Abstract
Sex difference involving chromosomes and gene expression has been extensively documented. In this study, the gender difference in the sweetpotato whitefly Bemisia tabaci was investigated using Illumina-based transcriptomic analysis. Gender-based RNAseq data produced 27 Gb reads, and subsequent de novo assembly generated 93,948 transcripts with a N50 of 1,853 bp. A total of 1,351 differentially expressed genes were identified between male and female B. tabaci, and majority of them were female-biased. Pathway and GO enrichment experiments exhibited a gender-specific expression, including enriched translation in females, and enhanced structural constituent of cuticle in male whiteflies. In addition, a putative transformer2 gene (tra2) was cloned, and the structural feature and expression profile of tra2 were investigated. Sexually dimorphic transcriptome is an uncharted territory for the agricultural insect pests. Molecular understanding of sex determination in B. tabaci, an emerging invasive insect pest worldwide, will provide potential molecular target(s) for genetic pest control alternatives.
Gender is a strong determinant of transcript levels in comparison to animals' ontogeny1, age, and genotype2. Animals from a broad range of taxa show sex differences in development time, lifespan3, body size4, sex-biased gene and protein expression5,6, and sex chromosomes7. Sex-biased gene expression has been documented in multiple insect species including Drosophila2,8,9,10, Anopheles gambiae11,12,13, Tribolium castaneum14, Daphnia pulex15, and vespid wasp16. Among them, a rapid evolution at transcription level is evident in Drosophila and An. Gambiae5, in which a large proportion of the transcriptome is sex-biased2,8,17. Expression of male-biased genes had significantly faster rates of evolution than that in female-biased or unbiased genes in D. melanogaster18. Also, specific gene families were preferentially expressed in unique tissues. The takeout family, for example, was enriched in the head of male D. melanogaster19.
The sweetpotato whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), is an emerging agricultural pest that causes serious damage through feeding and vectoring plant viruses in protected crops and field crops worldwide20. More than 34 morphologically indistinguishable cryptic species have been identified through various techniques21,22,23,24. Among them, Bemisia tabaci B (also referred to as the Middle East-Asia Minor 1) has been determined as the most invasive and destructive biotype in many parts of the world22. In China, B. tabaci was first recorded in the late 1940s. However, B. tabaci did not cause serious losses in vegetable, fruit and ornamental crops until the introduction and establishment of invasive B. tabaci B in the mid-1990s25,26.
Bemisia tabaci has a haplo-diploid life cycle. The males, derived from unfertilized eggs, are haploid, and females are diploid27. The estimated nuclear DNA content of male and female B. tabaci B was 1.04 and 2.06 pg, respectively28. Regarding the biological traits, male and female B. tabaci are different in longevity29, body size30,31,32, secondary symbionts33, and efficiency of transmitting plant viruses34,35,36,37,38,39. For squash leaf curl virus, however, female and male B. tabaci has the same efficiency40. There were no differences in response between male and female whitefly when they were exposed to thiamethoxam, a widely used neonicotinoid insecticide41. Molecular basis of sex determination has been traditionally focused on model insects with readily available genomes, including D. melanogaster42,43, Apis mellifera44,45, Nasonia46, and T. castaneum47. A suite of genes, including transformer (tra), transformer2 (tra2), feminize (fem), complementary sex determination (csd) and a key switch gene doublesex (dsx), are determined to play important role in sex determination. In other non-model insects, especially the ones without genome, such as haplo-diploid B. tabaci, molecular basis of sex determination remains largely unknown.
To reveal the gender differences at the transcription level, we carried out the largest de novo transcriptome sequencing in B. tabaci up-to-date (336 million reads, 27 Gb). Based on comparative analyses of eight B. tabaci transcriptomes generated from male and female whiteflies reside in four different host plants, genes and gene networks putatively involved in sex divergence were investigated. We also summarized the function and sequence divergence of genes with sexually enriched expressions to determine which genes, if any, are rapidly evolving and potentially involved in sex determination.
Results
Illumina sequencing and de novo assembly
We constructed 8 cDNA libraries derived from B-biotype (Middle East-Asia Minor 1) B. tabaci female or male adults reared on four different host plants: cabbage, cucumber, tomato and cotton. Each sample resulted in a library containing an average of 42 million reads equal to 3.4 Gb and a total data of 27 Gb were generated (Table 1). For further analysis, two different strategies (separate or pool assembly) were carried out to guarantee the amount of sequence data for each assembly. First, eight libraries were separately assembled by using Trinity with default parameters48, and 34,055–77,065 long transcripts were generated with lengths longer than 200 bp (Table 2). Second, pooled reads covering all 8 libraries were assembled with the same method above; the length distribution and length statistic were shown in Table 2. Pooled assembly was used as the final result, and it generated 93,948 long transcripts with a length longer than 200 bp (Table 2). The total length of assembled long transcripts was approximately 79.7 Mb and the N50 size was 1,853 bp with 21,273 sequences longer than 1 kb. In comparison to previously published transcriptome data of B. tabaci49,50,51, pooled assembly was better in data volume and in length (Table 2).
Table 1. Illumina sequencing metrics.
| Caf* | Cuf | Tof | Cof | Cam | Cum | Tom | Com | |
|---|---|---|---|---|---|---|---|---|
| Total Reads | 36,616,870 | 43,576,274 | 41,959,788 | 65,050,162 | 53,477,310 | 19,373,358 | 40,522,552 | 41,192,154 |
| Total Lengths (bp) | 2,929,349,600 | 3,486,101,920 | 3,356,783,040 | 5,204,012,960 | 4,278,184,800 | 1,549,868,640 | 3,241,804,160 | 3,295,372,320 |
“*”:Ca, Cu, Co, and Tom are abbreviation for the respective host plants including Cabbage, Cucumber, Cotton and Tomato, while m and f denote males and females.
