Background: Ribosomal protein S6 has been known to be a key downstream effector of the TOR signaling pathway.
Results: We demonstrated that ribosomal protein interacts with a histone deacetylase and binds to rRNA gene promoter.
Conclusion: The TOR signaling controls rRNA synthesis via interaction of RPS6 to rRNA genes.
Significance: This study links the environmental signals via TOR kinase to control growth of an organism by regulating ribosome biogenesis.
Keywords: Epigenetics, Histone Deacetylase, Ribosomal RNA (rRNA), Stress Response, TOR
Abstract
The target of rapamycin (TOR) kinase pathway regulates various biological processes, including translation, synthesis of ribosomal proteins, and transcription of rRNA. The ribosomal protein S6 (RPS6) is one of the well known downstream components of the TOR pathway. Ribosomal proteins have been known to have diverse functions in regulating cellular metabolism as well as protein synthesis. So far, however, little is known about other possible role(s) of RPS6 in plants, besides being a component of the 40 S ribosomal subunit and acting as a target of TOR. Here, we report that RPS6 may have a novel function via interaction with histone deacetylase 2B (AtHD2B) that belongs to the plant-specific histone deacetylase HD2 family. RPS6 and AtHD2B were localized to the nucleolus. Co-expression of RPS6 and AtHD2B caused a change in the location of both RPS6 and AtHD2B to one or several nucleolar spots. ChIP analysis suggests that RPS6 directly interacts with the rRNA gene promoter. Protoplasts overexpressing both AtHD2B and RPS6 exhibited down-regulation of pre-18 S rRNA synthesis with a concomitant decrease in transcription of some of the ribosomal proteins, suggesting their direct role in ribosome biogenesis and plant development. This is consistent with the mutation in rps6b that results in reduction in 18 S rRNA transcription and decreased root growth. We propose that the interaction between RPS6 and AtHD2B brings about a change in the chromatin structure of rDNA and thus plays an important role in linking TOR signaling to rDNA transcription and ribosome biogenesis in plants.
Introduction
Ribosome biogenesis is central to the growth and development of eukaryotic cells and organisms. Accordingly, rapidly growing cells invest most of the cells' transcriptional/translational capacities into the syntheses of rRNAs and the ribosomal proteins (1, 2). Ribosome biogenesis in eukaryotic cells involves coordinated syntheses of the four ribosomal RNAs (rRNAs) and more than 70 ribosomal proteins, and transcription/translation of each component is tightly regulated in response to the physiological status of the cell (3). Transcription of ribosomal DNA (rDNA) depends on multiple signaling pathways responding to the external environmental cues, including, stress, nutrients, hormones, and mitogens (1, 4).
Three of the four ribosomal RNAs (18 S, 5.8 S, and 25 S) are transcribed as a single precursor (pre-rRNA)4 by RNA polymerase I (pol I), which is an important rate-limiting step in the biogenesis of ribosomes. Processing of pre-rRNA begins at the 5′ external transcribed spacer (ETS). Subsequent cleavages occur at the 5′ end of the 18 S rRNA and the internal transcribed spacer 1 (ITS1) to generate 18 S rRNA and a precursor containing the 5.8 S and 25 S rRNAs. Final cleavage in the ITS2 and the 3′-ETS generates mature 5.8 S and 25 S rRNAs (5, 6).
Target of rapamycin (TOR) kinase (7, 8) signaling coordinates many cellular metabolic activities under varying energy and stress conditions (3). In yeast, the pol I-dependent transcription of 35S rRNA precursor is directly controlled by TOR, which binds to the rDNA promoter via its helix-turn-helix motif (9). More recently, association of mammalian TOR and the Arabidopsis TOR to their respective rDNA promoters has also been reported, and the binding of mammalian TOR was shown to be sensitive to rapamycin treatment (10, 11). TOR has also been implicated in the transcriptional activation of a number of ribosomal protein genes that is mediated by the activities of its downstream effector kinase (ribosomal protein S6 kinase) and the c-Myc transcription factor (Sch9 in animals and Sfp1 in yeast, respectively) (3). Activation of the pol I-mediated transcription by TOR is indirectly controlled by ribosomal protein S6 kinase, impinging on the general transcription factor UBF1 (Hmo1 in yeast) (12). Evidence suggests that the activity of TOR is required in derepressing the epigenetic silencing of the rDNA promoter (13, 14), and a possible role of histone deacetylases has been suggested in epigenetic silencing of the rRNA genes (15).
Ribosomal protein S6 (RPS6), a component of the 40 S ribosomal subunit, has been known to be a key downstream effector of the TOR signaling pathway, which is conserved among yeast, mammals, insects, and plants (16, 17). The phosphorylation status of RPS6, which reflects the activity of S6K, has been recognized as a hallmark of actively proliferating cells (18–20). The phosphorylation of RPS6 plays a role in the translational up-regulation of mRNAs containing the 5′-terminal oligopyrimidine tract (5′-TOP), which are found in many mRNAs encoding the proteins involved in ribosome biogenesis (21). However, the RPS6 phosphorylation-defective cells did not show a dramatic reduction in global protein translation as well as in translation of the 5′-TOP mRNAs (19). Thus, the exact role of RPS6 in the regulation of ribosome biogenesis and the identities of the factors involved in this process remain a subject of scrutiny.
To obtain a better insight into the possible role of RPS6 in the mechanism of regulation of ribosome biogenesis in plants, we attempted to identify novel interacting partners of RPS6 from Arabidopsis by GST pulldown followed by LC/MS protein identification. A plant-specific histone deacetylase AtHD2B (also known as HDT1) was identified as one of the interacting partners of RPS6. Here, we present evidence for a specific interaction of RPS6 with AtHD2B and demonstrate a possible role of this complex in transcriptional regulation of rRNA genes. We propose a new paradigm for controlling rDNA transcription in plants in which TOR may control a silencing mechanism of the rDNA transcription via its downstream signaling component RPS6, the mechanism of which involves interaction of the RPS6 with AtHD2B. Such an interaction can provide a direct link between stress signals and the regulation of translation and transcription (particularly rDNA) machineries controlling plant growth.
EXPERIMENTAL PROCEDURES
Plant Materials and Hormone Treatments
The Arabidopsis thaliana ecotype Columbia or Columbia-0 was used in BiFC and protoplast transformation assay and in mutant analysis of rps6b, respectively. Seeds were sterilized by gentle shaking in 70% ethanol for 5 min, followed by treatment with 50% hypochlorite and 0.01% Triton X-100 for 15 min and then with distilled water. Sterile seeds were grown on aseptic solid media (0.8% plant agar) containing ½ MS salts (Sigma) and 1% sucrose in a growth chamber at 22 °C with a 16-h photoperiod. For root elongation assays, wild-type, mutant, and transgenic seeds were germinated on MS agar media, and seedlings were transferred to plates with or without kinetin after 4 days and grown vertically. Root size was measured after an additional 10 days of growth. To apply plant hormones, 14-day-old seedlings were transferred to sterile liquid growth medium containing hormones and salt and harvested after 24 h. The concentrations of hormones used were as follows: 20 μm indole-3-acetic acid, 20 μm kinetin, 20 μm abscisic acid, and 300 mm NaCl.
Plasmid Construction for Transient Expression Studies
To make various constructs of RPS6-GFP fusion (S6FL-GFP, S6CT-GFP, or S6NT-GFP), the full-length and C- or N-terminal regions of RPS6 were amplified by PCR with Pfu DNA polymerase (Takara, Japan) using forward and reverse primers (see supplemental Table S1) with a SacI site and a BamHI site, respectively, and fused in-frame with GFP of the 326-smGFP vector.
