Background: Plasmodium falciparum merozoite surface protein 3 (MSP3) oligomerizes and binds heme.
Results: MSP3 forms amyloid-like structures that bind ∼35 heme molecules, and these filamentous structures are also present on the surface of merozoites.
Conclusion: Amyloid formation by MSP3 is related to heme binding.
Significance: The presence of MSP3 fibrils on merozoite surface and significant heme binding suggest its functional role during erythrocytic stages of P. falciparum.
Keywords: Amyloid, Erythrocyte, Heme, Infectious Diseases, Malaria
Abstract
Merozoite surface protein 3 of Plasmodium falciparum, a 40-kDa protein that also binds heme, has been biophysically characterized for its tendency to form highly elongated oligomers. This study aims to systematically analyze the regions in MSP3 sequence involved in oligomerization and correlate its aggregation tendency with its high affinity for binding with heme. Through size exclusion chromatography, dynamic light scattering, and transmission electron microscopy, we have found that MSP3, previously known to form elongated oligomers, actually forms self-assembled filamentous structures that possess amyloid-like characteristics. By expressing different regions of MSP3, we observed that the previously described leucine zipper region at the C terminus of MSP3 may not be the only structural element responsible for oligomerization and that other peptide segments like MSP3(192–196) (YILGW) may also be required. MSP3 aggregates on incubation were transformed to long unbranched amyloid fibrils. Using immunostaining methods, we found that 5–15-μm-long fibrillar structures stained by anti-MSP3 antibodies were attached to the merozoite surface and also associated with erythrocyte membrane. We also found MSP3 to bind several molecules of heme by UV spectrophotometry, HPLC, and electrophoresis. This study suggested that its ability to bind heme is somehow related to its inherent characteristics to form oligomers. Moreover, heme interaction with a surface protein like MSP3, which does not participate in hemozoin formation, may suggest a protective role against the heme released from unprocessed hemoglobin released after schizont egress. These studies point to the other roles that MSP3 may play during the blood stages of the parasite, in addition to be an important vaccine candidate.
Introduction
Malaria, caused by recurrent cycles of growth of pathogen Plasmodium in erythrocytes, leads to severe anemia and cerebral malaria (1). Merozoite surface proteins of Plasmodium falciparum are regarded as suitable candidates for developing vaccine against malaria mainly because they are relatively more exposed to the host immune system during the erythrocytic stage of parasite development (2). Surface location also suggests the possible role of these proteins in the invasion of red cells; several of these are also being investigated for their potential as vaccine candidates and as mediators of invasion of erythrocytes (3). Some of the major surface proteins of P. falciparum like MSP1, MSP4, MSP5, MSP8, and MSP10 are displayed to the parasite membrane through a glycosylphosphatidylinositol (GPI)5 anchor (2). Other surface proteins, including MSP3, MSP6, MSP7, and MSP9 (also known as ABRA), are soluble proteins and are present on the merozoite surface as a protein complex, possibly through protein-protein interactions (4). Of these proteins, MSP3 is of particular interest, both as a vaccine candidate antigen as well as for its peculiar structural characteristics (5–7).
MSP3 is a target of antibody response to P. falciparum infection, and anti-MSP3 antibodies were found to mediate antibody-dependent cellular inhibition of the parasite (5). Immunization with recombinant forms of MSP3 protected monkeys against malaria infection (8). A vaccine based on MSP3 N-terminal fragment is already in human trials (9, 10). In addition to being a potential vaccine candidate, two other characteristics of MSP3 stand out as follows: first, MSP3, although a soluble protein, forms oligomers, and second, it can bind to heme, although the significance of these characteristics is not well understood (7, 11).
During its blood stage development, the malaria parasite crucially depends on degradation of hemoglobin to utilize it as a major source of amino acids for its own protein synthesis (11). This inevitably releases large amounts of heme, a molecule highly toxic to the parasite (12). Most heme, but not all, released upon hemoglobin degradation is converted into a nontoxic polymeric form, hemozoin, in food vacuole within the infected erythrocyte (13, 14). Also, some amount of heme is still released from the unprocessed hemoglobin during the merozoite egress (12, 15). Parasite produces several heme-binding proteins that may be involved in detoxification of heme, in addition to hemozoin formation. For example, histidine-rich proteins I, II, and III, heme detoxification protein, and Maurer's associated protein have been characterized as heme-binding proteins (16–18). Although heme binding of MSP3 has been described, the mode and extent to which it binds heme remains unclear (18). MSP3 localized on the merozoite surface does not have any structural features, i.e. high histidine/cysteine content, etc., that would explain its heme binding properties (5). However, MSP3 contains three domains of alanine heptad repeats, a glutamic acid-rich region and a C-terminal leucine zipper-like motif (19). The presence of a specific 40-residue sequence in the leucine zipper region has been implicated in the formation of dimers and tetramers of MSP3 (16, 17). Whether these two characteristics of oligomerization and binding to heme are related to each other has not been investigated. Here, we have attempted to characterize oligomerization properties of MSP3 and investigated if its heme binding ability in some way is related to oligomerization.
EXPERIMENTAL PROCEDURES
Expression and Purification of MSP3 Recombinant Proteins
The full-length P. falciparum MSP3 (MSP3F) and a large N-terminal fragment MSP3(21–238) (MSP3N) from the 3D7 strain of P. falciparum were expressed in Escherichia coli and purified by immobilized metal affinity chromatography, followed by ion exchange chromatography, as described previously (20). The C-terminal hexahistidine-tagged polypeptides MSP3(191–312) (MSP3C), MSP3(219–312) (MSP3Cb), and MSP3(288–354) (MSP3C) were amplified from the MSP3F plasmid using NcoI and XhoI restriction sites and cloned in pET28a vector. These plasmids were transformed in BL21 (DE3) and expressed as soluble proteins. These recombinant proteins were purified under similar conditions as described previously (20). The purified proteins were dialyzed in 20 mm Tris, 150 mm NaCl, pH 8.0, and stored at −80 °C for further experiments.
Analytical Size-exclusion Chromatography
50 μg of MSP3 proteins in 20 mm Tris and 150 mm NaCl at pH 8.0 were loaded onto an analytical Superdex200 HR 10/30 size exclusion column with a loading volume of 1% of the column volume and at a flow rate of 0.3 ml/min. Theoretical molecular mass of proteins in different peak fractions was obtained by comparison with retention times of calibrant globular proteins that were chromatographed under identical conditions. Samples of all major peak fractions were analyzed by SDS-PAGE.
Dynamic Light Scattering (DLS) Studies
Light scattering studies were performed in Photocor complex using the multiple tau digital correlator. All studies were done at room temperature. A DLS experiment was carried out at an angle of 90° using a 632-nm laser using freshly prepared recombinant MSP3 proteins in a solution of 20 mm Tris, 150 mm NaCl, pH 8.0. Correlation data obtained were fitted using the DynaLS software to derive average apparent hydrodynamic radius (Rh). To monitor the inhibition of large aggregates of MSP3F by ammonium nitrate salt in 100 mm Tris buffer, pH 8.5, DLS was performed on the samples containing 0.5 mg/ml MSP3F with 0, 0.050, 0.1, 0.250, 0.5, 1, 2, and 2.5 m ammonium nitrate. The three protein populations observed were plotted against the intensity percentage along with the calculated diameter (in nanometers).
Blue Native-PAGE
For Blue Native-PAGE, native gradient gel was prepared from 4–20% in a cast as reported earlier with minor modifications (21). Cathode buffer (100 mm l-histidine, 0.02% Coomassie Blue (G-250), pH 8.0) and anode buffer (100 mm Tris, pH 8.8) were used to run the Blue Native gel at 4 °C for 18 h at 20 V. 1× native dye was used as sample buffer. 1 μl of native marker (prepared by addition of ferritin (880 and 440 kDa), aldolase (200 kDa), BSA (132 and 66 kDa), and β-lactoglobulin (18.4 kDa)) were used to analyze the protein size separated on the Blue Native-PAGE.
TANGO Algorithm for Predicting Aggregation Propensity
TANGO software that predicts a specific amino acid sequence in the protein that has the propensity to participate in aggregation was used for the study (22). Full-length MSP3 sequence was subjected to the algorithm at 25 °C, pH 7.4, to identify the regions responsible for aggregation.
Peptide Synthesis
The pentapeptide sequence from MSP3 protein (YILGW) determined as oligomerization motif by TANGO algorithm was synthesized by the methods described previously (23). Peptide was synthesized on Rink-amide 4-methylbenzhydrylamine resin (0.75 mmol/g) using Fmoc methodology. Couplings were performed by using carbodiimide. Fmoc deprotection was performed with piperidine (20% in dimethylformamide). After addition of the final residue, the resin was rinsed with dimethylformamide/dichloromethane/methanol and dried. The final peptide deprotection and cleavage from the resin were achieved with 10 ml of trifluoroacetic acid/triisopropylsilane/H2O/phenol (88:2:5:5) for 2 h. The crude peptide was precipitated with cold ether, filtered, lyophilized, and purified by preparative reverse phase HPLC. The purified fractions were pooled, lyophilized, and stored at −20 °C as dry powder. Peptide identity was confirmed by mass spectrometry.
