ABSTRACT
Known proteins associated with the cell-adhesion protein E-cadherin include catenins and proteins involved in signaling, trafficking and actin organization. However, the list of identified adherens junction proteins is likely to be incomplete, limiting investigation into this essential cell structure. To expand the inventory of potentially relevant proteins, we expressed E-cadherin fused to biotin ligase in MDCK epithelial cells, and identified by mass spectrometry neighboring proteins that were biotinylated. The most abundant of the 303 proteins identified were catenins and nearly 40 others that had been previously reported to influence cadherin function. Many others could be rationalized as novel candidates for regulating the adherens junction, cytoskeleton, trafficking or signaling. We further characterized lipoma preferred partner (LPP), which is present at both cell contacts and focal adhesions. Knockdown of LPP demonstrated its requirement for E-cadherin-dependent adhesion and suggested that it plays a role in coordination of the cell–cell and cell–substrate cytoskeletal interactions. The analysis of LPP function demonstrates proof of principle that the proteomic analysis of E-cadherin proximal proteins expands the inventory of components and tools for understanding the function of E-cadherin.
KEY WORDS: E-cadherin, Adherens junction, Proteomics, Biotin ligase
INTRODUCTION
The ability of cells to adhere to each other and to extracellular matrix depends on cell-specific adhesive proteins as well as cytoplasmic proteins that regulate signaling and actin cytoskeletal dynamics (Lecuit and Lenne, 2007). Given that both cell–cell and cell–substrate adhesion must be dynamic (Guillot and Lecuit, 2013; Wolfenson et al., 2013), to allow tissue growth and remodeling, and stable, to provide mechanical strength, these interactions are highly regulated. Numerous proteins have been identified that link adhesive proteins to the cytoplasmic components (Guillot and Lecuit, 2013; Wolfenson et al., 2013; Zaidel-Bar, 2013). In the present study, we used a relatively new biotin-tagging method (Roux et al., 2012) to expand the list of proteins that are proximal to the cell adhesion protein E-cadherin.
E-cadherin is the principal molecule supporting epithelial cell–cell adhesion at the adherens junction and is required for initiating the cell polarity program, normal morphogenesis and epithelial barrier formation (Niessen et al., 2011; Oda and Takeichi, 2011). It is a single-spanning transmembrane protein composed of an adhesive extracellular domain, transmembrane domain and relatively small (150 amino acids) intracellular domain (Takeichi, 1988). E-cadherin forms cis- and trans-homophilic clusters concentrated at the adherens junction and variably localized along the lateral cell membranes; these clusters are stabilized by their interactions with the actin cytoskeleton (Gomez et al., 2011; Yonemura, 2011). The interactions with actin are indirect and occur through extensively studied catenin proteins that bind to the intracellular domain of E-cadherin, namely β-catenin and α-catenin, and also through several catenin-associated actin-binding proteins, including vinculin, formin-1 and VASP (reviewed by Meng and Takeichi, 2009). Many proteins have been localized to adherens junctions by biochemical and microscopic techniques (Smith et al., 2011; Zaidel-Bar, 2013), but there have been relatively few attempts at global proteomic analysis.
With the goal of identifying more proteins that might be involved in E-cadherin function, we took advantage of a recently published technique (Roux et al., 2012) to identify proximal proteins in living cells. In this method, cells are transfected with a fusion protein, in this case, E-cadherin, attached to an engineered biotin ligase with decreased substrate specificity. E-cadherin directs the biotin ligase to adherens junctions; when biotin is added to the medium, the ligase releases highly reactive BioAMP which biotinylates primary amines on neighboring proteins. These proximal biotinylated proteins can be purified on streptavidin resin and subjected to proteomic analyses.
Similar to what we had found using this method with ZO-1 (Van Itallie et al., 2013), our results suggest that E-cadherin–biotin-ligase (EcadBL) identifies a large number of known functionally relevant proteins as well as proteins previously not known to be near E-cadherin and which might provide novel insights about E-cadherin function. The most abundant proteins identified were catenins, including α-E-catenin, β-catenin, p120 catenin and plakoglobin, and unexpectedly, α-N-catenin and α-T-catenin. Many proteins involved in cytoskeletal interactions and trafficking were also identified as proximal proteins. In addition were several proteins that are known to play a role not only at cell–cell interactions, but also at cell–substrate interactions. One of these proteins, lipoma preferred partner (LPP), a LIM-domain containing protein that is a member of the zyxin family (Petit et al., 2000) was chosen for further study. Its role at the adherens junction is not well studied and we previously observed it to be highly tagged and therefore presumably very close to ZO-1 (Van Itallie et al., 2013). ZO-1 associates with E-cadherin at initial cell contacts but moves under the adjacent tight junction as the apical junction complex matures (Ikenouchi et al., 2007). We report here that knockdown of LPP in epithelial cells both diminishes E-cadherin-dependent cell–cell adhesion, resulting in compromised tight junction assembly, and increases cell–substrate adhesion, suggesting that it functions to balance adhesion between these sites. Our findings further validate the utility of using biotin ligase fusion proteins to identify proximal proteins.
RESULTS
EcadBL fusion protein localizes to cell–cell contacts in MDCK cells
To determine how well EcadBL localized to cell contacts, we compared its distribution in stable, inducible cells MDCK II cell lines (Fig. 1A, middle panel) with that of endogenous E-cadherin (Fig. 1A, left panel) and found the fluorescent signals largely overlapped (Fig. 1A, right panel). Similarly, biotinylated proteins detected with fluorescent streptavidin colocalized with EcadBL in cells incubated overnight with biotin (Fig. 1B, bottom panels) but were not evident in cells without biotin treatment Fig. 1B, top panels). We conclude that the fusion protein localizes well, heavily tags proteins at the adherens junction and might reliably report on proteins near E-cadherin at the adherens junction and along the lateral membrane.
Fig. 1.
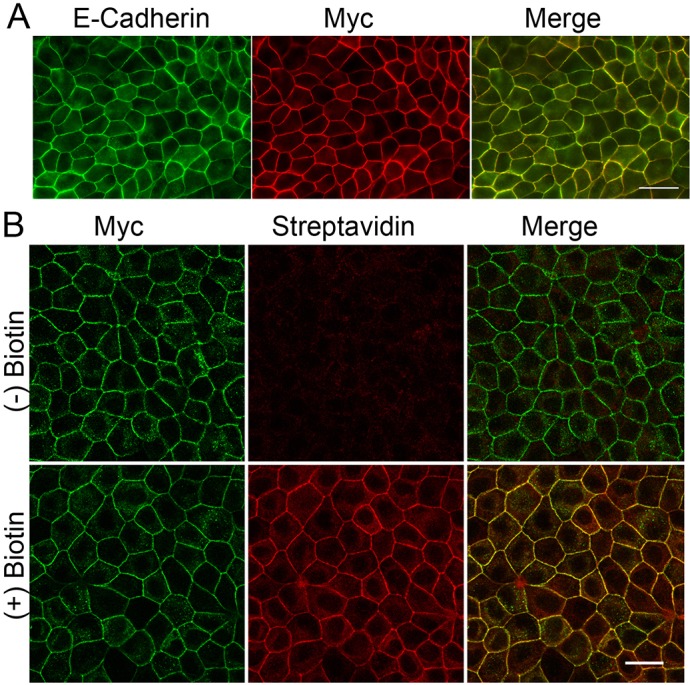
The E-cadherin–biotin-ligase fusion protein and biotinylated proteins colocalize with endogenous E-cadherin. (A) Myc-tagged EcadBL (Myc, middle) colocalized with endogenous E-cadherin (left and right, merge) detected using a canine-specific E-cadherin antibody that does not recognize the human EcadBL fusion protein. (B) In the presence (bottom panels) but not the absence (top panels) of exogenous biotin, fluorescent streptavidin signal (middle and right panels) colocalizes with the EcadBL fusion protein (Myc, left and right panels). Scale bars: 20 µm.