Table 2. Bemisia tabaci transcriptome resources used in this study.
| B. tabaci B50 | B. tabaci Q51 | B. tabaci B49 | Cafa | Cuf | Tof | Cof | Cam | Cum | Tom | Com | Pooled | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length distribution (bp) | ||||||||||||
| 0.1–0.5 k | 151,375 | 151,360 | 40,254 | 30,416 | 31,089 | 32,175 | 31,152 | 44,478 | 23,187 | 41,068 | 25,051 | 55,584 |
| 0.5–1.0 k | 22,546 | 12,949 | 13,007 | 11,259 | 12,885 | 12,599 | 11,234 | 15,864 | 6,934 | 13,808 | 6,127 | 17,091 |
| 1.0 k–2.0 k | 4,046 | 3,929 | 4,070 | 7,622 | 8,935 | 7,905 | 6,060 | 9,829 | 3,204 | 8,390 | 2,195 | 10,992 |
| >2.0 k | 702 | 662 | 410 | 6,443 | 7,144 | 4,684 | 2,967 | 6,894 | 890 | 5,384 | 682 | 10,281 |
| Total | 178,669 | 168,900 | 57,741 | 55,740 | 60,053 | 57,363 | 51,413 | 77,065 | 34,215 | 68,650 | 34,055 | 93,948 |
| Sequencing statistics (bp) | ||||||||||||
| Total (Mb) | 62.9 | 42.9 | 26.4 | 48.4 | 53.5 | 43.0 | 33.8 | 62.0 | 17.8 | 49.8 | 15.9 | 79.7 |
| Mean | 368 | 266 | 478 | 911 | 934 | 786 | 689 | 843 | 543 | 761 | 490 | 889 |
| Median | 327 | 172 | 355 | 444 | 478 | 435 | 403 | 417 | 331 | 388 | 335 | 392 |
| Min | 102 | 100 | 200 | 201 | 201 | 201 | 201 | 201 | 201 | 201 | 201 | 201 |
| Max | 5,872 | 7,297 | 5,936 | 16,744 | 19,356 | 16,173 | 17,748 | 24,983 | 12,022 | 19,970 | 9,551 | 27,456 |
| N50 | 423 | 331 | 552 | 1,757 | 1,710 | 1,294 | 1,023 | 1,503 | 723 | 1,320 | 561 | 1,853 |
| Comparative analysis | ||||||||||||
| Platform | 454 | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina |
| Assembly | gsAssembler | SOAPdenovo | SOAPdenovo | Trinity | Trinity | Trinity | Trinity | Trinity | Trinity | Trinity | Trinity | Trinity |
| Hitb | 72.5% | 77.7% | 93.7% | |||||||||
“a”: Ca, Cu, Co, and Tom are abbreviation for the respective host plants including Cabbage, Cucumber, Cotton and Tomato, while m and f denote males and females.
“b”: Blastn against the pooled transcriptome dataset using an e-value cutoff of 1e-5.
Functional annotation
For annotation, the pooled assembled unigenes were searched using blastx against the non-redundant (nr) NCBI protein database, SWISS-PROT, COG and KEGG all with an e-value cut-off of 10−5. As shown in Table 3, 25,554 genes (27.2% of all unigenes sequences) in nr, 20,003 genes in SWISS-PROT, 1656 genes in COG, and 5,548 genes in KEGG returned above the cut-off BLAST hits. This annotation rate is consistent with the previously reported B-type whitefly49,50 and Q-type whitefly51. Gene Ontology terms were assigned by Blast2GO through a search of NR database. InterPro was searched by InterProScan. Using this approach, in total, 12,222 unigenes were assigned to one or more Gene Ontology (GO) terms. The Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway analysis revealed that 5,548 unigenes could be assigned to 774 predicted pathways using Acyrthosiphon pisum as KEGG organism.
Table 3. Annotation of a pooled assembly, representing male and female B. tabaci transcriptomes on different host plants.
| Database | Number of Transcriptsa |
|---|---|
| Nr | 25,554 |
| SWISS-PROT | 20,003 |
| COG | 1,656 |
| KEGG | 5,548 |
| InterPro | 23,466 |
| GO | 12,222 |
“a”: Transcripts with a length greater than 200 bp were subjected to annotation.
Cluster correlation analysis
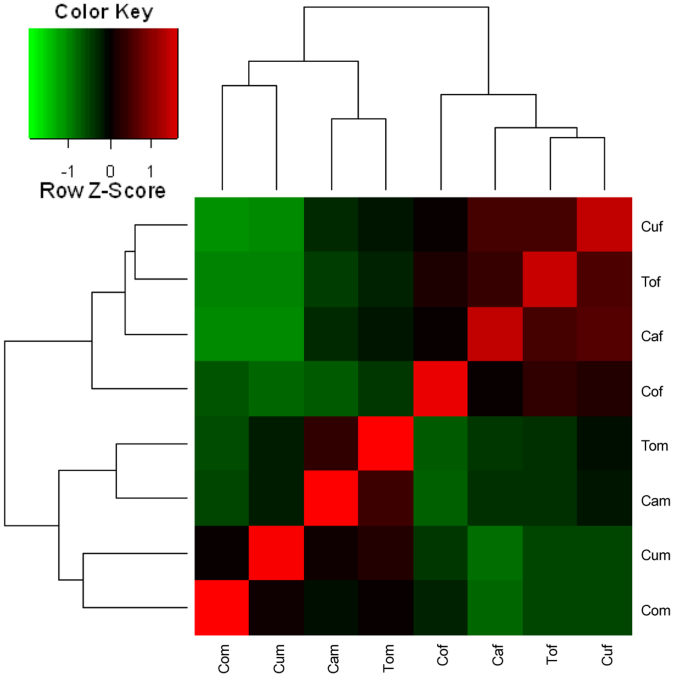
To measure the correlation of any two samples in all eight libraries, read number of each sample was used and calculated according to the Spearman correlation coefficient. In this study, gene expression differences between the sexes are much greater than those among the host plants (Figure 1). On the other hand, we analyzed the possible reciprocal best matched sequences (RBMs) pairs between transcripts from each separated and pooled transcriptomes using bidirectional best hit49,52,53. With this method, we identified 31658, 35692, 31560, 36318 and 42150, 17929, 26169, 17110 pairs of putative orthologs, respectively, in female B. tabaci on cabbage (Caf), cucumber (Cuf), tomato (Tof), cotton (Cof), and male tabaci on cabbage (Cam), cucumber (Cum), tomato (Tom), and cotton (Com) corresponding to the pooled transcripts (Table S3).
Figure 1. Pairwise correlation between all of the samples based on the Spearman correlation coefficient.
Based on the read counts measurements, the heatmap exhibits a hierarchical clustering of 8 transcriptome samples (sex*host plant). The inserted color key depicts the value of Spearman correlation coefficient, ranging from green (normalized expression of −1) to red (normalized expression value of 1).
