Background: Exosomes are small vesicles in the tumor microenvironment containing nucleic acids and proteins with the capacity to influence cell behavior.
Results: Exosomes contain double-stranded genomic DNA.
Conclusion: Exosomes have the capacity to carry and transport genomic DNA spanning all chromosomes with KRAS and p53 mutations.
Significance: Exosomes can aid in identifying genomic mutations in patients with pancreatic cancer.
Keywords: Chromosomes, DNA, Exosomes, p53, Pancreatic Cancer, Serum, KRAS, Double-stranded genomic DNA, Mutations
Abstract
Exosomes are small vesicles (50–150 nm) of endocytic origin that are released by many different cell types. Exosomes in the tumor microenvironment may play a key role in facilitating cell-cell communication. Exosomes are reported to predominantly contain RNA and proteins. In this study, we investigated whether exosomes from pancreatic cancer cells and serum from patients with pancreatic ductal adenocarcinoma contain genomic DNA. Our results provide evidence that exosomes contain >10-kb fragments of double-stranded genomic DNA. Mutations in KRAS and p53 can be detected using genomic DNA from exosomes derived from pancreatic cancer cell lines and serum from patients with pancreatic cancer. In addition, using whole genome sequencing, we demonstrate that serum exosomes from patients with pancreatic cancer contain genomic DNA spanning all chromosomes. These results indicate that serum-derived exosomes can be used to determine genomic DNA mutations for cancer prediction, treatment, and therapy resistance.
Introduction
In addition to direct cell-to-cell contact via soluble factors such as cytokines and chemokines, there is emerging evidence that exosomes play a pivotal role in intercellular communication (1). Exosomes are small membrane vesicles with a size of 50–150 nm (2, 3). They are secreted by many different cell types such as cancer cells, mesenchymal cells, and thrombocytes (1, 4, 5). The first step in exosome biogenesis involves the inward budding from the limiting membrane of late endosomes (6). During this process, exosomes are packed with RNA molecules and proteins from the parental cells (3, 6). After the release into the extracellular space, tumor-derived exosomes can transfer proteins and RNAs with oncogenic activity to recipient cells (7–9). Because exosomes are very stable under different conditions, they can protect the biological cargo against degradation and denaturation in the extracellular environment (10). Exosomes are found in all body fluids of cancer patients such as serum, saliva, and ascites (9, 11, 12). Therefore, exosomes are recognized as promising diagnostic and predictive biomarkers in cancer. However, whether exosomes contain DNA is largely unknown.
It has been reported that exosomes from astrocytes and glioblastoma cells carry mitochondrial DNA (13). Furthermore, it has been shown that exosomes from glioblastoma cell lines contain small amounts of single-stranded DNA as well as high levels of transposable elements (14). In the current study, we investigated whether exosomes contain double-stranded genomic DNA. To test this hypothesis, we extracted DNA from exosomes derived from pancreatic cancer cell lines and serum from pancreatic cancer patients. We show that exosomes from human serum samples, which span all chromosomes and contain DNA with mutated KRAS and p53, contain genomic DNA.
EXPERIMENTAL PROCEDURES
Patient Samples and Tissue Collection
Collection of serum samples was approved by the Ethics Committee of the University of Heidelberg. A written informed consent for the serum sampling was obtained preoperatively from all patients with disclosure of planned analyses regarding potential prognostic markers. No neoadjuvant radiotherapy or chemotherapy was provided prior to surgical resection tumors in the patients. On the day of surgery, 10-ml serum separator tubes were used to collect blood samples through a central venous catheter immediately before surgical incision. To prevent dilution with blocking saline, the first 5–7 ml of the drawn blood were discarded. The blood samples were then centrifuged at 2500 × g for 10 min to extract the serum, and the serum was stored at −80 °C until analysis.
Cell Lines
The following human cell lines were used: Panc-1 cells (American Type Culture Collection (ATCC)) and T3M-4 cells (Cell Bank, RIKEN BioResource Centre). Panc-1 and T3M4 cells were maintained in RPMI 1640 (Sigma), supplemented with 10% (v/v) fetal bovine serum (FBS), 100 units/ml penicillin, amphotericin B, and 100 μg/ml streptomycin. Both cell lines were cultured in a humidified atmosphere of 5% CO2 at 37 °C.
