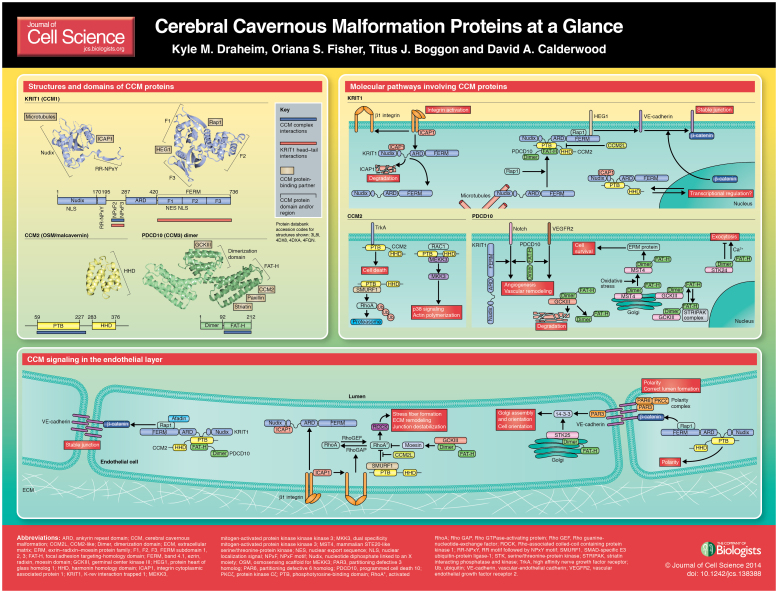
ABSTRACT
Loss-of-function mutations in genes encoding KRIT1 (also known as CCM1), CCM2 (also known as OSM and malcavernin) or PDCD10 (also known as CCM3) cause cerebral cavernous malformations (CCMs). These abnormalities are characterized by dilated leaky blood vessels, especially in the neurovasculature, that result in increased risk of stroke, focal neurological defects and seizures. The three CCM proteins can exist in a trimeric complex, and each of these essential multi-domain adaptor proteins also interacts with a range of signaling, cytoskeletal and adaptor proteins, presumably accounting for their roles in a range of basic cellular processes including cell adhesion, migration, polarity and apoptosis. In this Cell Science at a Glance article and the accompanying poster, we provide an overview of current models of CCM protein function focusing on how known protein–protein interactions might contribute to cellular phenotypes and highlighting gaps in our current understanding.
KEY WORDS: Cerebral cavernous malformations, CCM, Cell signaling, KRIT1, PCDC10, Rho Signaling, Vascular biology
Introduction
Cerebral cavernous malformations (CCMs) consist of clusters of enlarged endothelial channels (‘caverns’) that are arranged back-to-back to form densely packed sinusoids with little or no intervening brain parenchyma (reviewed in Cavalcanti et al., 2012; Fischer et al., 2013). These lesions lack smooth muscle and elastic tissue, so the vessel walls are thin, leaky and lack sub-endothelial support and an intact basal lamina. Ultra-structural analysis has revealed ruptures in the luminal endothelium (probably due to damaged intercellular junctions), endothelial detachment from the basal lamina and decreased numbers of pericytes (Tanriover et al., 2013). CCMs have been reported in up to 0.5% of the population and, although they are primarily found within the neurovasculature of the central nervous system (i.e. brain, spinal cord, retina), where they result in increased risk for stroke, seizures and focal neurological deficits, they are also seen in the skin and liver (Cavalcanti et al., 2012; Fischer et al., 2013). Currently, the only treatment for CCM is surgical resection.
CCMs are associated with loss-of-function mutations in any one of the three CCM genes: KRIT1 (Krev interaction trapped 1, also known as CCM1), CCM2 or PDCD10 (programmed cell death 10, also known as CCM3) (Cavalcanti et al., 2012) (see Box 1), suggesting that there is an essential pathway involving all three proteins (Stahl et al., 2008). This hypothesis is bolstered by the knowledge that all three proteins can be found in the same complex within the cell (see below). However, PDCD10 might also act separately from KRIT1 and CCM2, as its mutation often results in a more severe form of the disease (Denier et al., 2006; Zhu et al., 2011). Despite extensive analysis of CCM-knockout animals (see supplementary material Table S1) and recent advances in identifying binding partners and determining the structures of KRIT1, CCM2 and PDCD10, there still is limited knowledge of the mechanisms through which the loss of each of these proteins leads to CCM formation. Here, we review the current information on CCM protein signaling. In light of the CCM disease phenotype, we focus primarily on potential roles in vascular cells, but because all three CCM proteins are widely expressed, they might also have roles outside of the vasculature.
Box 1. The Genetics of CCM.
Familial CCM accounts for only ∼20% of cases but tends to be more severe than sporadic CCM (Cavalcanti et al., 2012), with patients exhibiting multiple lesions and increased hemorrhage rates. Familial CCM is associated with a heterozygous germline loss-of-function mutation in KRIT1, CCM2 or PDCD10 (Cavalcanti et al., 2012). Malformation development appears to require a local second hit to remove the remaining wild-type copy of the CCM gene (Akers et al., 2009; Dammann et al., 2013; Pagenstecher et al., 2009; Stahl et al., 2008), but other factors in the neurovasculature microenvironment could potentially also have a role in lesion formation (Boulday et al., 2011; Dammann et al., 2013). Local mutations in CCM genes also cause sporadic CCM (Limaye et al., 2009), and there is a growing list of disease-causing mutations, most of which are nonsense mutations leading to a loss of function. In 19 out of 20 of familial cases and two thirds of sporadic cases with multiple lesions, mutation in at least one of the CCM genes has been identified (Cavalcanti et al., 2012; Riant et al., 2013). In lesions with identified mutations, KRIT1 is mutated in 65% of cases, CCM2 in 19%, and PDCD10 in 16% (Riant et al., 2013; Stahl et al., 2008).
