Abstract
Phenotypic plasticity is the ability of a single genotype to yield distinct phenotypes in different environments. The molecular mechanisms linking phenotypic plasticity to the evolution of heritable diversification, however, are largely unknown. Here, we show that insulin/insulin-like growth factor signalling (IIS) underlies both phenotypic plasticity and evolutionary diversification of ovariole number, a quantitative reproductive trait, in Drosophila. IIS activity levels and sensitivity have diverged between species, leading to both species-specific ovariole number and species-specific nutritional plasticity in ovariole number. Plastic range of ovariole number correlates with ecological niche, suggesting that the degree of nutritional plasticity may be an adaptive trait. This demonstrates that a plastic response conserved across animals can underlie the evolution of morphological diversity, underscoring the potential pervasiveness of plasticity as an evolutionary mechanism.
Keywords: ovariole, ovary, Drosophila sechellia, Drosophila erecta, phenotypic plasticity, insulin receptor
1. Introduction
Phenotypic plasticity is the ability of a single genotype to yield distinct phenotypes in different environments. Phenotypic plasticity may play an important role in evolutionary diversification, as it is capable of generating striking examples of biodiversity, including differences in morphology, behaviour, life history and species interactions [1]. However, whether or not phenotypic plasticity promotes or impedes evolutionary diversification is still unclear, and has been under debate for decades [2]. One hypothesis is that common molecular mechanisms underlie both plasticity and interspecific variation in a trait, which would allow plasticity to promote diversification by providing a range of phenotypes whose underlying genetic variation can be subject to selection by genetic accommodation, genetic assimilation or other means [3]. The molecular underpinnings of plasticity within a single species are known for several systems [4–7], and there is also evidence that plasticity contributes to species differentiation [8]. However, specific examples that functionally demonstrate the molecular basis for both the plasticity and interspecies divergence of the same trait are lacking. We therefore sought to provide such an example, by examining the molecular basis of the evolutionary divergence and of the phenotypic plasticity of a single trait.
Reproductive traits are particularly relevant models for investigating the molecular mechanisms of phenotypic plasticity and evolutionary changes, because they affect the number of offspring, and hence fitness. Here, we examine one such trait: insect ovariole number. Ovarioles are egg-producing structures of insect ovaries. At the anterior end of each ovariole is the germarium, where germ-line stem cells (GSCs), supported within their somatic niche, self-renew and also differentiate to ultimately yield the mature oocyte and supporting germ cells. Posterior to the germarium, progressively maturing oocytes are arranged in an anterior to posterior progression within each ovariole.
Ovariole number spans three orders of magnitude across insects [9]. Several lines of evidence suggest that ovariole number is adaptive. First, ovariole number is a strong determinant of reproductive capacity, and thus is positively correlated to female fecundity and fitness [10–13]. Second, ovariole number is heritable and lineage-specific. Quantitative and developmental genetic analyses suggest that inter- and intraspecies variations in ovariole number are controlled through multiple loci [14–21]. Third, ovariole number shows latitudinal and altitudinal clinal variation on multiple continents [22,23]. In two cosmopolitan Drosophila species, D. melanogaster and D. simulans, ovariole number is greater in temperate populations than in tropical populations [24]. Finally, ovariole number is correlated with species ecology. Low ovariole numbers commonly evolve among ecological specialists, whereas generalists, or insects with more heterogeneous food sources, tend to evolve higher ovariole numbers [25–28].
Ovariole number exhibits strong phenotypic plasticity in response to larval rearing environment, particularly nutrition [29,30] and temperature [31]. Previous attempts to relate genetically fixed variation in and phenotypic plasticity of ovariole number in Drosophila concluded that different developmental mechanisms were responsible for species-specific ovariole number and ovariole number plasticity [29]. However, the underlying molecular mechanisms remained unknown. Many developmental genetic details underlying ovariole number determination have since emerged [32–34], allowing for molecular investigations of the basis for ovariole number determination and divergence. In the following paragraph, we describe the essential cellular behaviours involved in ovariole morphogenesis. These developmental events suggest specific candidate processes and molecular mechanisms that may underlie the evolution of variation in ovariole number.
Ovary morphogenesis in Drosophila begins with the specification of somatic gonad precursor cells in late embryogenesis [35]. Unlike most larval tissues in Drosophila, somatic ovarian cells proliferate continuously throughout larval life with no dramatic cell death [36]. Ovariole morphogenesis begins with the stacking of somatic ovarian cells into structures called terminal filaments (TFs) in the anterior of the larval ovary [37]. TF number at the larval–pupal (LP) transition stage directly determines adult ovariole number [29], and the number and morphogenesis of TF cells at LP stage determines TF number [32]. Somatic ovarian cells are then specified as anterior versus posterior cells, and a constant percentage of the anterior cells become TF cells [33]. Insulin and ecdysone signalling regulates TF cell number through modulating somatic ovarian cell proliferation, differentiation and morphogenesis [33,38,39]. This suggests that variation in hormonal signalling could underlie one or both of species-specific ovariole number and the phenotypic plasticity of ovariole number.
Here, we examine the role of insulin/insulin-like growth factor signalling (IIS) in the determination of mean ovariole number and the phenotypic plasticity of ovariole number in Drosophila. Furthermore, we use a comparison of two Drosophila species, D. melanogaster and D. sechellia, to investigate the hypothesis that the same molecular mechanism regulates both species-specific values and phenotypic plasticity of the same trait, ovariole number.
2. Methods
(a). Drosophila strains, culture conditions and diet manipulations
The following strains were used as wild-type strains for species comparisons: D. melanogaster OregonR-C (Bloomington Drosophila Stock Center (BDSC) no. 5, gift of the Hartl laboratory, Harvard University); D. sechellia Robertson strain (UC San Diego Drosophila Species Stock Center (DSSC) no. 14021-0248.25, gift of the Hartl laboratory); D. simulans (DSSC no. 14021-0251.194); and D. erecta (DSSC no. 14021-0224.01). To evaluate the amount of intraspecies variation in ovariole number, we counted adult ovariole number in isofemale lines of D. melanogaster and D. sechellia. Both tropical and temperate populations of D. melanogaster were considered. Tropical D. melanogaster isofemale lines, established from a population in Zambia, were a gift of the Flatt Laboratory (University of Lausanne). North American D. melanogaster isofemale lines (derived from females collected in Nevada, Catalina Island, CA, Santa Fe, NM and Raleigh, NC) were a gift of the DePace laboratory (Harvard Medical School). D. sechellia isofemale lines were a gift of the Hartl laboratory.