Isolation of Exosomes from Cells
Cells were grown in T225-cm2 flasks for 2–3 days until they reached a confluency of 60–70%. Next, cells were cultured in serum-free medium for 48 h. The media were collected and centrifuged at 1000 rpm for 5 min followed by a centrifugation step of 3000 rpm for 10 min to discard cellular detritus. Afterward, the medium was filtered using a 0.22-μm pore filter (Thermo Fisher Scientific). A total of 225 ml of conditioned medium was collected and ultracentrifuged at 4 °C for 2 h. The supernatant was collected and stored on ice until further processing on the same day. An additional 225 ml of conditioned, filtered medium was ultracentrifuged at 4 °C for 2 h. The exosome pellets of each ultracentrifugation step were pooled in 500 μl of PBS and incubated with 10 μl of DNase I (1 unit/μl, catalog number M6101, Promega) at 37 °C for 30 min. Subsequently, 50 μl of DNase stop solution (catalog number M199A, Promega) were added, and the samples were heated at 65 °C in a water bath for 5 min. Next, the pooled exosome pellet was washed in 30 ml of PBS, and a second step of ultracentrifugation was performed at 150,000 × g at 4 °C for 2 h. After aspiration of the supernatant, the exosome pellet was suspended in 200 μl of PBS. Five μl of this sample were obtained and diluted at 1:100 in PBS and stored at −20 °C for further analysis with NanoSight® LM10 (NanoSight Ltd., Minton Park, Amesbury, UK). Exosomes validation included flow cytometry analysis (FACS)4 and immunoblotting with exosomal markers as well as electron microscopy (see sections below).
Exosome Isolation from Human Serum Samples
After cell-free serum samples were thawed on ice, 500 μl of serum (5 ml of serum in the case of Bioanalyzer analysis) were diluted in 11 ml of 1× PBS, filtered through a 0.22-μm pore filter (syringe filter, catalog number 6786-1302, GE Healthcare), and ultracentrifuged at 150,000 × g at 4 °C overnight. Afterward, the exosome-depleted serum (∼11 ml) was collected and stored on ice until further processing on the same day. The exosome pellet was incubated with 1 μl of DNase I in 200 μl of PBS (1 unit/μl, catalog number M6101, Promega) at 37 °C for 30 min. Subsequently, 5 μl of DNase stop solution (catalog number M199A, Promega) were added, and the samples were heated at 65 °C in a water bath for 5 min. Next, the exosome pellet was washed in 11 ml of 1× PBS, and a second step of ultracentrifugation was performed at 150,000 × g at 4 °C for 2 h. The supernatant of the second step was discarded, and the pellet was suspended in 200 μl of PBS. Five μl of this sample were obtained and diluted 1:100 and stored at −20 °C for further analysis using NanoSight® LM10. The supernatant of this first run was filtered by using a fiber filtration system with a pore size of 70 kDa (catalog number C02-E070-05-N, Spectrum Laboratories, Inc.). NanoSight measurements using NanoSight® LM10 confirmed the depletion of exosomes.
DNA Extraction
The DNA was extracted using the DNeasy blood and tissue kit (catalog number 69506, Qiagen, Hilden, Germany) according to the manufacturer's instructions. Finally, the DNA was eluted in 50 μl of AE buffer and stored at −20 °C until processing. Double-stranded DNA was analyzed using an Agilent DNA 7500 reagent kit (catalog number 5067-1507, Agilent Technologies). To conduct DNA extraction on the whole exosome-depleted serum supernatant (∼10 ml), 1 ml of proteinase K and 10 ml of lysis buffer were added. After heat inactivation at 56 °C in a water bath, 10 ml of ethanol (100%) were supplemented. Subsequently, the whole volume was sequentially centrifuged in one spin column. After the washing steps had been performed according to the manufacturer's instructions, 50 μl of AE buffer were pipetted directly onto the column membrane, incubated at room temperature for 1 min, and then centrifuged for 1 min at 6000 × g (8000 rpm) to elute. The eluate was stored at −20 °C until processing.
FACS
Exosomes were attached to 4-μm aldehyde/sulfate latex beads (Invitrogen) by mixing ∼30 μg of exosomes in a 100-μl volume of beads for 2 h at room temperature. This suspension was diluted to 1 ml with PBS, and the reaction was stopped with 100 mm glycine and 2% BSA in PBS. Exosome-bound beads were washed in PBS/1% BSA, blocked with 10% BSA, and stained for FACS with CD9 (Abcam, 1:400, ab92726), TSG101 (Abcam; 1:400; anti-ab83), and CD63 antibodies (Santa Cruz Biotechnology, 1:400; sc-15363). Secondary antibodies with Alexa Fluor 488 (Life Technologies) were used.