CCM proteins
KRIT1
KRIT1 is the largest of the CCM proteins. It contains an N-terminal Nudix domain followed by a stretch of three NPxY/F motifs (Liu et al., 2013), a predicted ankyrin-repeat domain and a C-terminal FERM (band 4.1, ezrin, radixin, moesin) domain (Gingras et al., 2013; Li et al., 2012) (see poster). No catalytic activity has been reported for KRIT1, and it is thought to signal through its many binding partners (see below). KRIT1 is ubiquitously expressed in early embryogenesis with pronounced endothelial expression in large vessels (Guzeloglu-Kayisli et al., 2004). KRIT1 is observed in many different cellular compartments and is actively shuttled between the cytoplasm and the nucleus (Zawistowski et al., 2005). A polybasic sequence within a KRIT1 Nudix domain loop is responsible for the targeting of KRIT1 to microtubules, although the mode of interaction is not yet completely understood (Béraud-Dufour et al., 2007). KRIT1 also localizes to endothelial cell boundaries or cell–cell junctions through its FERM domain (Glading et al., 2007; Zawistowski et al., 2005).
KRIT1 was initially identified in a yeast two-hybrid screen as a Rap1-binding protein (Serebriiskii et al., 1997). Recent crystallographic analysis revealed a novel mode of interaction between the KRIT1 FERM domain and the small GTPase Rap1 that represents a paradigm for how small G-proteins can bind and recognize FERM domains (Gingras et al., 2013; Li et al., 2012). Rap1 binding inhibits the binding of KRIT1 to microtubules (Béraud-Dufour et al., 2007), thereby enabling the relocalization of KRIT1 and the stabilization of cell–cell junctions (Liu et al., 2011). The molecular mechanisms by which this occurs are not yet known, but KRIT1 binds Rap1 and microtubules through different domains, suggesting that there is a conformational regulation controlling the subcellular localization of KRIT1 (discussed below). The KRIT1 FERM domain can also bind to the membrane anchor protein heart of glass 1 (HEG1), a protein essential for KRIT1 junction localization. The functional importance of these interactions is highlighted in zebrafish studies, as KRIT1 mutants that are unable to bind either Rap1 or HEG1 do not rescue the KRIT1-null (santa) phenotype, which is associated with defects in cardiovascular development (Gingras et al., 2012; Liu et al., 2011) (see supplementary material Table S1).
Another important binding partner of KRIT1 is integrin cytoplasmic domain associated protein-1 (ICAP1), a phosphotyrosine binding (PTB)-domain-containing protein that negatively regulates β1 integrin activation (Liu et al., 2013; Millon-Frémillon et al., 2008). ICAP1 binds KRIT1 in a bidentate mode, recognizing two regions: the highly-conserved RR region and the first of the three KRIT1 NPxY/F motifs (Liu et al., 2013). Importantly, ICAP1 uses the same binding site to interact with either the KRIT1 NPxY motif or the cytoplasmic tail of integrin β1. As ICAP1 cannot inhibit integrin activation when it is bound to KRIT1, increased integrin activation is observed when increasing amounts of KRIT1 are available to bind to ICAP1 (Liu et al., 2013). In endothelial cells, KRIT1 also appears to stabilize the ICAP1 protein, so KRIT1 loss leads to decreased ICAP1 levels and consequently increased β1 integrin activation (Faurobert et al., 2013). Thus, the exact role of KRIT1 in modulating ICAP1-mediated regulation of integrin activation in endothelial cells is complex and is still being discerned.
KRIT1 has also been linked to several other important signaling pathways. KRIT1 overexpression leads to increased expression of HEY1 and DLL4 (indicative of Notch activation) and, conversely, silencing of KRIT1 diminishes Notch signaling (Wüstehube et al., 2010). Notch signaling is linked to phosphoinositol 3-kinase (PI3K)/AKT activation and the repression of ERK1/2. Consistent with this, loss of KRIT1 in endothelial cells leads to increased ERK1/2 phosphorylation, and CCM lesions show increased ERK1/2 phosphorylation (Wüstehube et al., 2010). Loss of KRIT1 also reduces the expression of the reactive oxygen species (ROS)-scavenging enzyme SOD2 with consequent increases in the steady state levels of ROS, AKT phosphorylation, and AKT-dependent FOXO1 phosphorylation (and thus inactivation) (Goitre et al., 2010). Another KRIT1 interactor, the long isoform of Nd1 (Nd1-L), an actin cytoskeleton-stabilizing protein, was recently identified and data suggests that its co-expression with KRIT1 supports SOD2 expression (Guazzi et al., 2012). Collectively, KRIT1 is involved in multiple pathways that prevent cell death.
The conformational organization of KRIT1 appears to be important for its subcellular localization and consequent signaling. KRIT1 is thought to harbor intramolecular binding sites, i.e. its N-terminus can interact with its FERM domain (Béraud-Dufour et al., 2007; Francalanci et al., 2009). It is therefore possible that KRIT1, like other FERM proteins, adopts both open and closed conformations through a ‘head-to-tail’ interaction. The head–tail interaction probably occurs though the recognition of the KRIT1 NPxY/F motifs by the FERM domain, although the specificity of this interaction is still unclear (Faurobert and Albiges-Rizo, 2010). Changes in the conformation of KRIT1 are thought to regulate its localization; for example, microtubule binding is associated with a presumed ‘closed’ conformation (Francalanci et al., 2009) and ICAP1 binding is associated with a presumed ‘open’ conformation (Béraud-Dufour et al., 2007). The functional and signaling importance of KRIT1 conformational changes has not yet been adequately explored.
CCM2
CCM2 is a scaffolding protein with no enzymatic activity and an expression pattern very similar to that of KRIT1, including in arterial endothelial cells of multiple tissues (Petit et al., 2006; Seker et al., 2006). It acts as the hub of the CCM complex by simultaneously binding both KRIT1 and PDCD10, in addition to a number of other signaling proteins (Hilder et al., 2007a). CCM2 contains a predicted PTB domain at its N-terminus (Liquori et al., 2003) and has recently been shown to contain a helical domain at its C-terminus termed the harmonin-homology domain (HHD) (Fisher et al., 2013) (see poster).