Transgenic Plants with P35S-AtHD2B or PrDNA-GUS
The DNA fragment of AtHD2B-HA or PrDNA-GUS were transferred into pK7WGF2.0. The resulting fusion constructs were introduced into the Agrobacterium tumefaciens strain GV3101, and Arabidopsis plants were transformed by the floral dip method (22). To make a construct of the rDNA minimal promoter fused to a GUS reporter gene, a 500-bp upstream region of rDNA was amplified using the following primers: forward, 5′-AGAATTCGTCGACCAGGACGGCGGAAC-3′, and reverse, 5′-AGACTCCCTCAACACCCACCCCCCTATA-3′.
Real Time Quantitative PCR
Real time (RT)-quantitative PCR was set up with SYBR Green PCR master mix (Takara, Japan) using Rotor-Gene 3000 (Qiagen, Germany). All reactions were normalized using ACTIN2 gene as an internal control. Primers used for various fragments are listed in supplemental Table S1.
Identification and Complementation of the T-DNA Insertion of RPS6B
Seeds of a T-DNA insertion line SALK_012147 of RPS6B were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). To confirm the insertion of T-DNA of RPS6B, genomic PCR was performed, and the PCR product was sequenced. Primers used were as follows: T-DNA left border-specific forward primer (LBb1), 5′-GCGTGGACCGCTTGCTGCAACT-3′; RPS6B forward primer (S6Bf), 5′-CAATGACCAAGTTAAGAACAGACAGGTCA-3′, and RPS6B reverse primer (S6Br), 5′-CTGCGTTGGTCTGATATATAACCAGTTC-3′. To search whether RPS6B expression is decreased in the T-DNA insertion rps6b mutant, RT-quantitative PCR was performed to examine the RPS6B expression in the mutant. For complementation of the mutant, the RPS6B promoter was fused with a full-length RPS6B cDNA. The recombinant plasmid was introduced into a homozygous mutant, rps6b-3, by the floral dip method (22).
Protein Pulldown Assay
GST protein was immobilized onto the CNBr-activated Sepharose resin and used for pre-clearing the cell extract. Then 100–300 mg of Arabidopsis total soluble proteins were applied to the columns of different GST fusion protein substrates arranged in tandem as outlined in supplemental Fig. S1. The protein sample passed through the columns was re-circulated at least three times to facilitate specific interaction between the GST fusion proteins and the cellular proteins. The columns were then washed with 1000 volumes of wash buffer A (50 mm Tris·Cl, pH 7.5, 125 mm NaCl, 5 mm benzamidine, 1 mm PMSF, 1 mm DTT, 10% glycerol), 100 volumes of wash buffer B (50 mm Tris·Cl, pH 7.5, 500 mm NaCl, 5 mm benzamidine, 1 mm PMSF), and then 500 volumes of wash buffer A. For the samples to be eluted by thrombin digestion, the wash step was completed with an additional 500 volumes of wash buffer C (50 mm Tris·Cl, pH 7.5, 125 mm NaCl, 1 mm DTT) to remove the traces of the protease inhibitors from the resins before they were subjected to thrombin digestion. Isolated proteins were visualized by Coomassie staining after SDS-PAGE, and the protein band identification by peptide mass fingerprinting by MALDI-TOF mass spectrometry was conducted at the proteomics facility at Ohio State University and at Genomine Inc. His-fused proteins were purified as described above and incubated with total soluble proteins extracted from 3HA-RPS6 transgenic plant for 16 h. After incubation, His-AtHD2B and 3HA-RPS6 were washed and resolved by SDS-PAGE.
BiFC Assay
The BiFC assay was performed according to the method described by Hu et al. (23). Full-length or the N-terminal region of AtHD2B and full-length cDNA of N35 were cloned into the binary BiFC vectors, p2YN and p2YC, respectively. The resulting fusion constructs were verified by sequencing and introduced into Arabidopsis protoplasts by transfection (24). After 16 h of incubation, fluorescence and DAPI staining were visualized by Olympus fluorescence microscope.
Chromatin Immunoprecipitation (ChIP)
The ChIP experiments were performed as described (11) with minor modifications. 1.5 g of 12-day-old Arabidopsis seedlings were fixed with 37 ml of 1% formaldehyde solution for 10 min until seedlings turn translucent for cross-linking proteins to DNA. The reaction was stopped by the addition of 2.5 ml of 2 m glycine. The chromatin was isolated and fragmented by sonication followed by immunoprecipitation with HA antibody (Santa Cruz Biotechnology). The chromatin solution was precleared with protein-agarose beads with sheared salmon sperm DNA. Co-precipitated DNA was amplified using Ex-Tag polymerase (Takara, Japan) and the primers listed in supplemental Table S2.
RESULTS
Identification of Proteins Interacting with the RPS6 C Terminus
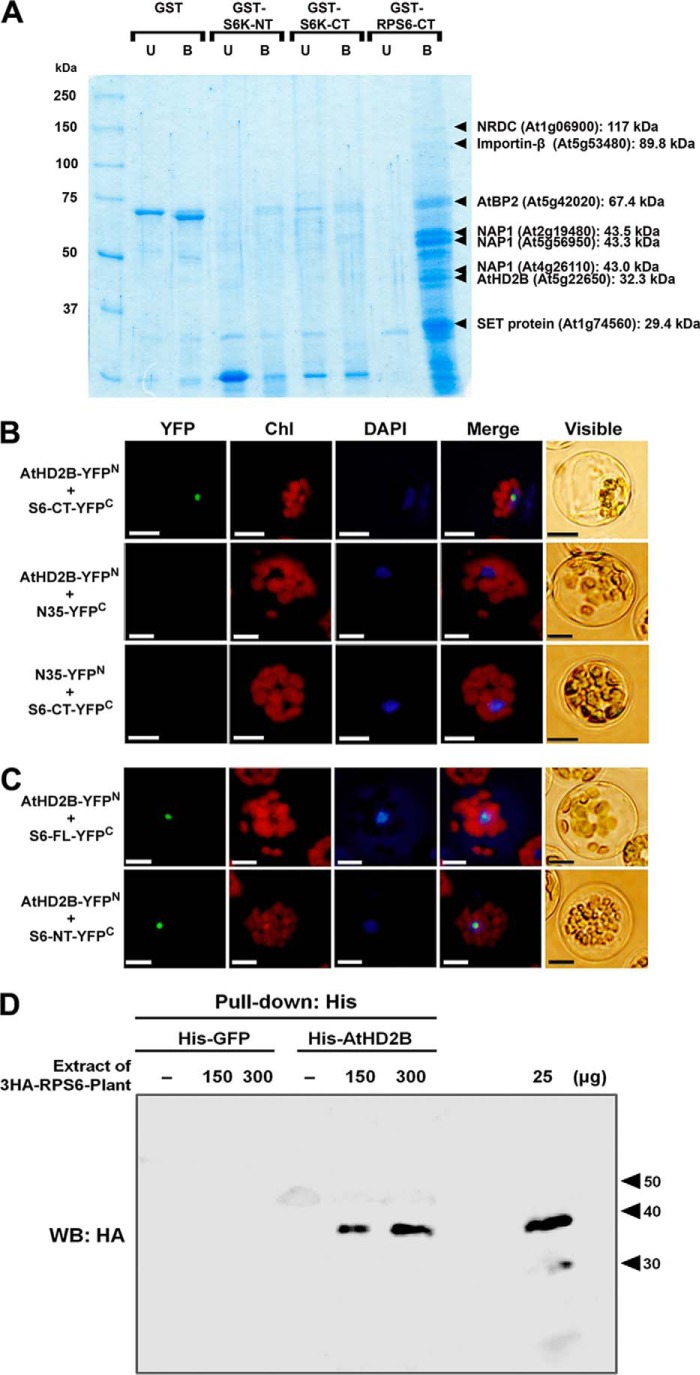
To isolate potential candidates for the interacting partners of RPS6, total soluble proteins extracted from Arabidopsis suspension cells were applied to an affinity column containing GST-fused RPS6 protein (used as an affinity bait). To reduce nonspecific interaction and focus on the candidates that are relevant to the TOR signaling, only the C-terminal 100 amino acids of the RPS6 (RPS6-CT), which includes the region of the putative phosphorylation sites by S6 kinase (19), was used as a bait. The GST-fused AtS6K1 N-terminal fragment (GST-S6K-NT) and the C-terminal fragment (GST-S6K-CT) were also prepared and used as controls together with the full-length of GST protein, in affinity purification (supplemental Fig. S1). The experiment was repeated several times, and in each case, about 100–300 mg of total soluble proteins extracted from actively growing Arabidopsis suspension culture cells were applied to the columns containing glutathione-agarose resin to which GST fusion proteins were bound by affinity interaction. After rigorous washing, the affinity-bound proteins were released by in-column thrombin digestion. Following size-exclusion fractionation to remove the GST fusion proteins co-eluted during thrombin digestion, the proteins were visualized by SDS-PAGE (Fig. 1A) and two-dimensional gel electrophoresis (data not shown). In several repeated experiments, GST fusion protein of RPS6-CT consistently pulled down a number of specifically interacting proteins (indicated by arrowheads in Fig. 1A) that were subjected to identification through both MALDI-TOF and LC-MS spectrometry analyses.