Western Blot and Antibodies
To monitor exosomal expression of TSG101 and CD63, exosomes were harvested in 8 m urea/2.5% SDS, 5 μg ml−1 leupeptin, 1 μg ml−1 pepstatin, and 1 mm phenylmethylsulfonyl fluoride buffer. Samples were loaded according to Bradford quantification and analyzed using acrylamide gels. Wet electrophoretic transfer was used to transfer the proteins in the gel onto PVDF membranes (Immobilon P). The protein blot was blocked for 1 h at room temperature with 5% nonfat dry milk in PBS/0.05% Tween and incubated overnight at 4 °C with the following primary antibodies against TSG101 (Abcam; 1:400 anti-ab83) and CD63 (Santa Cruz Biotechnology, 1:400; sc-15363). Afterward, secondary antibodies were incubated for 1 h at room temperature. Washes after antibody incubations were done on an orbital shaker, four times at 10-min intervals, with 1× PBS/0.05% Tween 20. Blots were developed with chemiluminescent reagents from Pierce.
PCR
The amount of DNA from cells and cell medium-derived exosomes was quantified using a Nanodrop® 1000 spectrophotometer (Thermo Fisher Scientific). The amount of DNA from human serum-derived exosomes was quantified using PicoGreen® (Quant-iTTM PicoGreen® dsDNA assay kit, catalog number P11496, Life Technologies). PCR was performed in a 25-μl reaction tube consisting of 10 μl of template DNA, 1 μm of each primer, 2.5 mm dNTP, 2.5 mm 10× PCR buffer, 25 mm magnesium solution, 0.5 μl of H2O, and 2.5 μl of Taq polymerase. Amplification was carried out in a T100 Thermocycler (Bio-Rad) under the following conditions: 94 °C for 1 min, 2 cycles of 94 °C for 10 s, 67 °C for 30 s, 70 °C for 30 s; 2 cycles of 94 °C for 10 s, 64 °C for 30 s, 70 °C for 30 s; 2 cycles of 94 °C for 10 s, 61 °C for 30 s, 70 °C for 30 s; 35 cycles of 94 °C for 10 s, 59 °C for 30 s, 70 °C for 30 s; endless 4 °C. KRAS analysis was performed using the following primers: forward 5′-AAGGCCTGCTGAAAATGACTG-3′, 5′-TCACAATACCAAGAAACCCAT-3′. p53 analysis was performed using the following primers: p53 Exon 7-8p (609 bp), forward 5′-TCCTAGGTTGGCTCTGAC-3′, reverse 5′-CCTGCTTGCTTACCTCGCT-3′ and p53 Exon 5-8 (1564 bp), forward 5′-TTCCTCTTCCTACAGTACTCC-3′, reverse 5′-CCTGCTTGCTTACCTCGCT-3′. PCR products were purified using the QIAquick PCR purification kit (Qiagen). Subsequently, a sequencing reaction was performed using BigDye terminator kit (v3.1, Life Technologies) according to the manufacturer's instructions.
For sequencing, the following primers were used: KRAS 5′-AAGGCCTGCTGAAAATGACTG-3′, 5′-AGAATGGTCCTGCACCAGTAA-3′; p53 Exon 5-8 forward 5′-TCTTCCTACAGTACTCCCCT-3′, reverse 5′-GCTTGCTTACCTCGCTTAGT-3′; p53 Exon 7-8 forward 5′-TAGGTTGGCTCTGACTGT-3′, reverse 5′-GCTTGCTTACCTCGCTTAGT-3′. Sequencing products were separated on an ABI 3730 automated sequencer (Life Technologies).
Electronic Microscopy
Samples were placed on 400 mesh Formvar-coated copper grids treated with poly-l-lysine for 1 h. Excess samples were blotted with filter paper and then negatively stained with Millipore paper-filtered aqueous 1% uranyl acetate for 1 min. Stain was blotted dry from the grids with filter paper, and samples were allowed to dry. Samples were then examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using the AMT imaging system (Advanced Microscopy Techniques Corp., Danvers, MA).
Whole Genome Shotgun Sequencing
Whole genome sequencing was performed using the ThruPLEX-FD library prep technology (catalog number R40048, Rubicon Genomics, Ann Arbor, MI) in combination with the Illumina HiSeq2000 sequencing platform, paired-end 2× 51 bp, to a coverage depth of 4× in exosomes and matched tumor samples. To assess copy number profile and gain additional insights into structural rearrangements, an algorithm called BIC-seq was utilized (15).