To examine IIS function in D. melanogaster, the following lines were used: the InR339 hypomorphic allele [40,41], a gift of the Hafen laboratory (ETH Zurich); the InRGC25 inversion allele ([42], BDSC no. 9554); and the Df(3R)Exel6186 deficiency allele (BDSC no. 7647).
To determine the role of systemic IIS from brain-derived peptides, we genetically ablated the principal insulin-producing cells of the brain. We used the dilp2-Gal4 driver [43], which is expressed specifically in the paired small clusters of medial neurosecretory cells that are known to produce Drosophila insulin-like peptides. We crossed this driver to the UAS-rpr (BDSC no. 5824) line to drive expression of the proapoptotic gene reaper.
To determine the responsiveness of somatic ovarian cells to IIS, we altered expression of the Drosophila insulin-like receptor InR specifically in the ovary by using the c587-GAL4 driver, which is expressed specifically in somatic ovarian cells beginning in the third larval instar ([44], gift of the Drummond-Barbosa laboratory, Johns Hopkins University). We crossed this driver to the following UAS lines to alter InR activity: UAS-InRExel (BDSC no. 8262), UAS-InRK1409A (BDSC no. 8259) and UAS-InRRNAi (BDSC no. 31037).
All adult ovariole counts and LP transition stage TF counts were performed as previously described [32]. Tibia length (adult females) was used as an adult body size proxy, as it has been previously demonstrated to correlate positively with body mass, which is indicative of overall body size [45].
Flies were maintained on standard laboratory diet (32 g Torula yeast, 60.5 g corn meal, 128 g dextrose, 9.2 g agar per litre). In all diet manipulation experiments, flies were raised from egg through to adult on the specified diet. Rich diet for all analyses consisted of standard laboratory diet supplemented with active dry yeast. Poor diet consisted of standard laboratory diet diluted with 3% agar in a ratio of 1 : 3 (25% final concentration standard laboratory diet) with no dry yeast supplementation. Wortmannin (EMD Millipore) was dissolved in 100% methanol and added to standard laboratory diet at 1% v/v. All rearing and experiments were performed at 25°C and 60–70% humidity.
(b). Quantitative PCR
As one measure of IIS pathway activity, we measured levels of Thor transcript [46]. Total RNA was extracted from ten biological replicates of five whole wandering third-instar females that were grown on rich diet. RNA was extracted using Trizol (Invitrogen), treated with TURBO DNase-I (Ambion, Life Technologies), and phenol–chloroform extracted. cDNA was prepared using oligo-dT primers and 0.5 μg RNA per reaction with Superscript III First Strand Synthesis Kit (Invitrogen). qPCR was performed using PerfeCta SYBR Green SuperMix, Low Rox (Quanta Biosciences). gapdh1 was used to normalize RNA levels and rp49 was used as expression control. Primer pairs were designed for use with both species templates. Primers were verified by performing species-specific standard curves for each primer pair, and showed less than 2.5% difference in amplification efficiency between species. Primer pairs were as follows: gapdh1-f, AGCCGAGTATGTGGTGGAGT, gapdh1-r, GGCTGTAGGCGTCCAGGTTA; Thor-f, AGCTAAGATGTCCGCTTCACC, Thor-r, TTTGGTGCCTCCAGGAGTGG; rp49-f, TGCTAAGCTGTCGCACAAATG, rp49-r, TTCTTGAATCCGGTGGGCAG.
(c). Immunohistochemistry, confocal imaging and analysis
Immunostaining was carried out as previously described [32]. The following primary antibodies were used: mouse 4D9 anti-Engrailed (1 : 40, Developmental Studies Hybridoma Bank); rabbit anti-Vasa (1 : 500, gift of P. Lasko, McGill University); and rabbit anti-phospho-Drosophila Akt (Ser505) (1 : 200, Cell Signaling Technology no. 4054). Secondary reagents used were Hoechst 33342 (Sigma, 1 : 1000 of 10 mg ml−1 stock solution), goat anti-mouse Alexa 488, goat anti-guinea pig Alexa 488 and donkey anti-rabbit Alexa 555 (1 : 500, Invitrogen). Samples were mounted in Vectashield (Vector laboratories) and imaged using a Zeiss LSM 780 confocal microscope.
Phosphorylated Akt (phospho-Akt) staining was quantified by measuring mean fluorescence signal intensity from maximum projection images composed of an equal number of confocal z-slices for each ovary. Secondary-only controls (figure 1b,d) indicate that the staining detected (figure 1a,c) and measured (figure 2b) is not background signal. A standard area of specifically anterior somatic ovarian cells, the cells from which TF precursor cells are specified, was analysed. Phospho-Akt intensity was normalized to mean DNA (Hoechst 33342) staining intensity to control for potential differences due to specific immunostaining experiments. Images were analysed with ImageJ v. 1.45I.
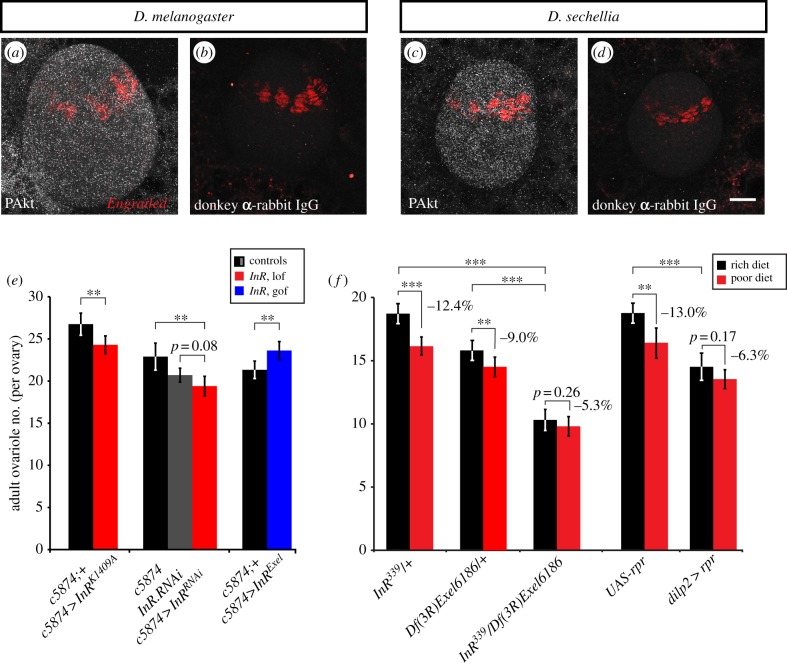
Figure 1.