Proteomic analysis of biotinylated proteins isolated from EcadBL-expressing cells reveals expected and unexpected proximal proteins
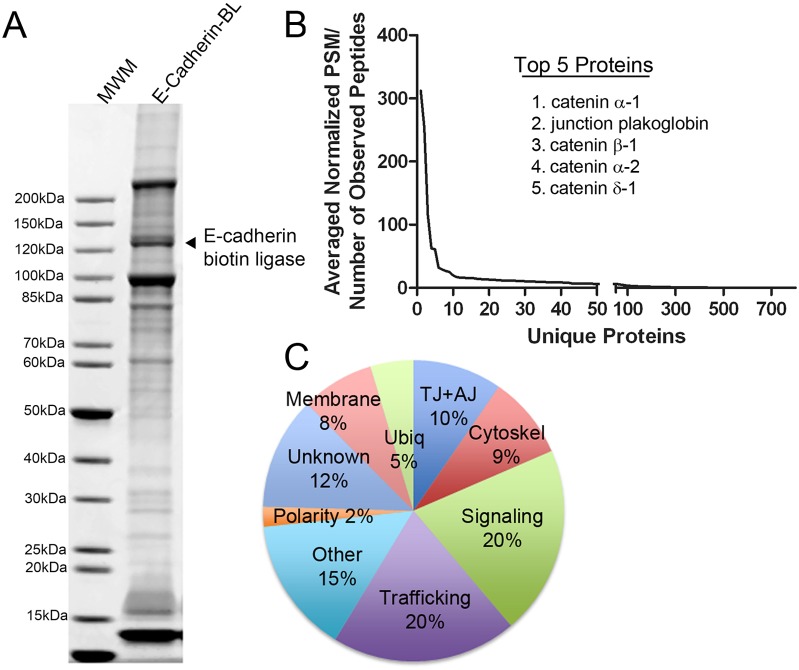
EcadBL-expressing MDCK cells were incubated for 15 hours with biotin, lysed and biotinylated proteins were purified on streptavidin beads, separated by SDS-PAGE and stained with Coomassie Brilliant Blue (Fig. 2A); a prominent band at ∼135 kDa was detected with an anti-E-cadherin antibody and is likely the EcadBL fusion protein. Unlike the results obtained with ZO-1 biotin ligase fusions (Van Itallie et al., 2013), the E-cadherin fusion protein was not the most abundant biotinylated protein in these cells, possibly because the E-cadherin intracellular domain available for self-biotinylation contains only five lysines. Three similar Coomassie-stained protein gel lanes from independent labeling experiments and streptavidin purifications were each cut into 12 bands (top to bottom) and used for proteomic analysis by mass spectrometry. Triplicate analysis identified 688 unique proteins in at least two out of three of the analyses. The relative abundance of these proteins was then compared with biotinylated proteins recovered from cells expressing biotin ligase alone. Ribosomal proteins, which are probably biotinylated during biosynthesis in the 15 hour labeling period, were sorted into a separate list; 303 proteins were identified as being present at levels that were at least twofold higher in the EcadBL-expressing cells compared with biotin ligase alone (supplementary material Table S1).
Fig. 2.
Proteomic analysis of proteins proximal to EcadBL. (A) Coomassie-Blue-stained SDS-PAGE of biotinylated proteins from EcadBL-expressing MDCK cells exposed to biotin overnight eluted from streptavidin beads; an aliquot of this sample and two other independent isolates were used for proteomic analysis. (B) Graph of relative abundance of proteins identified by mass spectrometry. The y-axis is calculated as follows: PSMs from each of the three isolations were normalized (PSM for each proteins/total PSMs for that isolation), these normalized PSMs were averaged between the three runs and then divided by the number of theoretical peptides falling in the size range detectable by MS (Pisitkun et al., 2012) and this value multiplied by 1000. Proteins were ordered by this value (largest to smallest); points on the x-axis indicate individual unique proteins identified using the Canis familiaris Ref Seq database (688 total). The top five most abundant proteins (all catenins) are listed. (C) Functional analysis of the 250 most abundant proteins identified as proximal to EcadBL. Cytoskel, cytoskeletal proteins; Ubiq, ubiquitin-related proteins.
Comparison of the relative abundance of the identified proteins purified from the EcadBL-expressing cells revealed that although many different proteins are recovered by this method, only a few are recovered in great abundance (Fig. 2B). In terms of relative abundance, the top five proteins identified are all adherens junction proteins (Fig. 2B); one of these, catenin α-2 (α-N-catenin) is generally thought to be restricted to the nervous system (Abe et al., 2004), although it was identified in a proteomic screen in A431 cells (Smith et al., 2011). A third α-catenin isoform, catenin α-3 (α-T-catenin), was also identified by EcadBL-dependent biotinylation (rank 10 in abundance); this catenin is enriched in heart and testes and has not been previously described in MDCK cells (Janssens et al., 2001).
With the caveat that recovery of relevant proteins requires that they contain lysines accessible for biotinylation, we predict that the most abundant proteins recovered are likely to be the most functionally relevant. Categorizing these proteins according to a combination of UniProt (The UniProt Consortium, 2013) and literature searches, we found that the majority of these proteins can be divided into proteins localized to adherens or tight junctions, proteins involved in trafficking and signaling, or cytoskeletal proteins (Fig. 2C).
LPP, a LIM-domain-containing member of zyxin family, is identified as an abundant proximal protein
One protein, lipoma preferred partner (LPP, rank 30) was of particular interest because it was also among the more abundant proteins tagged by the biotin ligase ZO-1 fusion protein (rank 36; Van Itallie et al., 2013). E-cadherin is essential not only in adherens junctions, but is also required for normal tight junction formation (Capaldo and Macara, 2007). We speculated that LPP, because it was identified as proximal to both ZO-1 and E-cadherin, might be an essential component of both tight and adherens junction organization. Along with LPP, a related family member, thyroid receptor-interacting protein 6 (TRIP6) was tagged by EcadBL (rank 67); in addition, zyxin, a third member of the same family, is biotinylated by E-cadherin and ZO-1 but at a lower level (rank 107). The relatively high level of LPP tagging compared with the other zyxin family members suggested that of its family, it might play a particularly important role at cell contacts.
LPP, like its zyxin family relatives, has been reported to localize to cell–cell contacts, to focal adhesions and to the nucleus (reviewed by Grunewald et al., 2009). Using MDCK cells, we verified localization of LPP to cell contacts, where it colocalizes with E-cadherin (Fig. 3, top panels) and to focal adhesions (Fig. 3, bottom panels), but we failed to see significant nuclear staining in normal cells. To verify proximity, we performed an in situ proximity ligation assay (PLA); this assay results in the production of a fluorescent signal when antibodies to two different antigens are close enough to allow ligation and amplification of oligonucleotides coupled to modified secondary antibodies (Söderberg et al., 2006). As a negative control, we stained first with antibodies against laterally distributed E-cadherin and the apical protein podocalyxin (Meder et al., 2005) (Fig. 4, top left); this combination failed to produce any fluorescent signal, confirming assay specificity. By contrast, the combination of E-cadherin and catenin delta-1 antibodies (p120 catenin) gave a strong fluorescent signal (Fig. 4, top right), as would be expected from their previously demonstrated biochemical interactions and close subcellular localization (Meng and Takeichi, 2009). E-cadherin and LPP antibodies also produced significant fluorescent signal in the PLA assay (Fig. 4, bottom left), consistent with the biotin ligase tagging results and with the colocalization visualized by conventional immunofluorescence (Fig. 3). As expected, a second negative control, that of LPP antibody alone incubated with PLA reagent also failed to generate significant fluorescence (Fig. 4, bottom right).
Fig. 3.
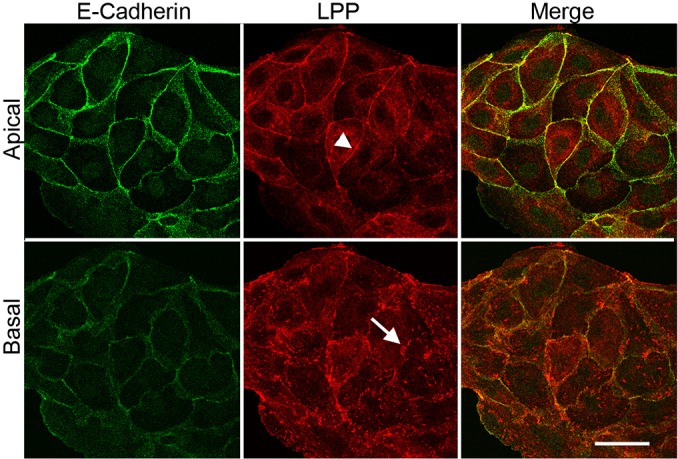
Immunofluorescent localization of LPP to cell contacts and focal adhesions. Top panels are confocal microscopic sections of the apical region of MDCK cells, which reveal that LPP (middle panel, red) is concentrated at cell contacts (white arrowhead) with E-cadherin (left panel, green and merge, right panel, yellow). Bottom panels are basal sections of the same cells, showing that LPP (middle panel, red) but not E-cadherin (left panel, green) is also localized to focal adhesions (white arrow). Scale bar: 20 µm.