Congo Red and Thioflavin-T Binding Assays
For the Congo Red assay, samples containing 200 μg/ml MSP3F and 2.5 μm Congo Red in PBS were incubated for 5 min before being scanned at wavelengths between 200 and 600 nm using a Cary 1E UV-visible spectrophotometer. For the thioflavin T (ThT) assay, 50 μg/ml MSP3F and 25 μm ThT in PBS was scanned at wavelengths between 200 and 600 nm. Emission spectra were collected with an excitation wavelength of 417 nm using a luminescence spectrometer model LS 50 B (PerkinElmer Life Sciences). For kinetics experiments, solution samples of filtered (0.2-μm membrane filter) MSP3F in 2-ml volume reactions in duplicates were incubated with 30 μm ThT in PBS in the dark. ThT fluorescence was assayed at 0.1-s intervals following agitations (20 s) at excitation and emission wavelengths of 443 and 484 nm, respectively.
Transmission Electron Microscopy
Time-dependent morphological evolution in MSP3 samples was viewed under transmission electron microscopy (TEM) (Tecnai 12 BioTWIN, FEI, Netherlands) using 1% uranyl acetate negative staining. MSP3 samples dialyzed in PBS, pH 7.4, were incubated for 72 h at room temperature. MSP3 samples were visualized after 0, 30, 50, and 72 h. Photomicrographs were digitally recorded using a Megaview II (SIS, Germany) digital camera. Image analysis to measure dimensions was carried using an Analysis II (Megaview, SIS, and Germany) software package.
Immunofluorescence Assay
P. falciparum 3D7 was utilized for isolation of merozoites for immunostaining assays. Parasite culture was synchronized at schizont-infected erythrocytes as described earlier (24). Invasive merozoites were purified from mature schizont stage on a Percoll/sorbitol gradient at 10,000 × g as described previously (25). Briefly, the cultures were monitored after 30 min for start of rupture of erythrocytes and the release of merozoites. Released merozoites along with fully mature schizont-infected erythrocytes that were ready to rupture were collected by passing the parasites through a 1.2-μm filter. They were passed a second time through the filter to remove any contaminating schizont-infected RBCs. Thin smears of the above isolated merozoites were fixed with chilled methanol for confocal microscopy and imaged under confocal microscopy as described earlier (26). Slides were co-immunostained with MSP3 polyclonal sera (20) at dilutions 1:1000 along MSP3 monoclonal Abs, Rh2b (1:100) as rhoptry marker, EBA-175 (1:100) and AMA-1 (1:500) as microneme marker, MSP5 (1:500) as merozoite surface marker, in blocking buffer for 1 h (27). For another set of experiments, freshly isolated merozoites were treated with cytochalasin D followed by addition of pre-warmed fresh erythrocytes. This preparation was used for fixing thin smears on the slide as described above for immunostaining assays with anti-MSP3 Abs.
Quantitative Invasion Assay
Invasion assay was performed as described previously (28, 29). Normal erythrocytes at 2% hematocrit in culture medium were inoculated with early schizonts of P. falciparum 3D7 to give a parasitemia of 0.3% and a hematocrit of 2% in a volume of 90 μl. MSP3F, MSP3Cb, and the pentapeptide YILGW were added to a final concentration of 0.5 and 1 mg/ml to a final volume of 100 μl. After incubation with proteins for 36 h, parasitemia of each well was determined by flow cytometry of SYBR Green 1 (Invitrogen)-stained trophozoite stage parasites using a FACSCalibur (BD Biosciences). For each well, ∼105 cells were counted. Growth of parasite was expressed as the percentage of parasitemia for the mean of duplicate wells for each protein concentration. Two independent assays were performed, and data were plotted as an average of the two in bar graphs for each condition.
MSP3-Heme Interaction Studies
MSP3-heme interactions were measured UV-spectrophotometrically as described earlier (17). In brief, for difference spectra, 10 μm hemin from a stock of 1 mm was added simultaneously in both test and reference cuvettes. The test cuvette contained 10 μm protein (MSP3F, MSP3N, MSP3Ca, MSP3Cb, MSP3Cc, or polyhistidine-tagged MSP19 (control)) in 20 mm Tris, 150 mm NaCl, pH 8.0, and only buffer was added in reference cuvette.
The HPLC heme quantitation method was used as an independent measure of a number of heme-binding sites as described previously (17). Briefly, 100 m excess heme was added to MSP3F in 20 mm Tris, 150 mm NaCl, pH 8.0, which was subjected to a desalting column to remove excess unbound heme. Simultaneously, a similar MSP3F amount was passed through the desalting column to determine protein concentration in the heme-bound sample. Heme concentration in the MSP3-heme complex was determined using an HPLC column using myoglobin standards with detection at 400 nm absorbance (17, 30).
ABTS Assay
To monitor the peroxidase-like activity of heme at pH 7.4, samples (200 μl) containing 2 mm ABTS and 10 μm heme with or without the protein (MSP3F; 2 μm) were added in triplicate to the wells of a 96-well plate. The reaction was initiated by the addition of 10 μl of 40 mm H2O2, and plates were incubated at 37 °C for 30 min in the dark. Peroxidase activity in each well was examined at 600 nm due to increase in the oxidative product.
RESULTS
Characterization of Oligomeric Aggregates of MSP3
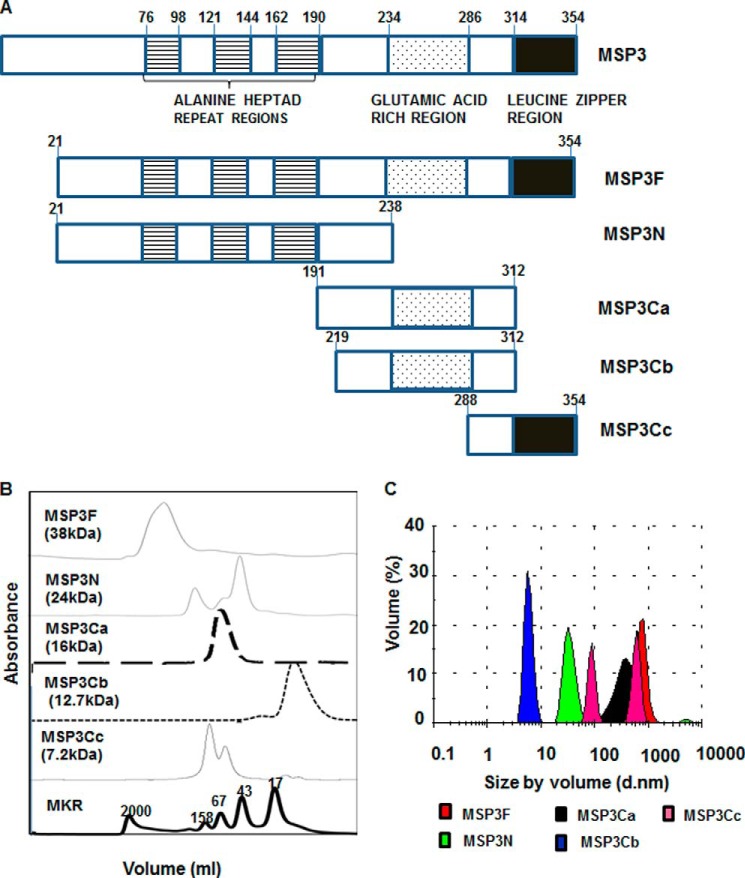
A full-length MSP3 (MSP3F), a 24-kDa heptad repeat domain (MSP3N), a 16-kDa middle fragment, including the glutamic acid stretch (MSP3Ca), a 12.7-kDa middle fragment, excluding the predicted aggregation motifs (MSP3Cb), and a 7.2-kDa C-terminal fragment possessing leucine zipper region (MSP3Cc) were expressed as soluble proteins and purified to homogeneity (Fig. 1A and supplemental Fig. S1A). Analytical SEC revealed that MSP3F, MSP3N, MSP3Ca, and MSP3Cc eluted at molecular masses that were several times higher than their calculated molecular masses (Fig. 1B). MSP3F migrated as a single oligomer species right after the blue dextran as reported previously (7). MSP3N migrated in two populations at ∼48 and 160 kDa, respectively. MSP3Ca, with a calculated molecular mass of 16 kDa, migrated as a single species at ∼50 kDa, possibly as a trimer. MSP3Cc migrated in two populations both with high molecular mass species (7). However, the truncated middle fragment MSP3Cb migrated as a single major population at its predicted molecular mass. Oligomerization was further studied by the DLS method, which showed MSP3F, MSP3N, MSP3Ca, and MSP3Cc to exhibit highly polydispersed species with a large hydrodynamic radii ranging from 50 to 1000 nm as percentage by volume, thereby suggesting the presence of high molecular mass oligomeric species (Fig. 1C). However, MSP3Cb that migrated as a monomer in SEC also showed the presence of monodispersed species with a hydrodynamic radius of 8 nm by DLS (Fig. 1C). Similarly, Blue Native-PAGE that allows analysis of protein aggregation in native conditions further confirmed the presence of high molecular weight species formed by these polypeptides. MSP3Cb, however, migrated as a single major band at its expected size (6.7 kDa) (supplemental Fig. S1B).
FIGURE 1.