Global expression profiles between sexes, and sex biased genes
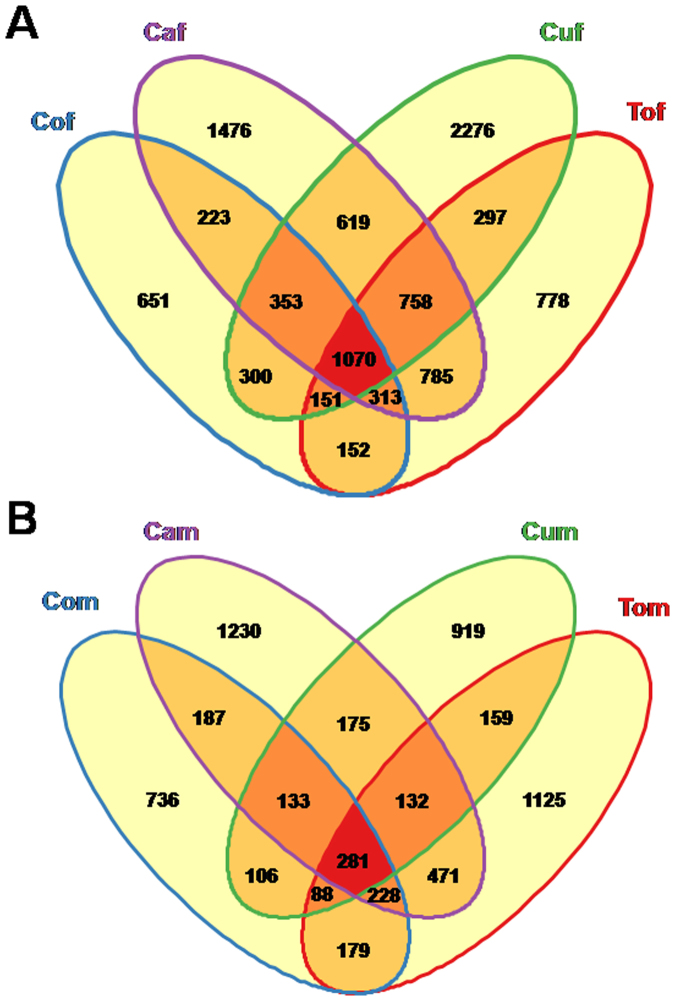
To specifically identify sex-biased genes that might also affect the sex difference process, we performed a series of expression profiling to examine gene activity changes between sexes. By comparing female to male, differentially expressed transcripts (DETs) (q-value < 0.05 and log2(Fold change) > 1or log2(Fold change) < −1) were generated in four host strains (Table S4). This resulted in 8,434 differentially expressed transcripts between Caf and Cam, 7817 between Cuf and Cum, 6967 between Tof and Tom and 5151 between Cof and Com (Table S4). These results suggested that majority of the differentially expressed transcripts between sexes were female-biased. To reduce the DETs between female and male whiteflies, intersection elements of female (Figure 2A) and male (Figure 2B) specific genes were determined. Among them, 1070 female and 281 male specific genes were annotated (Table S1 and S2). The analysis of the transcriptome dynamics was shown in Figure 3. Both the female and male biased genes were enriched with members of distinct KEGG pathways and GO categories. Pathway enrichment tests revealed that pyrimidine metabolism, ribosome, mRNA surveillance pathway, ribosome biogenesis in eukaryotes, RNA polymerase, RNA degradation, DNA replication and RNA transport were enriched in female whitely. In the case of male whitefly, starch and phototransduction, lysosome, nitrogen metabolism, glycerophospholipid metabolism and phagosome were enriched (Figure S1 and S2). The enriched GO analysis for female and male biased genes were shown in Figure S3, which indicated that many enriched GO categories in female included only extracellular region while structural constituent of cuticle and hormone activity were predominant in male (Figure S3).
Figure 2. Venn diagram demonstrating sex-specific gene expression in B. tabaci.
(A). Female specific gene expression in different host plants, including cabbage (Caf), cotton (Cof), cucumber (Cuf), and tomato (Tof). (B). Male specific gene expression in different host plants, including cabbage (Cam), cotton (Com), cucumber (Cum), and tomato (Tom).
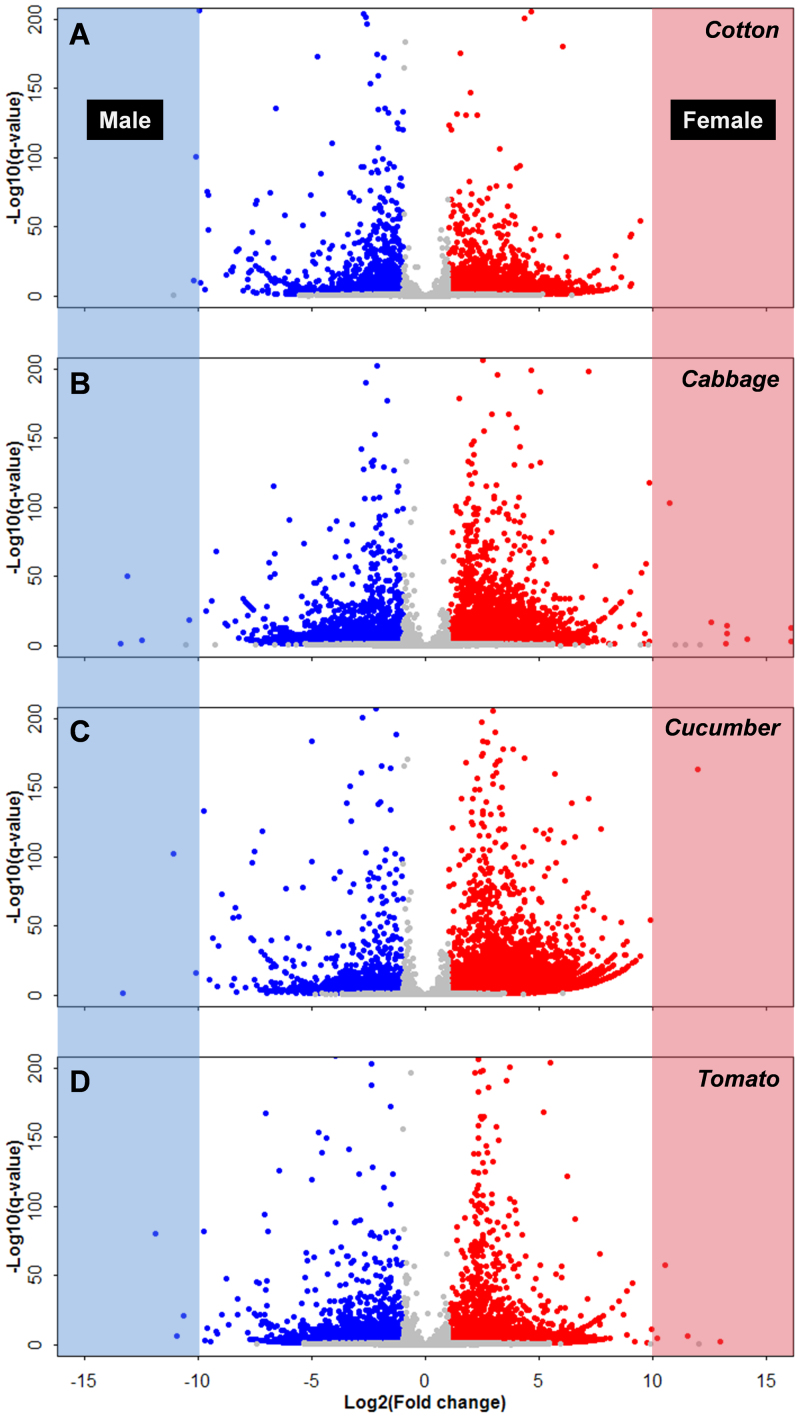
Figure 3. Volcano plot illustrating B. tabaci gender differences at transcription level.