FIGURE 1.
In vivo interaction of AtHD2B and RPS6. A, protein profiles after thrombin elution of the pulldown products. The cellular proteins bound with GST fusion protein substrates were released by thrombin digestion and run on SDS-polyacrylamide gels. For U lanes, the substrate resin was not incubated with the cellular proteins; for B lanes, substrate pulldown resins were incubated with the cellular proteins. Protein bands potentially representing the product of specific interaction with RPS6 were marked with arrowheads and were further identified. B, BiFC analyses of interaction between RPS6-CT and AtHD2B as follows: co-expression of P35S-AtHD2B-YFPN and 35S-S6-CT-YFPC (top panels); co-expression of P35S-AtHD2B-YFPN and P35S-N35-YFPC (middle panels); co-expression of P35S-N35-YFPN and P35S-S6-CT-YFPC (bottom panels). C, BiFC analyses of interaction between RPS6-NT and AtHD2B as follows: co-expression of P35S-AtHD2B-YFPN and P35S-S6-FL-YFPC (top panels); co-expression of P35S-AtHD2B-YFPN and P35S-S6-NT-YFPC (bottom panels). Transfected protoplasts were stained with DAPI to visualize the nucleus. These experiments were replicated three times with similar results. Bar, 10 μm. D, interaction between AtHD2B and RPS6 by a pulldown assay with His tag followed by a Western blot with anti-3HA antiserum. Protein extracts from plants expressing 3HA-RPS6 were incubated with different amounts (150 and 300 μg) of His-GFP or His-AtHD2B fusion proteins, respectively. WB, Western blot.
The identities of the proteins (and their corresponding gene accession numbers in parentheses) are shown in Fig. 1A. All of the identified proteins, with the exception of protease subtilisin homologue, have been functionally implicated in the chromatin-related activities, including the nucleosome-mediated regulation of gene expression. Of particular interest among these was the presence of a histone deacetylase, AtHD2B, a paralogue of which (AtHD2A) had been implicated in silencing of rDNA transcription in Arabidopsis (25). Because the physical interaction of AtHD2B with RPS6 presents an interesting perspective with regard to the role of RPS6 phosphorylation in the activation of translation and possibly ribosome biogenesis, we focused on uncovering the functional significance of the RPS6-AtHD2B interaction in vivo.
In Vivo Interaction of AtHD2B with RPS6
AtHD2A belongs to the plant-specific HD2 family of histone deacetylases that have been suggested to be involved in rDNA gene silencing and nucleolar dominance (15, 25). This means that AtHD2B might also be involved in the regulation of rDNA transcription. To confirm the physical interaction between the RPS6-CT and AtHD2B in vivo, we tested the formation of this complex using BiFC analysis using Arabidopsis protoplasts. The C-terminal regions of AtHD2B and RPS6 were fused to the complementary N- and C-terminal fragments of YFP (YFPN and YFPC), respectively. Soybean nodulin-35 (N35) (26) fused to either the N- or the C-terminal fragment of YFP was used as a negative control. Whereas no fluorescence was observed in protoplasts expressing pairs of N35 + AtHD2B or N35 + RPS6 proteins (Fig. 1B, middle and bottom panels), protoplasts transfected with AtHD2B-YFPN and RPS6-CT-YFPC constructs showed fluorescence signal that was concentrated into a smaller region within the nucleus, resembling the nucleolus (Fig. 1B, middle and bottom panels). These data suggest that AtHD2B specifically forms a complex with RPS6 in the nucleolar region of plant cells, and such a complex may be involved in the transcription of rDNA or processing of rRNAs.
To determine whether the interaction between the RPS6 and AtHD2B only occurs through the C-terminal region of the RPS6, the N-terminal fragment of RPS6 (RPS6-NT) that was excluded in the original GST pulldown experiment, by which AtHD2B was identified as an interacting partner, was also tested for interaction using the BiFC assay. Because fluorescence in protoplasts co-expressing a pair of AtHD2B-YFPN and RPS6-NT-YFPC was also detected in the nucleolus (Fig. 1C, bottom panels), these results suggest that more than one region of RPS6 might be associated with AtHD2B, forming a complex. To confirm that AtHD2B and AtRPS6 interact with each other, we performed an in vitro pulldown assay using His-AtHD2B-fused and RPS6-GST-fused proteins isolated from Escherichia coli. However, no interaction using proteins from E. coli was observed. Subsequently, transgenic plants expressing HA-tagged RPS6 driven by the CaMV-35S promoter was used for pulldown assay to test the possibility that AtHD2B may interact indirectly with RPS6 forming a complex. Proteins from the 3HA-RPS6-expressing plants were incubated with His-AtHD2B, which was used as a bait, and HA-RPS6 was found to interact with His-AtHD2B (Fig. 1D). These results suggest that AtHD2B and RPS6 may associate indirectly with each other, forming a multiprotein complex.
Interaction of RPS6 and AtHD2B Facilitates Their Nucleolar Localization
In contrast to the results obtained from the BiFC analyses described above, we observed that both GFP-tagged AtHD2B and RPS6 were localized in the nucleus when they were overexpressed alone in protoplasts (Fig. 2A, top two panels). The possibility of interaction-dependent nucleolar translocation of these proteins was also tested by co-expressing them in Arabidopsis protoplasts. Indeed, co-expression of RPS6-GFP with AtHD2B caused a change in localization of RPS6 to one or several putative nucleolar spots (Fig. 2A, bottom two panels). Although both the N-terminal (RPS6-NT) and the C-terminal (RPS6-CT) fragments of RPS6 were shown to interact with AtHD2B in the BiFC analyses, this nucleolar translocation was not observed when the GFP fusion construct of RPS6-NT was co-expressed with AtHD2B (Fig. 2B, bottom two panels). The interaction-dependent nucleolar translocation of AtHD2B was also confirmed by reciprocally monitoring the GFP fluorescence after overexpressing RPS6 in protoplasts prepared from transgenic Arabidopsis expressing GFP-AtHD2B (data not shown). These results indicate that formation of the RPS6-AtHD2B protein complex is critical for these proteins to be targeted to the nucleolus and also suggest that the protein motif present in the RPS6-CT may provide a tighter interaction with AtHD2B.
FIGURE 2.