CCM2 is found throughout the cell and can shuttle in and out of the nucleus, probably through its interaction with KRIT1 (Zhang et al., 2007). However, CCM2 binding has also been implicated in sequestration of the KRIT1–ICAP1 complex in the cytosol (Faurobert and Albiges-Rizo, 2010). CCM2 localization to endothelial cell–cell junctions is lost following the loss of KRIT1 localization to cell-cell junctions, suggesting that it is targeted there by KRIT1. Indeed, when re-expressed in CCM2-knockdown cells, wild-type CCM2 localizes to cell–cell junctions, but a mutant that cannot bind KRIT1 (CCM2-F217A) does not, implicating an interaction of the CCM2 PTB domain with one of the KRIT1 NPxY/F motifs in junctional targeting (Stockton et al., 2010).
The CCM2 PTB domain also binds TrkA, a receptor tyrosine kinase found on nerve cells (Harel et al., 2009). This interaction induces cell death in neuroblastoma or medulloblastoma, probably through the recruitment of a complex between PCDC10 and STK25, a member of the germinal center kinase III (GCKIII) group of serine/threonine kinases (Costa et al., 2012). How it induces cell death, and whether this pathway contributes to the CCM phenotype, remains unclear.
CCM2 is also known as osmosensing scaffold for MEKK3 (OSM) because it binds the mitogen-activated protein kinase (MAPK) kinase kinase MEKK3. It is required for hyperosmolar-induced p38 MAPK activation, and upon osmotic shock a significant portion of CCM2 relocalizes to membrane ruffles where CCM2 is thought to scaffold RAC1 and MEKK3 in the p38 MAPK cascade (Uhlik et al., 2003; Zawistowski et al., 2005). CCM2 binds F-actin in in vitro binding assays, suggesting that it organizes a complex that is capable of linking RAC1-dependent actin reorganization to p38 activity (Hilder et al., 2007b). Recent work confirms that osmotic stress elicits a response of the CCM2–RAC1 pathway, but indicates that the signaling might occur through phospholipase C (PLC)γ1 (Zhou et al., 2011). Conversely, another study suggests that CCM2 loss does not affect the p38 MAPK pathway, but rather affects JNK and MAPK kinase (MKK) signaling (Whitehead et al., 2009). The role of CCM2 in MAPK pathways is clearly complex, and a better understanding will require further study.
A significant recent advance in the field was the identification of CCM2-like (CCM2L), a protein with high sequence identity to CCM2 that is selectively expressed in activated endothelial cells (Zheng et al., 2012). Loss of zebrafish ccm2l phenocopies the ccm2-null (valentine) phenotype (supplementary material Table S1) and can be partially rescued by overexpression of CCM2 (Rosen et al., 2013). However, there is not complete overlap in functions, as CCM2L competes with CCM2 for binding to KRIT1, but not PDCD10, and functionally blocks CCM2-mediated junctional stability (Zheng et al., 2012). Whether CCM2L is linked to human disease remains to be determined.
PDCD10
PDCD10 is ubiquitously expressed and contains an N-terminal dimerization domain (Kean et al., 2011; Li et al., 2010) and a C-terminal focal adhesion targeting-homology (FAT-H) domain (Li et al., 2010) (see poster). It binds a variety of proteins including CCM2 (Hilder et al., 2007a; Voss et al., 2007), the GCKIII serine/threonine kinases (Fidalgo et al., 2010; Xu et al., 2013; Zhang et al., 2013a), paxillin (through its FAT-H domain) (Li et al., 2011), FAP-1 (also known as PTPN13) (Voss et al., 2007), protocadherin-γ (Lin et al., 2010), VEGFR (He et al., 2010), UNC13D (Zhang et al., 2013b) and striatin (through its FAT-H domain) (Goudreault et al., 2009; Kean et al., 2011). It also binds phosphotidylinositides (Dibble et al., 2010; Ding et al., 2010) (see poster).
The PDCD10 FAT-H domain interacts with CCM2 (Li et al., 2010), and PDCD10 is the third component in the heterotrimeric KRIT1–CCM2–PDCD10 complex (Hilder et al., 2007a). Its function in the CCM complex is still being explored, but PDCD10 also has roles outside of this complex. The best characterized of these involves dimerization-domain-mediated interactions with the GCKIII group of protein kinases, MST4/MASK, STK24/MST3 and STK25/YSK1/SOK1 (Sugden et al., 2013). PDCD10 predominately resides within the striatin interacting phosphatase and kinase (STRIPAK) complex where it binds to GCKIII kinases (Fidalgo et al., 2010). PDCD10 also binds striatin directly and thus complexes with protein phosphatase 2 (PP2) and other STRIPAK components indirectly, thereby linking GCKIII kinases to phosphatase PP2 (Goudreault et al., 2009). Alternatively, PDCD10–GCKIII localizes to the cis face of the Golgi complex through a GCKIII interaction with GOLGA2, an interaction mutually exclusive to the PDCD10–GCKIII interaction with STRIPAK. The PDCD10–GCKIII interaction is important for GCKIII localization as loss of PDCD10 shifts the binding of GCKIII kinases from the STRIPAK complex to the Golgi (Kean et al., 2011), leading to a loss of STK25 kinase activity, decreased protein stability and ultimately Golgi disassembly. Cells depleted of PDCD10 are impaired in repositioning both the Golgi complex and the centrosome towards the leading edge, which impairs cell migration (Fidalgo et al., 2010). Conversely, overexpression of PDCD10 or PDCD10–MST4 coexpression leads to increased cell migration, whereas mutants that are unable to bind one another do not (Zhang et al., 2013a). As directed migration is essential for blood vessel formation, its perturbation could lead to the vascular malformations found in CCM. PDCD10 also plays a role in exocytosis, and recent work shows that loss of either PDCD10 or STK24 increases neutrophil exocytosis owing to a loss of interaction with UNC13D, a regulator of vesicle fusion (Zhang et al., 2013b). This potentially implicates changes in exocytosis in the defective tubular morphology observed in CCM disease.