Schematic representations of P. falciparum merozoite surface protein 3 and its different deletion constructs. A, schematic of full-length MSP3 showing alanine heptad repeats (stripes), glutamic acid-rich regions (dotted), and leucine zipper-like motifs (black). MSP3 polypeptides MSP3(21–238) (MSP3N), MSP3(191–312) (MSP3Ca), MSP3(219) (MSP3Cb), and MSP3(288–354) (MSP3Cc) were expressed as soluble hexahistidine-tagged recombinant proteins. B, analytical size exclusion chromatography demonstrating oligomerization of MSP3 proteins. All chromatograms are plotted on the same x axis and offset in the y axis for comparison. MSP3 full-length and its corresponding fragments are denoted on each chromatogram along with their calculated molecular masses. MSP3Cb remained as a monomer and eluted at appropriate size corresponding to 17-kDa globular marker. The molecular mass (kDa) globular protein markers shown at the bottom are as follows: blue dextran (2000), aldolase (158), conalbumin (75), ovalbumin (43), and equine myoglobin (17). C, analysis of oligomeric size distribution of recombinant MSP3 proteins by dynamic light scattering experiments. DLS spectra of oligomers of MSP3F, MSP3N, MSP3Ca, and MSP3Cc exhibited high oligomeric particle diameters above 10 nm. MSP3Ca demonstrates a major single population diameter of 10 nm.
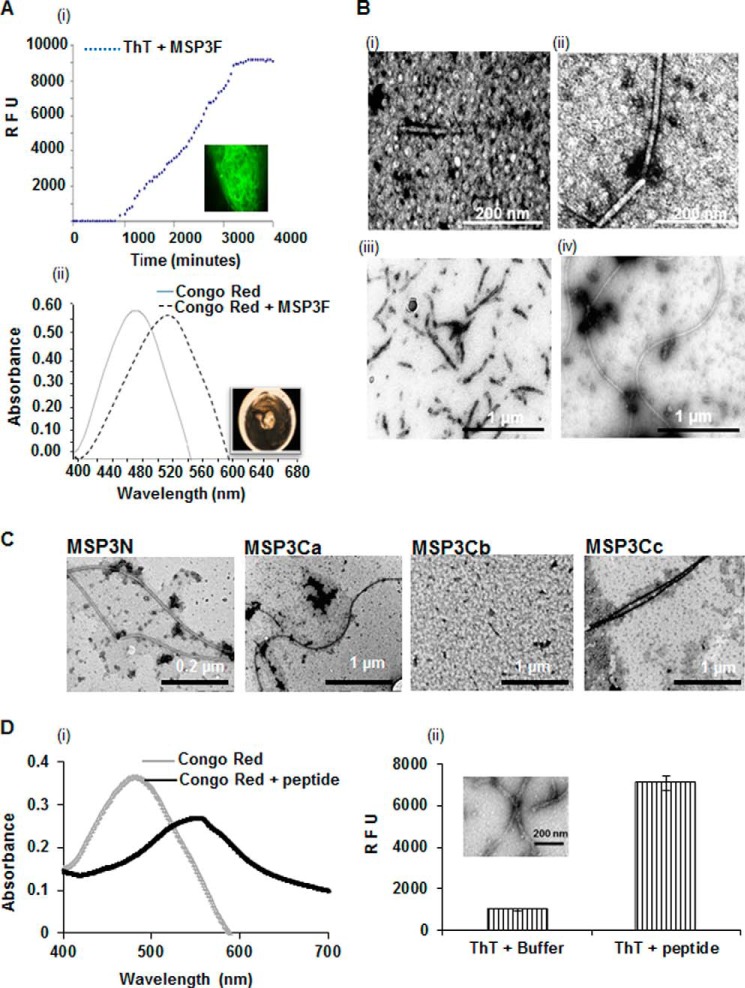
Aggregation behavior of MSP3F was further investigated by histochemical dyes ThT and Congo Red. Conformational transition in MSP3F in solution was monitored by ThT kinetics that demonstrated a substantial lag phase (∼40 h) followed by an exponential increase in fluorescence intensity indicative of a typical nucleation-dependent polymerization (Fig. 2A, panel i) (31). After 48 h, no further increase in ThT fluorescence was observed suggesting attainment of a stationary phase. MSP3F aggregates were also positive to Congo Red showing a red shift in the maximum absorbance from 484 to 584 nm and apple-green birefringence when examined under polarized light (Fig. 2A, panel ii).
FIGURE 2.
Morphological characterization of MSP3 aggregates after different time points as determined by electron micrographs. A, kinetics of MSP3F fibril formation assessed by ThT fluorescence and Congo Red assay. ThT fluorescence for 0.5 mg/ml solution of MSP3F incubated at 25 °C shows an increase in fluorescence at lag phase followed by the exponential phase. Fluorescence images after completion of ThT kinetics of MSP3F showed fibrillar assemblies. Polymeric MSP3-bound Congo Red demonstrated maximum absorbance red-shifting from 488 to 540 nm and showing birefringence under polarized light. RFU, relative fluorescence unit. B, dependence on time of TEM images of 0.5 mg/ml solution of MSP3F, which was incubated at 25 °C in phosphate-buffered saline, pH 7.4. and sampled at t = 1 h (panel i), 30 h (panel ii), 55 h (panel iii), and 72 h (panel iv). Spherical intermediates observed initially transformed to truncated fibrils and finally long elongated unbranched fibrils. C, electron micrographs show fibril formation by MSP3N, MSP3Ca, and MSP3Cc under similar conditions. MSP3Cb showed no visible ordered morphology. D, amyloid characterization of synthetic peptide YILGW corresponding to MSP3(192–196) predicted as an aggregation-prone motif in MSP3 sequence. Peptide was positive in ThT binding assay and when incubated with Congo Red showed a characteristic red shift for Congo Red (panels i and ii). TEM micrographs (inset) depict the fibril-like morphology (panel ii).
To understand the nucleation and progressive growth of soluble oligomers of MSP3, we used TEM to examine the negatively stained preparations at different time points. Initial TEM images (t = 1 h) of MSP3F showed the presence of some spherical aggregates of 15–30 nm in diameter resembling amylospheroids (32), and on further incubation (t = 30 h), homogeneous spheres were the dominant species along with some filamentous structures of ∼15 nm diameter (Fig. 2B, panels i and ii). Transitions from spheroids to the heterogeneous species of immature fibrils and short filamentous fragments were observed at t = 50 h (Fig. 2B, panel iii). At t = 72 h, long unbranched filamentous structures of 15–20 nm diameter and several microns in length were the dominant species along with some amorphous aggregates (Fig. 2B, panel iv). Aggregation characteristics of other MSP3-based polypeptides, including MSP3N, MSP3Ca, and MSP3Cc, were also examined under similar conditions as for MSP3F. TEM showed that although MSP3N, MSP3Ca, and MSP3Cc also formed long filamentous structures, MSP3Cb, lacking both the leucine zipper region and the predicted aggregation prone motif, MSP3(192–196) (YILGW), did not form any noticeable filamentous structures (Fig. 2C). Synthetic pentapeptide, YILGW, was also examined for its aggregating behavior. Both ThT and Congo Red binding assays showed that YILGW strongly interacted with the dyes as seen for MSP3F. Also, upon incubation for 72 h in PBS, pH 7.4, fibrillar assemblies similar to those formed by MSP3F were observed under TEM (Fig. 2D, panels i and ii).
Association of Fibril-like Structures of MSP3 with Merozoites
To explore if MSP3 fibrils were present in the blood stage cultures of P. falciparum, isolated merozoites were stained with MSP3 Abs along with known markers for micronemes (EBA-175 and AMA-1) and rhoptry (RH2B) and visualized under a confocal microscope. Both polyclonal and monoclonal anti-MSP3F Abs stained free merozoites with the characteristic surface staining. Interestingly, along with the surface staining with MSP3, several micron size long fibril-like structures attached with merozoite were observed (Fig. 3A). These thin structures were uniformly present in all fields examined and were positive to amyloid-specific dye ThT (supplemental Fig. S2A). Polyclonal sera against other parasite proteins like AMA-1, EBA-175, and Rh2b did not detect any extracellular fibril-like structures in co-immunostaining assays (Fig. 3A). To investigate the role of these fibrillar assemblies in the erythrocytic life cycle of the parasite, fresh erythrocytes were incubated with cytochalasin D-treated merozoites followed by addition of anti-MSP3F Abs. Cytochalasin D, a potent inhibitor of actin polymerization, blocks junction complex movement once the merozoite attaches to the erythrocytes. Confocal images of cytochalasin D-treated merozoites revealed association of the 8–15-μm-long fibrillar structures with erythrocytes. These thread-like structures, attached to the merozoite surface, appeared to wrap around erythrocytes (Fig. 3B). Individual merozoites attaching themselves to distant erythrocytes through these fibrillar structures were also observed in several fields (Fig. 3B).
FIGURE 3.