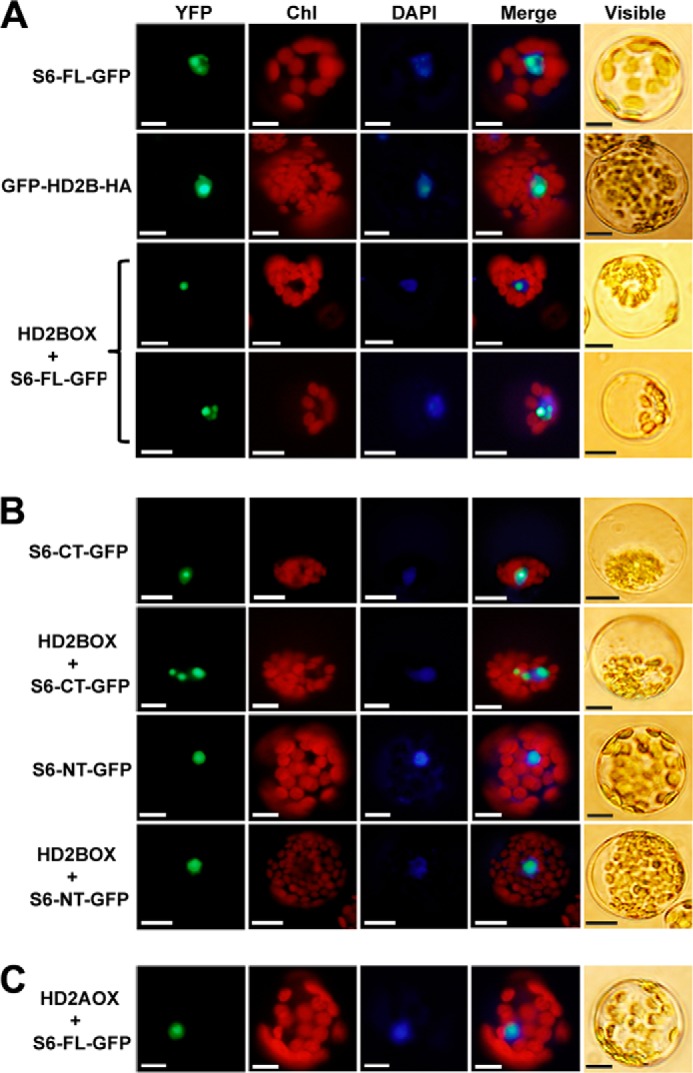
Interaction-dependent localization of AtHD2B and RPS6. A, subcellular localization of full-length RPS6 and AtHD2B. In the top panels, protoplasts were transfected with full-length RPS6 fused to GFP (S6FL-GFP). The 2nd panels show protoplasts isolated from transgenic plants with GFP-AtHD2B fusion (GFP-HD2B-HA). In the two bottom panels, transgenic protoplasts with S6FL-GFP were transfected with P35S-AtHD2B-NOS. B, subcellular localization of C- and N-terminal fragments of RPS6 and AtHD2B. Protoplasts were transfected with truncated RPS6 fused to GFP (S6CT-GFP or S6NT-GFP) or co-transfected with truncated RPS6 and AtHD2B-expressing plasmids (S6CT-GFP or S6NT-GFP and HD2BOX). C, subcellular localization of RPS6 in AtHD2A-expressing protoplasts. Protoplasts were co-transfected with RPS6 and AtHD2A-expressing plasmids (S6FL-GFP and HD2AOX). Transfected protoplasts were stained with DAPI to visualize the nucleus. These experiments were replicated three times with similar results. Bar, 10 μm.
The Arabidopsis genome contains four isoforms of the HDT-type histone deacetylases, namely AtHD2A, AtHD2B, AtHD2C, and AtHD2D, among which AtHD2A and AtHD2B share the highest sequence homology and the apparent functional similarities (27–29). To test the specificity of interaction between RPS6 and the AtHD2B, GFP fusion construct of a full-length RPS6 (S6FL-GFP) was co-expressed with AtHD2A in protoplasts (Fig. 2C). In this case, localization of the GFP fluorescence was not altered by co-expression of AtHD2A, supporting the idea that RPS6 may play a role as a specific functional component of the AtHD2B silencing complex, in regulating the synthesis and/or processing of rRNAs in plant nucleolus.
Effect of RPS6/AtHD2B Overexpression on rDNA Transcription
Several plant histone deacetylases have been reported to be involved in the silencing of rDNA genes (25, 30). We tested the possible effect of the RPS6-AtHD2B complex on rDNA gene transcription by examining pre-rRNA transcription levels in AtHD2B- or RPS6-expressing protoplasts. Arabidopsis pre-rRNA transcript contains the 18 S, 5.8 S, and 25 S rRNAs as well as the 5′ and 3′ external transcript spacer (ETS) and two internal transcript spacers between the three rRNAs (ITS1 and ITS2). Mature 18 S, 5.8 S, and 25 S rRNAs are generated after processing pre-rRNA (5, 6). Total RNA was isolated from the protoplasts transfected with AtHD2A-, AtHD2B-, or RPS6-overexpressing constructs, and DNA contamination was removed with DNase I digestion. Real time RT-PCR was performed with pre-18 S rRNA forward primer (position in 5′-ETS) and pre-18 S rRNA reverse primer (position in 18 S ribosomal RNA). AtHD2A-expressing protoplasts were used as a positive control for the silencing effect, as it has been reported that AtHD2A is associated with the silencing of rRNA genes (25).
These results directly demonstrated that an increase in the AtHD2A expression caused a decreased pre-rRNA transcript level (Fig. 3A). Co-expression of both AtHD2A and RPS6 in the protoplasts resulted in a more dramatic suppression of the pre-18 S rRNA transcript level (Fig. 3A), raising the possibility of a functional association of RPS6 with AtHD2A in the regulation of pre-rRNA transcription. However, as shown in Fig. 2, RPS6 might be specifically associated with AtHD2B, but not AtHD2A. Thus, the elevated inhibition of pre-18 S rRNA transcription observed in RPS6/AtHD2A-overexpressing protoplasts is likely to be caused by an independent synergistic effect of AtHD2A and RPS6 on rDNA transcription. Similar to the inhibitory effect observed from AtHD2A overexpression, protoplasts overexpressing AtHD2B, RPS6, or both AtHD2B and RPS6 constructs all exhibited down-regulation of pre-18 S rRNA synthesis with a concomitant decrease in some of the ribosomal protein transcriptions (Fig. 3B). This indicates that both AtHD2B and RPS6 are negatively involved in regulating rDNA transcription, probably as a single functional entity-protein complex. Co-expression of both AtHD2B and RPS6 did not result in an additional decrease in transcript levels of pre-18 S rRNA compared with those in AtHD2B- or RPS6-expressing protoplasts.
FIGURE 3.