Insulin signalling regulates ovariole number determination and plasticity in Drosophila. (a–d) IIS activity in larval ovaries of both D. melanogaster and D. sechellia visualized by phosphorylated Akt (pAkt: white; maximum projection of optical sections through whole ovary). Engrailed (red) marks terminal filament precursor cells. (b,d) Secondary antibody-only controls. Scale bar, 20 µm. (e) Adult ovariole number in females with ovary-specific expression of InR alleles driven by the c587-GAL4 driver. n ≥ 20 ovaries for all genotypes. For InRK1409A and InRExel, controls are siblings carrying a balancer chromosome (black bars). lof, loss of function; gof, gain of function. (f) Adult ovariole number in females with loss of InR function (InR339/Df(3R)Exel6186) or with dILP-producing neurons ablated (dilp2-GAL4>UAS:rpr), reared on rich or poor diets. n ≥ 20 for all genotypes. In (e,f), error bars show 95% CI of means. Student's t-test: ***p < 0.001, **p < 0.01.
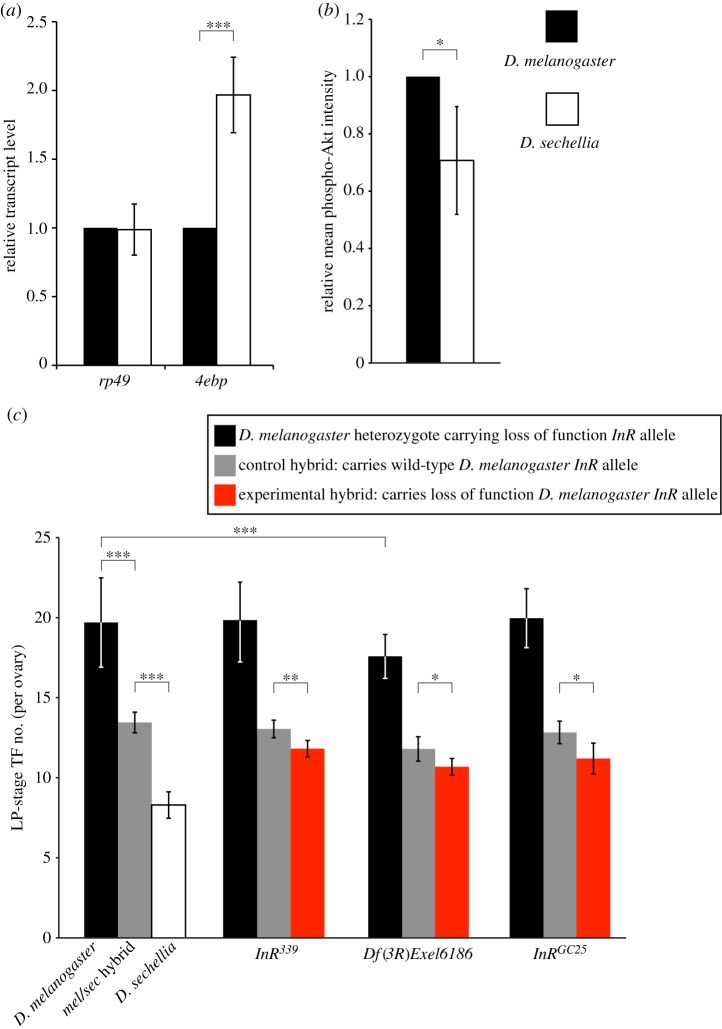
Figure 2.
Differential IIS activity exists between D. melanogaster and D. sechellia. Quantified levels of (a) larval expression of the growth attenuator Thor (normalized to gapdh1) and the ribosomal protein gene rp49 and (b) phospho-Akt intensity in wandering third-instar larval ovaries of D. melanogaster compared with D. sechellia. (c) Left-most set of bars shows TF number in D. melanogaster (black bar), D. melanogaster/D. sechellia hybrid (grey bar) and D. sechellia (white bar) females. Remaining sets show adult ovariole number in D. melanogaster females heterozygous for InR loss of function mutation (black bars), and final TF number in D. melanogaster InR*/D. sechellia hybrids (where InR* is any of three different InR loss of function alleles; red bars) compared with D. melanogaster InR+/D. sechellia control hybrids (grey bars). Controls are sisters carrying a wild-type copy of D. melanogaster InR. n ≥ 10 for all genotypes. Error bars show 95% CI of means. Student's t-test in (a,c); Mann–Whitney (Wilcoxon) test in (b): ***p < 0.001, **p < 0.005, *p < 0.05.
(d). Statistical analyses
Student's t-test was used for all pairwise comparisons of differences in means unless otherwise noted. Bonferroni adjustment for multiple comparisons was performed as appropriate. Mann–Whitney (Wilcoxon) test was used to evaluate differences in phospho-Akt staining intensity. To evaluate differences in interpopulational variation in ovariole number, Bartlett's test was used, as this test does not assume homogeneity of the variance of species-specific variances. Homo-/heterogeneity of species-specific variances was tested with Welch ANOVA (Welch t) to account for differences in mean values. Correlations, where noted, were evaluated by least-squares linear regression of mean values for each genotype. Statistical analyses were performed in Excel and JMP Pro v. 11.
3. Results
(a). Role of systemic IIS in determining ovariole number
We previously showed that loss of function of the Drosophila insulin-like receptor (InR) in D. melanogaster significantly reduces TF number by reducing both the number of somatic gonad precursor cells and the subsequent somatic cell proliferation rate throughout larval life [33]. To determine whether TF number reduction in InR mutants is due to autonomous IIS activity in somatic ovarian cells rather than through an indirect mechanism, we first asked whether IIS is active in ovarian cells at the relevant developmental time. Phosphorylated Akt (phospho-Akt) protein, an indicator of active IIS, was detectable at levels above background in wandering third larval instar ovaries, the time at which TF cells are proliferating and TFs are forming (figure 1a–d; compare a with b, and c with d). Phospho-Akt was also detected at above-background levels in the fat body, however at lower levels than in the ovary (figure 1a–d). We then used the somatic ovary-specific driver c587-GAL4 to abrogate or increase IIS specifically in the ovary. When IIS was decreased in the ovary either with the dominant negative InR allele K1409A, or with an InR RNAi construct, ovariole number was significantly decreased (p < 0.01 for InRK1409A, p < 0.01 and p = 0.08 for c587-GAL4 and UAS:InRRNAi parental controls, respectively; figure 1e). Conversely, when IIS was increased with overexpression of wild-type InR (InRExel), ovariole number was significantly increased (p < 0.01; figure 1e). As expected owing to the use of an ovary-specific GAL4 driver, these changes in ovariole number were not simple consequences of changes in body size (see electronic supplementary material, appendix S1). Finally, to determine whether systemic IIS from brain-derived insulin-like peptides (dILPs) regulates ovariole number determination, we genetically ablated insulin-producing neurons by using a dilp2-GAL4 driver to overexpress the proapoptotic gene reaper (rpr). Adult ovariole number was significantly reduced in dilp2 > rpr females compared with UAS:rpr control females (figure 1f; p < 0.001). Taken together, these results show that systemic IIS from brain-derived dILPs controls autonomous somatic ovarian cell proliferation, and modulation of IIS leads to changes in ovariole number.