RESULTS
Exosomes Contain >10-kb Fragments of Double-stranded Genomic DNA
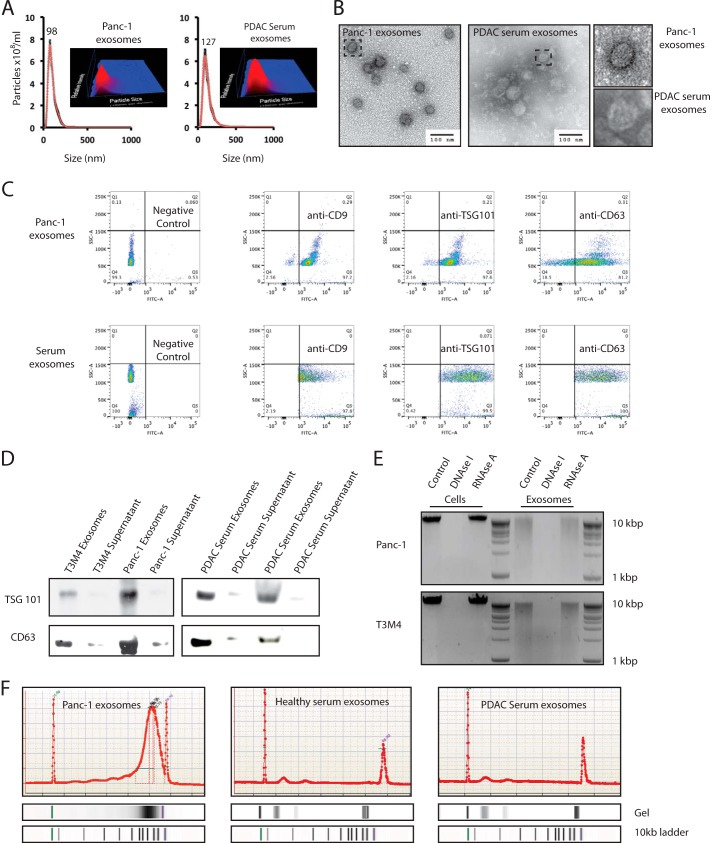
Exosomes were isolated from two human pancreatic cancer cell lines (Panc-1 and T3M4), two serum samples of healthy donors, and patients with pancreatic cancer (16, 17). To reduce external DNA contamination, prior to DNA extraction, exosomes were treated extensively with DNase I as described previously (14). The presence of exosomes and their concentration from both cancer cell lines and serum samples were confirmed using the NanoSight® LM10 (Fig. 1A and supplemental movie). Moreover, exosomes were identified as homogeneous population by electron microscopy (Fig. 1B) and by the expression of exosome markers TSG101, CD9, and CD63 (Fig. 1, C and D). Additionally, after extraction of exosomal DNA from cancer cell lines, the eluate was subjected to RNase A to exclude RNA. Subsequently, we analyzed the pretreated eluate on a 2% agarose gel (Fig. 1E). Our assessment revealed the presence of long fragments of DNA in exosomes without RNA. By using a double-stranded DNA detection kit, we show that exosomes from pancreatic cancer cells and serum samples contain genomic double-stranded DNA (Fig. 1F).
FIGURE 1.
Exosomes contain >10-kb fragments of double-stranded genomic DNA. A and B, the presence and concentration of exosomes from human pancreatic cancer cell lines and human serum samples from patients with pancreatic cancer were determined by using a NanoSight® LM10 (A) and electron microscopy (B). C and D, exosomes were characterized by the exosome-specific expression of CD9, TSG101, and CD63 by FACS analysis (C) and the exosome-specific expression of TSG101 and CD63 by Western blotting (D). E, to exclude RNA contamination after exosome lysis and DNA extraction, the DNA eluate from two cell lines (Panc-1 and T3M4) and the DNA eluate from corresponding exosomes were treated with DNase I and RNase A. Subsequently, the eluate was run on a 2% agarose gel. F, the presence of double-stranded DNA from cellular Panc-1 exosomes and human serum exosomes was confirmed by a double-stranded detection kit (representative figure for exosomal DNA from Panc-1, one healthy donor, and one patient with pancreatic cancer). PDAC, pancreatic ductal adenocarcinoma. SSC-A, side scatter detector A.