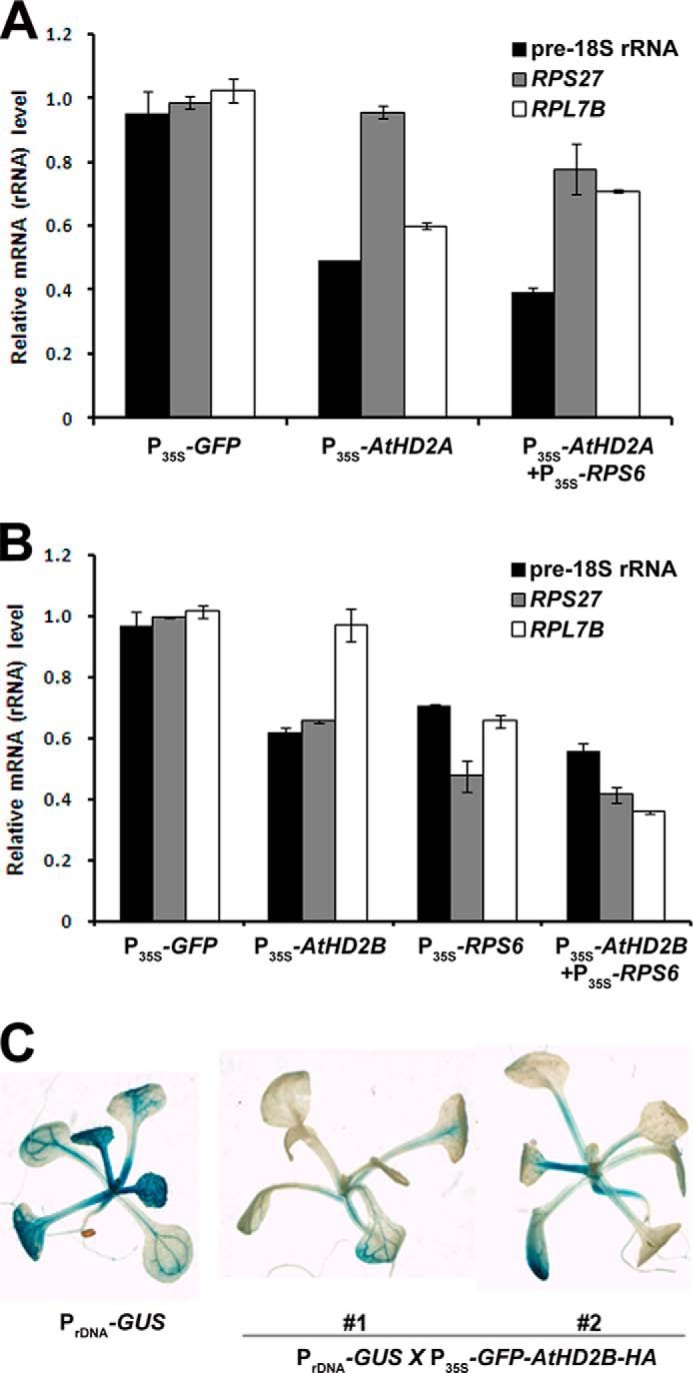
rDNA transcription regulated by AtHD2s and RPS6. A, transcript levels of rDNA and a few ribosomal protein genes in AtHD2A-expressing Arabidopsis protoplasts. Protoplasts were transfected with control plasmid (P35S-GFP), AtHD2A-expressing plasmid (P35S-AtHD2A), or both AtHD2A- and RPS6-expressing plasmids (P35S-AtHD2A and P35S-RPS6). Error bars represent standard deviations (n = 3). B, transcript levels of rDNA and a few ribosomal protein genes in AtHD2B-expressing Arabidopsis protoplasts. Protoplasts were transfected with control plasmid (P35S-GFP), AtHD2B-expressing plasmid (P35S-AtHD2B), RPS6-expressing plasmid (P35S-RPS6), or both AtHD2B- and RPS6-expressing plasmids (P35S-AtHD2B and P35S-RPS6).RNAs were extracted from the transfected protoplast, and then real time RT-PCRs were performed. Expression level of ACTIN2 was used as an internal control. Error bars represent standard deviations (n = 3). C, GUS expression in transgenic plants with PrDNA-GUS and in F2 progeny of a cross between PrDNA-GUS transgenic plants and P35S-AtHD2B-expressing plants.
Transcription of an mRNA gene driven by rDNA promoter has been successfully demonstrated previously in frog oocyte (31). To establish a convenient assay system for the transcriptional activity of rDNA in plants, we made transgenic plants expressing the PrDNA-GUS construct, which contains a 500-bp fragment, including the minimal Arabidopsis rDNA promoter region (32) fused with the GUS reporter. Transcription of this chimeric construct was confirmed to produce mRNA of GUS coding sequences with a poly(A) tail (supplemental Fig. S2C). To test if GUS expression driven by PrDNA-GUS is correlated to an endogenous rRNA transcription level, we made a comparison between GUS expression in PrDNA-GUS transgenic plants and the transcription level of endogenous pre-18 S rRNA after the treatment with several plant hormones and salt stress. The endogenous pre-18 S rRNA level was about two times higher after auxin, cytokinin, or both auxin and cytokinin treatments, but it was significantly lower with abscisic acid or NaCl treatments (supplemental Fig. S2). GUS expression in PrDNA-GUS transgenic plants was also increased in the presence of auxin or cytokinin but was decreased by abscisic acid. This means that PrDNA-GUS transgenic plants can be used to monitor transcription of rRNA and related genes. To further confirm whether rDNA transcription is negatively regulated by AtHD2B in Arabidopsis utilizing this system, PrDNA-GUS transgenic plants were crossed with transgenic plants containing GFP-tagged AtHD2B construct. Consistent with the results in AtHD2B-overexpressing protoplasts, the GUS expression level of PrDNA-GUS construct was significantly decreased in GFP-HD2B-HA-containing F2 transgenic plants compared with that of wild type (Fig. 3C). These results further support the negative role of the AtHD2B/RPS6 interaction in the regulation of rDNA transcription.
Effect of RPS6 Expression on rDNA Transcription and Root Growth
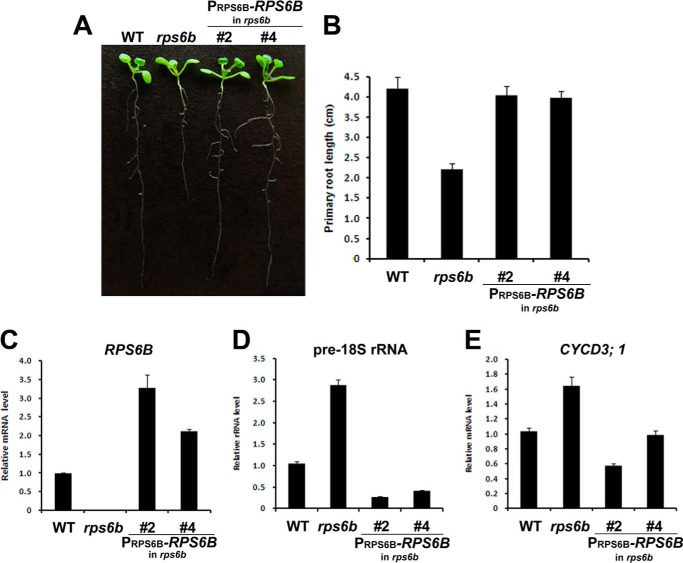
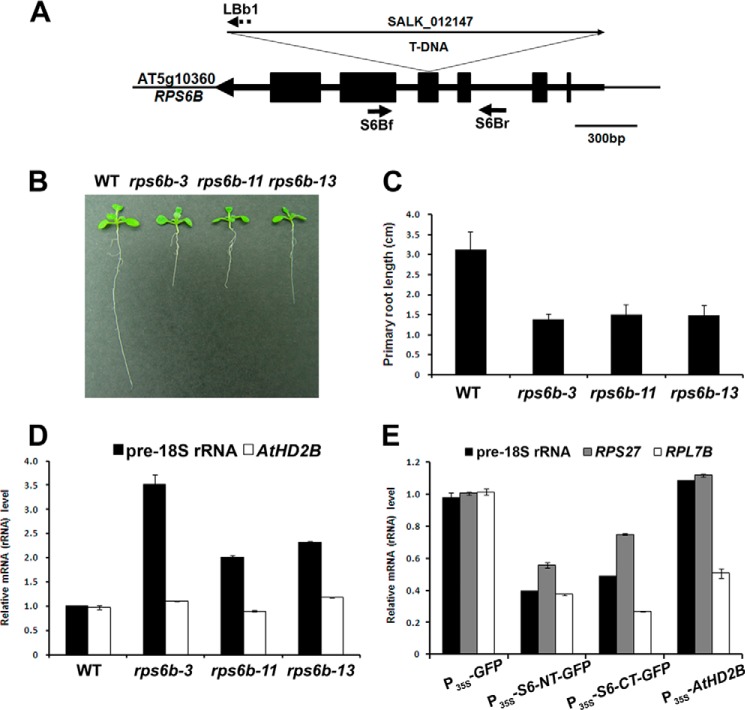
There are two copies of nearly identical ribosomal protein S6 genes, RPS6A (AT4g31700) and RPS6B (AT5g10360), in Arabidopsis (33). The deduced amino acid sequences of RPS6A and RPS6B predict 251 and 250 amino acids, respectively, with 95% sequence identity between them at the amino acid level, which strongly suggest the possible interaction of AtHD2B with RPS6A as well. To evaluate the function of the RPS6 gene in Arabidopsis development, we used a T-DNA insertion mutant having the decreased expression of RPS6B. The T-DNA insertion line was obtained from ABRC, and the position of the T-DNA insertion in this line was confirmed by genomic PCR and RT-PCR (Fig. 5C and data not shown). Based on genomic PCR and sequencing of PCR products, the rps6b mutant line was found to have T-DNA inserted in exon 4 of the RPS6B gene and was confirmed to be a homozygous line (Fig. 4A). RT-PCR analysis revealed that the expression level of RPS6B was dramatically reduced in three rps6b mutant lines compared with the wild-type Columbia-0 (data not shown). Although no apparent abnormality was observed in leaves and shoot apices of rps6b mutant seedlings, significant differences in root length were found in 14-day-old seedlings (Fig. 4B). The average root length of rps6b seedlings was about 50% shorter than that of WT (Fig. 4C). These results suggest that down-regulation of RPS6B significantly inhibits root growth apparently affecting ribosome biogenesis.
