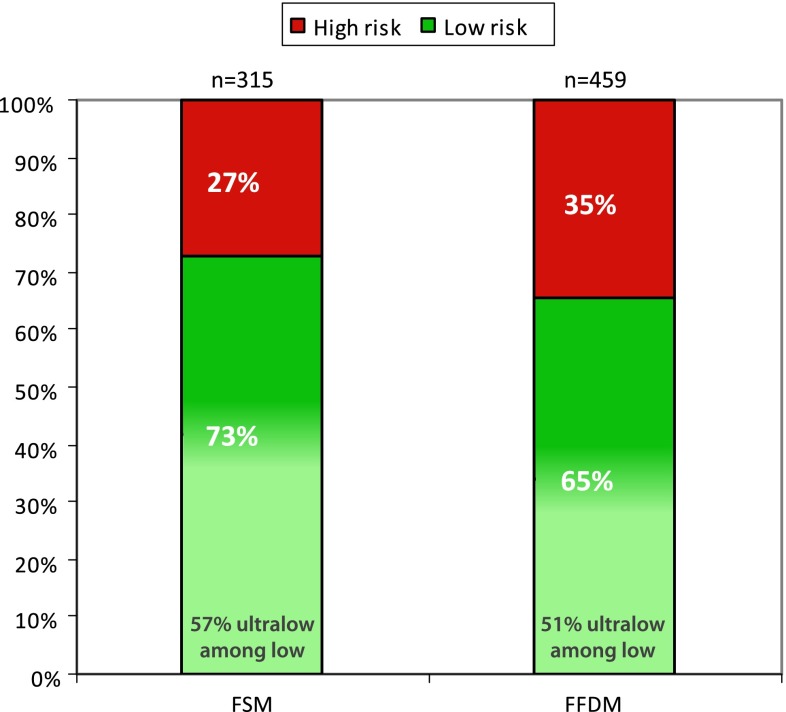
Between 2007 and 2011, a transition was seen in screening-modality used for the last screening before diagnoses. Supplementary Fig. 1 displays this transition in this cohort over time. Among the screen-detected cancers, 41 % were detected using FSM (n = 315) and 59 % were detected using FFDM (n = 459). FSM detected 27 % high-risk and 73 % low-risk tumors of whom 57 % could be defined as ultralow-risk (42 % of total). This is significantly different compared to cancers detected using FFDM (p X 2 test = 0.011), which detected 35 % high-risk and 65 % low-risk tumors of whom 51 % could be defined ultralow-risk (34 % of total) (Fig. 2; Table 3). Aside from a difference in tumor biology in tumors detected by FSM versus FFDM, there is also a difference in nodal involvement. For tumors detected by FSM, 8 % had one or more positive lymph nodes, while for tumors detected by FFDM 15 % had one or more positive lymph nodes (p X 2 test = 0.002). For other patient- and tumor-characteristics, such as age, size, histological type, grade, ER, PR, and HER2 status, no significant differences were seen between the two screening-modalities (Table 3). The association of nodal status with FFDM was at least partly attributable to the amendment of the MINDACT study in 2008, which allowed patients with 1–3 positive nodes to be included in the trial. This leads to an increase of nodal positive patients over the years (data not shown), however, nodal status was not associated with the 70-gene signature result (Supplementary Table 1).
Fig. 2.
Screen-detected cancers using film-screen versus digital mammography
Table 3.