(b). Role of IIS in nutritional plasticity of ovariole number
Systemic IIS is nutritionally controlled [47]. To test whether IIS mediates the nutritional plasticity of ovariole number, we reared flies with wild-type or modulated levels of IIS on rich or poor diets (see Methods). Like in wild-type flies [32], ovariole numbers were significantly reduced by poor diet in heterozygotes for InR loss of function mutations or UAS:rpr controls (figure 1f; p < 0.001 and p < 0.01, respectively). Body size of flies with altered IIS levels showed a more variable response to poor diet than wild-type flies, and body size was not a reliable predictor of ovariole number across genotypes (see electronic supplementary material, appendix S2). However, InR loss of function mutant and dilp2 > rpr females showed no statistically significant change in ovariole number on rich versus poor diet (figure 1f; p = 0.39 and p = 0.17, respectively). These results show that IIS is a molecular mediator of nutritional plasticity of ovariole number in D. melanogaster.
(c). IIS activity and sensitivity in D. melanogaster and D. sechellia
The melanogaster subgroup species D. melanogaster and D. sechellia, which diverged only five million years ago, have remarkably divergent mean ovariole numbers of 18.2 and 7.6, respectively. We previously showed that the heritable ovariole number difference between these species is caused by differences in somatic gonad precursor cell specification and somatic ovarian cell proliferation rate throughout larval life, and that InR loss of function mutants in D. melanogaster phenocopy both these differences [33]. We therefore hypothesized that IIS activity is reduced in D. sechellia compared with D. melanogaster. To test this hypothesis, we measured transcript expression of the growth attenuator Thor, which is negatively regulated by IIS [46]. 4E-BP, the protein product of the Thor transcript, is a known negative regulator of cell number in Drosophila [46]. We found that Thor expression in D. sechellia was significantly greater than in D. melanogaster (figure 2a; p < 0.001). In addition, we quantified the levels of phospho-Akt in the larval ovary of both species, and found that these levels were significantly higher in ovaries of D. melanogaster than of D. sechellia (figure 2b; p < 0.05). Taken together, these assessments of IIS activity indicate that IIS operates at higher levels in D. melanogaster than in D. sechellia. Consistent with these results, body size of D. sechellia is significantly smaller than that of D. melanogaster (see electronic supplementary material, appendix S4; p < 0.001). This suggests that evolutionary changes in IIS contribute to the divergence in ovariole number between these two species.
To further test for species-specific differences in IIS-mediated control of TF number between D. melanogaster and D. sechellia, we used interspecies hybrid complementation tests. Previous quantitative genetics analysis suggested that the InR locus may contribute to interspecies variation in ovariole number [15]. Therefore, we crossed D. melanogaster females carrying InR loss of function mutations with D. sechellia males, and counted TF number in resulting melanogaster/sechellia hybrids. Hybrids carrying mutant InR alleles from D. melanogaster had significantly reduced body size (see electronic supplementary material, appendix S3) and TF number compared with control hybrids carrying a wild-type D. melanogaster InR allele (figure 2c; p < 0.05 for InRGC25 and Df(3R)Exel6186; p < 0.01 for InR339). This suggests that the wild-type D. melanogaster InR allele may confer a higher level of IIS than the D. sechellia allele.
Because D. sechellia InR mutants are not available, we could not test this hypothesis directly by creating hybrids carrying a loss of function D. sechellia InR allele and a wild-type D. melanogaster InR allele. However, if our interpretation is correct, then the decrease in ovariole number caused by loss of one functional D. melanogaster InR allele should be less severe in D. melanogaster heterozygotes than in melanogaster/sechellia hybrids. Consistent with our hypothesis, adult ovariole number in D. melanogaster InR loss of function heterozygotes was not significantly different from wild-type (Oregon R) for two different InR alleles, InRGC25 and InR339 (figure 2c; p = 0.20, 0.34, respectively). For a third D. melanogaster InR loss of function allele, Df(3R)Exel6186, adult ovariole number was significantly lower than wild-type (figure 2c; p < 0.001), but the degree of reduction in ovariole was somewhat lower than that seen in the melanogaster/sechellia hybrid for the same InR allele (12.5% versus 13.6% reduction in ovariole number; figures 1f and 2c). In summary, with these experiments, we have compared the decrease in ovariole number caused by heterozygosis for a loss of function D. melanogaster InR allele in D. melanogaster heterozygotes versus D. melanogaster/D. sechellia hybrids, and shown that the ovariole number decrease is higher in the interspecies hybrids (figure 2c). Overall, these results are consistent with our hypothesis that the wild-type D. melanogaster InR allele confers a higher level of IIS than the wild-type D. sechellia allele, consistent with IIS activity being higher in D. melanogaster compared with D. sechellia. Taken together, these data demonstrate that IIS activity differs between D. melanogaster and D. sechellia, and that this activity difference contributes to species-specific ovariole number.
IIS sensitivity controls differential plastic response to nutrition in several insect species [48–50]. To determine how IIS activity difference could influence nutrition-dependent plasticity of ovariole number, we fed flies food containing Wortmannin, a specific inhibitor of PI3K [51], in a graded concentration series. Consistent with the results of genetic manipulation of IIS (figures 1e,f and 2c), body size (see electronic supplementary material, appendix S4) and ovariole number (figure 3a) were reduced in a dose-dependent manner in both species when grown on food containing Wortmannin. However, at all tested concentrations of Wortmannin, ovariole number was more significantly reduced in D. melanogaster than in D. sechellia (figure 3a). This indicates that ovariole number is more sensitive to changes in IIS in D. melanogaster than in D. sechellia. Together with our finding of evolved differences in IIS between the two species, this also shows that higher IIS activity in D. melanogaster is correlated with higher sensitivity to changes in IIS compared with D. sechellia.