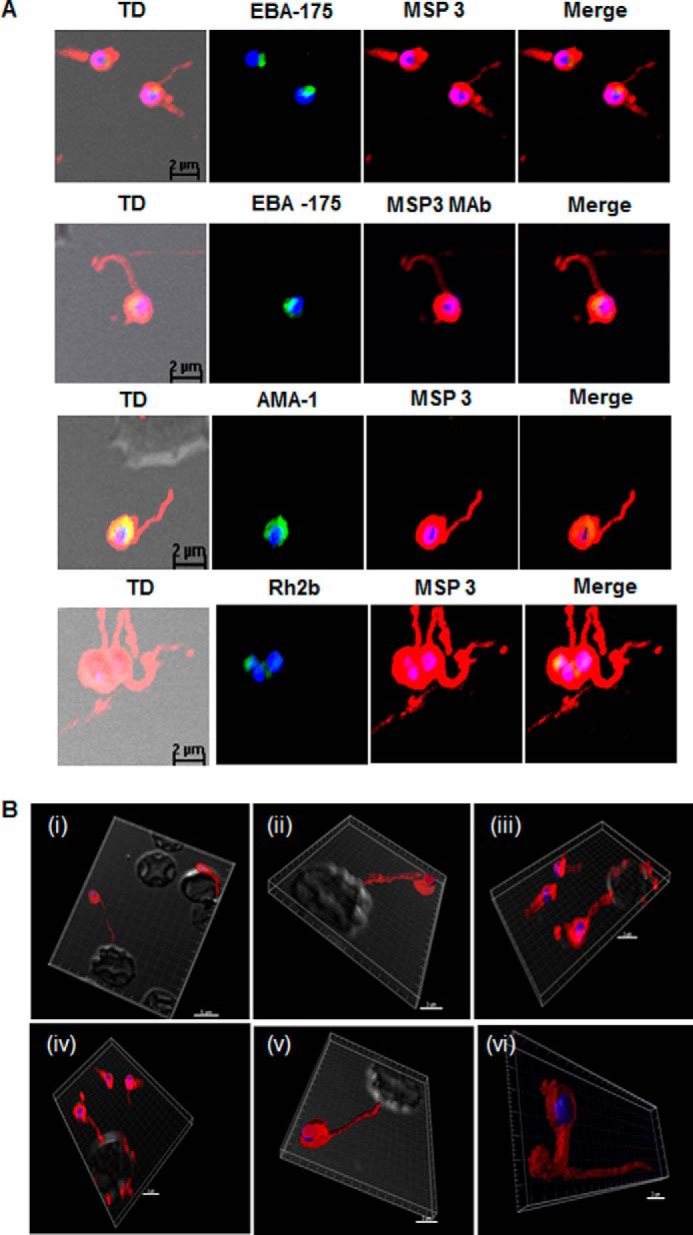
Detection of fibrillar structures in P. falciparum merozoites by immunofluorescence assay. A, co-immunostaining of free merozoites with anti-MSP3 Abs along with Abs against EBA-175, monoclonal MSP3 Abs (MSP3 MAb), AMA-1, Rh2b. and MSP5. In all the slides. long fibrous structure attached to free merozoite was detected by anti-MSP3 Abs. Monoclonal MSP3 antibodies also stained these fibrillar structures associated with merozoites, but other marker protein did not detect fibrillar assemblies. TD, transmission differential interference contrast/ B, association of fibril-like assemblies with erythrocytes. Cytochalasin D-treated merozoites when incubated with fresh RBC showed the staining of the fibril-like structures possessing MSP3 associated with merozoites and RBC membrane. Images from panels i–v are the various fields depicting similar association. Panel vi shows single free merozoite with long fibril-like structure stained with anti-MSP3 Ab.
To examine if exogenous addition of MSP3 polypeptides can influence the invasion and growth of the parasite, FACS-based quantitative invasion assay was performed. MSP3F that forms the amyloid MSP3Cb, which remains a monomer, and the aggregation prone pentapeptide YILGW were added to a synchronized culture of P. falciparum. No significant difference in parasitemia was observed in the wells in the presence of MSP3 polypeptides as compared with the control wells (supplemental Fig. S3).
Characterization of MSP3-Heme Complex
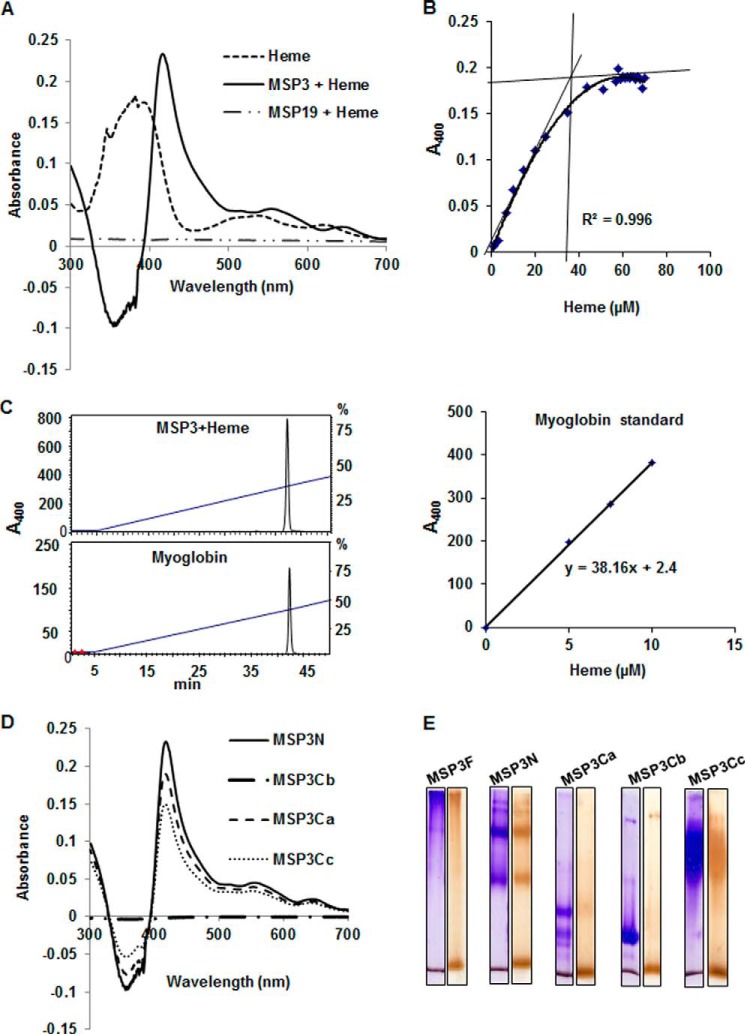
Heme binding with MSP3F and other MSP3 fragments was carried out by UV spectrophotometry. The broad Soret absorption 383-nm peak of free heme sharpened and shifted to 417 nm on incubation with MSP3F. Similarly, the p and q bands at 493 and 616 nm of free heme were found to red-shift at 560 and 646 nm in the heme-MSP3F complex (Fig. 4A). The number of heme-binding sites on MSP3 was determined by heme titration experiments. Difference absorption spectra were recorded at varying heme concentrations, and heme binding curves were generated by plotting ΔA417 nm versus heme concentration as shown (Fig. 4B). Fitting this heme binding curve yielded MSP3-heme binding stoichiometry of 35 ± 5 heme/MSP3. Independently, the number of heme-binding sites in MSP3 was also assessed by the HPLC method (17). Using myoglobin as a standard to calculate the concentration of heme, the stoichiometry calculated was 40 mol of heme/mol of MSP3 (Fig. 4C). Red-shift in the Soret band at 383–415 nm along with p and q bands at 545 and 646 nm for heme were also observed when it was incubated with MSP3N, MSP3Ca, and MSP3Cc. However, no red shift was observed in the difference spectrum of heme when incubated with MSP3Cb, suggesting absence of heme interaction with this fragment of MSP3 (Fig. 4D).
FIGURE 4.
MSP3 can bind several molecules of heme in vitro. A, UV spectrophotometry difference spectrum showed significant red shift of Soret peak of heme at 417 nm from 383 nm when MSP3F is scanned in the presence of 10 μm heme. MSP1(19) used as a control does not show absorption spectrum at 415 nm indicating absence of heme binding. B, determination of number of heme-binding sites on MSP3F by difference spectra recorded from heme titration assay and heme binding curve generated by plotting ΔA417 nm versus heme concentration showed 1:35 stoichiometry of MSP3 and heme. C, HPLC-based characterization of the MSP3F-bound heme using horse heart myoglobin and pure hemin as controls showed 1:40 stoichiometry of MSP3 and heme. D, UV spectrophotometry difference spectra showed significant red shift of Soret peak of heme at 415 nm from 383 nm when MSP3 fragments MSP3N, MSP3Cb, and MSP3Cb were recorded in the presence of 10 μm heme. MSP3Cb did not show any red shift in the presence of heme suggesting the absence of interaction. E, native PAGE analysis of heme binding by peroxidase staining. MSP3F and other polypeptides show heme interaction as evident by brown bands in peroxidase-stained native PAGE as compared with Coomassie-stained PAGE run in parallel. MSP3Cb did not show any significant binding to heme as evident by absence of brown band in peroxidase staining compared with Coomassie gel. Unbound heme is stained at the bottom of all the gel lanes.
Formation of heme complex with MSP3F and its fragments was also analyzed qualitatively by peroxidase-stained native PAGE. As shown in Fig. 4D, MSP3F, MSP3N, and MSP3Cc migrated as high molecular mass species. In comparison with Coomassie-stained native PAGE, peroxidase-stained gel demonstrated heme bound to these larger oligomeric species. MSP3Cb that migrated mainly as a single band at the lower end of the gel was not stained with peroxidase. Free heme in each sample was stained as a broad band at the bottom of the gel.