Breast cancer patients with screen-detected cancers
| Film-screen mammography (n = 315) | Ful-field digital mammography (n = 459) | p valuea | |
|---|---|---|---|
| 70-Gene signature | |||
| High risk | 85 (27 %) | 159 (35 %) | 0.04 |
| Low risk | 98 (31 %) | 143 (31 %) | |
| Ultralow risk | 132 (42 %) | 157 (34 %) | |
| Age (years) | |||
| 49–54 yrs | 77 (24 %) | 130 (28 %) | 0.633 |
| 55–59 yrs | 83 (26 %) | 110 (24 %) | |
| 60–64 yrs | 81 (26 %) | 119 (26 %) | |
| 65–69 yrs | 74 (24 %) | 96 (21 %) | |
| Tumor size | |||
| T1 (<20 mm) | 254 (81 %) | 358 (78 %) | 0.633 |
| T2 (20–50 mm) | 60 (19 %) | 100 (22 %) | |
| T3 (>50 mm) | 1 (0.3 %) | 1 (0.2 %) | |
| Lymph node status | |||
| Negative | 290 (92 %) | 389 (85 %) | 0.002 |
| Positive | 25 (8 %) | 70 (15 %) | |
| Histological type | |||
| Ductal | 269 (85 %) | 373 (81 %) | 0.406 |
| Lobular | 28 (9 %) | 48 (11 %) | |
| Mixed | 11 (4 %) | 17 (4 %) | |
| Other | 7 (2 %) | 20 (4 %) | |
| Grade | |||
| I | 111 (35 %) | 133 (29 %) | 0.160 |
| II | 143 (45 %) | 213 (46 %) | |
| III | 61 (19 %) | 112 (24 %) | |
| Unknown | 0 | 1 | |
| ER status | |||
| Negative | 28 (9 %) | 49 (11 %) | 0.415 |
| Positive | 287 (91 %) | 410 (89 %) | |
| PR status | |||
| Negative | 79 (25 %) | 109 (24 %) | 0.169 |
| Positive | 233 (74 %) | 339 (74 %) | |
| Unknown | 3 | 11 | |
| HER2 status | |||
| Negative | 271 (86 %) | 409 (89 %) | 0.242 |
| Positive | 43 (14 %) | 50 (11 %) | |
| Unknown | 1 | 0 | |
Patient and tumor characteristics stratified by film-screen or digital mammography
ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor 2
aChi square test
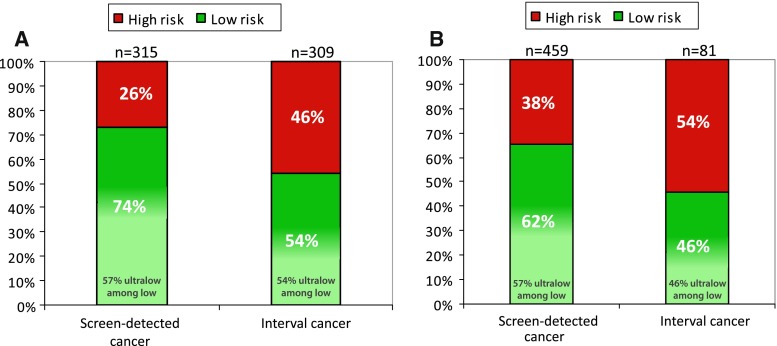
Overall, the proportion of interval cancers among the screened women within the Dutch MINDACT cohort was 33 % (390/1164). In the FSM-screened population (n = 624), the proportion of interval cancers was 49.5 % (309/624), while for the FFDM-screened population (n = 540) the interval rate was 15 % (81/540). Among the FSM interval cancers which became symptomatic within 30 months after a negative FSM (n = 309), 46 % had a high-risk and 54 % had a low-risk tumor of whom 54 % had an ultralow-risk tumor (Fig. 3a). Among the FFDM interval cancers, which became symptomatic within 30 months after a negative FFDM (n = 81), 54 % had a high-risk and 46 % had a low-risk tumor of whom 46 % an ultralow-risk tumor (Fig. 3b). Odd’s ratios for FSM and FFDM are shown in Table 4. There was no effect modification of screening-modality in the association between the 70-gene signature and screen-detected versus interval cancers.
Fig. 3.
70-Gene signature proportions among screen-detected and interval cancers after a film-screen mammography or b full-field digital mammography
Table 4.