Exosomes Contain Mutated KRAS and p53 DNA
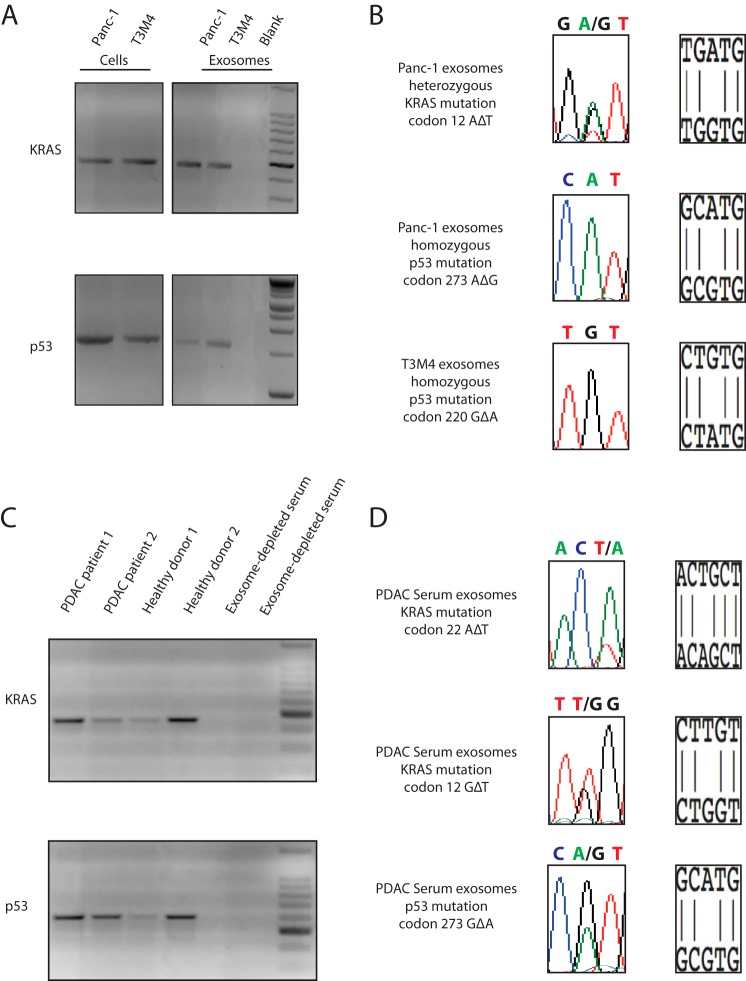
KRAS and p53 are the most frequently mutated genes in pancreatic ductal adenocarcinoma (18). We amplified a 466-bp fragment of KRAS encoding exon 2 and partial intron 2, as well as a 1564-bp fragment of p53 spanning from exon 5 to exon 8, including introns 5, 6, and 7, from both cell lines and DNA isolated from exosomes derived from the same cell lines (Fig. 2A). KRAS and p53 mutations in Panc-1 and T3M4 have been described previously (19). Panc-1 displays a heterozygous KRAS mutation in codon 12 (glycine to aspartate) and a homozygous p53 mutation in codon 273 (arginine to histidine) (19). T3M4 cells contain wild type sequence of KRAS, but display a homozygous p53 mutation in codon 220 (tyrosine to cysteine) (19). By Sanger sequencing of the PCR amplified DNA, we were able to detect identical KRAS and p53 mutations in the DNA isolated from exosomes derived from Panc-1 cells and the identical p53 mutation in the DNA from exosomes derived from the T3M4 cells (Fig. 2B). Mutation in the KRAS DNA was not detected in the T3M4 cells and the exosomes isolated from them (data not shown).
FIGURE 2.
Exosomes contain mutated KRAS and p53 DNA. A, by PCR, we amplified a 466-bp fragment of KRAS spanning exon 2 and intron 2 and a 1564-bp fragment of p53 spanning 4 exons and 3 introns. B, Sanger sequencing of genomic DNA from Panc-1 cells and corresponding exosomes revealed the same heterozygous mutation of KRAS on codon 12 (GGT to GAT) and the similar homozygous mutation of p53 on codon 273 (CGT to CAT). T3M4 cells and corresponding exosomes displayed the same homozygous mutation of p53 on codon 220 (TAT to TGT). C, PCR amplification provided evidence for long fragments of DNA in circulating exosomes from two healthy donors and two patients with pancreatic cancer. We could retrieve a 466-bp fragment of KRAS DNA and 609-bp fragment of p53 DNA spanning exons 7 and 8 and intron 7. When serum samples depleted of exosomes were subjected to PCR, no KRAS or p53 amplicon could be detected. PDAC, pancreatic ductal adenocarcinoma. D, Sanger sequencing of serum exosome-derived DNA was able to detect DNA with a KRAS mutation on codon 22. In a second patient, Sanger sequencing revealed a KRAS mutation on codon 12 and a p53 mutation on codon 273.