CCM lesions have poor endothelial integrity. Appropriate regulation of cell death is essential for maintenance of endothelial integrity and, as its name suggests, PDCD10 is thought to play a role in cell death. However, a specific role for PDCD10 in cell survival is not clear as both pro-survival and pro-apoptopic effects have been reported. PDCD10 was originally discovered to be upregulated during granulocyte apoptosis (Wang et al., 1999) and has been linked to several different signaling pathways involved in survival. For example, PDCD10 complexes and colocalizes with VEGFR2, and loss of PDCD10 leads to decreased stability of the VEGFR2 protein (He et al., 2010). PDCD10 also appears to induce VEGFR2 endocytosis after VEGF stimulation (He et al., 2010) and is required for the relocation of MST4 to the cell periphery after oxidative stress, where it phosphorylates and activates ERM proteins, thereby promoting cell survival (Fidalgo et al., 2012). Conversely, PDCD10 expression has been linked to cell death (Chen et al., 2009; Lin et al., 2010; Zhu et al., 2010) and loss of PCDC10 has been reported to increase survival and proliferation, possibly through reduced Notch signaling, enhanced VEGF signaling, or increased ERK activity (Louvi et al., 2011; You et al., 2013; Zhu et al., 2010). How these conflicting observations can be reconciled remains to be determined.
The role of CCM proteins in RhoA–ROCK signaling
The first indication that RhoA dysregulation might contribute to CCM pathology came from the observation of increased stress fiber formation (a sign of activated RhoA) after RNA-interference-mediated knockdown of any of the CCM proteins (Crose et al., 2009; Glading et al., 2007; Stockton et al., 2010; Whitehead et al., 2009; Zheng et al., 2010). Consistent with this, activated (GTP-bound) RhoA is increased in KRIT1-, CCM2- or PDCD10-deficient endothelial cells. One of the primary effectors of activated RhoA is the serine/threonine kinase ROCK, which increases actomyosin contractility by phosphorylating and inhibiting the myosin light chain (MLC) phosphatase. Knockdown of KRIT1, CCM2 or PDCD10 increases the amount of phosphorylated MLC, whereas treatment with ROCK-inhibitors reverses this increase and the stress fiber accumulation (Borikova et al., 2010; Stockton et al., 2010; Whitehead et al., 2009), confirming a role for RhoA–ROCK in CCM signaling.
How CCM proteins influence RhoA has still not been fully elucidated, but a recent report implicates β1 integrin signaling (Faurobert et al., 2013). In addition, CCM2 appears to direct the degradation of RhoA through a CCM2 PTB-domain-mediated interaction with the E3 ubiquitin ligase Smad ubiquitin regulatory factor 1 (SMURF1) (Crose et al., 2009). Notably, loss of CCM2 leads to increases in RhoA, but not of other SMURF1 substrates, suggesting that CCM2 might selectively promote SMURF1-mediated RhoA degradation. Increased RhoA activity is also seen when the PDCD10-binding partner STK25 (a GCKIII serine/threonine kinase) is knocked down (Zheng et al., 2010), suggesting that membrane-localized actin-associated complexes that are regulated by the PDCD10–STK25–ERM pathway might ultimately control RhoA activation. Cells that lack KRIT1, CCM2 or PDCD10 are defective in migration, invasion, three-dimensional tube formation and maintenance of a monolayer permeability barrier. Each of these functions can be rescued by ROCK inhibition (Borikova et al., 2010; Stockton et al., 2010; Whitehead et al., 2009). ROCK inhibitors also rescue lipopolysaccharide-induced vascular leak in KRIT1- and CCM2-deficient mice (Stockton et al., 2010) and inhibit the formation of vascular lesions in other mouse models (McDonald et al., 2012). Targeting RhoA–ROCK signaling is therefore one potential therapeutic strategy for CCM (Li and Whitehead, 2010).
Vascular polarity and permeability in CCMs
In animal models, KRIT1, CCM2 and PDCD10 are essential for cardiovascular development (supplementary material Table S1). Loss of either KRIT1 or PDCD10 leads to an induction of angiogenesis by impaired Delta–Notch signaling, and PDCD10 might be essential for venous endothelial cell differentiation (Wüstehube et al., 2010; You et al., 2013; Zheng et al., 2010). Additionally, KRIT1 deficiency disrupts the junctional localization of the TIAM–PAR3–PKCζ polarity complex (Lampugnani et al., 2010), impairing directed migration and vascular lumen formation. PDCD10 is also important for endothelial cell polarization in directed migration through its effects on Golgi positioning (Fidalgo et al., 2010), further linking PDCD10 with vascular development.
The leaky vasculature in CCM lesions is explained by their weak and disordered cell–cell junctions (see poster). The importance of KRIT1 in cell–cell junctions is underscored by its association with the junctional proteins VE-cadherin, α-catenin, β-catenin, AF6 (afadin, also known as MLLT4) and p120-catenin (Glading et al., 2007). Loss of KRIT1 reduces β-catenin and VE-cadherin at cell–cell junctions, leading to increased nuclear β-catenin and upregulation of its transcriptional targets (Glading and Ginsberg, 2010). Activation of Rap1 (which stabilizes KRIT1 in junctions) inhibits β-catenin transcription in a KRIT1-dependent manner. Interestingly, loss of CCM2 also leads to loss of KRIT1 from cell–cell junctions and their destabilization, a phenotype that is rescued by a mutant CCM2 that cannot bind KRIT1 (Schneider et al., 2011; Stockton et al., 2010). Recent data suggest that extracellular matrix (ECM) remodeling could also contribute to CCM pathology (Faurobert et al., 2013). As noted above, loss of KRIT1, CCM2 or PDCD10 increases RhoA-dependent contractility; this results in abnormal remodeling of ECM, altered integrin signaling and further increases in cell tension, which increases destabilization of cell–cell junctions. This may explain the abnormal ECM patterns seen in CCM lesions.