FIGURE 5.
Transgenic complementation of the rps6b mutant. The rps6b plants were transformed with PRPS6B-RPS6B, a complementation construct, to check if the mutant phenotype was caused by defective expression of RPS6B. A, root length of wild type (WT), rps6b mutant, and transformants with PRPS6B-RPS6B. B, comparison of primary root lengths of wild type, rps6b mutant, transformants with PRPS6B-RPS6B shown in A. Error bars represent standard deviations (n ≥ 10). C–E, expression of RPS6B (C), pre-18 S rRNA (D), and CYCD3;1 (E) in seedlings of wild type (WT), rps6b mutant, transformants with PRPS6B-RPS6B, respectively. The results shown are representative of more than three independent experiments. Data shown represent the mean ± S.D. (n > 3).
FIGURE 4.
Root development in rps6b mutant. A, scheme of the T-DNA insertion in the RPS6B gene. Exons are indicated by filled boxes. Gene-specific primers (S6Bf and S6Br) and a T-DNA border primer (LBb1) are indicated by arrows and a dotted arrow, respectively. B, root growth in 14-day-old seedlings of wild type and those of rps6b mutant lines. C, quantitative comparison of primary root lengths of wild type and rps6b mutants shown in B. Error bars represent standard deviations (n ≥ 10). D, expression of pre-18 S rRNA and AtHD2B in wild-type and rps6b seedlings examined by real time RT-PCR. The results shown are representative of more than three independent experiments. Data shown represent the mean ± S.D. (n > 3). E, expression of pre-18 S rRNA, RPS27, and RPL7B in Arabidopsis protoplasts with RPS6-expressing plasmids examined by real time RT-PCR. Arabidopsis protoplasts were isolated from rps6b mutant plants and then transfected with control plasmid (P35S-GFP), RPS6-expressing plasmids (P35S-S6NT-GFP and P35S-S6CT-GFP), or AtHD2B-expressing plasmid (P35S-AtHD2B). The results shown are representative of more than three independent experiments. Data shown represent the mean ± S.D. (n > 3).
To determine whether the lack of expression of RPS6B was directly responsible for the rps6b mutant phenotype, the rps6b mutant was transformed with a full-length RPS6B cDNA under the control of native promoter (PRPS6B-RPS6B). The full-length RPS6B cDNA rescued the root length to wild type in T2 generation plants carrying PRPS6B-RPS6B construct in a T-DNA insertion mutant of rps6b (Fig. 5, A and B), indicating that a functional copy of RPS6B can complement the observed rps6b phenotype.
A possible mechanism by which RPS6 could control rRNA gene transcription was explored by measuring the expression level of pre-18 S rRNAs in rps6b mutants. Increased transcript level of pre-18 S rRNA was observed in three rps6b mutants (Fig. 4D), providing the evidence that RPS6 participates in rRNA biogenesis in plants. However, the expression level of AtHD2B was not affected in the rps6b mutant indicating that RPS6B did not have any effect on AtHD2B transcription. Consistent with the phenotypic complementation in the rps6b mutant background, the full-length RPS6B cDNA restored the transcript level of pre-18 S rRNA (Fig. 5D).
To test if the increased level of pre-18 S rRNA in the rps6b mutant resulted from down-regulation of RPS6B, protoplasts isolated from rps6b mutant were transfected with the RPS6 N-terminal region or RPS6 C-terminal region-expressing plasmids. As expected, RPS6-NT or RPS6-CT overexpression resulted in decreased pre-18 S rRNA and some ribosomal protein gene expressions as compared with those in the rps6b mutant. However, in the rps6b mutant, AtHD2B overexpression did not result in decreased rRNA transcript levels (Fig. 4E), which suggests that functional RPS6 may be necessary for exerting the negative effect of AtHD2B on rDNA transcription. The expression levels of pre-18 S rRNA and CYCD3.1, both of which are known to be induced by cytokinin, were found to be increased in the rps6b mutant (supplemental Fig. S3). This is consistent with the above phenotypic complementation by RPS6B, which also reduced the transcript levels of pre-18 S rRNA and CYCD3:1 (Fig. 5, D and E).
Binding of RPS6 to rDNA Promoter
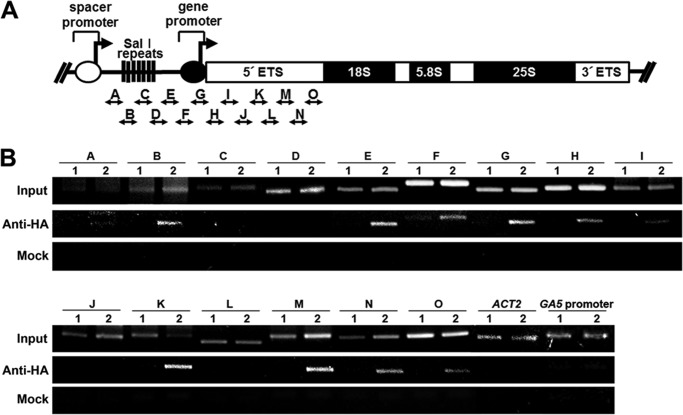
To determine whether the down-regulation of rDNA transcription by the RPS6-AtHD2B complex is mediated through a physical interaction of the complex with the rDNA promoter, thereby regulating rDNA transcription (9–11), chromatin immunoprecipitation (ChIP) was performed using chromatin from transgenic plants expressing HA-tagged RPS6B. Eighteen primer pairs covering the 45 S rRNA promoter and 5′-ETS regions (32, 34) were used in amplifying chromatin fragments after isolating DNA associated with RPS6 by immunoprecipitation with anti-HA antibody (Fig. 6A). Regions B, E–I, K, and M–O were significantly amplified using chromatin fragments extracted from 3HA-RPS6-expressing transgenic plants, whereas no specific band was observed using those from wild-type control (Fig. 6B, middle section). A mock experiment without adding anti-HA antibody in ChIP reaction also did not produce any specific band (Fig. 6B, bottom section). In addition, nonspecific binding of RPS6 to an actin gene (ACT2) or GA5 promoters was not observed, ensuring specificity of the RPS6 binding to the rDNA promoter. These results suggest that RPS6 is involved in the control of rDNA transcription by binding to the promoter and 5′-ETS of the ribosomal RNA gene in Arabidopsis. The binding of RPS6 with the rDNA promoter and specific interaction of RPS6 with HDA2B may bring HD2B in close contact with the rDNA chromatin affecting the acetylation state of histones and thus controlling transcriptional activation of these genes.