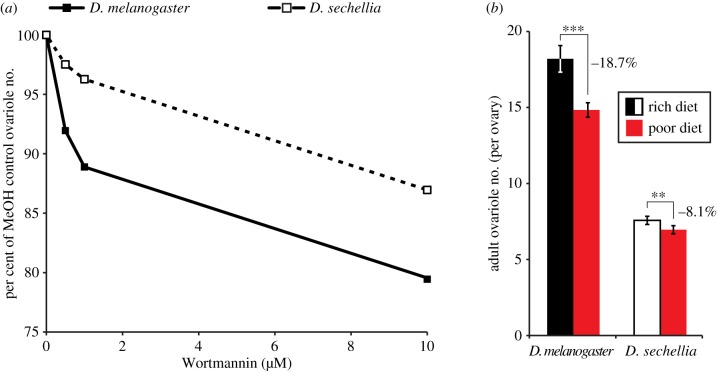
Figure 3.
Differential IIS sensitivity between D. melanogaster and D. sechellia correlates with species-specific nutritional plasticity of ovariole number. (a) Dose-dependent Wortmannin-induced decrease in adult ovariole number in D. melanogaster and D. sechellia, shown as percentage decrease relative to flies reared on control food containing methanol (Wortmannin solvent). n = 20 for each species at all concentrations except D. sechellia at 10 µM, n = 6 (owing to low eclosion rate of D. sechellia at high Wortmannin concentrations). (b) Adult ovariole number in D. melanogaster and D. sechellia reared on poor and rich diets. Error bars show 95% CI of means. n ≥ 20 for all genotypes and conditions. Student's t-test: ***p < 0.001, **p < 0.005. (Online version in colour.)
(d). Correlation between IIS sensitivity and nutritional plasticity
To test whether evolved differences in IIS activity levels and sensitivity could yield differences in nutritional plasticity between species, we measured ovariole number nutritional plasticity for D. sechellia. As in D. melanogaster [32], poor diet reduced ovariole number in D. sechellia, but only by 8.1%, in contrast to 18.7% in D. melanogaster (figure 3b). Body size was significantly reduced by poor diet in D. melanogaster (p < 0.001), whereas in D. sechellia body size was reduced numerically but not significantly (p = 0.08; electronic supplementary material, appendix S5). These data demonstrate that evolutionary change in IIS underlies the divergence of both mean ovariole number and the nutritional plasticity of ovariole number between these two Drosophila species.
(e). Interpopulational variation in ovariole number
If plasticity promotes diversification by providing a range of phenotypes whose underlying genetic variation can be subject to selection, then modulating the degree of plasticity may lead to differences in interpopulational divergence. Having observed that the degree of nutritional plasticity in ovariole number has diverged between D. melanogaster and D. sechellia, we asked whether interpopulational variation in ovariole number also differs between these species. We measured mean ovariole number for multiple isofemale lines from both species, and observed greater between-population variation for ovariole number in D. melanogaster compared with D. sechellia (figure 4a and the electronic supplementary material, appendix S6; p < 0.001). Although D. sechellia occupies an exclusively tropical habitat whereas D. melanogaster is distributed worldwide (figure 4b), even when considering variation within a tropical D. melanogaster population, variation is significantly greater in D. melanogaster compared with D. sechellia (figure 4a, p < 0.05). Genetic variation in D. sechellia is known to be lower than that of other melanogaster group species [52], and it is possible that this contributes to its reduced interpopulational variation in ovariole number. However, we argue that IIS-dependent plasticity provides a proximate molecular mechanism for the evolutionary divergence of ovariole number. Our data are consistent with the idea that plasticity plays a central role in diversifying ovariole number not only between species, but also within species.
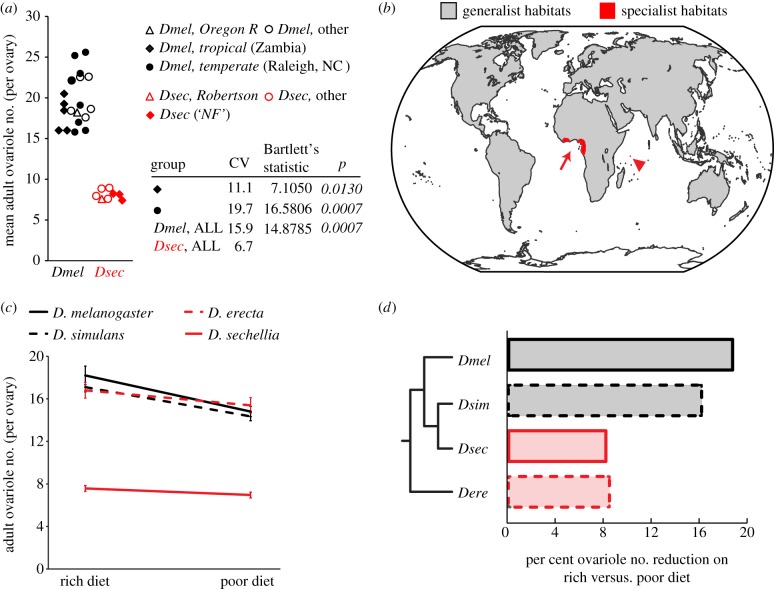
Figure 4.
Degree of plasticity correlates with relative interpopulational variation and ecological niche. (a) Range of ovariole number in different populations of D. melanogaster and D. sechellia. CV = coefficient of variation. Bartlett's test was used to compare variance of all D. sechellia populations with variance of indicated D. melanogaster groups. p-Values indicating a significant difference are italicized. (b) Global species distributions of the generalists D. melanogaster and D. simulans (worldwide distributions: grey); the specialists D. erecta (red arrow: specializes on fruits of the genus Pandanus in west Africa) and D. sechellia (red arrowhead: specializes on the fruit M. citrifolia in the Seychelles, which is toxic to other Drosophila species) have limited habitats. (c) Reaction norm of ovariole number on rich versus poor diet in four Drosophila species. Poor diet reduces ovariole number more in cosmopolitan generalist species (black lines) than in specialist species (red lines). Error bars show 95% CI of means. n ≥ 20 for all genotypes and conditions. (d) The phylogenetic relationship between the four species analysed is shown to the left of a bar graph showing per cent ovariole number reduction on different diets.