Fig. 4.
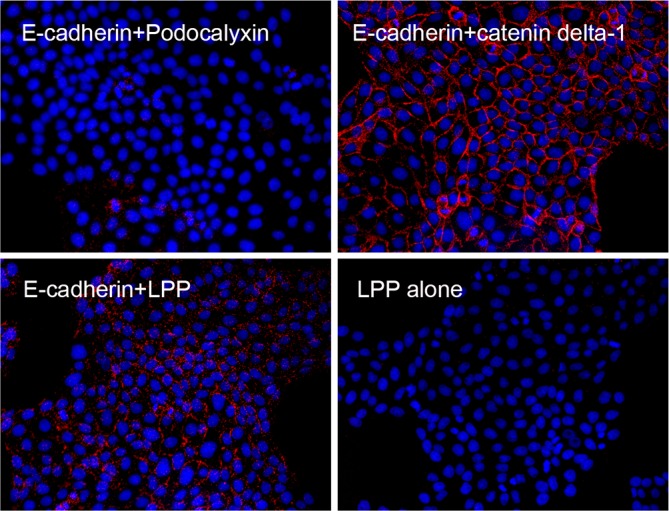
Proximity ligation assay demonstrates proximity of E-cadherin and LPP at adherens junctions. MDCK cells plated on coverslips were incubated with E-cadherin (rabbit) and podocalyxin (mouse) antibodies (top left), E-cadherin (rabbit) and catenin delta-1 (mouse) antibodies (top right), E-cadherin (rabbit) and LPP (mouse) antibodies (bottom left) and LPP (mouse) antibody alone (bottom right). Cells were then incubated with anti-mouse minus and anti-rabbit plus PLA reagents followed by ligation and far-red amplification reagents; nuclei were stained with Hoechst 33342 reagent during the last three washes. Fluorescent signal corresponding to successful amplification is only evident in the positive control, E-cadherin with catenin delta-1 antibodies (top right) and in the cells incubated with E-cadherin and LPP antibodies (bottom left).
Because LPP had been identified as a protein proximal to both E-cadherin and ZO-1, and further because it has a C-terminal PDZ binding motif (Petit et al., 2005a) that might interact with ZO-1 or ZO-2 PDZ domains, we tested to see whether LPP localization to cell contacts was dependent on the presence of these proteins. In both ZO-1 and ZO2 double-knockdown and in E-cadherin-knockdown cells, LPP was still found at cell contacts (supplementary material Fig. S1), suggesting that its recruitment to this site does not require an interaction with any of these proteins. In addition, LPP failed to coimmunoprecipitate with E-cadherin (not shown), which is consistent with the lack of a direct interaction.
LPP-knockdown cells show diminished E-cadherin-dependent adhesion
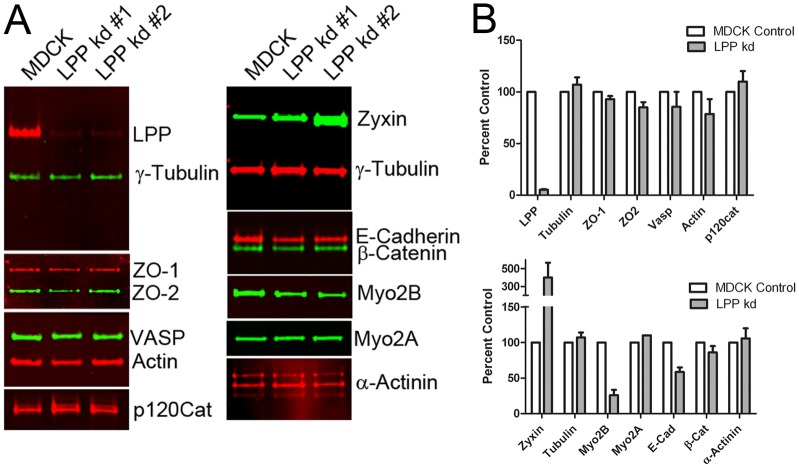
We hypothesized that LPP, like its relative, zyxin (Nguyen et al., 2010), might be important in modulating the strength of E-cadherin-dependent adhesion. To test this, we made MDCK cells lines stably depleted of endogenous LPP. Immunoblot analysis of equal amounts of control MDCKs and two different LPP knockdown cell lines confirmed LPP depletion of greater than 95% (Fig. 5A,B). The levels of the tight junction proteins ZO-1 and ZO-2 were not changed (Fig. 5A,B), nor was that of occludin (not shown). However, the level of E-cadherin was slightly but significantly decreased by about 30%. The levels of β-catenin (Fig. 5A,B), p120 catenin (Fig. 5A,B) and α-E-catenin (not shown) were unchanged in knockdown cells compared with control MDCK cells.
Fig. 5.
Immunoblot analysis of control and LPP-knockdown cells. (A) MDCK and two independent knockdown cell lines were probed for expression of (left, top panel), LPP (red) and γ-tubulin (green), (left, second panel), ZO-1 (red) and ZO-2 (green), (left, third panel), VASP (green) and actin (red), (left, bottom panel), p120 catenin (red), (right, top panel), zyxin (green) and γ-tubulin (red), (right, second panel), e-cadherin (red) and β-catenin (green), (right, third panel), myosin 2B (green), (right, fourth panel), myosin 2A (green) and (right, bottom panel) α-actinin (red). (B) Quantification of replicate immunoblots reveals increased levels of zyxin in the LPP-knockdown cells (300% of control values) and decreased levels of LPP (less than 5% of control values), myosin 2B (30% of control values) and slightly decreased levels of E-cadherin (70%) of control levels. The levels of most other proteins were unaltered in the LPP knockdowns. Values are means ± s.d.
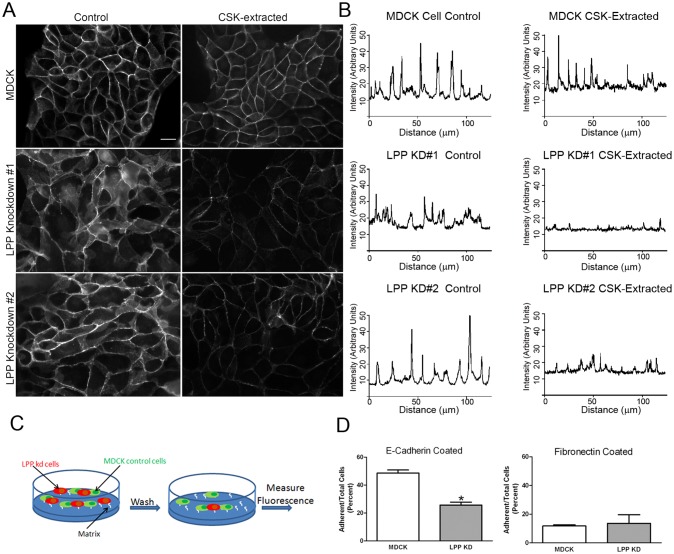
Stability of E-cadherin to extraction with Triton X-100 is associated with the maturity of cell–cell contacts (Shore and Nelson, 1991). To determine whether LPP is required to create or maintain mature E-cadherin-dependent cell contact, we extracted MDCK control monolayer cultures and LPP knockdown cells with standard Triton-containing cytoskeleton (CSK) buffer (Fig. 6A) before fixation and immunofluorescent staining for E-cadherin. In unextracted cells, approximately equal amounts of E-cadherin were present at cell–cell contacts in all lines, as determined by confocal line scans of the E-cadherin fluorescent signal (Fig. 6B, left). CSK extraction resulted in only minor loss of membrane-associated E-cadherin in control MDCK cells, but had a much greater effect on E-cadherin at cell contacts in two different LPP-knockdown cell lines (Fig. 6B, right). These results suggest that knockdown of LPP compromises junction stabilization and E-cadherin-dependent adhesion.
Fig. 6.