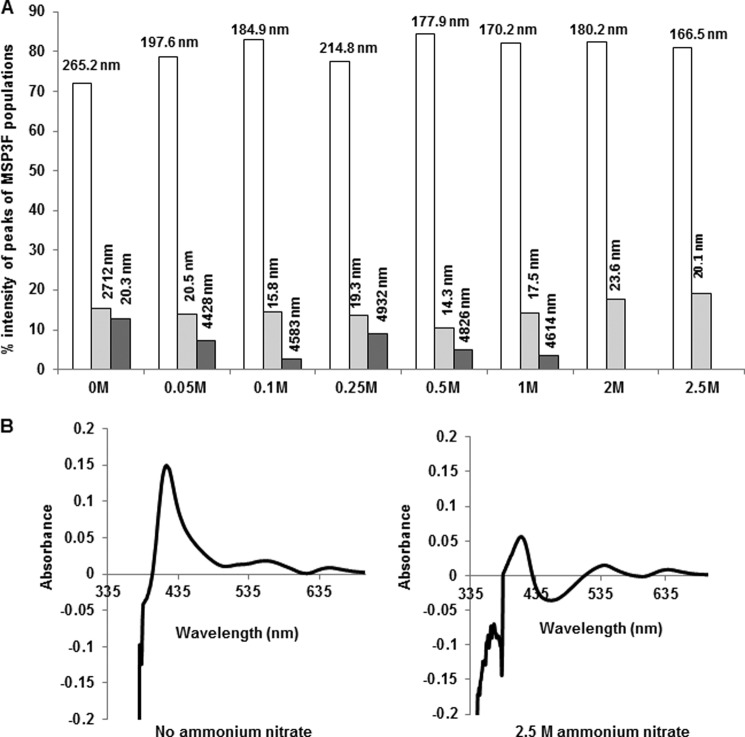
Aggregation of MSP3F was also examined under several different buffer conditions containing various inorganic salts (data not shown). ThT and Congo Red assays were negative in one of the screened buffers (100 mm Tris, 2.5 m ammonium nitrate), thereby suggesting the absence of amyloid aggregates in the protein sample. Furthermore, analysis by DLS showed that increasing concentration of ammonium nitrate in the buffer led to disintegration of larger aggregates of MSP3F, and at a concentration of 2.5 m ammonium nitrate, small oligomers of hydrodynamic diameter of 166.5 and 20.1 nm were mostly present (Fig. 5A). Under these conditions, there was no significant observable red shift of the Soret band of heme indicating the absence of interaction of heme with MSP3F (Fig. 5B).
FIGURE 5.
Effect of ammonium nitrate on aggregation and heme complex formation of MSP3. A, graph depicting ammonium nitrate concentration versus percentage of intensity peak detected by DLS. The readings above each column show the calculated diameter (in nanometers) of each population detected in DLS spectra for MSP3F. With increasing ammonium nitrate concentration, the percentage of larger aggregates is decreased and finally disappears at 2.5 m ammonium nitrate concentration. B, comparison of UV spectrophotometry analysis of MSP3 heme binding (50 μg) in the absence and addition of ammonium nitrate. Heme interaction is significantly reduced in the presence of ammonium nitrate.
MSP3-Heme Complex Possesses Peroxidase Activity
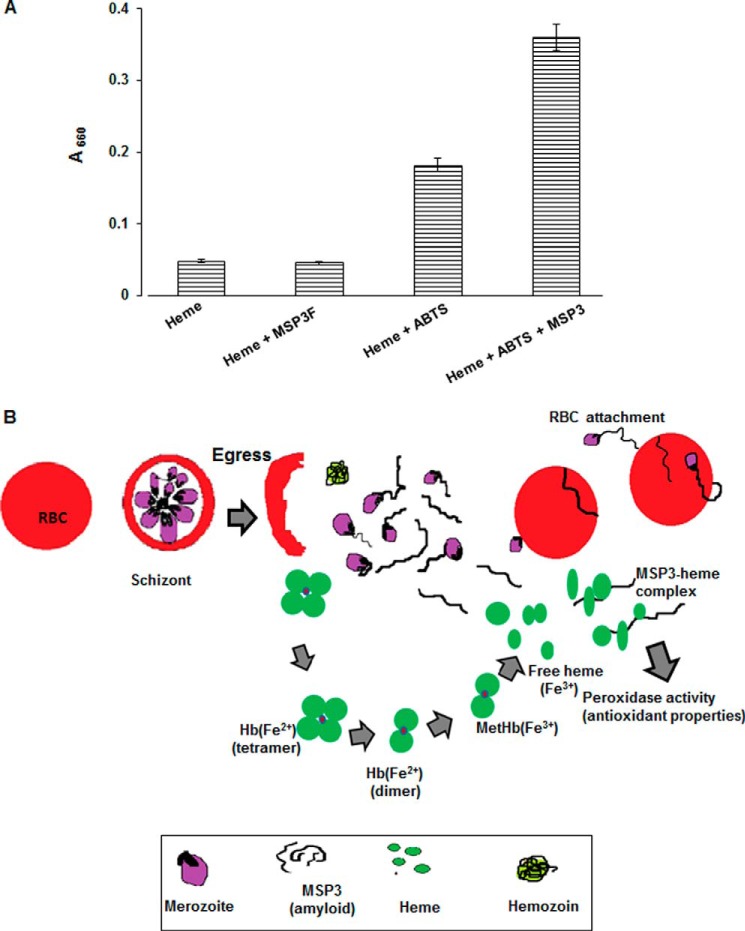
As heme is reported to possess peroxidase-like activity (33), we investigated its capability to catalyze the oxidation of the model substrate ABTS in the presence of MSP3. We found that MSP3F enhances a 2-fold increase in the H2O2-induced oxidation of ABTS as compared with heme alone (Fig. 6A).
FIGURE 6.
MSP3-heme complex possesses peroxidase activity. A, ABTS assay detected 2-fold increase in absorption at 600 nm for MSP3-heme complex as compared with heme. No significant increase at 600 nm was observed for heme or MSP3F alone suggesting MSP3-heme to be an active species. B, model proposing MSP3 to act as a possible heme scavenger in its amyloid morphology. After completion of erythrocytic asexual life cycle by P. falciparum, unprocessed hemoglobin (Hb) along with merozoites are released in blood plasma. Released merozoites with attached MSP3 fibrillar structures reinvade red blood cells by associating RBC membrane with these structures. Heme iron in ferrous state (Fe2+) has high affinity for oxygen molecule that leads to formation of oxyhemoglobin. Autoxidation of oxyhemoglobin leads to production of ferric (Fe3+) hemoglobin (methemoglobin), ferryl hemoglobin, free heme, and several reactive oxygen species. They possess a potential threat as a possible cause of tissue damage and cell destruction. Haptoglobulin, a high affinity Hb binder along with α1-microglobulins, transferrin, albumins, hemopexins, and antioxidants such as vitamin E and ascorbic acid, cooperatively entrap free heme during hemolysis. However, in pathological conditions of malarial infection, these systems are not sufficient to combat oxidative stress to both host and parasite. Parasite expresses merozoite surface protein3 (MSP3) in abundance along with other unknown proteins that form amyloid fibril-like structures. These nascent heme molecules are entrapped by soluble MSP3 fibrils that modulate its peroxidase activity that might have antioxidant properties of some biological relevance.
DISCUSSION
PfMSP3, a 40-kDa non-GPI-anchored surface protein, is abundantly present on the surface of the merozoite and is being developed as a blood stage vaccine candidate antigen (9). Oligomerization and heme binding properties described earlier have remained largely unexplored (16). In this study, we have attempted to further explore and understand the aggregating properties of MSP3 and its correlation, if any, with its ability to bind heme, a molecule potentially toxic to the parasite.
While preparing recombinant MSP3F and MSP3N for immunological characterization studies, we observed that these proteins formed oligomers of high molecular masses (20). The presence of large molecular mass species of MSP3 has been described previously and attributed to the presence of a leucine zipper-like sequence in the C terminus of the protein (7). However, aggregates formed by MSP3N, which is devoid of the leucine zipper region, as shown by SEC and DLS, prompted us to explore aggregation characteristics of MSP3 further. Based on the predicted high propensity aggregating pentapeptide sequence MSP3(192–196) (YILGW), and earlier described role of the C-terminal leucine zipper region in oligomerization, three more MSP3 fragments, i.e. MSP3Ca, MSP3Cb, and MSP3Cc, were designed and produced as soluble recombinant polypeptides. Biophysical characterization of MSP3 proteins showed consistent oligomerization behavior. MSP3F showed high molecular mass oligomeric species in SEC, DLS, and Blue Native-PAGE. Other shorter fragments followed similar aggregating patterns except for MSP3Cb, which demonstrated a single population of ∼10 kDa, corresponding to its monomeric form.
MSP3F was further examined for any conformational transitions under near physiological conditions. Electron micrographs of a sample of MSP3F on the 1st day showed the presence of homogeneous spherical structures that were similar to the self-assemblies (amylospheroid) and intermediate species formed by Aβ-derived diffusible ligands (32, 34). At 50 h, these spherical structures were primarily transformed to short fibrillar assemblies resembling intermediate species formed during amyloidogenesis of Aβ peptide (35), whereas at 72 h clear amyloid fibrils were noticeable along with few amorphous aggregates. The role of amorphous aggregates has been studied in detail in a kinetic study of a prion protein-based peptide in which the aggregates were shown to have a dual role for releasing monomeric species as well as to act as a site for initiation of amyloid growth (36). Kinetics of amyloid formation of MSP3F was also studied by using ThT assay. As expected, the initial lag phase did not show any ThT binding, whereas the exponential phase, during which time MSP3 underwent fibril formation, demonstrated increasing ThT binding. This was followed by a stationary phase during which no further binding of ThT was observed. These results were similar to the amyloid formation by several self-aggregating proteins, including recombinant prion proteins, α-synuclein, and amyloid-β polypeptide (31, 37–39).