Unadjusted and adjusted Odd’s ratios of the tumor biology among interval versus screen-detected cancers for film-screen and digital mammography
| Unadj. OR (95 % CI) | p valuea | Adj. ORb (95 % CI) | p valuea | |
|---|---|---|---|---|
| Last screen FSM | ||||
| 70-Gene signature ultralow vs. low | 1.59 (1.07–2.38) | 0.023 | 1.27 (0.84–1.94) | 0.308 |
| 70-Gene signature ultralow vs. high | 2.84 (1.93–4.20) | <0.0001 | 1.45 (0.88–2.38) | 0.261 |
| Last screen FFDM | ||||
| 70-Gene signature ultralow vs. low | 1.29 (0.65–2.56) | 0.464 | 1.15 (0.57–2.32) | 0.695 |
| 70-Gene signature ultralow vs. high | 2.56 (1.40–4.66) | 0.002 | 1.68 (0.82–3.45) | 0.158 |
aLogistic regression model
bAdjusted for grade, estrogen receptor status, and tumor size
These proportions in tumor biology remained the same for FSM and FFDM when only including those patients that were diagnosed after the amendment. Sensitivity analyses in the period when FFDM screening was implemented in at least half of the population and potentially 2 years had passed for women with a negative FFDM screen in order for interval cancers to become manifest, i.e., 2009 and 2010 showed similar proportions of high-risk tumors among FSM- and FFDM-screened patients (26.2 % FSM and 33.0 % FFDM).
Discussion
The effectiveness of breast cancer screening is extensively debated, particularly regarding the estimated proportion of overdiagnosed cancers [3, 22]. Identification of these overdiagnosed screen-detected cancers is challenging. Screen-detected cancers have shown to have more favorable clinico-pathological factors and better outcome compared to interval cancers [7]. Our results also show that the majority of the cancers detected in screening (68 %) are biologically low risk and over half of the low-risk tumors are even ultralow-risk. This indicates that knowledge of the biological background may help to identify those screen-detected breast cancers at such a low risk of recurrence that concerns about overdiagnosis can be raised. Especially for this subgroup of patients, overtreatment with chemotherapy should be avoided. To determine whether the group with screen-detected ultralow-risk tumors is indeed overdiagnosed, a randomized controlled trial would provide further insight.
Mammographic screening on the other hand, has proven to be an effective way to detect breast cancer at an early stage [23]. Our results confirm that screening also detects cancers with poor prognosis tumor biology, which are at a high risk of recurrence. Almost one-third of the patients with a tumor detected in the screening program had a high-risk 70-gene signature result. The 70-gene signature is likely to be a useful tool to separate patients at a high-risk from those at a low- or even an ultralow risk of recurrence. Patients with a screen-detected cancer are two-times more likely to have an ultralow-risk tumor compared to interval cancers. Even when adjusting for other prognostic factors with a substantial association with method of detection (in our population tumor size, grade, and ER status), the 70-gene signature remained an important prognostic factor. Previous analyses showed that the proportion of low- and ultralow-risk tumors among screen-detected cancers is higher compared to symptomatic cancers diagnosed before the introduction of screening [2]. Our current results validate this finding in a larger cohort, showing 68 % low risk among screen-detected cancers of whom 54 % had an ultralow risk. In literature, it is still debated whether the prevalent screen-detected cancers should be included when analyzing screen-detected cancers [23]. In this study, we aimed to look at screen-detected cancers from a different, more biologically oriented perspective to evaluate the type of tumors that are detected in screening programs. Since prevalent cancers are also screen-detected and a substantial proportion of overdiagnosis may be present in this subgroup, they were included in our analyses. Good prognosis for prevalent cancers has been suggested by others [1], and our observation on the biological level supports that notion, albeit not significant. The number of prevalent cancers in this cohort is low and in univariate analyses the screening round was not a significant prognostic factor.
The recent transition from FSM to FFDM resulted in a larger proportion of high-risk tumors among the screen-detected cancers, which may indicate that the introduction of FFDM leads to the detection of more aggressive cancers with a worse prognosis. It may also indicate that breast cancer screening using FFDM is even more effective than when using solely FSM. Given the possibility that high-risk tumors that used to be missed in screening are now detected with FFDM, the introduction of FFDM might be responsible for an increase in the proportion of high-risk tumors among the screen-detected cancers and decrease in the number of interval cancers. The fact that the proportion of interval cancers among FFDM-screened patients was low (15 %) may therefore be a result of more sensitive screening, but can also explained by the fact that the accrual of women to FFDM was in transition from 2008 till 2010. Hence, for many women insufficient time had passed after a negative FFDM for the development of interval cancers. Thus, the ratio between the number of women at risk for a screen-detected tumor versus an interval cancer is lower for FSM compared to FFDM. Therefore, no conclusions regarding the relative amount of interval cancers for FFDM versus FSM can be drawn based on the data presented here. Since the Dutch screening program is still collecting data on the effect of the transition from FSM to FFDM, we were not able compare our result to those of the entire screened population in the Netherlands. Of note is that the MINDACT trial currently only has available data of the tumor samples provided by the local pathology departments. Tumor characteristics, especially grade, may change after central review of the samples. A limitation is the possibility of selection bias in the MINDACT trial itself. The novelty of gene-signatures and the limited experience of doctors with this new prognostic tool may have resulted in the inclusion of patients with more favorable tumor characteristics in the beginning of the trial.