Based on our observations using cell lines, we speculated that human circulating serum exosomes might also contain KRAS and p53 DNA. We amplified a 466-bp fragment of KRAS encoding exon 2 and a portion of intron 2 in two samples from patients with pancreatic cancer and two samples from healthy donors (Fig. 2C). Subsequently, we also isolated a 609-bp DNA fragment of p53 overlapping exons 7 and 8 and intron 7 in all human samples (Fig. 2C). To evaluate the presence of DNA in serum samples depleted of exosomes, we also performed a PCR for KRAS and p53. However, we could not detect any amplified PCR product of KRAS or p53 (Fig. 2C) in the exosome-depleted serum. The PCR amplicons from the DNA isolated from exosomes were subjected to Sanger sequencing. Sanger sequencing detected DNA with a KRAS mutation in serum samples of patients with pancreatic cancer (Fig. 2D). One KRAS mutation was located on codon 12 and was characterized by a base exchange of GGT to TGT. The second KRAS mutation was found on codon 22 with a base exchange from CAG to CTG. Additionally, in one patient with pancreatic cancer, we were able to detect a p53 mutation on codon 273 with a base exchange from CGT to CAT (Fig. 2D).
Circulating Exosomes from the Peripheral Blood of Pancreatic Ductal Adenocarcinoma Patients Contain Double-stranded Genomic DNA Spanning All Chromosomes
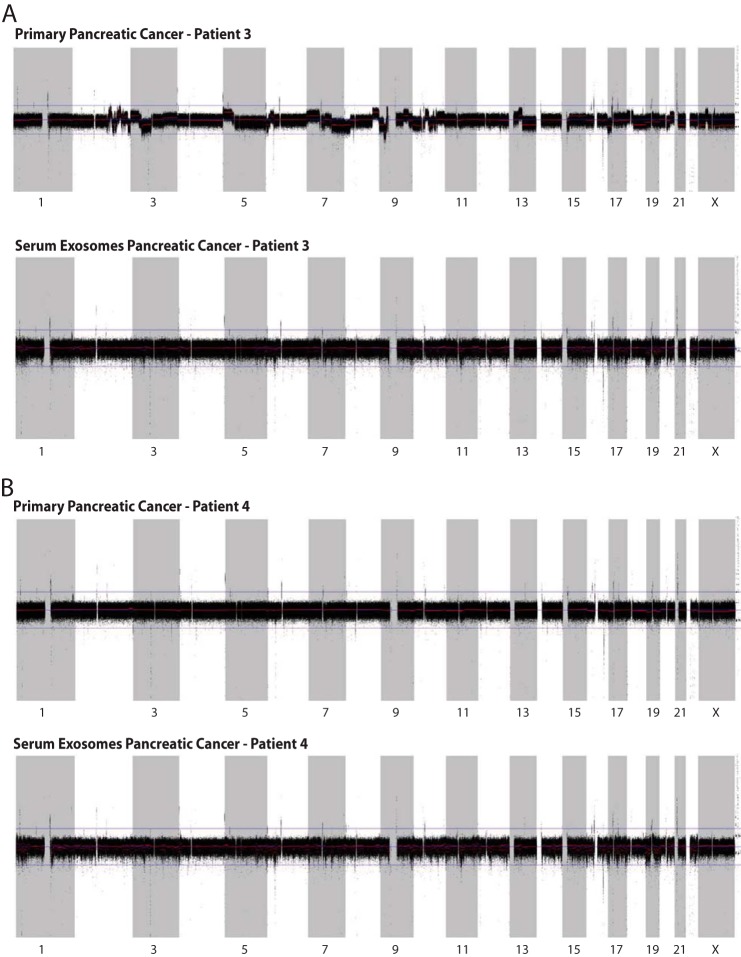
Two pancreatic cancer samples were investigated using paired serum exosomal DNA and matched tumor sample. A 4× whole genome sequence coverage was achieved with inferred insert size of libraries ∼160 bp. The percentage of reads mapped to the human genome was around 96%. The properly paired percentage read ∼92% between tumor genomic DNA and exosomal genomic DNA. Sequence complexity as a number of unique reads was over 9 × 108 in all samples. Our analysis revealed that a bulk of serum-derived exosomes contain DNA spanning uniformly across all chromosomes resembling nuclear genomic DNA (Fig. 3, A and B).
FIGURE 3.