The x-axis represents the relative expression of transcript for female and male B. tabaci, respectively. The color-shaded regions depict significantly female-biased genes [red, Log2(fold change) > 1, q-value < 0.05], and male-biased genes [blue, Log2(fold change) < −1, q-value < 0.05] among cotton (A), cabbage (B), cucumber (C), and tomato (D).
qRT-PCR validation
Only 52% (33/64) of the genes exhibited significantly different expression profiles between female and male whiteflies (Table S6). For the 48 female enriched genes, 18 of them were over-expressed in the female compared to male (expression ratio of female/male > 1 and significant based on t test), whereas 15 out of 16 were over-expressed in the male compared to female. The comparative analysis of qRT-PCR and RNA sequencing of selected sex enriched genes revealed that the qRT-PCR results were, for the most part, consistent with the sequencing analysis results, especially to the male enriched genes. For instance, among the differentially expressed genes from qRT-PCR analysis, 85% (28/33) exhibited significantly difference at a fold change of above 2 (female enriched genes) or below 2 (male enriched genes) (Table S6).
Characterization of Bt-tra2 gene
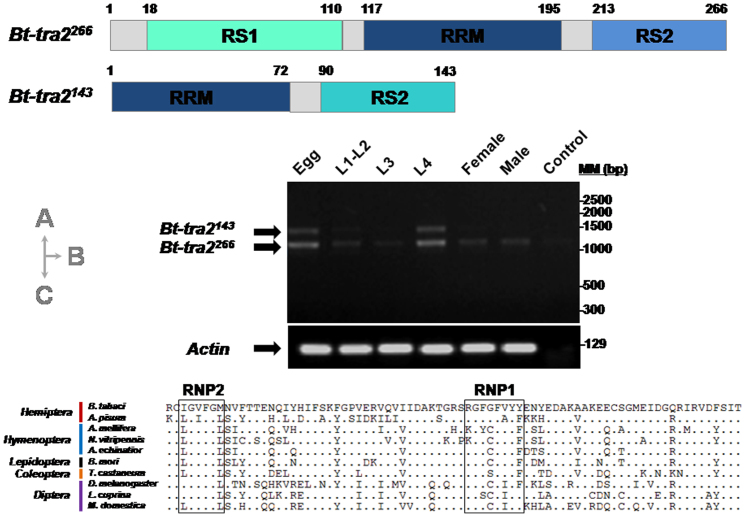
Two different splice variants (Bt-tra2266 and Bt-tra2143) were identified that were not sex-specific in embryos, larvae or adults by verifying their amplicons in all developmental stages by sequencing (Figure 4A, 4B). High expression abundance in eggs and L4 compared to L1-L2, L3 and adults stages was also found by semi-quantitative transcriptional profile analysis of Bt-tra2 transcripts at different developmental stages (Figure 4B). Because of evolutionary constrain in RRM (RNA binding domain or RNA-recognition motif), we only unambiguously aligned the amino acid sequences with those of hymenopteran (N. vitripennis, A. mellifera and A. echinatior), lepidopteran (B. mori), coleopteran (T. castaneum) and dipteran (D. melanogaster, L. cuprina and M. domestica) that shared structural features for RRM domain in above insects (Figure 4C). Results indicated that the RRM amino acid sequence diverges in relation to phylogenetic distance. The RRM domains of the hymenopterans (A. mellifera, N. vitripennis and A. echinatior) show a pairwise sequence identity of 81 to 85%, and 68 to 80% for the dipterans (L. cuprina, M. domestica and D. melanogaster), whereas RRMs of B. tabaci and hymenopterans or dipterans have pairwise sequence identity of 70 to 78%, and 61 to 68%, respectively. Within the RRM of Bt-Tra2 protein, there are two putative RNP (ribonucleoprotein consensus peptide) elements (Figure 4C), which can directly interact with the dsx pre-mRNAs in D. melanogaster. In comparison to D. melanogaster, three amino acid differences were indentified in the eight-amino acids-long RNP1 sequence element of B. tabaci, whereas, only one amino acid differences exhibited in the six-amino-acid long RNP2 sequence element.
Figure 4. Characterization of Bt-tra2, a putative sex determination gene in B. tabaci.
(A). The schematic drawing shows structural features of the two predicted Bt-tra2 isoforms (RS, arginine/serine rich domain; RRM, RNA binding domain). (B). Semi-quantitative transcriptional profile of Bt-tra2 transcripts throughout different developmental stages, including L1–L2 (1-2rd instar larvae), L3 (3rd instar larvae), L4 (4rd instar larvae), female and male adults. The actin was used as a loading control. (C). The phylogenetic relationship of Tra2 RRM domains among different insect orders, including Hemiptera (B. tabaci and Acyrthosiphon pisum),Hymenoptera (Apis mellifera, Nasonia vitripennis, and Acromyrmex echinatior), Lepidoptera (Bombyx mori), Coleoptera (Tribolium castaneum), and Diptera (Drosophila melanogaster, Lucilia cuprina, and Musca domestica). Dots indicate amino acids identical to the predicted Bt-Tra2 RRM of the whitefly. RNP sequence elements (RNP1 and RNP2) are highlighted in boxes.