In conclusion, screen‐detection was found to be associated with a higher likelihood of a 70-gene signature biologically low-risk tumor, which prospectively validates our previous analyses [2]. Half of all screen‐detected low-risk tumors even had an ultralow-risk of distant metastases. Especially for this screen-detected patient group, the use of tools to differentiate breast cancers by risk of recurrence may minimize overtreatment. Second, the transition from FSM to FFDM resulted in the detection of a larger proportion of high-risk tumors, which may indicate that FFDM is a more effective screening-modality than FSM.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We acknowledge the contribution of the European Organization for Research and Treatment of Cancer (EORTC) and the TransBig Consortium. We thank the Dutch Screening Facilities, Frank Yntema in particular, for providing the screening data used in this study. We thank Annuska Glas from Agendia for providing value information on the 70-gene signature results. We are indebted to all the Dutch women who participated in the MINDACT trial. This work was supported by the EORTC Breast Cancer Group (type 3 Grant 2011/2012), the Dutch Cancer Society (NKI 2009-4363), BBMRI-NL (NWO 184.021.007, complementation project 45), and the Dutch Genomics Initiative “Cancer Genomics Centre.” The funding sources had no role in the study design, data collection, data analysis, data interpretation, in writing the report, or in the decision to submit for publication.
Conflict of interest
LvtV is named inventor on the patent for the 70-gene signature used in this study. LvtV reports being shareholder in and employed by Agendia Inc, the commercial company that markets the 70-gene signature as MammaPrint™.
References
- 1.Nagtegaal ID, Allgood PC, Duffy SW, Kearins O, Sullivan EO, Tappenden N, Wallis M, Lawrence G. Prognosis and pathology of screen-detected carcinomas: how different are they? Cancer. 2011;117:1360–1368. doi: 10.1002/cncr.25613. [DOI] [PubMed] [Google Scholar]
- 2.Esserman LJ, Shieh Y, Rutgers EJ, Knauer M, Retel VP, Mook S, Glas AM, Moore DH, Linn S, van Leeuwen FE, van ‘t Veer LJ. Impact of mammographic screening on the detection of good and poor prognosis breast cancers. Breast Cancer Res Treat. 2011;130:725–734. doi: 10.1007/s10549-011-1748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esserman LJ, Thompson IM, Reid B. Overdiagnosis and overtreatment in cancer: an opportunity for improvement. JAMA. 2013;310(8):797–798. doi: 10.1001/jama.2013.108415. [DOI] [PubMed] [Google Scholar]
- 4.Lundgren B, Helleberg A. Single oblique-view mammography for periodic screening for breast cancer in women. J Natl Cancer Inst. 1982;68:351–355. [PubMed] [Google Scholar]
- 5.Puliti D, Duffy SW, Miccinesi G, de Koning H, Lynge E, Zappa M, Paci E. Overdiagnosis in mammographic screening for breast cancer in Europe: a literature review. J Med Screen. 2012;19(Suppl 1):42–56. doi: 10.1258/jms.2012.012082. [DOI] [PubMed] [Google Scholar]
- 6.de Gelder R, Heijnsdijk EA, van Ravesteyn NT, Fracheboud J, Draisma G, De Koning HJ. Interpreting overdiagnosis estimates in population-based mammography screening. Epidemiol Rev. 2011;33:111–121. doi: 10.1093/epirev/mxr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mook S, van’t Veer LJ, Rutgers EJ, Ravdin PM, van de Velde AO, van Leeuwen FE, Visser O, Schmidt MK. Independent prognostic value of screen detection in invasive breast cancer. J Natl Cancer Inst. 2011;103:585–597. doi: 10.1093/jnci/djr043. [DOI] [PubMed] [Google Scholar]