Serum-derived exosomes contain genomic DNA spanning all chromosomes. A and B, whole genome sequencing was conducted on serum-derived, exosomal DNA and corresponding primary tumor from two patients. BIC-seq control-free log2 copy-number profile across all human chromosomes, bin size 1000 bp; RAW profile, black; segmented, red line. Profiles demonstrate somatic chromosomal gains (upper) and losses (lower), as well as normal polymorphism. In the second case (B), a lack of structural chromosomal rearrangement expected for pancreatic ductal adenocarcinoma is explained due to the possible low number of cancer cells in the sample. Sequencing revealed that circulating exosomes contain genomic DNA spanning all chromosomes.
DISCUSSION
There is emerging evidence that the exosome-mediated cell-to-cell communication might have importance in health and disease. Because tumor-derived exosomes contain a wide range of RNA molecules and proteins, they have been considered to be promising candidates for discovery of diagnostic biomarkers in cancer and other diseases (1, 20). However, the presence of DNA in exosomes is largely undetermined (21). Two previous studies have shown that exosomes contain mitochondrial DNA, single-stranded DNA, and transposable elements (13, 14). Our study provides evidence that exosomes can carry large fragments (>10 kb) of double-stranded genomic DNA, apart from mitochondrial DNA and small amounts of single-stranded DNA (13, 14). Moreover, we show that exosomes from human serum samples contain genomic DNA spanning all chromosomes. This novel finding contradicts the current opinion that circulating DNA is highly fragmented with an estimated length of only 60–100 bp (22). Intriguingly, although we were able to extract >10-kb fragments of DNA from serum exosomes, we could not detect any PCR product in the exosome-depleted serum. These data require further validation but suggest that the majority of circulating DNA isolated from the serum samples may come from inside the exosomes and are not present as free floating circulating DNA. Our studies suggest a new source of circulating DNA in addition to potential cell-free DNA derived from cell lysis or apoptosis (23).
As a proof of concept, we were able to demonstrate that exosome-derived DNA carry mutations identical to their parental cancer cells or tumors. Additionally, our preliminary whole genome sequencing attempt led to the identification of significant genomic DNA in the exosomes covering all chromosomes. We speculate that our inability to detect mutation via our low coverage whole genomic sequencing effort may be due to extensive contribution of exosomes from non-cancer cells in the serum samples. Future studies can attempt deep coverage sequencing or identify methods to specifically isolate cancer cell-specific exosomes. Nevertheless, our study provides unequivocal evidence for the presence of double-stranded genomic DNA in the exosomes that spans across all the chromosomes. Whether our discovery will enable the use of genomic DNA from circulating exosomes to sequence a significant portion of human genome for diagnostic and therapeutic purpose remains to be determined. Exosomes may turn out to be a powerful DNA diagnostic tool and likely also provide new methods to predict the prognosis of cancer patients and improve treatment via a personalized medicine approach.
Acknowledgments
The Metastasis Research Center at the MD Anderson Cancer Center and Cancer Prevention and Research Institute of Texas were supported by Institutional Core Grant CA16672 for High Resolution Electron Microscopy Facility.
This work was supported, in whole or in part, by National Institutes of Health Grants CA-125550, CA-155370, CA-151925, DK 081576, and DK 055001 (to R. K.).

This article contains a supplemental movie.
- FACS
- flow cytometry analysis.