LPP-knockdown cells show diminished E-cadherin-dependent adhesion. (A) MDCK control and two separate LPP-knockdown cell lines were cultured on glass coverslips for 3 days and either extracted (right panels) or not (left panels) with CSK buffer before fixation and immunofluorescence staining for E-cadherin. Scale bar: 20 µm. (B) Representative line scan analysis of images as shown in C demonstrates little loss of E-cadherin fluorescence in control cells after CSK extraction (top panels). By contrast, there is greater loss of signal from E-cadherin fluorescence in LPP-knockdown cells after CSK extraction (right two bottom panels) compared with unextracted cells (left two bottom panels). (C) Schematic of modified assay to measure E-cadherin-dependent adhesion. MDCK and LPP-knockdown cells were separately labeled with fluorescent dyes, mixed and incubated for 60 minutes on E-cadherin- or fibronectin-coated plates. Wells were washed to remove loosely adhering cells and the relative fluorescent signals from MDCK and LPP-knockdown cells were compared with unwashed wells. (D) MDCK control cells adhered twice as well as LPP-knockdown cells to E-cadherin-coated wells; there was no difference in adhesion to fibronectin in the control versus knockdown cells. Values are means ± s.d.; *P<0.005 by unpaired Student's t-test.
To test a role for LPP in adhesion directly, control and LPP-knockdown cells were differentially labeled with green or red fluorescent CellTracker, mixed and allowed to attach to E-cadherin or fibronectin-coated wells (Fig. 6C). Non-adherent cells were removed by washing and the ratio of adherent MDCK cells compared with LPP-knockdown cells was determined by fluorescent quantification on a flat-bed fluorescent imager. Control MDCK cells adhered to the E-cadherin plates about twice as well as the LPP-knockdown cells (Fig. 6D). There was no statistically significant difference between control and knockdown cells in adherence to fibronectin (Fig. 6D).
E-cadherin-dependent adhesion is required for tight junction assembly after calcium removal and replacement (Gumbiner et al., 1988). To test whether the observed defects in E-cadherin-dependent adhesion might also impair tight junction barrier formation, we measured the transepithelial electrical resistance (TER) in control and LPP-knockdown cells during recovery after calcium replacement following overnight incubation in low calcium (Fig. 7A). Baseline TER was similar in both control and knockdown cells (not shown). MDCK control cells demonstrated the previously described TER early overshoot during recovery (Gonzalez-Mariscal et al., 1985), whereas LPP-knockdown cells failed to generate any appreciable barrier function in the first 15 hours following calcium replacement. Immunofluorescent analysis of ZO-1 to follow morphologic barrier re-formation showed a parallel delay in tight junction continuity in the LPP-knockdown cells compared with control cells (Fig. 7B). Staining for ZO-1 (and occludin, not shown) is fully continuous in MDCK controls after 4 hours of recovery (Fig. 7B, top panels), but is still largely discontinuous in LPP-knockdown cells 8 hours after calcium replacement (Fig. 7B, bottom panels), consistent with the delay in recovery of TER. A similar loss of the early peak in TER is also observed when E-cadherin is depleted (Capaldo and Macara, 2007) and in ZO-1 and ZO-2 double-knockdown MDCK cells (Fanning et al., 2012).
Fig. 7.
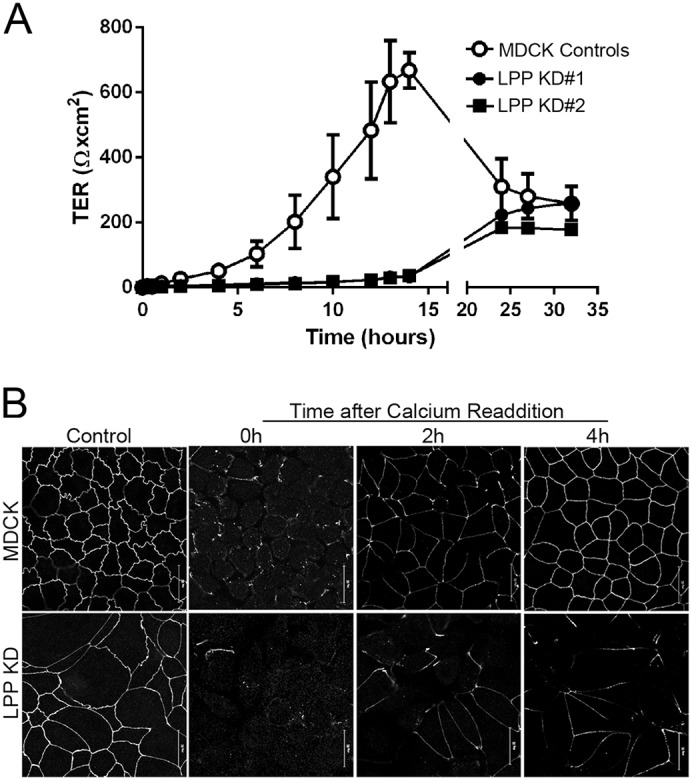
LPP knockdown slows barrier recovery after calcium removal. Confluent MDCK and LPP cell monolayers were exposed to calcium-free media for 18 hours. At t = 0 hours, calcium-free medium was replaced with normal calcium-containing media and (A) TER was measured on duplicate filters at the indicated intervals. Unlike MDCK cell controls, LPP-knockdown cells failed to recover appreciable TER over the first 15 hours after cells were placed back into normal calcium-containing medium. By 24 hours, TER values in the knockdown cells were similar to control MDCK cells. (B) Filters with MDCK controls and LPP-knockdown cells from the above time points were collected, and recovery of ZO-1 at tight junctions monitored by immunofluorescence. By 4 hours, ZO-1 staining has become continuous in control cells but not in LPP-knockdown cells. Scale bars: 20 µm.
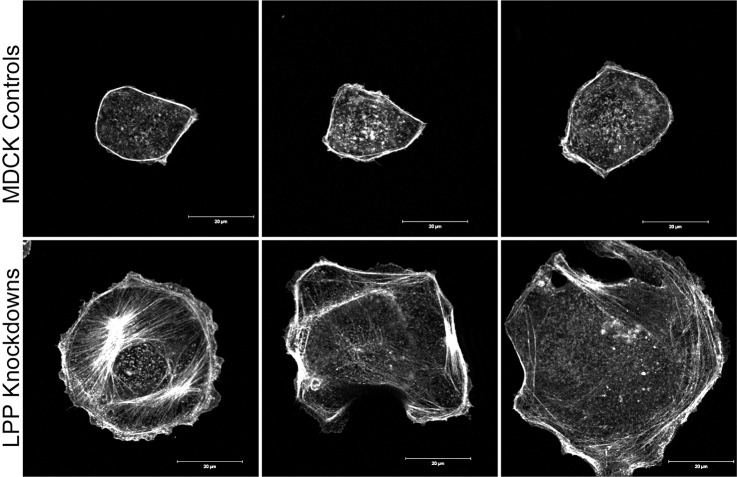
LPP-knockdown cells tended to be irregular in shape compared with control MDCK cells and were often larger and flatter (Figs 7B, 8 and 9); in addition, BrdU incorporation assays demonstrate a slower proliferative rate (45±2% labeled nuclei in control cells after 90 minutes, compared with 29±2% in LPP-knockdown cells, P = 0.0003). Along with the alteration in E-cadherin dependent interactions, the irregular cell shapes suggested that there might be differences in actin or myosin organization at cell contacts. Rhodamine-phalloidin staining of freshly plated, subconfluent cells revealed striking differences in the actin filament organization between control MDCK cells and LPP-knockdown cells (Fig. 8 and Fig. 9A). Four hours after trypsinization and replating on glass coverslips, actin is organized in a tight cortical ring in control cells (Fig. 8, top panels). By contrast, in the LPP-knockdown cells, much less of the cellular actin is organized at the cortex and it is often present in a loose array (Fig. 8, bottom panels). The difference in actin organization is most evident in isolated cells; in confluent monolayers, cortical actin organization is similar between control and knockdown cells (supplementary material Fig. S2) although cell shape differences make the actin organization and density difficult to compare. The localization of apical myosin 2A and 2B is also similar between control and LPP-knockdown cells (supplementary material Fig. S2), in spite of the decreased myosin 2B levels as determined by immunoblotting (Fig. 5A,B).
Fig. 8.