Electron micrographs of both MSP3N and MSP3Ca that do not include the previously described C-terminal 40 amino acids responsible for oligomerization (40) also showed the presence of self-assembled fibril-like structures. However, MSPCb did not exhibit any self-assembled structures under similar conditions. The main difference between MSP3Ca that formed fibrils and MSP3Cb that did not is the presence of extra amino acid residues, including the predicted aggregation prone sequence YILGW at the N terminus of MSP3Ca. To examine if YILGW peptide sequence was indeed prone to aggregation, we synthesized this pentapeptide and studied its aggregation and found that YILGW also readily formed fibrils. Interestingly, MSP3Cc that contains the leucine zipper region but does not include the aggregation-prone pentapeptide also formed fibrillar structures. These results indicate that both the leucine zipper region as well as the YILGW can independently be responsible for forming fibril-like structures in MSP3 polypeptides.
Merozoites immunostained with anti-MSP3 Abs revealed novel long fibrillar assemblies associated with the surface, possessing characteristics often attributed to amyloid-like structures, including fibril morphology and binding to ThT (supplemental Fig. S2A). However, these fibril-like structures were not observed at the ring or the trophozoite stages of the parasite. Because MSP3 is expressed only during schizogony (5), it is quite likely that the major content of these elongated fibrils is MSP3 (5). This notion was further supported by the fact that these fibrillar assemblies in parasite culture were not stained by Abs specific for AMA-1, EBA-175, or Rh2b. The results of immunostaining with cytochalasin D-treated merozoites showed remarkable association of merozoites with freshly incubated erythrocytes. In the presence of cytochalasin D that disrupts actin polymerization, merozoite attaches to RBCs and apically reorients for initiation of invasion (25). The long fibril-like structures attached at one end with the merozoite surface were consistently seen to associate with distant erythrocytes as well as wrap around them. An earlier study with Plasmodium knowlesi has described the presence of an amyloid-like coat on the merozoite surface leading to the adhesion to erythrocyte (41). The amyloid coat on the merozoite surface was described as patches of regular denticulation in early schizogony and later formed long bristles and filaments that appeared thicker at apices than their stems and thus suggested the presence of a complex formation (42). Interestingly, similar filamentous extracellularly extended structures have also been reported in many eukaryotic and prokaryotic cells termed adhesins, extracellular appendages, fibrils, and filaments (43–46). However, the components of merozoite's amyloid coat have remained unexplored so far. It may be that the 5–15-μm sized fibrils attached to the end of the merozoite surface as observed in this study somehow assist in anchoring erythrocytes. However, we also found that exogenous addition of MSP3 proteins or YILGW peptide to the culture did not affect the growth of the parasite at all. On the contrary, an earlier study (47) using iodinated synthetic peptides based on the MSP3 sequence showed erythrocytic binding as well as invasion inhibition. Erythrocytic invasion, a complex phenomenon involving several proteins, is initiated by contact with the erythrocyte followed by apical reorientation by the merozoite that results in formation of a tight junction (48, 49). Considerable functional redundancy in malaria protein as well as alternate invasion pathways used by the parasite are well known. Although our results clearly showed the presence of MSP3 fibrils on merozoite surface and their attachment with erythrocyte, it may not be possible to ascribe any definite role(s) of MSP3 in the invasion process.
Another vaccine candidate merozoite surface protein MSP2, which is a GPI-anchored ∼30-kDa highly abundant protein, was also shown to form amyloid-like fibrils under physiological conditions as observed for MSP3 in this study (50, 51). Structurally, MSP2 has two conserved domains at the N and C termini but is highly polymorphic in the central region. The core sequence responsible for fibril formation was shown to be located in the conserved N-terminal domain, and the peptides based on sequences from this region also formed fibrils similar to the full-length MSP2 (52). However, it was also observed that regions of MSP2 outside the conserved N-terminal domain were also important for fibril formation by MSP2. Interestingly, failure of mAb 6D8 in situ that reacted with the N-terminal conserved region of MSP2 was taken as indirect evidence of the presence of MSP2 on the merozoite surface as a homo-oligomer; no direct evidence for MSP2 fibril has been reported (53). A major difference between these two highly abundant blood stage proteins with regard to their intrinsic propensity to form fibrils is that although MSP2 is anchored to the merozoite surface via the GPI anchor, MSP3 is supposed to be present on the merozoite surface through association with other surface proteins (47, 48). Oligomerization of MSP3, as a soluble protein, is facile and likely inherent to its structure that includes the presence of leucine zipper-like sequences at the C terminus (40). In both the proteins, although small peptides based on the domains involved in fibril formation also form fibrils like the full-length proteins themselves, the sequences outside the fibril forming domains are also involved and/or assist in fibril formation by the two proteins (52, 53). Results described in this study, however, clearly show that MSP3-based fibrils are present on the merozoite surface and seem to interact with erythrocytes. It will be interesting to explore if other merozoite surface proteins, indeed other malarial proteins present in specific organelles within the parasite, form fibril structures, and if this property is related to their functional activities. However, association of fibril structures of secretory non-GPI-anchored MSP3 on the surface of the merozoite may well be due to its interactions with other proteins present on the merozoite surface (54, 55).
Another intriguing property of MSP3 is that it binds heme, although it does not have any structural features generally known for heme binding (16). Heme, an iron protoporphyrin complex, serves as a prosthetic group of numerous hemoproteins responsible for several biological reactions such as oxygen transport, respiration, drug detoxification, and signal transduction (56, 57). However, free heme released due to hemolysis during pathological states, like sickle cell disease, ischemia reperfusion, and malaria, leads to undesirable toxicity that damages lipids, protein, and DNA through generation of reactive oxygen species (58–60). To further characterize the nature of the heme-MSP3 complex, we first quantitatively measured heme binding by two independent methods (17) and found that MSP3 can bind 35–40 mol of hemin/mol of protein. Such multiple heme-binding sites present in a protein sequence are often attributed to high histidine content or due to the presence heme regulatory motifs (61–63). For example, histidine-rich protein II that has high histidine content can bind 35–50 mol of heme/mol of protein (17). Similarly, as many as 35 heme molecules bind to a 93-kDa protein (HBP.93) isolated from rabbit serum that possesses 16.6 and 9.9% of proline and histidine content (63, 64). Results of heme binding and native peroxidase staining experiments showed that all MSP3 polypeptides used in this study bound heme except for MSP3Cb. Because MSP3Cb stays as a monomer and does not form amyloid structures, we wondered whether the MSP3-heme interaction is indeed due to oligomerization of MSP3. To probe this further, we carried out MSP3-heme interaction experiments in a buffer containing ammonium nitrate that did not favor aggregation of MSP3 as was shown by ThT or Congo Red assays (data not shown). A derivative of ammonium nitrate is a well known solute that disrupts aggregation of proteins (65). In the presence of ammonium nitrate, binding of heme was significantly reduced suggesting that heme binding somehow depends on the aggregation of the protein.
During malarial infections, a large amount of free heme released during intra-erythrocytic growth of the parasite is detoxified by the molecular mechanism of hemozoin formation evolved by the parasite (16). However, the remaining 40% unprocessed hemoglobin when released in plasma after schizont egress releases nascent heme upon enzymatic processing (12, 66). We also observed during confocal experiments on purified merozoites using anti-MSP3 Abs that hemozoin crystals, which were released along with merozoites, were consistently enveloped by MSP3 (supplemental Fig. S2B).
Ferriprotoporphyrin IX or heme can react with H2O2 to form ferryl intermediates that contribute in various enzyme-like reactions, including peroxidase-like activity that reduces H2O2 at the expense of organic substrate (18, 33). Malarial heme-binding proteins like histidine-rich protein II and MAHRP-1 that are present in the erythrocytic cytosol as well as in plasma and MSP3-heme complex also catalyzed H2O2-mediated oxidation of ABTS (18, 67). This enhanced peroxidase-like activity of heme may suggest some role of MSP3 in heme detoxification and a protective role for the parasite. Amyloids, earlier associated with diseases, have been reported to be present on the microbial surface of several organisms and may have a functional role in adapting the microbe to the environment by providing adhesion, mechanical invasion, and cell to cell communication (68, 69). Interestingly, several studies have also shown that amyloids interact with red blood cells as well as heme (70–73).
In conclusion, we have shown that MSP3 not only forms oligomers but also readily forms amyloid-like structures. Also, the C-terminal leucine zipper in MSP3, although clearly involved in oligomerization, may not be essentially required for fibril formation of MSP3. Using a number of MSP3 fragments, we also show that heme binding of MSP3 may be related to its ability to form aggregated structures. Whether binding of heme to a major malaria protein like MSP3 has any physiological relevance in malaria infection would need further investigation.