Exciting new data show that the loss of KRIT1 or PDCD10 leads to increased expression of mesenchymal markers in endothelial cells. This endothelial–mesenchymal transition (EndMT) is due to increased BMP6–SMAD signaling (Maddaluno et al., 2013), a pathway that is also upregulated in patient samples with KRIT1 or CCM2 mutations. EndMT is characterized by loss of polarity, increases in cell proliferation or migration and changes in cell–cell junctions, characteristics frequently seen in CCM lesions, suggesting that EndMT might contribute to CCM initiation and progression. It will be interesting to see whether BMP6 or TGFβ inhibitors will be effective therapeutics for patients with CCM.
Conclusions
Despite significant progress in determining the genetics of CCM and the functions of CCM proteins, many questions remain. The relevant signaling pathways in which CCM proteins participate in endothelial cells have not been fully established and the significance of the trimeric CCM protein complex remains controversial. Because of the disease phenotype, much work has focused on the roles of CCM proteins in the vasculature, but CCM proteins are widely expressed and why their loss results predominantly in a neurovasculature phenotype requires further study. It is clear, however, that further studies are necessary before we can link the known functions of CCM proteins to CCM formation.
Supplementary Material
Acknowledgments
Space limitations preclude detailed discussion of all reported CCM interactions and we apologize to colleagues whose work is not cited.
Footnotes
Competing interests
The authors declare no competing interests.
Funding
K.M.D. is supported by an American Cancer Society post-doctoral fellowship. O.S.F. is funded by a National Science Foundation Graduate Research Fellowship. T.J.B. and D.A.C. are funded by the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.138388/-/DC1
Cell science at a glance
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.138388/-/DC2.
References
- Akers A. L., Johnson E., Steinberg G. K., Zabramski J. M., Marchuk D. A. (2009). Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 18, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béraud-Dufour S., Gautier R., Albiges-Rizo C., Chardin P., Faurobert E. (2007). Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. FEBS J. 274, 5518–5532 10.1111/j.1742-4658.2007.06068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borikova A. L., Dibble C. F., Sciaky N., Welch C. M., Abell A. N., Bencharit S., Johnson G. L. (2010). Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J. Biol. Chem. 285, 11760–11764 10.1074/jbc.C109.097220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulday G., Blécon A., Petit N., Chareyre F., Garcia L. A., Niwa-Kawakita M., Giovannini M., Tournier-Lasserve E. (2009). Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis. Model. Mech. 2, 168–177 10.1242/dmm.001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulday G., Rudini N., Maddaluno L., Blécon A., Arnould M., Gaudric A., Chapon F., Adams R. H., Dejana E., Tournier-Lasserve E. (2011). Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J. Exp. Med. 208, 1835–1847 10.1084/jem.20110571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti D. D., Kalani M. Y., Martirosyan N. L., Eales J., Spetzler R. F., Preul M. C. (2012). Cerebral cavernous malformations: from genes to proteins to disease. J. Neurosurg. 116, 122–132 10.3171/2011.8.JNS101241 [DOI] [PubMed] [Google Scholar]
- Chan A. C., Drakos S. G., Ruiz O. E., Smith A. C., Gibson C. C., Ling J., Passi S. F., Stratman A. N., Sacharidou A., Revelo M. P. et al. (2011). Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J. Clin. Invest. 121, 1871–1881 10.1172/JCI44393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tanriover G., Yano H., Friedlander R., Louvi A., Gunel M. (2009). Apoptotic functions of PDCD10/CCM3, the gene mutated in cerebral cavernous malformation 3. Stroke 40, 1474–1481 10.1161/STROKEAHA.108.527135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B., Kean M. J., Ast V., Knight J. D., Mett A., Levy Z., Ceccarelli D. F., Badillo B. G., Eils R., König R. et al. (2012). STK25 protein mediates TrkA and CCM2 protein-dependent death in pediatric tumor cells of neural origin. J. Biol. Chem. 287, 29285–29289 10.1074/jbc.C112.345397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crose L. E., Hilder T. L., Sciaky N., Johnson G. L. (2009). Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J. Biol. Chem. 284, 13301–13305 10.1074/jbc.C900009200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K., Uchida Y., O'Donnell E., Claudio E., Li W., Soneji K., Wang H., Mukouyama Y. S., Siebenlist U. (2011). Conditional deletion of Ccm2 causes hemorrhage in the adult brain: a mouse model of human cerebral cavernous malformations. Hum. Mol. Genet. 20, 3198–3206 10.1093/hmg/ddr225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann P., Hehr U., Weidensee S., Zhu Y., Gerlach R., Sure U. (2013). Two-hit mechanism in cerebral cavernous malformation? A case of monozygotic twins with a CCM1/KRIT1 germline mutation. Neurosurg. Rev. 36, 483–486 10.1007/s10143-013-0456-z [DOI] [PubMed] [Google Scholar]
- Denier C., Labauge P., Bergametti F., Marchelli F., Riant F., Arnoult M., Maciazek J., Vicaut E., Brunereau L., Tournier-Lasserve E. Société Française de Neurochirurgie(2006). Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann. Neurol. 60, 550–556 10.1002/ana.20947 [DOI] [PubMed] [Google Scholar]
- Dibble C. F., Horst J. A., Malone M. H., Park K., Temple B., Cheeseman H., Barbaro J. R., Johnson G. L., Bencharit S. (2010). Defining the functional domain of programmed cell death 10 through its interactions with phosphatidylinositol-3,4,5-trisphosphate. PLoS ONE 5, e11740 10.1371/journal.pone.0011740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Wang X., Li D. F., Hu Y., Zhang Y., Wang D. C. (2010). Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation. Biochem. Biophys. Res. Commun. 399, 587–592 10.1016/j.bbrc.2010.07.119 [DOI] [PubMed] [Google Scholar]