Actin in subconfluent LPP-knockdown cells is less well organized than in MDCK control cells. Confocal immunofluorescent localization of F-actin with Rhodamine-phalloidin in freshly plated (4 hours) MDCK (top panels) reveals a narrow cortical band of actin in isolated cells. By contrast (bottom panels), actin in LPP-knockdown cells is loosely organized in actin cables that are found both near cell borders and elsewhere in the cells. Scale bars: 20 µm.
Fig. 9.
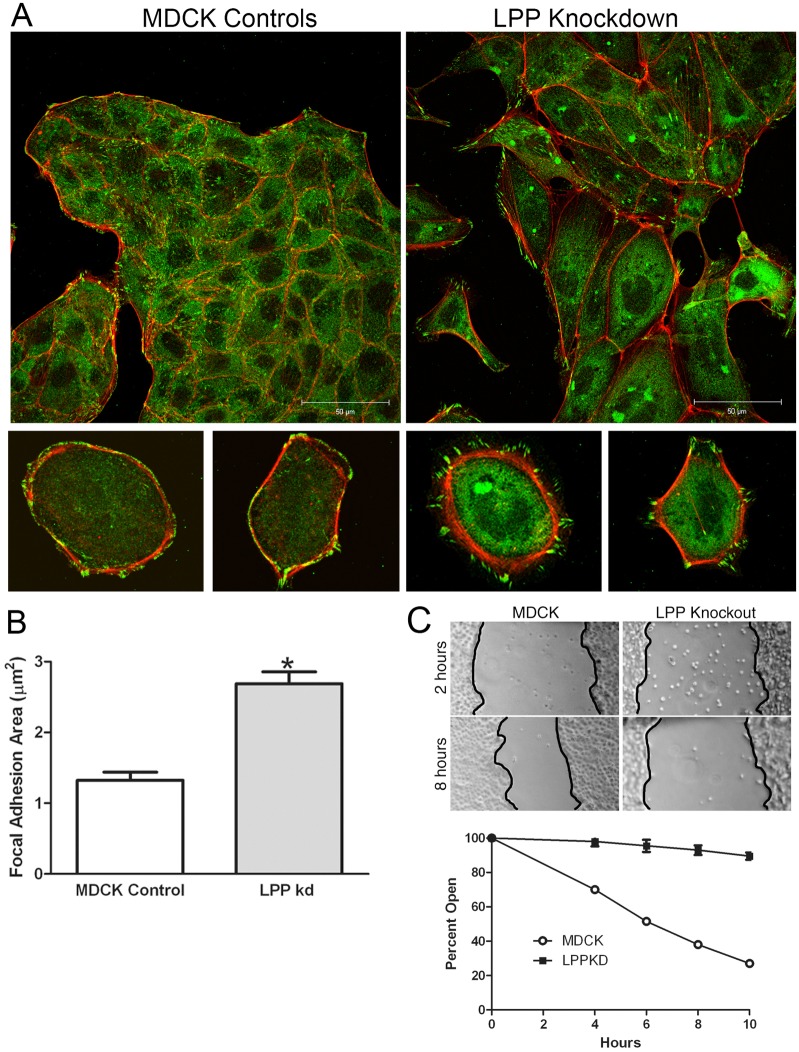
LPP-knockdown cells have larger, less numerous focal adhesions and decreased migration in wound-healing assays. (A) MDCK control cells (left panels) and LPP-knockdown cells (right panels) were stained for paxillin (green) and actin (red). Both in cell islands and in isolated cells, there were larger and fewer focal adhesions than in MDCK control cells and cortical actin appeared slightly less organized in knockdown cells. (B) Focal adhesion size as defined by paxillin staining was measured in control MDCK and LPP-knockdown cells using ImageJ, n = 133 and 219 focal adhesions, respectively for control and knockdown cells. Values are means ± s.d.; P<0.001, unpaired Student's t-test. (C) Confluent monolayers of MDCK and LPP-knockdown cells were wounded using a plastic pipette tip and (top panel) wound edges photographed at indicated intervals up to 10 hours post wounding. Distances between wound edges at predetermined places were measured using ImageJ (bottom panel); at least three measurements were made at each time point. Similar results were seen with three LPP-knockdown cell lines. Scale bar: 50 µm.
How LPP modulates cortical actin organization is unclear. LPP is known to bind directly to the actin regulatory proteins α-actinin (Li et al., 2003) and VASP (Petit et al., 2000), but neither the levels (Fig. 5A,B) nor subcellular localization of these proteins appears significantly changed in the LPP knockdown cells compared with MDCK controls (supplementary material Fig. S3), suggesting that the observed actin changes might not be mediated by these proteins. However, it is known that activity of both VASP (Thomson et al., 2011) and α-actinin (Feng et al., 2013) can be influenced by phosphorylation, so essential changes in post-translational modifications are possible in knockdown cells. In addition, although there are marked increases in zyxin levels in the knockdown cells compared with controls (Fig. 5A,B), which could also act to alter actin organization (Beckerle, 1997), there is no clear change in zyxin distribution at cell–cell contacts (supplementary material Fig. S3, bottom panels). The immunofluorescence data suggest that amount of zyxin at this site is not reciprocally regulated by the loss of LPP, although zyxin staining of focal adhesions is more prominent in knockdown cells compared with control cells (supplementary material Fig. S3).
LPP-knockdown cells have larger focal adhesions and decreased migration
Because LPP is associated not only with cell contacts but also with focal contacts, we next asked whether there were changes in cell-substrate adhesions in the LPP-knockdown cells. We found that in both cell islands (Fig. 9A, top panels) and in isolated cells (Fig. 9A, bottom panels) that focal adhesions, as visualized with paxillin immunofluorescence (Fig. 9A, green) were larger and less numerous in the knockdown cells than in MDCK control cells (Fig. 9B). Because large focal adhesions have been negatively associated with cell motility (Fincham and Frame, 1998), we determined the effects of LPP knockdown in wound-healing assays (Fig. 9C). LPP-knockdown cells showed markedly lower migration rates in this assay than did MDCK cell controls (Fig. 9C and quantified in Fig. 9D).
DISCUSSION
The use of E-cadherin fused to biotin ligase as a probe to identify proteins proximal to the adherens junction has identified a large number of both expected and unexpected proteins. Among the most abundant proteins are catenins, which are the best studied cadherin-interacting proteins (reviewed by Franke, 2009). Numerous cytoskeletal proteins are also well tagged by E-cadherin, as are many trafficking and signaling proteins. Some apparently irrelevant proteins, including ribosomal proteins, are also tagged and are probably biotinylated during E-cadherin translation. In an extensive literature search, Zaidel-Bar (Zaidel-Bar, 2013) recently identified 175 proteins (including many tissue-specific proteins) as components of the ‘cadherin adhesome’; of this list, nearly 40 are also identified in the present proteomic screen. In contrast to the overlap between the ‘cadherin adhesome’ and the EcadBL results, there was less overlap (6/54; namely E-cadherin, alpha-E-catenin (catenin-alpha-1), nectin-2 (poliovirus-related receptor, protein 2), SNAP23 (synaptosomal-associated protein 23), desmoglein-2, integrin-beta-1 and ADP-ribosylation factor 1) between our findings (supplementary material Table S1) and the 54 proteins identified in a proteomic analysis performed on a biochemically enriched preparation of tight and adherens junctions (Yamazaki et al., 2008). However, this lack of overlap is probably due to differences in methodology, because their tissue preparation included lateral membrane material that was not necessarily proximal to E-cadherin.
In addition to those proteins identified as components of the ‘cadherin adhesome’, we also identify a number of different kinases and signaling molecules, which are thus likely candidates for further investigation. Easily rationalized proteins that were well tagged by EcadBL that are not identified in the ‘cadherin adhesome’ include members of the pleckstrin homology domain containing family A members 5 and 6, EH-domain binding protein 1-like, the coxsackie and adenovirus receptor, vang-like protein 1 and 4F2 cell surface antigen heavy chain; this last protein was previously identified as interacting with catenin delta-1 (Smith et al., 2011). Proteins that might play a role but have yet undescribed functions include sickle tail homolog and uncharacterized protein LOC100688057, which contains an actin binding/calponin homology domain. Also well tagged were the vesicular trafficking proteins synaptosomal-associated proteins 23 and 29, members of the rab5 family and proteins involved in clathrin-dependent endocytosis, which could play a role in cadherin recycling (Le et al., 1999). It is worth noting that in MDCK cells, E-cadherin is distributed not only at apical adherens junctions, but also along the lateral membrane, suggesting that some tagged identified proteins are not uniquely proximal to adherens junction components. In fact, the abundant lateral membrane transporter, the sodium/potassium transporter ATPase, was recovered in cells expressing EcadBL, but at a low level (rank 229), consistent with considerable specificity in this assay as we previously found for the N- and C-terminal of ZO-1 (Van Itallie et al., 2013).