- 8.Azim HA, Jr, Michiels S, Zagouri F, Delaloge S, Filipits M, Namer M, Neven P, Symmans WF, Thompson A, Andre F, Loi S, Swanton C. Utility of prognostic genomic tests in breast cancer practice: The IMPAKT 2012 Working Group Consensus Statement. Ann Oncol. 2013;24:647–654. doi: 10.1093/annonc/mds645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 10.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 12.Bueno-de-Mesquita JM, Linn SC, Keijzer R, Wesseling J, Nuyten DS, van Krimpen C, Meijers C, de Graaf PW, Bos MM, Hart AA, Rutgers EJ, Peterse JL, Halfwerk H, de Groot R, Pronk A, Floore AN, Glas AM, van’t Veer LJ, van de Vijver MJ. Validation of 70-gene prognosis signature in node-negative breast cancer. Breast Cancer Res Treat. 2009;117:483–495. doi: 10.1007/s10549-008-0191-2. [DOI] [PubMed] [Google Scholar]
- 13.Drukker CA, Bueno-de-Mesquita JM, Retel VP, van Harten WH, van Tinteren H, Wesseling J, Roumen RM, Knauer M, van’t Veer LJ, Sonke GS, Rutgers EJ, van de Vijver MJ, Linn SC. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer. 2013;133:929–936. doi: 10.1002/ijc.28082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Evaluation Team for Breast Cancer Screening in the Netherlands: NETB report 2012
- 15.Bluekens AM, Holland R, Karssemeijer N, Broeders MJ, den Heeten GJ. Comparison of digital screening mammography and screen-film mammography in the early detection of clinically relevant cancers: a multicenter study. Radiology. 2012;265:707–714. doi: 10.1148/radiol.12111461. [DOI] [PubMed] [Google Scholar]
- 16.Vinnicombe S, Pinto Pereira SM, McCormack VA, Shiel S, Perry N, Dos Santos Silva IM. Full-field digital versus screen-film mammography: comparison within the UK breast screening program and systematic review of published data. Radiology. 2009;251:347–358. doi: 10.1148/radiol.2512081235. [DOI] [PubMed] [Google Scholar]
- 17.Pisano ED, Gatsonis C, Hendrick E, Yaffe M, Baum JK, Acharyya S, Conant EF, Fajardo LL, Bassett L, D’Orsi C, Jong R, Rebner M. Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med. 2005;353:1773–1783. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 18.Kerlikowske K, Hubbard RA, Miglioretti DL, Geller BM, Yankaskas BC, Lehman CD, Taplin SH, Sickles EA. Comparative effectiveness of digital versus film-screen mammography in community practice in the United States: a cohort study. Ann Intern Med. 2011;155:493–502. doi: 10.7326/0003-4819-155-8-201110180-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardoso F, van’t Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26:729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 20.Rutgers E, Piccart-Gebhart MJ, Bogaerts J, Delaloge S, van‘t Veer LJ, Rubio IT, Viale G, Thompson AM, Passalacqua R, Nitz U, Vindevoghel A, Pierga JY, Ravdin PM, Werutsky G, Cardoso F. The EORTC 10041/BIG 03-04 MINDACT trial is feasible: results of the pilot phase. Eur J Cancer. 2011;47:2742–2749. doi: 10.1016/j.ejca.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 21.De Koning HJ, Fracheboud J, Boer R, Verbeek AL, Collette HJ, Hendriks JH, van Ineveld BM, de Bruyn AE, van der Maas PJ. Nation-wide breast cancer screening in The Netherlands: support for breast-cancer mortality reduction. National Evaluation Team for Breast Cancer Screening (NETB) Int J Cancer. 1995;60:777–780. doi: 10.1002/ijc.2910600608. [DOI] [PubMed] [Google Scholar]
- 22.Esserman L, Shieh Y, Thompson I. Rethinking screening for breast cancer and prostate cancer. JAMA. 2009;302:1685–1692. doi: 10.1001/jama.2009.1498. [DOI] [PubMed] [Google Scholar]
- 23.Marmot MG, Altman DG, Cameron DA, Dewar JA, Thompson SG, Wilcox M. The benefits and harms of breast cancer screening: an independent review. Br J Cancer. 2013;108:2205–2240. doi: 10.1038/bjc.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.