FIGURE 6.
Interaction of RPS6 with rDNA promoter and its 5′-ETS. A, structure of ribosomal RNA genes, including an intergenic spacer, the gene promoter, the ETS, and the coding region for the 18 S, 5.8 S, and 25 S rRNA. DNA fragments used in chromatin precipitation (ChIP) (fragments A–O) are shown. B, ChIP with anti-HA antiserum followed by PCRs. Input is chromatin before immunoprecipitation. Mock is ChIP product with no antibody. Lane 1, ChIP product using chromatin of wild type. Lane 2, ChIP product using chromatin of transgenic plants with P35S-3HA-RPS6.
DISCUSSION
Regulation of rRNA Genes via Histone Deacetylases
Multiple mechanisms have been implicated in the regulation of rRNA gene transcription (1, 4). Based on the result of this study, we suggest that histone deacetylase AtHD2B is involved in the regulation of rDNA transcription via the interaction of its promoter with the RPS6B protein. AtHD2B was identified as one of the interacting proteins using a pulldown assay (Fig. 1 and supplemental Fig. S1).
Maintenance of nucleolar dominance, a phenomenon in hybrids or allopolyploids, requires that only rDNA genes inherited from one parent are transcribed (34–37). Histone deacetylases and methylases control rDNA genes and thus regulate the epigenetic on/off switch (gene dosage), providing an important mechanism in the biogenesis of ribosomes (25, 27). It has been demonstrated that the activity of the yeast RPD3 is required for nucleolar reorganization and RNA pol I delocalization via its association with the rDNA chromatin, which can be antagonized by TOR (13). In Arabidopsis, two histone deacetylases, HD2A and HDA6 that are localized to the nucleolus, have been reported to directly interact with the rDNA promoter to repress rRNA transcription via epigenetic alteration of rDNA chromatin acetylation on histone H3, and knockdown of these histone deacetylases results in increased transcription of the rDNA genes (25, 30).
Our results identified AtHD2B as a new regulator of the rDNA transcription, in addition to the aforementioned two histone deacetylases. Of particular interest in this finding is that the regulatory activity of AtHD2B involves interaction with RPS6, a component of the 40 S ribosome and a substrate of the S6 kinase, AtS6K (Figs. 1–3). This observation was further substantiated by our ChIP results as shown in Fig. 6, which demonstrated specific binding of the RPS6 to the Arabidopsis rDNA promoter region and thereby suggested a possible role of RPS6 as a component of the repressor complex for the rDNA transcription. It is likely that the down-regulation of pre-18 S rRNA synthesis observed in RPS6-overexpressing protoplasts (Fig. 3) as well as in rps6b mutant plants complemented with transgene expression of RPS6b (Fig. 4), is a result of the RPS6 binding to the rDNA promoter, thereby recruiting AtHD2B to the proximity and thus repressing rDNA transcription. The ChIP assay data showed the binding footprints of RPS6 over multiple regions of the rDNA promoter at regular intervals, which is consistent with the proposed role of the RPS6-AtHD2B complex in chromatin modification. In this regard, determining how TOR is involved in this process would be a key to fully understand the mechanisms by which the TOR pathway controls rDNA transcription and ribosome biogenesis.
It is possible that a multimeric protein complex containing RPS6 recruits AtHD2B to rDNA promoter to repress rRNA transcription. Among the proteins identified as interacting proteins (Fig. 1A), it seems that nucleosome assembly protein 1 (NAP1) is a strong candidate for acting as a bridging factor between the RPS6 and AtHD2B. This is consistent with the fact that several isoforms of NAP1 were isolated as interacting proteins with RPS6 (Fig. 1A). In addition, NAP1 has been shown to serve as a bridge between Kap114p and histone H2A and H2B in mediating chromatin assembly (38). Interestingly, the HD2B protein, which is about 39 kDa, was originally identified in maize as a nucleolar targeted phosphoprotein and was purified as an enzyme with a native molecular mass of about 400 kDa (39), suggesting the nature of the RPS6-AtHD2B protein complex as a massive multiprotein complex. Thus, our working model for the RPS6-AtHD2B complex in the control of rDNA transcription illustrated in Fig. 7 portrays NAP1 acting as an adaptor protein bringing the RPS6 and AtHD2B together.
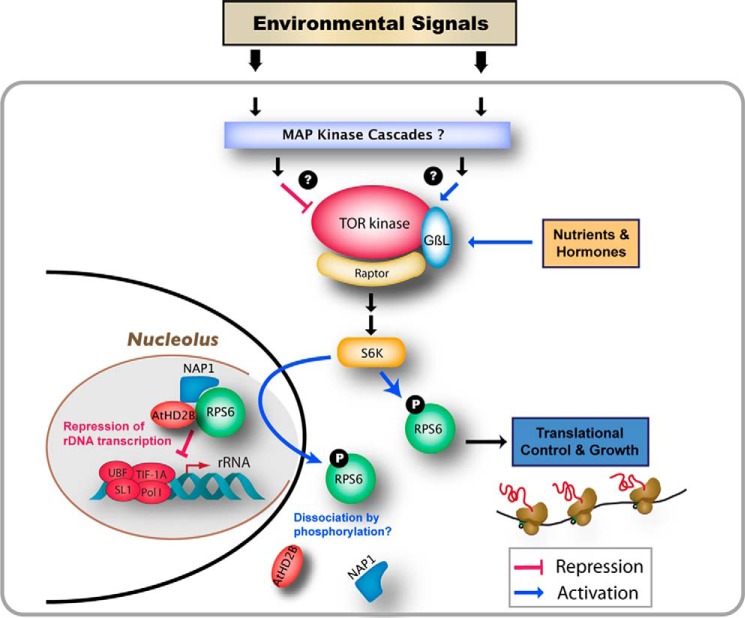
FIGURE 7.
Model for plant growth control by TOR complex via the interaction between AtHD2B and RPS6.
If TOR also binds to rDNA and protein phosphatase 2A (PP2A) is a part of the TOR complex (40), it may regulate the phosphorylation state of RPS6 under stress conditions. We have shown that one of the AtS6Ks is localized in the nucleolus (17), and RPS6 phosphorylation in the nucleolus occurs during active ribosome biogenesis. The phosphorylation of RPS6 in the cytoplasm promotes translation (41), and this phosphorylation state is directly linked to stress signals (17, 42). Phosphorylation of RPS6 by AtS6K may cause dissociation of this complex, directly linking rRNA biogenesis to the TOR signaling pathway. This is consistent with the negative effect of AtHD2B on the regulation of rDNA transcription that was abolished in the rps6b mutant, and it also offers a possible explanation on the correlations between dephosphorylation of RPS6 and repression of rDNA transcription observed in our previous work (17) and this study, suggesting that AtHD2B regulates rRNA transcription levels through the activity of RPS6, which may be dependent on its phosphorylation state.
Similar to yeast and animals (9, 10), binding of the Arabidopsis TOR to the rDNA promoter has recently been reported (11). However, our ChIP data suggest that the region of the rDNA promoter to which RPS6 binds is largely different from the binding sites for Arabidopsis TOR reported by Ren et al. (11). In contrast to the fact that the activity of TOR has been positively implicated in transcriptional regulation of rDNA in the study of Ren et al. (11), as well as in those of yeast and animals (9, 10), our results show an RPS6-dependent mechanism in transcriptional repression of the rDNA. However, it is possible that the RPS6 exists as a component of the nucleolar TOR complex and plays a negative regulator for the rDNA transcription, antagonizing the positive effect provided by the TOR kinase.