Along with other proteins previously identified as members of the ‘cadherin adhesome’ (Zaidel-Bar, 2013), EcadBL tagged several members of the zyxin family. As described above, LPP was the most heavily tagged of the zyxin family members in both the EcadBL and BL-ZO1 proteomics screens. LPP shares a similar domain organization with zyxin, including N-terminal binding sites for α-actinin, proline-rich ActA sites which interact with members of the Ena/VASP family and other proteins, leucine-rich nuclear export signals and three C-terminal LIM domains, also known to act as sites for protein interaction (Grunewald et al., 2009). Zyxin family members act as scaffolds regulating actin-interacting proteins and other proteins at cell adhesion sites; the presence of the LIM domains are characteristic of this and other protein families that can also shuttle in and out of the nucleus to influence gene transcription (reviewed by Hervy et al., 2006).
We confirmed the colocalization of LPP with E-cadherin and its localization to focal adhesions. In confluent MDCK cells, the cell-contact localization was much more obvious for LPP than it was for zyxin, although both proteins were equally easily visualized at focal adhesions. This observation, along with the heavier tagging of LPP than zyxin in the proteomic screens, suggested to us that it might play a different role in cell-contact regulation in MDCK cells than does zyxin. Although several studies have identified a role for zyxin at cell contacts (Hansen and Beckerle, 2006; Nguyen et al., 2010; Sperry et al., 2010), the importance of epithelial LPP is not well understood. Similar to zyxin-knockout animals (Hoffman et al., 2003), LPP-knockout mice develop normally with no obvious physiologic defects and morphologically normal adherens junctions (Vervenne et al., 2009). In one of the few sets of experiments in epithelial cells, Hansen and Beckerle (Hansen and Beckerle, 2006) expressed wild-type LPP and zyxin in MDCK cells and saw no alteration in cell adhesion as measured by a hanging-drop assay. However, these authors found expression of LPP or zyxin mutated in the ActA (VASP interacting) and/or LIM domains altered aggregation; further experiments suggested this modulation was dependent on interaction with VASP.
We demonstrate here that knockdown of LPP results in decreased cadherin-dependent cell adhesion, similar to what has been described for zyxin knockdown (Nguyen et al., 2010). However, the mechanism of this decrease in adhesion is unclear. Zxyin knockdown in MDCK cells is associated with loss of VASP from focal adhesions and cadherin-dependent cell contacts; this change in localization of VASP is implicated in the observed decrease in adhesion (Nguyen et al., 2010). By contrast, there are no obvious changes in VASP localization in LPP-knockdown cells. However, LPP contains only one functional VASP-binding domain, compared with the four functional domains found in zyxin, thus it is possible that other protein–protein interactions might predominate in the effects of LPP on cell adhesion (Drees et al., 2000; Petit et al., 2000; Li et al., 2003).
In the above study by Hansen and Beckerle, there were no obvious differences in the effects of wild-type LPP or zyxin on cell adhesion. This, with the findings that neither LPP- nor zyxin-knockout animals show obvious defects, has led to the suggestion that the proteins are functionally redundant in vivo (Hoffman et al., 2003; Vervenne et al., 2009). In fact, although there is no change in zyxin levels in LPP-knockout animals (Vervenne et al., 2009) or vice versa (Hoffman et al., 2003), zyxin levels are variably elevated in our LPP-knockdown cells. In spite of the increase in the level of zyxin protein, it was difficult to visualize any significant change in zyxin localization in knockdown cells.
Strikingly, LPP-knockdown cells show dramatic increases in the size of focal adhesions and changes in peripheral actin organization that are not evident in zyxin-knockdown cells. Consistent with the large focal adhesions, LPP knockdown results in decreased cell migration; decreased migration is also seen in mouse embryonic fibroblasts isolated from LPP-knockout animals (Vervenne et al., 2009). Again, this differs from zyxin knockdowns, because MDCK cells depleted of zyxin show normal migration in a wound-healing assay (Nguyen et al., 2010). Both normal E-cadherin-dependent adhesion (Li et al., 2012) and focal contact organization have been implicated in normal migratory behavior. The decrease in E-cadherin-dependent adhesion and increase in the size of focal adhesions suggests that LPP, unlike zyxin, regulates the balance between these adhesive sites. One possibility is that LPP titrates regulatory proteins at cell contacts and focal adhesions to coordinate actin organization. Two candidates among many include the LPP-interacting proteins paladin (Jin et al., 2007), which is an actin organizing protein, and Scrib (Petit et al., 2005b), which has been implicated in both cell migration and adhesion (Qin et al., 2005). However, the number of possible interacting regulatory proteins is large and their complete examination is outside the scope of this study. In any case, the changes in cell–cell and cell–substrate organization in the LPP-knockdown cells, along with the potential for changes in nuclear transcription suggest that this protein provides regulation and/or feedback in balancing cell movement with cell adhesion.
The proteins identified by EcadBL should provide a resource for further understanding the organization of adherens junctions; the results from investigation of just one of these proteins, LPP, support the utility of this approach. In addition, although the many cytoskeletal, trafficking and signaling proteins identified are unlikely to be unique to adherens junctions, their identification in this screen suggests that they could play important roles associated with this structure. Finally, comparison between proteins tagged by E-cadherin and those identified in our previous study with ZO-1 (Van Itallie et al., 2013), along with those of other tight junction proteins (work in progress), should eventually allow identification of a consensus set of tight and adherens junction proteins and their sub-junctional compartmentalization.
MATERIALS AND METHODS
Constructs and cell lines
Myc–biotin ligase plasmid (pcDNA3.1 mycBioID) was a gift from Kyle Roux (Addgene, Cambridge, MA, plasmid 35700 (Roux et al., 2012). The Myc–biotin ligase insert was excised and subcloned into pTRE2hyg (BD Biosciences, San Jose, CA) and the Myc tag moved by PCR to the C-terminal end of the biotin ligase, as previously described (Van Itallie et al., 2013). Full-length human E-cadherin was a gift from Jennifer Stow (Addgene plasmid 28009 (Miranda et al., 2001). The E-cadherin coding sequence (minus GFP) was cloned using the Infusion (Clontech, Mountain View, CA) protocol into the modified pTRE2hyg biotin ligase plasmid; the final fusion protein was (N-terminal) E-cadherin–biotin-ligase–Myc tag (C-terminal). MDCK II tet-off (Clontech) cells were transfected by nucleofection (Lonza, Walkersville, MD); stable cell lines expressing the fusion protein were selected with medium containing 0.25 mg/ml hygromycin. Cells were kept uninduced in the presence of doxycycline until plated for immunofluorescence or proteomic analysis.
Three different LPP-knockdown oligonucleotides (GATGCTGTATGATATGGAA, GGCCTACCTTTAATGTACA, GGTCGTTACTATGAAGCCT were cloned into pSuper (Oligoengine, Seattle, WA) and were cotransfected with pTKhyg into MDCK cells and stable lines selected as described above. Both the first and second antisense sequence resulted in excellent knockdown, so one clone from each was used in all studies.