- Faurobert E., Albiges-Rizo C. (2010). Recent insights into cerebral cavernous malformations: a complex jigsaw puzzle under construction. FEBS J. 277, 1084–1096 10.1111/j.1742-4658.2009.07537.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurobert E., Rome C., Lisowska J., Manet-Dupé S., Boulday G., Malbouyres M., Balland M., Bouin A. P., Kéramidas M., Bouvard D. et al. (2013). CCM1-ICAP-1 complex controls β1 integrin-dependent endothelial contractility and fibronectin remodeling. J. Cell Biol. 202, 545–561 10.1083/jcb.201303044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M., Fraile M., Pires A., Force T., Pombo C., Zalvide J. (2010). CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J. Cell Sci. 123, 1274–1284 10.1242/jcs.061341 [DOI] [PubMed] [Google Scholar]
- Fidalgo M., Guerrero A., Fraile M., Iglesias C., Pombo C. M., Zalvide J. (2012). Adaptor protein cerebral cavernous malformation 3 (CCM3) mediates phosphorylation of the cytoskeletal proteins ezrin/radixin/moesin by mammalian Ste20-4 to protect cells from oxidative stress. J. Biol. Chem. 287, 11556–11565 10.1074/jbc.M111.320259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Zalvide J., Faurobert E., Albiges-Rizo C., Tournier-Lasserve E. (2013). Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol. Med. 19, 302–308 10.1016/j.molmed.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Fisher O. S., Zhang R., Li X., Murphy J. W., Demeler B., Boggon T. J. (2013). Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus. FEBS Lett. 587, 272–277 10.1016/j.febslet.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francalanci F., Avolio M., De Luca E., Longo D., Menchise V., Guazzi P., Sgrò F., Marino M., Goitre L., Balzac F. et al. (2009). Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp. Cell Res. 315, 285–303 10.1016/j.yexcr.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Gingras A. R., Liu J. J., Ginsberg M. H. (2012). Structural basis of the junctional anchorage of the cerebral cavernous malformations complex. J. Cell Biol. 199, 39–48 10.1083/jcb.201205109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A. R., Puzon-McLaughlin W., Ginsberg M. H. (2013). The structure of the ternary complex of Krev Interaction Trapped 1 (KRIT1) bound to both the Rap1 GTPase and the heart of Glass (HEG1) cytoplasmic tail. J. Biol. Chem. 288, 23639–23649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A. J., Ginsberg M. H. (2010). Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis. Model. Mech. 3, 73–83 10.1242/dmm.003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A., Han J., Stockton R. A., Ginsberg M. H. (2007). KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J. Cell Biol. 179, 247–254 10.1083/jcb.200705175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitre L., Balzac F., Degani S., Degan P., Marchi S., Pinton P., Retta S. F. (2010). KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS ONE 5, e11786 10.1371/journal.pone.0011786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A. V., Lampugnani M. G., Dye L., Dejana E., Weinstein B. M. (2008). Combinatorial interaction between CCM pathway genes precipitates hemorrhagic stroke. Dis. Model. Mech. 1, 275–281 10.1242/dmm.000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I. et al. (2009). A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8, 157–171 10.1074/mcp.M800266-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guazzi P., Goitre L., Ferro E., Cutano V., Martino C., Trabalzini L., Retta S. F. (2012). Identification of the Kelch family protein Nd1-L as a novel molecular interactor of KRIT1. PLoS ONE 7, e44705 10.1371/journal.pone.0044705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzeloglu-Kayisli O., Amankulor N. M., Voorhees J., Luleci G., Lifton R. P., Gunel M. (2004). KRIT1/cerebral cavernous malformation 1 protein localizes to vascular endothelium, astrocytes, and pyramidal cells of the adult human cerebral cortex. Neurosurgery 54, 943–949, discussion 949 10.1227/01.NEU.0000114512.59624.A5 [DOI] [PubMed] [Google Scholar]
- Harel L., Costa B., Tcherpakov M., Zapatka M., Oberthuer A., Hansford L. M., Vojvodic M., Levy Z., Chen Z. Y., Lee F. S. et al. (2009). CCM2 mediates death signaling by the TrkA receptor tyrosine kinase. Neuron 63, 585–591 10.1016/j.neuron.2009.08.020 [DOI] [PubMed] [Google Scholar]
- He Y., Zhang H., Yu L., Gunel M., Boggon T. J., Chen H., Min W. (2010). Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci. Signal. 3, ra26 10.1126/scisignal.2000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilder T. L., Malone M. H., Bencharit S., Colicelli J., Haystead T. A., Johnson G. L., Wu C. C. (2007a). Proteomic identification of the cerebral cavernous malformation signaling complex. J. Proteome Res. 6, 4343–4355 10.1021/pr0704276 [DOI] [PubMed] [Google Scholar]
- Hilder T. L., Malone M. H., Johnson G. L. (2007b). Hyperosmotic induction of mitogen-activated protein kinase scaffolding. Methods Enzymol. 428, 297–312 10.1016/S0076-6879(07)28017-6 [DOI] [PubMed] [Google Scholar]
- Hogan B. M., Bussmann J., Wolburg H., Schulte-Merker S. (2008). ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum. Mol. Genet. 17, 2424–2432 10.1093/hmg/ddn142 [DOI] [PubMed] [Google Scholar]
- Kean M. J., Ceccarelli D. F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L. C., Derry W. B., Scott I. C., Pelletier L. et al. (2011). Structure-function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25065–25075 10.1074/jbc.M110.214486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B., Zheng X., Liu J. J., Blum Y., Tung J. J., Zou Z., Sweeney S. M., Chen M., Guo L., Lu M. M. et al. (2009). Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat. Med. 15, 169–176 10.1038/nm.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani M. G., Orsenigo F., Rudini N., Maddaluno L., Boulday G., Chapon F., Dejana E. (2010). CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 123, 1073–1080 10.1242/jcs.059329 [DOI] [PubMed] [Google Scholar]