Discussion
In this study, eight paired end libraries, each containing an average of 42 million reads, were constructed individually using the Trinity method48, representing the largest de novo transcriptome assembly in whiteflies (27 Gb)49,50,51.
Sexual dimorphism represents phenotypic differences between males and females of the same species. Differences between males and females can be resolved by the evolution of differential gene expression in the two sexes5. Features of B. tabaci make it particularly interesting for a comparative study of sex-biased genes. B. tabaci just like thrips (Thysanoptera), beetles (Coleoptera) and bees (Hymenoptera), have haplodiploid sex determination; males are haploid, develop from unfertilized eggs and only inherit a maternal genome, whereas females are diploid, develop from fertilized eggs and inherit a paternal and a maternal genome. Thus in B. tabaci, we expect to find the patterns of sex-biased gene expression common or specific to the other arthropods. Previous experiments have found that a large portion of the transcriptome is differentially expressed between the sexes11,14,54,55. However, most of these studies have focused on model organisms such as Drosophila, An. gambiae and T. castaneum, with few studies on non-model species due to the lack of genome sequences.
Our comparative transcriptome analyses result in 1351 sex biased genes, in which 1070 are female specific and 281 are male specific. It is worth noting that without replications, the intersection of four experiments involving gender and host plants was fairly conservative in detecting the differentially expressed genes. Nevertheless, we do find a large number of genes to be female-biased, which is consistent with Tribolium castaneum14 and Anopheles gambiae13. In contrast, Eads et al (2007)15 reported that majority of the sex-biased genes were male specific in Daphnia. The discrepancy reflects the species-specific nature of gender-associated genes. On the other hand, the intrinsic genetic differences between haploid male and diploid female might contribute to such inconsistency as well. Recent studies in other haploid/diploid organisms, including Nasonia, beetles and honeybee44,45,46,47, indicate that the transformer (tra) -doublesex (dsx) axis is conserved among all insects and plays a pivotal role in sex determination mechanisms.
Functional categories enriched in female B. tabaci were associated with translation, pyrimidine metabolism, ribosome function and DNA replication. Previous report indicated that proteins involved in translation and DNA packaging were predominantly female-biased in expressions among flies9, mosquitoes11 and Daphnia15. One of the enriched functional categories in B. tabaci is the structural constituent of cuticle, which is consistent with a previous report in Daphnia15, suggesting that structural difference (e.g., cuticle composition) and increased cuticle metabolism in males might be a result of gender differences. Previous report indicated that diet could affect lifespan and reproduction, and favor different type of nutrient intake in males and females56,57. During starvation, neurons from males are readily undergoing autophagy and dead, whereas neurons from females mobilize fatty acids, accumulate triglycerides, form lipid droplets, and survive longer58. Differential immune responses between the sexes were also observed in an autumnal moth, Epirrita autumnata59. In whiteflies, functional categories such as nutrient-related (nitrogen metabolism and glycerophospholipid metabolism) and immune-related (lysosome and phagosome) pathways were enriched in males.
The accuracy of the corresponding genes of most enriched pathway and GO terms was confirmed by the qRT-PCR analysis, with 52% of the selected ESTs showed significant differences. Similar to other microarray or RNA-seq analysis, this method tends to generate false positives. Consequently, results from both analyses need to be validated by qRT-PCR. Whitefly female and male biology difference were obvious in previous report including some virus transmission and biological characters and so on, but seemly are not distinctly and necessarily associated with sexual divergence in this study. The public release of the assembled whitefly genome, combined with genetic mapping studies, should provide new insight into the roles of sex-biased genes.
Genetic regulation of sex determination was focused initially on model insects D. melanogaster and Caenorhabditis elegans42, and recently expanded to other insects including Hymenoptera (A. mellifera and Nasonia) and Coleoptera (T. castaneum)46,47,60,61. In most insects, the sex-determining pathway consists of the basal switch doublesex that is acting as a conserved major switch at the downstream of the sex-determining cascade62. In Drosophila melanogaster, protein-protein interactions between the female-biased transformer (tra), the splicing regulator of doublesex, and transformer2 (tra2), a non-sex-specific auxiliary splicing factor, are essential to promote female sexual differentiation63. In hemipterans, such as aphid and whitefly, regulation mechanism of sex determination are largely unknown. In this study, a gene encoding transformer2 was cloned from the whitefly (Bt-tra2) and its expression profiles were documented. Not surprisingly, Bt-tra2 did not display any detectable sexual dimorphic differences in expression or splicing patterns (Figure 4). Sex-specific splicing of tra as an important regulatory mechanism was described in the fruitfly, wasp46 and red flour beetle47. However, non-sex-specifically spliced tra2 from honeybee also act as an essential regulator of the sex determination hierarchy45. In addition, sex-specific doublesex (Bt-dsx) was also found and cloned from our other EST database (unpublished data). Functional characterization studies are currently underway to test the hypothesis that Bt-tra2 affects embryonic viability and female splicing of Bt-dsx transcripts. In addition, a recent report showed that endosymbionts can actively manipulate the sex determination mechanism of their host, Ostrinia scapulalis64,65. This discovery can shed new light on the sex determination mechanism of lepidopteran insects and other herbivores. Thus, in the near future, a study may also be focused about the Bt-tra2 splicing patterns in females with different types and densities of the bacterial symbionts.
Methods
Insect rearing and sample preparation
The four B B. tabaci host strains (cabbage, cucumber, tomato and cotton) were the same populations as described previously66. For sample collection, only 2-day-old adults were collected from each host strain using a glass tube (5 × 0.5 cm) and the sex was determined under a stereo microscope. Then adults of the same sex were pooled into a plastic tube using an aspirator. Finally, these different host strain female and male samples were respectively collected, snap frozen in liquid nitrogen, and transferred to a −80°C freezer for the long-term storage (Figure S4). Before sample collection, the populations had been completely separated and reared for at least five years, and all identified as B-type (the Middle East-Asia Minor 1) using mtDNA COI marker in the laboratory every 2–3 generations.