(f). Correlation between ecology and nutritional plasticity of ovariole number
Finally, we asked whether nutrition-dependent plasticity of this critical reproductive trait was linked to broader ecological patterns of ovariole number diversity, which may indicate an adaptive value of nutritional plasticity. Mean ovariole number is correlated with nutritional host preference in many insect species from a range of global habitats [25–27]. Specifically, species that have a wide host preference (generalists) or feed on abundant food sources tend to have more ovarioles than species that feed on a restricted niche (specialists) or scarce food sources. This correlation has been used to support the idea of adaptive value of ovariole number in terms of r–K selection theory [11,25,28]. Briefly, higher ovariole numbers permitting a larger number of offspring (r-selection) would be favoured when host substrates are numerous and nutritionally rich, whereas restricted substrates would favour production of fewer offspring and hence decreased ovariole number (K-selection). Given our finding that mean ovariole number and nutritional plasticity of ovariole number are controlled by the same molecular mechanisms, we predicted that nutritional plasticity would also correlate with range of host preference. Consistent with our hypothesis, we found that the cosmopolitan generalist species D. melanogaster and D. simulans (figure 4b) show high ovariole number plasticity and moderate body size plasticity in response to nutrition, whereas the specialist species D. sechellia [53] and D. erecta [54] show low nutritional plasticity and no significant change in body size (figure 4c,d and the electronic supplementary material, appendix S5). Because all species were reared on standard laboratory medium rather than native diets, we cannot rule out the possibility that our observed ovariole numbers and associated phenotypes may be affected by the use of a standard, non-native diet that was necessary to allow us to make comparisons between species. We note, however, that in the case of D. sechellia, ovariole number reported here is the same as that reported when D. sechellia is reared on its host plant Morinda citrifolia [53], suggesting that it may indeed be the degree of food source specialization, rather than a specific food source, which is the relevant parameter influencing ovariole number and its plasticity. Furthermore, low nutritional plasticity in D. erecta, which specializes on the non-toxic Pandanus genus of plants, indicates that this effect is not an artefact of the toxicity of M. citrifolia to other Drosophila species. Our experiments thus demonstrate that Drosophila species differ in their sensitivity to nutritional input, and suggest that relative IIS activity level may mediate these sensitivity differences.
Differences in plasticity lead to different relative ovariole numbers, and hence different relative reproductive capacities, between species in different environments (figure 4c). These results imply that the degree of nutritional plasticity in ovariole number may be subject to selection, and has diverged across species in response to ecological niche. If specific nutritional plasticity is an adaptation to host preference range, then variation in IIS levels and sensitivity could provide a proximate mechanism for the observed correlation between mean ovariole number and host preference.
4. Discussion
Ovariole number is believed to be under stabilizing selection [17], and environmental changes cannot increase ovariole number beyond a lineage-specific maximum [10,55]. Evolution of reduced ovariole number has occurred convergently in many insect lineages [33], and is correlated with occupation of specialist ecological and nutritional niches [25–27]. Consistent with these observations, we suggest that nutritional plasticity and reproductive capacity may present a trade-off dependent on relative IIS activity: high IIS activity can increase mean ovariole number, but at the cost of strongly reducing ovariole number in poor nutritional conditions (figure 4c). Because increased IIS also correlates with shortened lifespan [56,57], it is also possible that evolution of low plasticity due to low IIS levels could confer the advantage of an increased lifespan that is relatively robust to changes in nutritional conditions. Although we cannot yet determine which of these traits is the target of selection, we suggest that evolutionary diversification of both ovariole number and its nutritional plasticity occurs through genetic changes that modulate IIS activity and sensitivity. Our data show that a functional consequence of evolutionary changes in IIS activity and sensitivity is modulation of plastic range between species, and that this range is correlated with interpopulational diversification. We previously showed that different developmental mechanisms, which are genetically separable, contribute to ovariole number evolution [33]. We hypothesize that these alternative mechanisms may be targets of evolution for generating population-specific ovariole number while maintaining species-specific plastic responses.
While we have demonstrated that IIS has diverged between Drosophila species, what remains to be elucidated are the specific loci responsible for this divergence. Our data, particularly the interspecies hybrid complementation results, are consistent with the hypothesis that evolutionary change at the InR locus contributes to interspecies variation in ovariole number. Cross-species transgenesis and in-depth genetic analysis of IIS differences between species will be necessary to address this problem. We note here that both coding and noncoding differences exist between D. melanogaster and D. sechellia at the InR locus, none of which suggest obvious candidates for functional divergence. The protein coding sequences are 97% identical between these two species, and none of the amino acid changes occur within the known kinase domain. This suggests that slight structural or non-kinase activity-related alterations in the InR protein could modulate signalling in such a way as to contribute to phenotypic change. Natural variation in a coding region indel polymorphism in InR among D. melanogaster populations is consistent with this hypothesis [58].
IIS in multicellular animals is a conserved mechanism that coordinates cellular growth and proliferation with physiological condition, particularly nutritional state. The regulation of IIS contributes to evolutionary change within invertebrate and vertebrate species [49,59]. We have now shown that the regulation of IIS can underlie evolutionary morphological diversity both within and between species. Interestingly, evidence from functional studies in D. melanogaster and in horned beetles suggests that both increasing [48] and decreasing [49] IIS can reduce nutritional plasticity. This suggests that IIS may be able to act as a nutritional stress response system that is either environment-sensitive or environment-insensitive. Ovariole number in Drosophila (this study) and ornament size in horned beetles [49] appear to be examples of environment-sensitive nutritional stress responses, allowing generation of more offspring or exaggerated ornaments when food is plentiful, and restricting investment in these traits when food is scarce. An example of environment-insensitive nutrient stress response may be external genitalia in Drosophila, which continue to devote resources to growth despite unfavourable environmental conditions [48]. Given the wide conservation of IIS-mediated growth response, this work suggests a potentially pervasive role of plasticity in generating adaptive diversity.
Acknowledgements
D.A.G. is supported by an NSF pre-doctoral fellowship and a Ford Foundation Dissertation Fellowship. This work was partially supported by NIH grant 1R01-HD073499. The authors declare no competing financial interests. D.A.G. and C.G.E. designed research; D.A.G. performed experiments and collected data; D.A.G. and C.G.E. analysed data and wrote the paper; C.E. obtained funding for the research. We thank Julien Ayroles, Ernst Hafen, Daniel Hartl, Paul Lasko, Peter Klepsatel, Thomas Flatt, Akhila Rajan, Norbert Perrimon, Sarah Saminadin-Peter and Angela DePace for reagents, the Extavour laboratory for discussion, and Yuichiro Suzuki and James Mallet for comments on the manuscript.