- Li D. Y., Whitehead K. J. (2010). Evaluating strategies for the treatment of cerebral cavernous malformations. Stroke 41 Suppl., S92–S94 10.1161/STROKEAHA.110.594929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang R., Zhang H., He Y., Ji W., Min W., Boggon T. J. (2010). Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J. Biol. Chem. 285, 24099–24107 10.1074/jbc.M110.128470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ji W., Zhang R., Folta-Stogniew E., Min W., Boggon T. J. (2011). Molecular recognition of leucine-aspartate repeat (LD) motifs by the focal adhesion targeting homology domain of cerebral cavernous malformation 3 (CCM3). J. Biol. Chem. 286, 26138–26147 10.1074/jbc.M110.211250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang R., Draheim K. M., Liu W., Calderwood D. A., Boggon T. J. (2012). Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1). J. Biol. Chem. 287, 22317–22327 10.1074/jbc.M112.361295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye N., Boon L. M., Vikkula M. (2009). From germline towards somatic mutations in the pathophysiology of vascular anomalies. Hum. Mol. Genet. 18 R1, R65–R74 10.1093/hmg/ddp002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Meng S., Zhu T., Wang X. (2010). PDCD10/CCM3 acts downstream of gamma-protocadherins to regulate neuronal survival. J. Biol. Chem. 285, 41675–41685 10.1074/jbc.M110.179895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori C. L., Berg M. J., Siegel A. M., Huang E., Zawistowski J. S., Stoffer T., Verlaan D., Balogun F., Hughes L., Leedom T. P. et al. (2003). Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am. J. Hum. Genet. 73, 1459–1464 10.1086/380314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. J., Stockton R. A., Gingras A. R., Ablooglu A. J., Han J., Bobkov A. A., Ginsberg M. H. (2011). A mechanism of Rap1-induced stabilization of endothelial cell – cell junctions. Mol. Biol. Cell 22, 2509–2519 10.1091/mbc.E11-02-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Draheim K. M., Zhang R., Calderwood D. A., Boggon T. J. (2013). Mechanism for KRIT1 release of ICAP1-mediated suppression of integrin activation. Mol. Cell 49, 719–729 10.1016/j.molcel.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A., Chen L., Two A. M., Zhang H., Min W., Günel M. (2011). Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. Proc. Natl. Acad. Sci. USA 108, 3737–3742 10.1073/pnas.1012617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably J. D., Mohideen M. A., Burns C. G., Chen J. N., Fishman M. C. (2003). heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138–2147 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- Mably J. D., Chuang L. P., Serluca F. C., Mohideen M. A., Chen J. N., Fishman M. C. (2006). santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development 133, 3139–3146 10.1242/dev.02469 [DOI] [PubMed] [Google Scholar]
- Maddaluno L., Rudini N., Cuttano R., Bravi L., Giampietro C., Corada M., Ferrarini L., Orsenigo F., Papa E., Boulday G. et al. (2013). EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 498, 492–496 10.1038/nature12207 [DOI] [PubMed] [Google Scholar]
- McDonald D. A., Shenkar R., Shi C., Stockton R. A., Akers A. L., Kucherlapati M. H., Kucherlapati R., Brainer J., Ginsberg M. H., Awad I. A. et al. (2011). A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum. Mol. Genet. 20, 211–222 10.1093/hmg/ddq433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D. A., Shi C., Shenkar R., Stockton R. A., Liu F., Ginsberg M. H., Marchuk D. A., Awad I. A. (2012). Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke 43, 571–574 10.1161/STROKEAHA.111.625467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millon-Frémillon A., Bouvard D., Grichine A., Manet-Dupé S., Block M. R., Albiges-Rizo C. (2008). Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J. Cell Biol. 180, 427–441 10.1083/jcb.200707142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagenstecher A., Stahl S., Sure U., Felbor U. (2009). A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum. Mol. Genet. 18, 911–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit N., Blécon A., Denier C., Tournier-Lasserve E. (2006). Patterns of expression of the three cerebral cavernous malformation (CCM) genes during embryonic and postnatal brain development. Gene Expr. Patterns 6, 495–503 10.1016/j.modgep.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Plummer N. W., Gallione C. J., Srinivasan S., Zawistowski J. S., Louis D. N., Marchuk D. A. (2004). Loss of p53 sensitizes mice with a mutation in Ccm1 (KRIT1) to development of cerebral vascular malformations. Am. J. Pathol. 165, 1509–1518 10.1016/S0002-9440(10)63409-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer N. W., Squire T. L., Srinivasan S., Huang E., Zawistowski J. S., Matsunami H., Hale L. P., Marchuk D. A. (2006). Neuronal expression of the Ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm. Genome 17, 119–128 10.1007/s00335-005-0098-8 [DOI] [PubMed] [Google Scholar]
- Riant F., Cecillon M., Saugier-Veber P., Tournier-Lasserve E. (2013). CCM molecular screening in a diagnosis context: novel unclassified variants leading to abnormal splicing and importance of large deletions. Neurogenetics 14, 133–141 10.1007/s10048-013-0362-0 [DOI] [PubMed] [Google Scholar]
- Rosen J. N., Sogah V. M., Ye L. Y., Mably J. D. (2013). ccm2-like is required for cardiovascular development as a novel component of the Heg-CCM pathway. Dev. Biol. 376, 74–85 10.1016/j.ydbio.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H., Errede M., Ulrich N. H., Virgintino D., Frei K., Bertalanffy H. (2011). Impairment of tight junctions and glucose transport in endothelial cells of human cerebral cavernous malformations. J. Neuropathol. Exp. Neurol. 70, 417–429 10.1097/NEN.0b013e31821bc40e [DOI] [PubMed] [Google Scholar]
- Seker A., Pricola K. L., Guclu B., Ozturk A. K., Louvi A., Gunel M. (2006). CCM2 expression parallels that of CCM1. Stroke 37, 518–523 10.1161/01.STR.0000198835.49387.25 [DOI] [PubMed] [Google Scholar]