RNA isolation, cDNA library construction, and illumina sequencing
RNA from each female or male sample group was extracted with Trizol reagent (Invitrogen) according to the manufacturer's instructions, and purity and degradation were checked on 1% agarose gels. RNA integrity was further confirmed using the 2100 Bioanalyzer (Agilent Technologies) with a minimum RNA integrated number value of 8. Poly (A)-containing RNA was then separated from total RNA using the Dynabeads mRNA purification kit (Invitrogen) and the quality was verified on a denaturing gel. Then, the eight paired-end cDNA libraries were all prepared using Illumina's protocols following manufacturer's recommendations and sequenced on the Illumina GAIIplatform. Briefly, the mRNA was fragmented into small pieces using divalent cations under elevated temperature and the cleaved RNA fragments were used for first strand cDNA synthesis using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA polymerase I and RNaseH. These cDNA fragments were subjected to end repair process and ligation of adapters. These products were purified and enriched with PCR to create the final cDNA library.
Sequence preprocessing, assembly, and functional annotation
The 8 cDNA library was separately sequenced on the Illumina sequencing platform (GAII). The insert size of each library is approximately 500 bp and both ends of the cDNA are sequenced. Image deconvolution and quality value calculations were performed using the Illumina GA pipeline 1.3. The raw reads were filtered by removing adaptor sequences, low quality sequences (containing reads with unknown sequences ‘N’ and lower than 10 of mean phred scores). The reads obtained from each library were assembled separately, and all pooled reads of 8 libraries were also assembled using Trinity (version r2011-08-20) with default parameters48. To annotate the pooled assembly transcriptome, we performed a BLAST search against the non-redundant (NR) database in NCBI, SWISS-PROT, KEGG and COG with an e-value cut-off of 1e-5. Gene Ontology terms were assigned by Blast2GO67 through a search of the NR database.
Identification of reciprocal best matched sequences (RBMs)
With each library as an independent sample, identification pipeline of RBMs genes was done similar to the methods described in Wang et al. (2011)49. Briefly, the bidirectional best hit method (BBH) was considered in BLAST search (blastn, e-value cutoff is 1e-10) to identify genes that are putatively pairs of reciprocal best matched sequences between the pooled assembly and 8 separate assembly results. Pairs of sequences that were each other's best hit and longer than 200 bp were retained.
Expression profiling pipeline
Gene expression value measurement and differentially expressed genes
Based on the number of assembled genes, trinity pooled transcripts and raw reads for each sample were analyzed using RSEM software (v1.1.15)68. Second, in order to calculate the normalized read counts for each library, RNA composition bias and normalization factors were taken into account and calculated by edger69. Read counts for each sample is also used to calculate spearman correlation coefficient of two selected samples with cor function in R package, and based that heat map of spearman correlation coefficient for all samples was generated with heatmap.2 in R package70. Then, the normalized count was directly used for subsequent differential expression gene analyses. DEGseq packages71 were used to identify differentially expressed genes using MARS model (Q-value < 0.05 and Fold Change > 2)71,72.
Enrichment analysis
GOSeq (Version 1.8.0)73 is used for GO enrichment analysis, which takes into account the gene selection bias due to difference in gene lengths and thus numbers of overlapping sequencing reads. Pathway mapping (used Acyrthosiphon pisum as KEGG organisms) and further KEGG enrichment analysis was carried out with local server KOBAS 2.074. The Q-value < 0.05 is chosen as significant cutoff for enriched GO and pathway.
Quantitative real time PCR (qRT-PCR) analysis
Sample preparation
Female adult (Caf) and male adult (Cam) from cabbage strain were determined as described previously66. A total of 300 newly emerged and virgin Caf and Cam, respectively, were collected, snap frozen in liquid nitrogen, and stored at −80°C for the subsequent qRT-PCR analysis.
qRT-PCR
Total RNA was extracted from Caf and Cam, respectively, using Trizol (Invitrogen) following the manufacturer's protocols. The total RNA obtained was re-suspended in nuclease-free water and the concentration was measured using Nanodrop (Thermo Scientific Nanodrop 2000). About 0.5 μg of total RNA was used as template to synthesize the first-strand cDNA using a PrimerScript RT reagent Kit (TaKaRa) following the manufacturer's protocols. The resultant cDNA was diluted to 0.1 μg/μl for further analysis in the qRT-PCR (ABI 7500) using an SYBR Green Realtime PCR Master Mix (TaKaRa).
qRT-PCR primers (Table S5) for the selected 64 unique genes, corresponding to the most enriched pathways, such as the pyrimidine metabolism in females (Figure S1) and the phototransduction in males (Figure S2), and the most enriched GO terms, including the structural constituent of ribosome, ribosome and translation in females, and the extracellular region, structural constituent of cuticle and hormone activity in males (Figure S3), were designed using the Primer Express 2.0 software. The cycling parameter was 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 62°C for 34 s, ending with melting curve analysis (65°C to 95°C in increments of 0.5°C every 5 s) to check for nonspecific product amplification. Relative gene expression was calculated by the 2−ΔΔCt method using housekeeping genes RPL2975 as the reference to eliminate sample-to-sample variations in the initial cDNA samples.
Characterization of Bt-tra2
To determine the entire sequences of the Bt-tra2 transcripts, we analyzed in detail the EST sequence (annotated transformer-2 sex-determining protein in Table S1) from the female biased genes relative to male. Based on this EST, combined with other insect EST annotated tra2 in NCBI, the complete open reading frame (ORF) sequence of Bt-tra2 was cloned and both-strands sequenced. We translated the mRNA sequences into the amino acid sequence, and we predicted the protein domains by the similarity to domains in the PROSITE database (http://www.expasy.org/prosite/). The GenBank accession numbers are KC333625 (Bt-tra2266) and KC333626 (Bt-tra2143).
Bt-tra2 distribution throughout development
Total RNA was extracted from male and female eggs, larvae (L1–L2, L3 and L4 instar larvae), female and male adult. We amplified cDNA fragments using oligonucleotides primers (forward primer, 5′-CCCCACTGAATCCTGTT-3′ and reverse primer, 5′-CGGTAATGAGTGGAGAAGA-3′) that span the complete open reading frame (ORF) of these two Bt-Tra2 splice variants. The identity of the amplicons of all developmental stages was verified by sequencing.
Phylogenetic analysis
We utilized nine Tra2 amino acid sequences from different insect species with ClustalW alignment through Molecular Evolutionary Genetic Analysis software version 5 (MEGA 5) to compare the phylogenetic and molecular relationships: Acyrthosiphon pisum (GenBank: XP_001944825), Apis mellifera (GenBank: NP_001252514), Nasonia vitripennis (GenBank: XP_001601106), Acromyrmex echinatior (GenBank: EGI70155), Bombyx mori (GenBank: NP_001119709), Tribolium castaneum (GenBank:XP_968550), Drosophila melanogaster (GenBank: NP_476765), Lucilia cuprina (GenBank: ACS34688), Musca domestica (GenBank: AAW34233).