Extraribosomal Function of RPS6 Links Stress Signals to Ribosome Biogenesis
It has been recognized that many ribosomal proteins have additional functions besides their role as components of the ribosome, namely the extraribosomal functions, including mRNA processing and translational control of their own genes (42, 43). In animal cells, RPS6 has also been implicated in the regulation of protein synthesis through association with 5′-TOP tract mRNA, such as RPL11 and RPS16 mRNAs, suggesting that RPS6 has a role of a negative regulator in the translation of 5′-TOP mRNAs (21). With the apparent lack of the 5′-TOP motif in plant mRNA structures, it is not certain whether the same mode of translational control by RPS6 is conserved in plants. Our findings revealed a novel extraribosomal function of RPS6 in regulating rRNA transcription via interaction with HD2B. Because the levels of rRNA transcripts and the amount of ribosomal proteins for ribosome assembly are tightly regulated according to the need for protein synthesis in eukaryotic cells, the mechanism proposed here may allow shutting down proliferative activities of cells to minimize wasting cellular resources in response to the environmental signals.
The data presented here provide evidence for the involvement of RPS6 in possible control of plant rRNA gene expression via its interaction with AtHD2B, although given that the catalytic activity of AtHD2B has not been verified unlike that of AtHD2A (25), it cannot be ruled out that the repression of rDNA by AtHD2B could be achieved through a mechanism not involving the chromatin modification. RPS6 has been identified as the first protein to be phosphorylated in response to various signals (44, 45). The presence of S6K in the nucleolus and demonstration that TOR phosphorylates RPS6 via S6K, which is sensitive to osmotic stress (17), and the direct binding of TOR to rDNA promoter (11), may control optimum transcription of rRNA according to the environmental conditions and gene dosage. Such a mechanism would allow environmental signals to be transduced via TOR to control not only translation but also transcription (particularly of rRNA) for optimum growth of the plant under a given environment (Fig. 7). AtHD2B may play a critical role in linking the translation and transcription machineries to the transduction of such signals. Further studies may resolve the exact roles of AtHD2B, S6K, and TOR in this process.
This work was supported by National Science Foundation Grant 0726284 (to D. P. S. V.), National Research Foundation of Korea Grant 2011-00300074 funded by the Korean Government (to Sookmyung MRC Center and C. I. C.), and National Research Foundation of Korea Grant 2010-A0604-C00031 (to S. K.).

This article contains supplemental Figs. S1–S3 and Table S1 and S2.
- pre-rRNA
- precursor rRNA
- pol
- polymerase
- TOR
- target of rapamycin
- BiFC
- bimolecular fluorescence complementation
- ETS
- external transcribed spacer
- 5′-TOP
- 5′-terminal oligopyrimidine tract
- N35
- nodulin-35.
REFERENCES
- 1. Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. (2007) A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lempiäinen H., Shore D. (2009) Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21, 855–863 [DOI] [PubMed] [Google Scholar]
- 3. Mayer C., Grummt I. (2006) Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25, 6384–6391 [DOI] [PubMed] [Google Scholar]
- 4. Russell J., Zomerdijk J. C. (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004) Plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24, 7284–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi D. Q., Liu J., Xiang Y. H., Ye D., Sundaresan V., Yang W. C. (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18 S ribosomal RNA biogenesis. Plant Cell 17, 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 8. Moss T., Stefanovsky V. Y. (2002) At the center of eukaryotic life. Cell 109, 545–548 [DOI] [PubMed] [Google Scholar]
- 9. Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F. (2006) Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 10. Tsang C. K., Liu H., Zheng X. F. (2010) mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle 9, 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ren M., Qiu S., Venglat P., Xiang D., Feng L., Selvaraj G., Datla R. (2011) Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155, 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger A. B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O. (2007) Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell. Biol. 27, 8015–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsang C. K., Bertram P. G., Ai W., Drenan R., Zheng X. F. (2003) Chromatin-mediated regulation of nucleolar structure and RNA pol I localization by TOR. EMBO J. 22, 6045–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha C. W., Huh W. K. (2011) Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 39, 1336–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bártová E., Horáková A. H., Uhlírová R., Raska I., Galiová G., Orlova D., Kozubek S. (2010) Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J. Histochem. Cytochem. 58, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hay N., Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 17. Mahfouz M. M., Kim S., Delauney A. J., Verma D. P. (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turck F., Zilbermann F., Kozma S. C., Thomas G., Nagy F. (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 134, 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. (2005) Ribosomal nucleolar accumulation of ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 19, 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiocchetti A., Zhou J., Zhu H., Karl T., Haubenreisser O., Rinnerthaler M., Heeren G., Oender K., Bauer J., Hintner H., Breitenbach M., Breitenbach-Koller L. (2007) Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 42, 275–286 [DOI] [PubMed] [Google Scholar]
- 21. Hagner P. R., Mazan-Mamczarz K., Dai B., Balzer E. M., Corl S., Martin S. S., Zhao X. F., Gartenhaus R. B. (2011) Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5′ terminal oligopyrimidine tract. Oncogene 30, 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clough S. J., Bent A. F. (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 23. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 24. Yoo S. D., Cho Y. H., Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 25. Lawrence R. J., Earley K., Pontes O., Silva M., Chen Z. J., Neves N., Viegas W., Pikaard C. S. (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13, 599–609 [DOI] [PubMed] [Google Scholar]
- 26. Suzuki H., Verma D. P. (1991) Soybean nodule-specific uricase (Nodulin-35) is expressed and assembled into a functional tetrameric holoenzyme in Escherichia coli. Plant Physiol. 95, 384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu K., Tian L., Zhou C., Brown D., Miki B. (2003) Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34, 241–247 [DOI] [PubMed] [Google Scholar]
- 28. Zhou C., Labbe H., Sridha S., Wang L., Tian L., Latoszek-Green M., Yang Z., Brown D., Miki B., Wu K. (2004) Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 38, 715–724 [DOI] [PubMed] [Google Scholar]
- 29. Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19, 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Earley K., Lawrence R. J., Pontes O., Reuther R., Enciso A. J., Silva M., Neves N., Gross M., Viegas W., Pikaard C. S. (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fleischer S., Grummt I. (1983) Expression of an mRNA coding gene under the control of an RNA polymerase I promoter. EMBO J. 2, 2319–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doelling J. H., Pikaard C. S. (1995) The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J. 8, 683–692 [DOI] [PubMed] [Google Scholar]
- 33. Creff A., Sormani R., Desnos T. (2010) The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol. Biol. 73, 533–546 [DOI] [PubMed] [Google Scholar]
- 34. Tucker S., Vitins A., Pikaard C. S. (2010) Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 22, 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grummt I., Pikaard C. S. (2003) Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4, 641–649 [DOI] [PubMed] [Google Scholar]
- 36. Santoro R., Grummt I. (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 25, 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preuss S., Pikaard C. S. (2007) rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta 1769, 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mosammaparast N., Ewart C. S., Pemberton L. F. (2002) A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lusser A., Brosch G., Loidl A., Haas H., Loidl P. (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91 [DOI] [PubMed] [Google Scholar]
- 40. Pracheil T., Thornton J., Liu Z. (2012) TORC2 signaling is antagonized by protein phosphatase 2A and the Far complex in Saccharomyces cerevisiae. Genetics 190, 1325–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fumagalli S., Thomas G. (2000) Translational Control of Gene Expression (Hershey J. W., Mathews M. B., Sonenberg N., eds) pp. 695–717, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Williams A. J., Werner-Fraczek J., Chang I.-F., Bailey-Serres J. (2003) Regulated phosphorylation of 40 S ribosomal protein S6 in root tips of maize. Plant Physiol. 132, 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue S., Barna M. (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warner J. R., McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franco R., Rosenfeld M. G. (1990) Hormonally inducible phosphorylation of a nuclear pool of ribosomal protein S6. J. Biol. Chem. 265, 4321–4325 [PubMed] [Google Scholar]