References
- 1.Moczek AP, Sultan S, Foster S, Ledon-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705–2713 (doi:10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West-Eberhard MJ. 2003. Developmental plasticity and evolution, p. 794 Oxford, UK: Oxford University Press [Google Scholar]
- 3.Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 3811, 563–565 (doi:10.1038/150563a0) [DOI] [PubMed] [Google Scholar]
- 4.Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. 1996. Development, plasticity and evolution of butterfly eyespot patterns. Nature 384, 236–242 (doi:10.1038/384236a0) [DOI] [PubMed] [Google Scholar]
- 5.Rutherford AL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (doi:10.1038/24550) [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y, Nijhout HF. 2006. Evolution of a polyphenism by genetic accommodation. Science (New York, NY) 311, 650–652 (doi:10.1126/science.1118888) [DOI] [PubMed] [Google Scholar]
- 7.Abouheif E, Wray GA. 2002. Evolution of the gene network underlying wing polyphenism in ants. Science 297, 249–252 (doi:10.1126/science.1071468) [DOI] [PubMed] [Google Scholar]
- 8.Bloom S, Ledon-Rettig C, Infante C, Everly A, Hanken J, Nascone-Yoder N. 2013. Developmental origins of a novel gut morphology in frogs. Evol. Dev. 15, 213–223 (doi:10.1111/ede.12035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büning J. 1994. The insect ovary: ultrastructure, previtellogenic growth and evolution, p. 400 London, UK: Chapman and Hall [Google Scholar]
- 10.Cohet Y, David JR. 1978. Control of the adult reproductive potential by preimaginal thermal conditions. Oecologia 36, 295–306 (doi:10.1007/BF00348055) [DOI] [PubMed] [Google Scholar]
- 11.Bouletreau-Merle J, Allemand R, Cohet Y, David JR. 1982. Reproductive strategy in Drosophila melanogaster: significance of a genetic divergence between temperate and tropical populations. Oecologia 53, 323–329 (doi:10.1007/BF00389008) [DOI] [PubMed] [Google Scholar]
- 12.David JR. 1970. Le nombre d'ovarioles chez Drosophila melanogaster: relation avec la fécondité et valeur adaptive. Arch. Zool. Exp. Gen. 111, 357–370 [Google Scholar]
- 13.Klepsatel P, Galikova M, De Maio N, Ricci S, Schlotterer C, Flatt T. 2013. Reproductive and post-reproductive life history of wild-caught Drosophila melanogaster under laboratory conditions. J. Evol. Biol. 26, 1508–1520 (doi:10.1111/jeb.12155) [DOI] [PubMed] [Google Scholar]
- 14.Bergland AO, Genissel A, Nuzhdin SV, Tatar M. 2008. Quantitative trait loci affecting phenotypic plasticity and the allometric relationship of ovariole number and thorax length in Drosophila melanogaster. Genetics 180, 567–582 (doi:10.1534/genetics.108.088906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orgogozo V, Broman KW, Stern DL. 2006. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 173, 197–205 (doi:10.1534/genetics.105.054098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wayne ML, Hackett JB, Mackay TFC. 1997. Quantitative genetics of ovariole number in Drosophila melanogaster. I. Segregating variation and fitness. Evolution 4, 1156–1163 (doi:10.2307/2411045) [DOI] [PubMed] [Google Scholar]
- 17.Wayne ML, Mackay TFC. 1998. Quantitative genetics of ovariole number in Drosophila melanogaster. II. Mutational variation and genotype–environment interaction. Genetics 148, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayne ML, Hackett JB, Dilda CL, Nuzhdin SV, Pasyukova EG, Mackay TF. 2001. Quantitative trait locus mapping of fitness-related traits in Drosophila melanogaster. Genet. Res. 77, 107–116 (doi:10.1017/S0016672300004894) [DOI] [PubMed] [Google Scholar]
- 19.Wayne ML, McIntyre LM. 2002. Combining mapping and arraying: an approach to candidate gene identification. Proc. Natl Acad. Sci. USA 99, 14 903–14 906 (doi:10.1073/pnas.222549199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telonis-Scott M, Bono L, McIntyre L, Wayne M. 2005. Analyses of the quantitative genetic architecture of ovariole number and body size in Drosophila melanogaster using diallels. In 46th Annual Drosophila Conf., San Diego, CA, 30 March–3 April 2005, p. 806B [Google Scholar]
- 21.Telonis-Scott M, McIntyre LM, Wayne ML. 2005. Genetic architecture of two fitness-related traits in Drosophila melanogaster: ovariole number and thorax length. Genetica 125, 211–222 (doi:10.1007/s10709-005-8549-4) [DOI] [PubMed] [Google Scholar]
- 22.Collinge JE, Hoffmann AA, McKechnie SW. 2006. Altitudinal patterns for latitudinally varying traits and polymorphic markers in Drosophila melanogaster from eastern Australia. J. Evol. Biol. 19, 473–482 (doi:10.1111/j.1420-9101.2005.01016.x) [DOI] [PubMed] [Google Scholar]
- 23.Wayne ML, Korol A, Mackay TF. 2005. Microclinal variation for ovariole number and body size in Drosophila melanogaster in ‘evolution canyon’. Genetica 123, 263–270 (doi:10.1007/s10709-004-5056-y) [DOI] [PubMed] [Google Scholar]
- 24.Capy P, Pla E, David JR. 1993. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and D. simulans. I. Geographic variations. Genet. Sel. Evol. 25, 517–536 (doi:10.1186/1297-9686-25-6-517) [Google Scholar]
- 25.Kambysellis MP, Heed WB. 1971. Studies of oogenesis in natural populations of Drosophilidae. I. Relation of ovarian development and ecological habitats of the Hawaiian species. Am. Nat. 941, 31–49 (doi:10.1086/282700) [Google Scholar]
- 26.Fitt G. 1990. Variation in ovariole number and egg size of species of Dacus (Diptera; Tephritidae) and their relation to host specialization. Ecol. Entomol. 15, 255–264 (doi:10.1111/j.1365-2311.1990.tb00807.x) [Google Scholar]
- 27.Leather S, Wellings P, Walters K. 1988. Variation in ovariole number within the Aphidoidea. J. Nat. Hist. 22, 381–393 (doi:10.1080/00222938800770271) [Google Scholar]
- 28.Montague JT, Mangan RL, Starmer WT. 1981. Reproductive allocation in the Hawaiian Drosophilidae: egg size and number. Am. Nat. 118, 865–871 (doi:10.1086/283877) [Google Scholar]