Acknowledgments
MSP3 RAM 1 monoclonal antibodies used for confocal experiments were obtained from the European Vaccine Initiative as part of their harmonization of the malaria vaccine assay. Dr. Dinkar Sahal (International Centre for Genetic Engineering and Biotechnology, New Delhi, India) was involved in technical discussions related to heme binding studies. Dr. Aseem Mishra and Dr. Jiban Jyoti Panda kindly assisted in EM studies. We thank Dr. Shaheena Parveen for synthesizing the YILGW peptide. Jitender Singh is acknowledged for assisting in PCR and cloning of MSP3 polypeptides.
This work was supported in part by the Department of Biotechnology.

This article contains supplemental Figs. S1–S3.
- GPI
- glycosylphosphatidylinositol
- SEC
- size exclusion chromatography
- DLS
- dynamic light scattering
- TEM
- transmission electron microscopy
- Ab
- antibody
- ThT
- thioflavin T
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- ABTS
- 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid).
REFERENCES
- 1. Miller L. H., Good M. F., Milon G. (1994) Malaria pathogenesis. Science 264, 1878–1883 [DOI] [PubMed] [Google Scholar]
- 2. Chauhan V. S., Yazdani S. S., Gaur D. (2010) Malaria vaccine development based on merozoite surface proteins of Plasmodium falciparum. Hum. Vaccin. 6, 757–762 [DOI] [PubMed] [Google Scholar]
- 3. Schwartz L., Brown G. V., Genton B., Moorthy V. S. (2012) A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar. J. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaur D., Mayer D. C., Miller L. H. (2004) Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int. J. Parasitol. 34, 1413–1429 [DOI] [PubMed] [Google Scholar]
- 5. Oeuvray C., Bouharoun-Tayoun H., Gras-Masse H., Bottius E., Kaidoh T., Aikawa M., Filgueira M. C., Tartar A., Druilhe P. (1994) Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood 84, 1594–1602 [PubMed] [Google Scholar]
- 6. Singh S., Soe S., Mejia J. P., Roussilhon C., Theisen M., Corradin G., Druilhe P. (2004) Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J. Infect. Dis. 190, 1010–1018 [DOI] [PubMed] [Google Scholar]
- 7. Burgess B. R., Schuck P., Garboczi D. N. (2005) Dissection of merozoite surface protein 3, a representative of a family of Plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J. Biol. Chem. 280, 37236–37245 [DOI] [PubMed] [Google Scholar]
- 8. Hisaeda H., Saul A., Reece J. J., Kennedy M. C., Long C. A., Miller L. H., Stowers A. W. (2002) Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J. Infect. Dis. 185, 657–664 [DOI] [PubMed] [Google Scholar]
- 9. Audran R., Cachat M., Lurati F., Soe S., Leroy O., Corradin G., Druilhe P., Spertini F. (2005) Phase I malaria vaccine trial with a long synthetic peptide derived from the merozoite surface protein 3 antigen. Infect. Immun. 73, 8017–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sirima S. B., Nébié I., Ouédraogo A., Tiono A. B., Konaté A. T., Gansané A., Dermé A. I., Diarra A., Ouédraogo A., Soulama I., Cuzzin-Ouattara N., Cousens S., Leroy O. (2007) Safety and immunogenicity of the Plasmodium falciparum merozoite surface protein-3 long synthetic peptide (MSP3-LSP) malaria vaccine in healthy, semi-immune adult males in Burkina Faso, West Africa. Vaccine 25, 2723–2732 [DOI] [PubMed] [Google Scholar]
- 11. Huy N. T., Serada S., Trang D. T., Takano R., Kondo Y., Kanaori K., Tajima K., Hara S., Kamei K. (2003) Neutralization of toxic heme by Plasmodium falciparum histidine-rich protein 2. J. Biochem. 133, 693–698 [DOI] [PubMed] [Google Scholar]
- 12. Francis S. E., Sullivan D. J., Jr., Goldberg D. E. (1997) Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 51, 97–123 [DOI] [PubMed] [Google Scholar]
- 13. Schneider E. L., Marletta M. A. (2005) Heme binding to the histidine-rich protein II from Plasmodium falciparum. Biochemistry 44, 979–986 [DOI] [PubMed] [Google Scholar]
- 14. Pagola S., Stephens P. W., Bohle D. S., Kosar A. D., Madsen S. K. (2000) The structure of malaria pigment β-haematin. Nature 404, 307–310 [DOI] [PubMed] [Google Scholar]
- 15. Balla J., Vercellotti G. M., Jeney V., Yachie A., Varga Z., Eaton J. W., Balla G. (2005) Heme, heme oxygenase, and ferritin in vascular endothelial cell injury. Mol. Nutr. Food Res. 49, 1030–1043 [DOI] [PubMed] [Google Scholar]
- 16. Campanale N., Nickel C., Daubenberger C. A., Wehlan D. A., Gorman J. J., Klonis N., Becker K., Tilley L. (2003) Identification and characterization of heme-interacting proteins in the malaria parasite, Plasmodium falciparum. J. Biol. Chem. 278, 27354–27361 [DOI] [PubMed] [Google Scholar]
- 17. Choi C. Y., Cerda J. F., Chu H. A., Babcock G. T., Marletta M. A. (1999) Spectroscopic characterization of the heme-binding sites in Plasmodium falciparum histidine-rich protein 2. Biochemistry 38, 16916–16924 [DOI] [PubMed] [Google Scholar]
- 18. Spycher C., Klonis N., Spielmann T., Kump E., Steiger S., Tilley L., Beck H. P. (2003) MAHRP-1, a novel Plasmodium falciparum histidine-rich protein, binds ferriprotoporphyrin IX and localizes to the Maurer's clefts. J. Biol. Chem. 278, 35373–35383 [DOI] [PubMed] [Google Scholar]
- 19. McColl D. J., Silva A., Foley M., Kun J. F., Favaloro J. M., Thompson J. K., Marshall V. M., Coppel R. L., Kemp D. J., Anders R. F. (1994) Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 68, 53–67 [DOI] [PubMed] [Google Scholar]
- 20. Imam M., Devi Y. S., Verma A. K., Chauhan V. S. (2011) Comparative immunogenicities of full-length Plasmodium falciparum merozoite surface protein 3 and a 24-kilodalton N-terminal fragment. Clin. Vaccine Immunol. 18, 1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 22. Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., Serrano L. (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 [DOI] [PubMed] [Google Scholar]
- 23. Pathak S., Chauhan V. S. (2011) Rationale-based, de novo design of dehydrophenylalanine-containing antibiotic peptides and systematic modification in sequence for enhanced potency. Antimicrob. Agents Chemother. 55, 2178–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haynes J. D., Diggs C. L., Hines F. A., Desjardins R. E. (1976) Culture of human malaria parasites Plasmodium falciparum. Nature 263, 767–769 [DOI] [PubMed] [Google Scholar]
- 25. Srinivasan P., Beatty W. L., Diouf A., Herrera R., Ambroggio X., Moch J. K., Tyler J. S., Narum D. L., Pierce S. K., Boothroyd J. C., Haynes J. D., Miller L. H. (2011) Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc. Natl. Acad. Sci. U.S.A. 108, 13275–13280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh S., Alam M. M., Pal-Bhowmick I., Brzostowski J. A., Chitnis C. E. (2010) Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 6, e1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller L. H., Aikawa M., Johnson J. G., Shiroishi T. (1979) Interaction between cytochalasin B-treated malarial parasites and erythrocytes. Attachment and junction formation. J. Exp. Med. 149, 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Persson K. E., McCallum F. J., Reiling L., Lister N. A., Stubbs J., Cowman A. F., Marsh K., Beeson J. G. (2008) Variation in use of erythrocyte invasion pathways by Plasmodium falciparum mediates evasion of human inhibitory antibodies. J. Clin. Invest. 118, 342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tham W. H., Wilson D. W., Reiling L., Chen L., Beeson J. G., Cowman A. F. (2009) Antibodies to reticulocyte binding protein-like homologue 4 inhibit invasion of Plasmodium falciparum into human erythrocytes. Infect. Immun. 77, 2427–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brandish P. E., Buechler W., Marletta M. A. (1998) Regeneration of the ferrous heme of soluble guanylate cyclase from the nitric oxide complex: acceleration by thiols and oxyhemoglobin. Biochemistry 37, 16898–16907 [DOI] [PubMed] [Google Scholar]
- 31. Baskakov I. V. (2004) Autocatalytic conversion of recombinant prion proteins displays a species barrier. J. Biol. Chem. 279, 7671–7677 [DOI] [PubMed] [Google Scholar]
- 32. Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., Sato K. (2003) Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3β. Proc. Natl. Acad. Sci. U.S.A. 100, 6370–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loria P., Miller S., Foley M., Tilley L. (1999) Inhibition of the peroxidative degradation of haem as the basis of action of chloroquine and other quinoline antimalarials. Biochem. J. 339, 363–370 [PMC free article] [PubMed] [Google Scholar]
- 34. Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedrich R. P., Tepper K., Rönicke R., Soom M., Westermann M., Reymann K., Kaether C., Fändrich M. (2010) Mechanism of amyloid plaque formation suggests an intracellular basis of Aβ pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 107, 1942–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lundberg K. M., Stenland C. J., Cohen F. E., Prusiner S. B., Millhauser G. L. (1997) Kinetics and mechanism of amyloid formation by the prion protein H1 peptide as determined by time-dependent ESR. Chem. Biol. 4, 345–355 [DOI] [PubMed] [Google Scholar]