REFERENCES
- 1. Kahlert C., Kalluri R. (2013) Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 91, 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan B. T., Teng K., Wu C., Adam M., Johnstone R. M. (1985) Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 101, 942–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trams E. G., Lauter C. J., Salem N., Jr., Heine U. (1981) Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 645, 63–70 [DOI] [PubMed] [Google Scholar]
- 4. Heijnen H. F., Schiel A. E., Fijnheer R., Geuze H. J., Sixma J. J. (1999) Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood 94, 3791–3799 [PubMed] [Google Scholar]
- 5. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 7. Kucharzewska P., Christianson H. C., Welch J. E., Svensson K. J., Fredlund E., Ringnér M., Mörgelin M., Bourseau-Guilmain E., Bengzon J., Belting M. (2013) Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. U.S.A. 110, 7312–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grange C., Tapparo M., Collino F., Vitillo L., Damasco C., Deregibus M. C., Tetta C., Bussolati B., Camussi G. (2011) Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 71, 5346–5356 [DOI] [PubMed] [Google Scholar]
- 9. Peinado H., Alečkoví M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor D. D., Gercel-Taylor C. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 11. Lau C., Kim Y., Chia D., Spielmann N., Eibl G., Elashoff D., Wei F., Lin Y. L., Moro A., Grogan T., Chiang S., Feinstein E., Schafer C., Farrell J., Wong D. T. (2013) Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 288, 26888–26897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi D. S., Park J. O., Jang S. C., Yoon Y. J., Jung J. W., Choi D. Y., Kim J. W., Kang J. S., Park J., Hwang D., Lee K. H., Park S. H., Kim Y. K., Desiderio D. M., Kim K. P., Gho Y. S. (2011) Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics 11, 2745–2751 [DOI] [PubMed] [Google Scholar]
- 13. Guescini M., Guidolin D., Vallorani L., Casadei L., Gioacchini A. M., Tibollo P., Battistelli M., Falcieri E., Battistin L., Agnati L. F., Stocchi V. (2010) C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res. 316, 1977–1984 [DOI] [PubMed] [Google Scholar]
- 14. Balaj L., Lessard R., Dai L., Cho Y. J., Pomeroy S. L., Breakefield X. O., Skog J. (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xi R., Hadjipanayis A. G., Luquette L. J., Kim T. M., Lee E., Zhang J., Johnson M. D., Muzny D. M., Wheeler D. A., Gibbs R. A., Kucherlapati R., Park P. J. (2011) Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc. Natl. Acad. Sci. U.S.A. 108, E1128–E1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., Buchanan M., Hosein A. N., Basik M., Wrana J. L. (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 [DOI] [PubMed] [Google Scholar]
- 17. Théry C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol., Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 18. Biankin A. V., Waddell N., Kassahn K. S., Gingras M. C., Muthuswamy L. B., Johns A. L., Miller D. K., Wilson P. J., Patch A. M., Wu J., Chang D. K., Cowley M. J., Gardiner B. B., Song S., Harliwong I., Idrisoglu S., Nourse C., Nourbakhsh E., Manning S., Wani S., Gongora M., Pajic M., Scarlett C. J., Gill A. J., Pinho A. V., Rooman I., Anderson M., Holmes O., Leonard C., Taylor D., Wood S., Xu Q., Nones K., Fink J. L., Christ A., Bruxner T., Cloonan N., Kolle G., Newell F., Pinese M., Mead R. S., Humphris J. L., Kaplan W., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chou A., Chin V. T., Chantrill L. A., Mawson A., Samra J. S., Kench J. G., Lovell J. A., Daly R. J., Merrett N. D., Toon C., Epari K., Nguyen N. Q., Barbour A., Zeps N., Australian Pancreatic Cancer Genome Initiative, Kakkar N., Zhao F., Wu Y. Q., Wang M., Muzny D. M., Fisher W. E., Brunicardi F. C., Hodges S. E., Reid J. G., Drummond J., Chang K., Han Y., Lewis L. R., Dinh H., Buhay C. J., Beck T., Timms L., Sam M., Begley K., Brown A., Pai D., Panchal A., Buchner N., De Borja R., Denroche R. E., Yung C. K., Serra S., Onetto N., Mukhopadhyay D., Tsao M. S., Shaw P. A., Petersen G. M., Gallinger S., Hruban R. H., Maitra A., Iacobuzio-Donahue C. A., Schulick R. D., Wolfgang C. L., Morgan R. A., Lawlor R. T., Capelli P., Corbo V., Scardoni M., Tortora G., Tempero M. A., Mann K. M., Jenkins N. A., Perez-Mancera P. A., Adams D. J., Largaespada D. A., Wessels L. F., Rust A. G., Stein L. D., Tuveson D. A., Copeland N. G., Musgrove E. A., Scarpa A., Eshleman J. R., Hudson T. J., Sutherland R. L., Wheeler D. A., Pearson J. V., McPherson J. D., Gibbs R. A., Grimmond S. M. (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moore P. S., Sipos B., Orlandini S., Sorio C., Real F. X., Lemoine N. R., Gress T., Bassi C., Klöppel G., Kalthoff H., Ungefroren H., Löhr M., Scarpa A. (2001) Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 439, 798–802 [DOI] [PubMed] [Google Scholar]
- 20. Kosaka N., Yoshioka Y., Hagiwara K., Tominaga N., Katsuda T., Ochiya T. (2013) Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front. Genet. 4, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 22. Mouliere F., Thierry A. R. (2012) The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert. Opin. Biol. Ther. 12, Suppl. 1, S209–S215 [DOI] [PubMed] [Google Scholar]
- 23. Pinzani P., Salvianti F., Pazzagli M., Orlando C. (2010) Circulating nucleic acids in cancer and pregnancy. Methods 50, 302–307 [DOI] [PubMed] [Google Scholar]