Author Contributions
Conceived and designed the experiments: W.X., L.T.G., X.G.Z. and Y.J.Z. Performed the experiments: W.X. and L.T.G. Analyzed the data: W.X. and L.T.G. Contributed reagents/materials/analysis tools: W.X., L.T.G., X.G.J., N.N.Y., X.Y., Q.J.W., S.L.W., X.G.Z. and Y.J.Z. Wrote the paper: W.X., L.T.G., Q.J.W., S.L.W., X.G.Z. and Y.J.Z. All authors reviewed the manuscript.
Additional Information
Data Deposition The Illumina reads of Bemisia tabaci of eight libraries were submitted to the NCBI Sequence Read Archive under the accession number SRA050082.1-SRA050089.1.
Supplementary Material
Supplementary Info File #1
Acknowledgments
This research was supported by the National Science Fund for Distinguished Young Scholars (31025020), the 973 Program (2012 CB017359), and Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China.
References
- Arbeitman M. N., Fleming A. A., Siegal M. L., Null B. H. & Baker B. S. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131, 2007–2021 (2004). [DOI] [PubMed] [Google Scholar]
- Jin W. et al. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29, 389–395 (2001). [DOI] [PubMed] [Google Scholar]
- Bonduriansky R., Maklakov A., Zajitschek F. & Brooks R. Sexual selection, sexual conflict and the evolution of ageing and lifespan. Funct. Ecol. 22, 443–453 (2008). [Google Scholar]
- Fairbairn D. J. Allometry for sexual size dimorphism: pattern and process in the coevolution of body size in males and females. Annu. Rev. Ecol. S. 28, 659–687 (1997). [Google Scholar]
- Ellegren H. & Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8, 689–698 (2007). [DOI] [PubMed] [Google Scholar]
- Viitaniemi H. M. & Leder E. H. Sex-biased protein expression in threespine stickleback, Gasterosteus aculeatus. J. Proteome Res. 10, 4033–4040 (2011). [DOI] [PubMed] [Google Scholar]
- Kaiser V. B. & Bachtrog D. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 44, 91–112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M. N. et al. Gene expression during the life cycle of Drosophila melanogaster. Science 297, 2270–2275 (2002). [DOI] [PubMed] [Google Scholar]
- Ranz J., Castillo-Davis C., Meiklejohn C. & Hartl D. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742–1745 (2003). [DOI] [PubMed] [Google Scholar]
- Chang P. L., Dunham J. P., Nuzhdin S. V. & Arbeitman M. N. Somatic sex-specific transcriptome differences in Drosophila revealed by whole transcriptome sequencing. BMC Genomics 12, 364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W. & Lanzaro G. C. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr. Biol. 15, 192–193 (2005). [DOI] [PubMed] [Google Scholar]
- Marinotti O. et al. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol. Biol. 15, 1–12 (2006). [DOI] [PubMed] [Google Scholar]
- Baker D. A. et al. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 12, 296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince E. G., Kirkland D. & Demuth J. P. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linkedgenes in the flour beetle. Genome Biol. Evol. 2, 336–346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads B. D., Colbourne J. K., Bohuski E. & Andrews J. Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex. BMC Genomics 8, 464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt B. G. & Goodisman M. A. Evolutionary variation in gene expression is associated with dimorphism in eusocial vespid wasps. Insect Mol. Biol. 19, 641–652 (2010). [DOI] [PubMed] [Google Scholar]
- Ranz J. M., Castillo-Davis C. I., Meiklejohn C. D. & Hartl D. L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742–1745 (2003). [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hambuch T. M. & Parsch J. Molecular Evolution of Sex-Biased Genes in Drosophila. Mol. Biol. Evol. 21, 2130–2139 (2004). [DOI] [PubMed] [Google Scholar]
- Vanaphan N., Dauwalder B. & Zufall R. A. Diversification of takeout, a male biased gene family in Drosophila. Gene 491, 142–148 (2012). [DOI] [PubMed] [Google Scholar]
- Jones D. R. Plant viruses transmitted by whiteflies. Eur. J. Plant. Pathol. 109, 195–219 (2003). [Google Scholar]
- Perring T. M. The Bemisia tabaci species complex. Crop Prot. 20, 725–737 (2001). [Google Scholar]
- Dinsdale A., Cook L., Riginos C., Buckley Y. M. & De Barro P. J. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 103, 196–208 (2010). [Google Scholar]
- Elbaz M., Lahav N. & Morin S. Evidence for pre-zygotic reproductive barrier between the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull. Entomol. Res. 100, 581–590 (2010). [DOI] [PubMed] [Google Scholar]
- De Barro P. J., Liu S. S., Boykin L. M. & Dinsdale A. Bemisia tabaci: A statement of species status. Ann. Rev. Entomol. 56, 1–19 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang L. P. et al. Analysis of genetic diversity among different geographical populations and determination of biotypes of Bemisia tabaci in China. J. Appl. Entomol. 129, 121–128 (2005). [Google Scholar]
- Chu D. et al. The introduction of the exotic Q biotype of Bemisia tabaci from the Mediterranean region China on ornamental crops. Fla. Entomol. 89, 168–174 (2006). [Google Scholar]
- Blackman R. L. & Cahill M. The karyotype of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull. Entomol. Res. 88, 213–215 (1998). [Google Scholar]
- Brown J. K., Lambert G. M., Ghanim M., Czosnek H. & Galbraith D. W. Nuclear DNA content of the whitefly Bemisia tabaci (Aleyrodidae: Hemiptera) estimated by flow cytometry. Bull. Entomol. Res. 95, 309–312 (2005). [DOI] [PubMed] [Google Scholar]
- Azab A. K., Megahed M. M. & El-Mirswami H. D. On the biology of Bemisia tabaci (Genn) (Homoptera: Aleyrodidae). Bull. Soc. Ent. Egypte. 55, 305–315 (1971). [Google Scholar]
- Mohanty A. K. & Basu A. N. Biology of whitefly vector Bemisia tabaci (Genn) on four host plants throughout the year. J. Ent. Res. 11, 15–18 (1987). [Google Scholar]
- Brar D. S., Aneja A. K., Singh J. & Mahal M. S. Biology of whitefly Bemisia tabaci (Gennadius) on American cotton Gossypium hirsutum Linnaeus. J. Insect Sci. 18, 48–59 (2005). [Google Scholar]
- Feng Y. T. et al. Fitness costs and morphological change of laboratory-selected thiamethoxam resistance in the B-type Bemisia tabaci (Hemiptera: Aleyrodidae). J. Appl. Entomol. 133, 466–472 (2008). [Google Scholar]
- Pan H. P. et al. Factors affecting population dynamics of maternally transmitted endosymbionts in Bemisia tabaci. Plos One 7, e30760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. & Nitzany F. E. Transmission and host range of the tomato yellow leaf curl virus. Phytopathology 56, 1127–1131 (1966). [Google Scholar]