- Serebriiskii I., Estojak J., Sonoda G., Testa J. R., Golemis E. A. (1997). Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene 15, 1043–1049 10.1038/sj.onc.1201268 [DOI] [PubMed] [Google Scholar]
- Stahl S., Gaetzner S., Voss K., Brackertz B., Schleider E., Sürücü O., Kunze E., Netzer C., Korenke C., Finckh U. et al. (2008). Novel CCM1, CCM2, and CCM3 mutations in patients with cerebral cavernous malformations: in-frame deletion in CCM2 prevents formation of a CCM1/CCM2/CCM3 protein complex. Hum. Mutat. 29, 709–717 10.1002/humu.20712 [DOI] [PubMed] [Google Scholar]
- Stockton R. A., Shenkar R., Awad I. A., Ginsberg M. H. (2010). Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207, 881–896 10.1084/jem.20091258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden P. H., McGuffin L. J., Clerk A. (2013). SOcK, MiSTs, MASK and STicKs: the GCKIII (germinal centre kinase III) kinases and their heterologous protein-protein interactions. Biochem. J. 454, 13–30 10.1042/BJ20130219 [DOI] [PubMed] [Google Scholar]
- Tanriover G., Sozen B., Seker A., Kilic T., Gunel M., Demir N. (2013). Ultrastructural analysis of vascular features in cerebral cavernous malformations. Clin. Neurol. Neurosurg. 115, 438–444 10.1016/j.clineuro.2012.06.023 [DOI] [PubMed] [Google Scholar]
- Uhlik M. T., Abell A. N., Johnson N. L., Sun W., Cuevas B. D., Lobel-Rice K. E., Horne E. A., Dell'Acqua M. L., Johnson G. L. (2003). Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 5, 1104–1110 10.1038/ncb1071 [DOI] [PubMed] [Google Scholar]
- Voss K., Stahl S., Schleider E., Ullrich S., Nickel J., Mueller T. D., Felbor U. (2007). CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics 8, 249–256 10.1007/s10048-007-0098-9 [DOI] [PubMed] [Google Scholar]
- Voss K., Stahl S., Hogan B. M., Reinders J., Schleider E., Schulte-Merker S., Felbor U. (2009). Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum. Mutat. 30, 1003–1011 10.1002/humu.20996 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu H., Zhang Y., Ma D. (1999). cDNA cloning and expression of an apoptosis-related gene, humanTFAR15 gene. Sci. China C Life Sci. 42, 323–329 10.1007/BF03183610 [DOI] [PubMed] [Google Scholar]
- Whitehead K. J., Plummer N. W., Adams J. A., Marchuk D. A., Li D. Y. (2004). Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development 131, 1437–1448 10.1242/dev.01036 [DOI] [PubMed] [Google Scholar]
- Whitehead K. J., Chan A. C., Navankasattusas S., Koh W., London N. R., Ling J., Mayo A. H., Drakos S. G., Jones C. A., Zhu W. et al. (2009). The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat. Med. 15, 177–184 10.1038/nm.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüstehube J., Bartol A., Liebler S. S., Brütsch R., Zhu Y., Felbor U., Sure U., Augustin H. G., Fischer A. (2010). Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl. Acad. Sci. USA 107, 12640–12645 10.1073/pnas.1000132107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wang X., Zhang Y., Wang D. C., Ding J. (2013). Structural basis for the unique heterodimeric assembly between cerebral cavernous malformation 3 and germinal center kinase III. Structure 21, 1059–1066 10.1016/j.str.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Yoruk B., Gillers B. S., Chi N. C., Scott I. C. (2012). Ccm3 functions in a manner distinct from Ccm1 and Ccm2 in a zebrafish model of CCM vascular disease. Dev. Biol. 362, 121–131 10.1016/j.ydbio.2011.12.006 [DOI] [PubMed] [Google Scholar]
- You C., Sandalcioglu I. E., Dammann P., Felbor U., Sure U., Zhu Y. (2013). Loss of CCM3 impairs DLL4-Notch signalling: implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J. Cell. Mol. Med. 17, 407–418 10.1111/jcmm.12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski J. S., Stalheim L., Uhlik M. T., Abell A. N., Ancrile B. B., Johnson G. L., Marchuk D. A. (2005). CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum. Mol. Genet. 14, 2521–2531 10.1093/hmg/ddi256 [DOI] [PubMed] [Google Scholar]
- Zhang J., Rigamonti D., Dietz H. C., Clatterbuck R. E. (2007). Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery 60, 353–359, discussion 359 10.1227/01.NEU.0000249268.11074.83 [DOI] [PubMed] [Google Scholar]
- Zhang M., Dong L., Shi Z., Jiao S., Zhang Z., Zhang W., Liu G., Chen C., Feng M., Hao Q. et al. (2013a). Structural mechanism of CCM3 heterodimerization with GCKIII kinases. Structure 21, 680–688 10.1016/j.str.2013.02.015 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tang W., Zhang H., Niu X., Xu Y., Zhang J., Gao K., Pan W., Boggon T. J., Toomre D. et al. (2013b). A Network of Interactions Enables CCM3 and STK24 to Coordinate UNC13D-Driven Vesicle Exocytosis in Neutrophils. Dev. Cell 27, 215–226 10.1016/j.devcel.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J. et al. (2010). CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest. 120, 2795–2804 10.1172/JCI39679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Xu C., Smith A. O., Stratman A. N., Zou Z., Kleaveland B., Yuan L., Didiku C., Sen A., Liu X. et al. (2012). Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev. Cell 23, 342–355 10.1016/j.devcel.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Izumi Y., Burg M. B., Ferraris J. D. (2011). Rac1/osmosensing scaffold for MEKK3 contributes via phospholipase C-gamma1 to activation of the osmoprotective transcription factor NFAT5. Proc. Natl. Acad. Sci. USA 108, 12155–12160 10.1073/pnas.1108107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wu Q., Xu J. F., Miller D., Sandalcioglu I. E., Zhang J. M., Sure U. (2010). Differential angiogenesis function of CCM2 and CCM3 in cerebral cavernous malformations. Neurosurg. Focus 29, E1 10.3171/2010.5.FOCUS1090 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wu Q., Fass M., Xu J. F., You C., Müller O., Sandalcioglu I. E., Zhang J. M., Sure U. (2011). In vitro characterization of the angiogenic phenotype and genotype of the endothelia derived from sporadic cerebral cavernous malformations. Neurosurgery 69, 722–731, discussion 731-732 10.1227/NEU.0b013e318219569f [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.