Immunoblotting and immunofluorescence
Immunoblotting and immunofluorescence were performed as previously described (Van Itallie et al., 2013); unless otherwise noted, fixation for immunofluorescence was 1% paraformaldehyde. Antibody sources as follows. Millipore (Billerica, MA): actin (MAB1501R, immunoblotting only); Sigma-Aldrich (St Louis, MO, USA): α-actinin (A5044), β-catenin (C2206), e-cadherin (U3254, immunofluorescence only) gamma tubulin (T5326) vinculin (V9131), BD Biosciences; afadin (610732), p120 catenin (610133), E-cadherin (610181, immunoblot only), paxillin (612405), Life Technologies (Grand Island, NY); α-catenin (71-1200), occludin (33-1500), ZO-2, ethanol fixation (38-9100), Cell Signaling Technology (Danvers, MA); LPP (3389), Myc tag (2276), VASP (3132) E-cadherin rabbit polyclonal antibody (3195), Covance (Chantilly, VA); myosin 2A (ethanol fixation, PRB-440P), myosin 2B (ethanol fixation, PRB-445P), Abcam (Cambridge, MA); gamma tubulin (ab11317), zyxin (ab71842). ZO-1 antibody was hybridoma 40.76, a gift from Bruce Stevenson. IR-dye secondary antibodies for immunoblots were from Rockland Immunochemicals (Gilbertsville, PA) and for immunofluorescence from Jackson Immunoresearch (West Grove, PA), except for Streptavidin 568 (Life Technologies); actin was localized with Rhodamine-phalloidin (Life Technologies). In experiments where nuclei were stained Hoechst 33342 reagent was included at a concentration of 0.1 µg/ml during the last three washes.
Proximity ligation assays (Söderberg et al., 2006) were performed as recommended by the manufacturer (Duolink, Sigma Chemical Company) using LPP monoclonal and E-cadherin rabbit polyclonal antibodies described above. E-cadherin rabbit polyclonal and p120 catenin antibodies were used as positive controls; rabbit E-cadherin and mouse gp135/podocalyxin (kindly provided by G. Ojakian, State University of New York Downstate Medical Center) antibodies and LPP antibody alone were used as negative controls.
Proteomic analyses
Purification of biotinylated proteins and mass spectrometry was carried out as previously described (Van Itallie et al., 2013). MASCOT database search was performed using a Canis familiaris RefSeq database. EcadBL identification was performed from three separate isolations. Criteria for inclusion in the final protein lists were appearance in at least two of the three sample runs. Relative abundance was determined by normalizing PSM (peptide spectral match) values within a single run (values for each protein/total PSM value for that run) and then averaging normalized PSM for two or three runs for that protein. Average normalized PSM values for each protein was divided by the number of theoretical tryptic peptides detectable by LC-MS/MS (Pisitkun et al., 2012) and this value multiplied by 1000 for convenience.
Adhesion assay
E-cadherin-dependent adhesion was extensively modified from a protocol described by Johnson et al. (Johnson et al., 2009). To coat wells with substrate, 75 μl/well of 10 μg/ml E-cadherin (R&D Systems recombinant human E-cadherin FC Chimera, Minneapolis, MN) or fibronectin (Life Technologies) was added to a 96-well plate and incubated overnight at 4°C; 4 hours before use, wells were blocked with 10% BSA.
To make single cells, MDCK control and knockdown cells were trypsinized, pelleted and plated at low (106 cells/100 mm dish) density one day prior to the assay. The next morning, trypsinization, pelleting and plating was repeated into duplicate 60 mm dish at 105 cells/dish. Cells were allowed to attach for 4 hours; after 3 hours, medium was removed and cells were incubated in 2.5 μM Cell Tracer Green CMFDA or Red CMPTX (Molecular Probes, Life Technologies) in serum-free Optimem (Life Technologies). Cells were incubated with dyes for 30 minutes and then changed back into normal serum-supplemented medium. 4 hours following trypsinization, cells were rinsed twice with PBS with calcium and magnesium and incubated in 0.05% trypsin in Hanks-buffered salt solution supplemented with 0.1 mM CaCl2. When cells were fully dissociated (after ∼55 minutes), they were collected, pelleted, resuspended (105/ml) and 100 μl of MDCK control (red or green) and LPP-knockdown (green or red) cells added together to E-cadherin-coated 96-well dishes. Six wells were plated for each mixture on each substrate and allowed to adhere for 60 minutes; wells (three each) were left unwashed or washed three times with medium and then fluorescence measured using a fluorescent imager (Typhoon, GE Healthcare, Piscataway, NJ). Data are present as the relative fluorescent signal in the washed and unwashed wells. The adhesion assay was repeated four times.
Calcium-switch assay
For the calcium-switch experiments, MDCK controls and LPP-knockdown cell lines were grown to confluence on 12 mm Transwell filters (Costar, Corning, NY, washed extensively with phosphate-buffered saline (PBS) without Ca2+ and Mg2+ and incubated overnight in S-MEM (Sigma-Aldrich) supplemented with 2% dialyzed fetal bovine serum and penicillin-streptomycin to dissociate cell–cell contacts. The low-Ca2+ medium was replaced the next day with normal growth medium (1.8 mM Ca2+) and TER (measured with an EVOM2 Ohm meter and Endohm-12 electrode (World Precision Instruments, Sarasota, FL) and the cells were prepared for immunostaining at indicated time points.
BrdU incorporation
BrdU incorporation was performed according to the manufacturer's instructions (BD Biosciences); MDCK control and LPP-knockdown cells labeled for 90 minutes were quantified by comparison of immunofluorescent signal from labeled nuclei (BrdU antibody Bu20a, Cell Signaling Technology) to DAPI staining (Life Technologies) using ImageJ (NIH). More than 300 MDCK controls and LPP knockdowns were analyzed in each of two separate experiments; statistical analyses were performed using the unpaired Student's t-test (GraphPad Prism).
CSK extraction
MDCK controls and LPP-knockdown cells were grown to confluence on coverslips, washed three times with PBS and extracted with CSK buffer (10 mM PIPES, pH 6.8, 50 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100, 300 mM sucrose (Shore and Nelson, 1991), washed, fixed with 4% paraformaldehyde and stained for e-cadherin. Line scans of the acquired images were acquired using ImageJ (NIH) and plotted using GraphPadPrism (La Jolla, CA) software.
Quantification of focal adhesion
Focal adhesion size was quantified using paxillin images of control and knockdown cells, values were thresholded and the area of focal adhesions quantified using ImageJ; at least 100 focal adhesions were measured in control and knockdown cell lines.
Wound-healing assay
Newly confluent MDCK and LPP-knockdown cells cultured on 60-mm dishes were scratched with a plastic pipette. Wounds were photographed at designated sites along the wound at the indicated times and the size of the wound at the sites (as measured by the distance between the edges) was calculated using ImageJ. The wounding experiment was repeated twice with at least two LPP-knockdown clones.
Supplementary Material
Acknowledgments
We thank Mark Knepper, Trairak Pisitkun and Jason Hoffert (Systems Biology Center, NHLBI, National Institutes of Health) and Christian Combs and Daniela Malide (Light Microscopy Core, NHLBI, National Institutes of Health) for invaluable help. We thank Ana Pasapera (Clare Waterman Laboratory, NHLBI, National Institutes of Health) for help with paxillin staining and interpretation.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
C.M.V, A.J.T., A.A., K.F. and M.G. performed experiments. J.M.A. assisted in interpreting data and contributed to writing the manuscript. A.S.F. assisted in interpreting data. C.M.V. conceived the study, performed experiments, supervised experimental work and wrote the manuscript.