- 29.Hodin J, Riddiford LM. 2000. Different mechanisms underlie phenotypic plasticity and interspeficic variation for a reproductive character in Drosophilds (Insecta: Diptera). Evolution 5, 1638–1653 [DOI] [PubMed] [Google Scholar]
- 30.Tu MP, Tatar M. 2003. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 2, 327–333 (doi:10.1046/j.1474-9728.2003.00064.x) [DOI] [PubMed] [Google Scholar]
- 31.Delpuech J-M, Noreteau B, Chiche J, Pla E, Vouidibio J, David JR. 1995. Phenotypic plasticity and reaction norms in temperate and tropical populations of Drosophila melanogaster: ovarian size and developmental temperature. Evolution 4, 670–675 (doi:10.2307/2410320) [DOI] [PubMed] [Google Scholar]
- 32.Sarikaya D, Assefa Belay A, Ahuja A, Green AD, Dorta A, Extavour CG. 2012. The roles of cell size and cell number in determining ovariole number in Drosophila. Dev. Biol. 363, 279–289 (doi:10.1016/j.ydbio.2011.12.017) [DOI] [PubMed] [Google Scholar]
- 33.Green DA, II, Extavour CG. 2012. Convergent evolution of a reproductive trait through distinct developmental mechanisms in Drosophila. Dev. Biol. 372, 120–130 (doi:10.1016/j.ydbio.2012.09.014) [DOI] [PubMed] [Google Scholar]
- 34.Bartoletti M, et al. 2012. Genetic basis for developmental homeostasis of germline stem cell niche number: a network of Tramtrack-Group nuclear BTB factors. PLoS ONE 7, e49958 (doi:10.1371/journal.pone.0049958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle M, DiNardo S. 1995. Specification, migration and assembly of the somatic cells of the Drosophila gonad. Development 121, 1815–1825 [DOI] [PubMed] [Google Scholar]
- 36.King RC. 1970. Ovarian development in Drosophila melanogaster, p. 227 New York, NY: Academic Press [Google Scholar]
- 37.Sahut-Barnola I, Godt D, Laski FA, Couderc J-L. 1995. Drosophila ovary morphogenesis: analysis of terminal filament formation and identification of a gene required for this process. Dev. Biol. 170, 127–135 [DOI] [PubMed] [Google Scholar]
- 38.Hodin J, Riddiford LM. 1998. The ecdysone receptor and ultraspriacle regulate the timing and progression of ovarian morphogenesis during Drosophila metamorphosis. Dev. Genes Evol. 208, 304–317 (doi:10.1007/s004270050186) [DOI] [PubMed] [Google Scholar]
- 39.Gancz D, Lengil T, Gilboa L. 2011. Coordinated regulation of niche and stem cell precursors by hormonal signaling. PLoS Biol. 9, e1001202 (doi:10.1371/journal.pbio.1001202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. 1995. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14, 3373–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 (doi:10.1016/S0960-9822(01)00068-9) [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Jack J, Garofalo RS. 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137, 846–856 [DOI] [PubMed] [Google Scholar]
- 43.Wu Q, Zhang Y, Xu J, Shen P. 2005. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl Acad. Sci. USA 102, 13 289–13 294 (doi:10.1073/pnas.0501914102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manseau L, et al. 1997. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209, 310–322 (doi:10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L) [DOI] [PubMed] [Google Scholar]
- 45.Catchpole RDJ. 1994. Wing length is not the best predictor of body size. Drosoph. Inf. Serv. 75, 84–86 [Google Scholar]
- 46.Puig O, Marr MT, Ruhf ML, Tjian R. 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 (doi:10.1101/gad.1098703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hietakangas V, Cohen SM. 2009. Regulation of tissue growth through nutrient sensing. Annu. Rev. Genet. 43, 389–410 (doi:10.1146/annurev-genet-102108-134815) [DOI] [PubMed] [Google Scholar]
- 48.Tang HY, Smith-Caldas MSB, Driscoll MV, Salhadar S, Shingleton AW. 2011. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 7, e1002373 (doi:10.1371/journal.pgen.1002373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337, 860–864 (doi:10.1126/science.1224286) [DOI] [PubMed] [Google Scholar]
- 50.Snell-Rood EC, Moczek AP. 2012. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS ONE 7, e34857 (doi:10.1371/journal.pone.0034857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yano H, Nakanishi S, Kimura K, Hanai N, Saitoh Y, Fukui Y, Nonomura Y, Matsuda Y. 1993. Inhibition of histamine secretion by wortmannin through the blockade of phosphatidylinositol 3-kinase in RBL-2H3 cells. J. Biol. Chem. 268, 25 846–25 856 [PubMed] [Google Scholar]
- 52.Legrand D, Tenaillon MI, Matyot P, Gerlach J, Lachaise D, Cariou M-L. 2009. Species-wide genetic variation and demographic history of Drosophila sechellia, a species lacking population structure. Genetics 182, 1197–1206 (doi:10.1534/genetics.108.092080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R'Kha S, Capy P, David JR. 1991. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetic analysis. Proc. Natl Acad. Sci. USA 88, 1835–1839 (doi:10.1073/pnas.88.5.1835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lachaise D, Carious M-L, David JR, Lemeunier F, Tsacas L, Ashburner M. 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Ecol. 22, 159–225 [Google Scholar]
- 55.Engstrom LE. 1971. Studies of the effects of two-way selection for ovariole number in Drosophila melanogaster. Urbana-Champaign, IL: University of Illinois at Urbana-Champaign [Google Scholar]
- 56.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107–110 (doi:10.1126/science.1057987) [DOI] [PubMed] [Google Scholar]
- 57.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104–106 (doi:10.1126/science.1057991) [DOI] [PubMed] [Google Scholar]
- 58.Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. 2010. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 19, 760–774 (doi:10.1111/j.1365-294X.2009.04508.x) [DOI] [PubMed] [Google Scholar]
- 59.Sutter NB, et al. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115 (doi:10.1126/science.1137045) [DOI] [PMC free article] [PubMed] [Google Scholar]