- 37. Naiki H., Gejyo F. (1999) Kinetic analysis of amyloid fibril formation. Methods Enzymol. 309, 305–318 [DOI] [PubMed] [Google Scholar]
- 38. Wood S. J., Wypych J., Steavenson S., Louis J. C., Citron M., Biere A. L. (1999) α-Synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson's disease. J. Biol. Chem. 274, 19509–19512 [DOI] [PubMed] [Google Scholar]
- 39. Harper J. D., Lansbury P. T., Jr. (1997) Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 40. Gondeau C., Corradin G., Heitz F., Le Peuch C., Balbo A., Schuck P., Kajava A. V. (2009) The C-terminal domain of Plasmodium falciparum merozoite surface protein 3 self-assembles into α-helical coiled coil tetramer. Mol. Biochem. Parasitol. 165, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bannister L. H., Butcher G. A., Mitchell G. H. (1977) Recent advances in understanding the invasion of erythrocytes by merozoites of Plasmodium knowlesi. Bull. World Health Organ. 55, 163–169 [PMC free article] [PubMed] [Google Scholar]
- 42. Bannister L. H., Mitchell G. H., Butcher G. A., Dennis E. D., Cohen S. (1986) Structure and development of the surface coat of erythrocytic merozoites of Plasmodium knowlesi. Cell Tissue Res. 245, 281–290 [DOI] [PubMed] [Google Scholar]
- 43. Dworkin M. (1999) Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. BioEssays 21, 590–595 [DOI] [PubMed] [Google Scholar]
- 44. Saarimaa C., Peltola M., Raulio M., Neu T. R., Salkinoja-Salonen M. S., Neubauer P. (2006) Characterization of adhesion threads of Deinococcus geothermalis as type IV pili. J. Bacteriol. 188, 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clark M. E., Edelmann R. E., Duley M. L., Wall J. D., Fields M. W. (2007) Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ. Microbiol. 9, 2844–2854 [DOI] [PubMed] [Google Scholar]
- 46. Walker J. R., Gnanam A. J., Blinkova A. L., Hermandson M. J., Karymov M. A., Lyubchenko Y. L., Graves P. R., Haystead T. A., Linse K. D. (2007) Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol. Microbiol. 63, 629–643 [DOI] [PubMed] [Google Scholar]
- 47. Rodríguez L. E., Curtidor H., Ocampo M., Garcia J., Puentes A., Valbuena J., Vera R., López R., Patarroyo M. E. (2005) Identifying Plasmodium falciparum merozoite surface antigen 3 (MSP3) protein peptides that bind specifically to erythrocytes and inhibit merozoite invasion. Protein Sci. 14, 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cowman A. F., Berry D., Baum J. (2012) The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J. Cell Biol. 198, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cowman A. F., Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 50. Zhang X., Adda C. G., Low A., Zhang J., Zhang W., Sun H., Tu X., Anders R. F., Norton R. S. (2012) Role of the helical structure of the N-terminal region of Plasmodium falciparum merozoite surface protein 2 in fibril formation and membrane interaction. Biochemistry 51, 1380–1387 [DOI] [PubMed] [Google Scholar]
- 51. Low A., Chandrashekaran I. R., Adda C. G., Yao S., Sabo J. K., Zhang X., Soetopo A., Anders R. F., Norton R. S. (2007) Merozoite surface protein 2 of Plasmodium falciparum: expression, structure, dynamics, and fibril formation of the conserved N-terminal domain. Biopolymers 87, 12–22 [DOI] [PubMed] [Google Scholar]
- 52. Yang X., Adda C. G., MacRaild C. A., Low A., Zhang X., Zeng W., Jackson D. C., Anders R. F., Norton R. S. (2010) Identification of key residues involved in fibril formation by the conserved N-terminal region of Plasmodium falciparum merozoite surface protein 2 (MSP2). Biochimie 92, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adda C. G., Murphy V. J., Sunde M., Waddington L. J., Schloegel J., Talbo G. H., Vingas K., Kienzle V., Masciantonio R., Howlett G. J., Hodder A. N., Foley M., Anders R. F. (2009) Plasmodium falciparum merozoite surface protein 2 is unstructured and forms amyloid-like fibrils. Mol. Biochem. Parasitol. 166, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Puentes A., Ocampo M., Rodríguez L. E., Vera R., Valbuena J., Curtidor H., García J., López R., Tovar D., Cortes J., Rivera Z., Patarroyo M. E. (2005) Identifying Plasmodium falciparum merozoite surface protein-10 human erythrocyte specific binding regions. Biochimie 87, 461–472 [DOI] [PubMed] [Google Scholar]
- 55. Mills K. E., Pearce J. A., Crabb B. S., Cowman A. F. (2002) Truncation of merozoite surface protein 3 disrupts its trafficking and that of acidic-basic repeat protein to the surface of Plasmodium falciparum merozoites. Mol. Microbiol. 43, 1401–1411 [DOI] [PubMed] [Google Scholar]
- 56. Kumar S., Bandyopadhyay U. (2005) Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 57. Ponka P. (1999) Cell biology of heme. Am. J. Med. Sci. 318, 241–256 [DOI] [PubMed] [Google Scholar]
- 58. Letarte P. B., Lieberman K., Nagatani K., Haworth R. A., Odell G. B., Duff T. A. (1993) Hemin: levels in experimental subarachnoid hematoma and effects on dissociated vascular smooth-muscle cells. J. Neurosurg. 79, 252–255 [DOI] [PubMed] [Google Scholar]
- 59. Vincent S. H. (1989) Oxidative effects of heme and porphyrins on proteins and lipids. Semin. Hematol. 26, 105–113 [PubMed] [Google Scholar]
- 60. Suliman H. B., Carraway M. S., Velsor L. W., Day B. J., Ghio A. J., Piantadosi C. A. (2002) Rapid mtDNA deletion by oxidants in rat liver mitochondria after hemin exposure. Free Radic. Biol. Med. 32, 246–256 [DOI] [PubMed] [Google Scholar]
- 61. Morgan W. T. (1985) The histidine-rich glycoprotein of serum has a domain rich in histidine, proline, and glycine that binds heme and metals. Biochemistry 24, 1496–1501 [DOI] [PubMed] [Google Scholar]
- 62. Kühl T., Wissbrock A., Goradia N., Sahoo N., Galler K., Neugebauer U., Popp J., Heinemann S. H., Ohlenschläger O., Imhof D. (2013) Analysis of Fe(III) heme binding to cysteine-containing heme-regulatory motifs in proteins. ACS Chem. Biol. 8, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 63. Aft R. L., Mueller G. C. (1984) Hemin-mediated oxidative degradation of proteins. J. Biol. Chem. 259, 301–305 [PubMed] [Google Scholar]
- 64. Tsutsui K., Mueller G. C. (1982) A protein with multiple heme-binding sites from rabbit serum. J. Biol. Chem. 257, 3925–3931 [PubMed] [Google Scholar]
- 65. Summers C. A., Flowers R. A., 2nd (2000) Protein renaturation by the liquid organic salt ethylammonium nitrate. Protein Sci. 9, 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pamplona A., Ferreira A., Balla J., Jeney V., Balla G., Epiphanio S., Chora A., Rodrigues C. D., Gregoire I. P., Cunha-Rodrigues M., Portugal S., Soares M. P., Mota M. M. (2007) Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat. Med. 13, 703–710 [DOI] [PubMed] [Google Scholar]
- 67. Mashima R., Tilley L., Siomos M. A., Papalexis V., Raftery M. J., Stocker R. (2002) Plasmodium falciparum histidine-rich protein-2 (PfHRP2) modulates the redox activity of ferri-protoporphyrin IX (FePPIX): peroxidase-like activity of the PfHRP2-FePPIX complex. J. Biol. Chem. 277, 14514–14520 [DOI] [PubMed] [Google Scholar]
- 68. Gebbink M. F., Claessen D., Bouma B., Dijkhuizen L., Wösten H. A. (2005) Amyloids–a functional coat for microorganisms. Nat. Rev. Microbiol. 3, 333–341 [DOI] [PubMed] [Google Scholar]
- 69. Austin J. W., Sanders G., Kay W. W., Collinson S. K. (1998) Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162, 295–301 [DOI] [PubMed] [Google Scholar]
- 70. Khodarahmi R., Naderi F., Mostafaie A., Mansouri K. (2010) Heme, as a chaperone, binds to amyloid fibrils and forms peroxidase in vitro: possible evidence on critical role of non-specific peroxidase activity in neurodegenerative disease onset/progression using the α-crystallin-based experimental system. Arch. Biochem. Biophys. 494, 205–215 [DOI] [PubMed] [Google Scholar]
- 71. Ravi L. B., Poosala S., Ahn D., Chrest F. J., Spangler E. L., Jayakumar R., Nagababu E., Mohanty J. G., Talan M., Ingram D. K., Rifkind J. M. (2005) Red cell interactions with amyloid-β(1–40) fibrils in a murine model. Neurobiol. Dis. 19, 28–37 [DOI] [PubMed] [Google Scholar]
- 72. Murali J., Koteeswari D., Rifkind J. M., Jayakumar R. (2003) Amyloid insulin interaction with erythrocytes. Biochem. Cell Biol. 81, 51–59 [DOI] [PubMed] [Google Scholar]
- 73. Atamna H., Boyle K. (2006) Amyloid-β peptide binds with heme to form a peroxidase: relationship to the cytopathologies of Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 103, 3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]