- Nateshan H. M., Muniyappa V., Swanson M. M. & Harrison B. D. Host range, vector relations and serological relationships of cotton leaf curl virus from southern India. Ann. Appl. Biol. 128, 233–244 (1996). [Google Scholar]
- Muniyappa V. et al. Tomato leaf curl virus from Bangalore (ToLCV-Ban4): sequence comparison with Indian ToLCV isolates, detection in plants and insects, and vector relationships. Arch. Virol. Arch. Virol. 145, 1583–1598 (2000). [DOI] [PubMed] [Google Scholar]
- Mann R. S. & Singh L. Retention of Cotton leaf curl virus (CLCUV) in its vector whitefly Bemisia tabaci (Gennadius). Ind. J. Entomol. 66, 96–98 (2004). [Google Scholar]
- Mann R. S. & Singh L. Studies on the relationship of Cotton leaf curl virus (CLCuV) with its vector, Bemisia tabaci (Gennadius). Ind. J. Plant Prot. 32, 140–141 (2004). [Google Scholar]
- Xie W., Xu Y. X., Jiao X. G. & Zhang Y. J. High efficient of females of B-type Bemisia tabaci as males in transmitting the whitefly-borne tomato yellow leaf curl virus to tomato plant with Q-PCR method confirmation. Commun. Integr. Biol. 5, 543–545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polston J. E., Al-Musa A., Perring T. M. & Dodds J. A. Association of the nucleic acid of squash leaf curl geminivirus with the whitefly Bemisia tabaci. Phytopathology 80, 850–856 (1990). [Google Scholar]
- Santillán-Ortega C. et al. Susceptibility of females and males of Bemisia tabaci (Gennadius) B-biotype and Trialeurodes vaporariorum (Westwood) to thiamethoxam. Southwest Entomol. 36, 167–176 (2011). [Google Scholar]
- Cline T. W. & Meyer B. J. Vive la différence: males vs females in flies vs worms. Annu. Rev. genet. 30, 637–702 (1996). [DOI] [PubMed] [Google Scholar]
- Erickson J. W. & Quintero J. J. Indirect effects of ploidy suggest X chromosome dose, not the X: A ratio, signals sex in Drosophila. PLoS Biol. 5, e332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe T. et al. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7, e1000222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen I., Müller M. & Beye M. The Am-tra2 gene is an essential regulator of female splice regulation at two levels of the sex determination hierarchy of the honeybee. Genetics 192, 1015–1126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., Beukeboom L. W. & van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328, 620–623 (2010). [DOI] [PubMed] [Google Scholar]
- Shukla J. N. & Palli S. R. Sex determination in beetles: production of all male progeny by parental RNAi knockdown of transformer. Sci. Rep. 2, 602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W. et al. Transcriptome analysis and comparison reveal divergence between two invasive whitefly cryptic species. BMC Genomics 12, 458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W. et al. Pyrosequencing the Bemisia tabaci transcriptome reveals a highly diverse bacterial community and a robust system for insecticide resistance. Plos One 7, e35181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W. et al. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics 11, 400–410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff A. M. & Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 5, e1000262 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto F. S., Moy G. W. & Burton R. S. Interpopulation patterns of divergence and selection across the transcriptome of the copepod Tigriopus californicus. Mol. Ecol. 20, 560–572 (2011). [DOI] [PubMed] [Google Scholar]
- McIntyre L. M. et al. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 7, 79 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small C. M., Carney G. E., Mo Q. X., Vannucci M. & Jones A. G. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: Evidence for masculization of the transcriptome. BMC Genomics 10, 579 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P. et al. Lifespan and reproduction in Drosophila:new insights from nutritional geometry. PNAS 105, 2498–2503 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov A. A. et al. Sex-specific fitness effects of nutrient intake on reproduction and lifespan. Curr. Biol. Jul. 18, 1062–1066 (2008). [DOI] [PubMed] [Google Scholar]
- Du L. et al. Starving neurons show sex difference in autophagy. J. Biol. Chem. 284, 2383–2396 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala M. J. & Roff D. A. Inbreeding and extreme outbreeding cause sex differences in immune defence and life history traits in Epirrita autumnata. Heredity (Edinb) 98, 329–336 (2007). [DOI] [PubMed] [Google Scholar]
- Saccone G., Pane A. & Polito L. C. Sex determination in flies, fruitflies and butterflies. Genetica 116, 15–23 (2002). [DOI] [PubMed] [Google Scholar]
- Sánchez L. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52, 837–56 (2008). [DOI] [PubMed] [Google Scholar]
- Oliveira D. C. et al. Identification and characterization of the doublesex gene of Nasonia. Insect Mol. Biol. 18, 315–324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L. & Beukeboom L. W. Insect sex determination: it all evolves around transformer. Curr. Opin. Genet. Dev. 20, 376–383 (2010). [DOI] [PubMed] [Google Scholar]
- Sugimoto T. N. & Ishikawa Y. A male-killing Wolbachia carries a feminizing factor and is associated with degradation of the sex-determining system of its host. Biol. Lett. 8, 412–415 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L. W. Microbial manipulation of host sex determination. Endosymbiotic bacteria can directly manipulate theirhost's sex determination towards the production of female offspring. Bioessays 34, 484–488 (2012). [DOI] [PubMed] [Google Scholar]
- Xie W. et al. Induction effects of host plants on insecticide susceptibility and detoxification enzymes of Bemisia tabaci (Hemiptera: Aleyrodidae). Pest Manag. Sci. 67, 87–93 (2011). [DOI] [PubMed] [Google Scholar]
- Aparicio G. et al. Blast2GO goes grid: developing a grid-enabled prototype for functional genomics analysis. Stud. Health Technol. Inform. 120, 194–204 (2006). [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E. & Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X. & Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- Storey J. D. & Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K. & Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, 14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids. Res. 39, 316–322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8, e53006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info File #1