Funding
This research was funded by the Office of the Director through the Intramural Research Program of the National Institutes of Health. A.S.F. is supported by grant number DK061397 from the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.140475/-/DC1
References
- Abe K., Chisaka O., Van Roy F., Takeichi M. (2004). Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat. Neurosci. 7, 357–363 10.1038/nn1212 [DOI] [PubMed] [Google Scholar]
- Beckerle M. C. (1997). Zyxin: zinc fingers at sites of cell adhesion. Bioessays 19, 949–957 10.1002/bies.950191104 [DOI] [PubMed] [Google Scholar]
- Capaldo C. T., Macara I. G. (2007). Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 18, 189–200 10.1091/mbc.E06-05-0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B., Friederich E., Fradelizi J., Louvard D., Beckerle M. C., Golsteyn R. M. (2000). Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 275, 22503–22511 10.1074/jbc.M001698200 [DOI] [PubMed] [Google Scholar]
- Fanning A. S., Van Itallie C. M., Anderson J. M. (2012). Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 23, 577–590 10.1091/mbc.E11-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Ngu H., Alford S. K., Ward M., Yin F., Longmore G. D. (2013). α-actinin1 and 4 tyrosine phosphorylation is critical for stress fiber establishment, maintenance and focal adhesion maturation. Exp. Cell Res. 319, 1124–1135 10.1016/j.yexcr.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham V. J., Frame M. C. (1998). The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 17, 81–92 10.1093/emboj/17.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke W. W. (2009). Discovering the molecular components of intercellular junctions—a historical view. Cold Spring Harb. Perspect. Biol. 1, a003061 10.1101/cshperspect.a003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G. A., McLachlan R. W., Yap A. S. (2011). Productive tension: force-sensing and homeostasis of cell-cell junctions. Trends Cell Biol. 21, 499–505 10.1016/j.tcb.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Chávez de Ramírez B., Cereijido M. (1985). Tight junction formation in cultured epithelial cells (MDCK). J. Membr. Biol. 86, 113–125 10.1007/BF01870778 [DOI] [PubMed] [Google Scholar]
- Grunewald T. G., Pasedag S. M., Butt E. (2009). Cell Adhesion and Transcriptional Activity - Defining the Role of the Novel Protooncogene LPP. Transl. Oncol. 2, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C., Lecuit T. (2013). Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340, 1185–1189 10.1126/science.1235249 [DOI] [PubMed] [Google Scholar]
- Gumbiner B., Stevenson B., Grimaldi A. (1988). The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 107, 1575–1587 10.1083/jcb.107.4.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. D., Beckerle M. C. (2006). Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J. Biol. Chem. 281, 16178–16188 10.1074/jbc.M512771200 [DOI] [PubMed] [Google Scholar]
- Hervy M., Hoffman L., Beckerle M. C. (2006). From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr. Opin. Cell Biol. 18, 524–532 10.1016/j.ceb.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Hoffman L. M., Nix D. A., Benson B., Boot-Hanford R., Gustafsson E., Jamora C., Menzies A. S., Goh K. L., Jensen C. C., Gertler F. B. et al. (2003). Targeted disruption of the murine zyxin gene. Mol. Cell. Biol. 23, 70–79 10.1128/MCB.23.1.70-79.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Umeda K., Tsukita S., Furuse M., Tsukita S. (2007). Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 176, 779–786 10.1083/jcb.200612080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens B., Goossens S., Staes K., Gilbert B., van Hengel J., Colpaert C., Bruyneel E., Mareel M., van Roy F. (2001). alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J. Cell Sci. 114, 3177–3188 [DOI] [PubMed] [Google Scholar]
- Jin L., Kern M. J., Otey C. A., Wamhoff B. R., Somlyo A. V. (2007). Angiotensin II, focal adhesion kinase, and PRX1 enhance smooth muscle expression of lipoma preferred partner and its newly identified binding partner palladin to promote cell migration. Circ. Res. 100, 817–825 10.1161/01.RES.0000261351.54147.de [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Winterwood N., DeMali K. A., Stipp C. S. (2009). Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J. Cell Sci. 122, 2263–2273 10.1242/jcs.045997 [DOI] [PubMed] [Google Scholar]
- Le T. L., Yap A. S., Stow J. L. (1999). Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219–232 [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., Lenne P. F. (2007). Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 10.1038/nrm2222 [DOI] [PubMed] [Google Scholar]
- Li B., Zhuang L., Reinhard M., Trueb B. (2003). The lipoma preferred partner LPP interacts with alpha-actinin. J. Cell Sci. 116, 1359–1366 10.1242/jcs.00309 [DOI] [PubMed] [Google Scholar]
- Li L., Hartley R., Reiss B., Sun Y., Pu J., Wu D., Lin F., Hoang T., Yamada S., Jiang J. et al. (2012). E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell. Mol. Life Sci. 69, 2779–2789 10.1007/s00018-012-0951-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder D., Shevchenko A., Simons K., Füllekrug J. (2005). Gp135/podocalyxin and NHERF-2 participate in the formation of a preapical domain during polarization of MDCK cells. J. Cell Biol. 168, 303–313 10.1083/jcb.200407072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Takeichi M. (2009). Adherens junction: molecular architecture and regulation. Cold Spring Harb. Perspect. Biol. 1, a002899 10.1101/cshperspect.a002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K. C., Khromykh T., Christy P., Le T. L., Gottardi C. J., Yap A. S., Stow J. L., Teasdale R. D. (2001). A dileucine motif targets E-cadherin to the basolateral cell surface in Madin-Darby canine kidney and LLC-PK1 epithelial cells. J. Biol. Chem. 276, 22565–22572 10.1074/jbc.M101907200 [DOI] [PubMed] [Google Scholar]
- Nguyen T. N., Uemura A., Shih W., Yamada S. (2010). Zyxin-mediated actin assembly is required for efficient wound closure. J. Biol. Chem. 285, 35439–35445 10.1074/jbc.M110.119487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen C. M., Leckband D., Yap A. S. (2011). Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91, 691–731 10.1152/physrev.00004.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H., Takeichi M. (2011). Evolution: structural and functional diversity of cadherin at the adherens junction. J. Cell Biol. 193, 1137–1146 10.1083/jcb.201008173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M. M., Fradelizi J., Golsteyn R. M., Ayoubi T. A., Menichi B., Louvard D., Van de Ven W. J., Friederich E. (2000). LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11, 117–129 10.1091/mbc.11.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M. M., Crombez K. R., Vervenne H. B., Weyns N., Van de Ven W. J. (2005a). The tumor suppressor Scrib selectively interacts with specific members of the zyxin family of proteins. FEBS Lett. 579, 5061–5068 10.1016/j.febslet.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Petit M. M., Meulemans S. M., Alen P., Ayoubi T. A., Jansen E., Van de Ven W. J. (2005b). The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 6, 1 10.1186/1471-2121-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T., Gandolfo M. T., Das S., Knepper M. A., Bagnasco S. M. (2012). Application of systems biology principles to protein biomarker discovery: urinary exosomal proteome in renal transplantation. Proteomics Clin. Appl. 6, 268–278 10.1002/prca.201100108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Capaldo C., Gumbiner B. M., Macara I. G. (2005). The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171, 1061–1071 10.1083/jcb.200506094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M., Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore E. M., Nelson W. J. (1991). Biosynthesis of the cell adhesion molecule uvomorulin (E-cadherin) in Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 266, 19672–19680 [PubMed] [Google Scholar]
- Smith A. L., Friedman D. B., Yu H., Carnahan R. H., Reynolds A. B. (2011). ReCLIP (reversible cross-link immuno-precipitation): an efficient method for interrogation of labile protein complexes. PLoS ONE 6, e16206 10.1371/journal.pone.0016206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G. et al. (2006b). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Sperry R. B., Bishop N. H., Bramwell J. J., Brodeur M. N., Carter M. J., Fowler B. T., Lewis Z. B., Maxfield S. D., Staley D. M., Vellinga R. M. et al. (2010). Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. J. Cell. Physiol. 222, 612–624 [DOI] [PubMed] [Google Scholar]
- Takeichi M. (1988). The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development 102, 639–655 [DOI] [PubMed] [Google Scholar]
- Thomson D. M., Ascione M. P., Grange J., Nelson C., Hansen M. D. (2011). Phosphorylation of VASP by AMPK alters actin binding and occurs at a novel site. Biochem. Biophys. Res. Commun. 414, 215–219 10.1016/j.bbrc.2011.09.059 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium(2013). Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids. Res. 41, D43–D47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Aponte A., Tietgens A. J., Gucek M., Fredriksson K., Anderson J. M. (2013). The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J. Biol. Chem. 288, 13775–13788 10.1074/jbc.M113.466193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervenne H. B., Crombez K. R., Delvaux E. L., Janssens V., Van de Ven W. J., Petit M. M. (2009). Targeted disruption of the mouse Lipoma Preferred Partner gene. Biochem. Biophys. Res. Commun. 379, 368–373 10.1016/j.bbrc.2008.12.074 [DOI] [PubMed] [Google Scholar]
- Wolfenson H., Lavelin I., Geiger B. (2013). Dynamic regulation of the structure and functions of integrin adhesions. Dev. Cell 24, 447–458 10.1016/j.devcel.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Okawa K., Yano T., Tsukita S., Tsukita S. (2008). Optimized proteomic analysis on gels of cell-cell adhering junctional membrane proteins. Biochemistry 47, 5378–5386 10.1021/bi8002567 [DOI] [PubMed] [Google Scholar]
- Yonemura S. (2011). Cadherin-actin interactions at adherens junctions. Curr. Opin. Cell Biol. 23, 515–522 10.1016/j.ceb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R. (2013). Cadherin adhesome at a glance. J. Cell Sci. 126, 373–378 10.1242/jcs.111559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.