Background: Diazaborine is the only known inhibitor of eukaryotic ribosome biogenesis, but its target is unknown.
Results: Diazaborine binds the AAA-ATPase Drg1 and inhibits its ATP hydrolysis, thereby blocking release of Rlp24 from pre-60S particles.
Conclusion: Diazaborine blocks ribosome biogenesis by inhibiting the physiological activity of Drg1.
Significance: Our results highlight the potential of the ribosome biogenesis pathway as target for novel inhibitors.
Keywords: ATPases, Drug Action, Enzyme Inhibitors, Protein-Drug Interactions, Ribosome Assembly, AAA-ATPases, Diazaborine, Small Molecule Inhibitor
Abstract
The drug diazaborine is the only known inhibitor of ribosome biogenesis and specifically blocks large subunit formation in eukaryotic cells. However, the target of this drug and the mechanism of inhibition were unknown. Here we identify the AAA-ATPase Drg1 as a target of diazaborine. Inhibitor binding into the second AAA domain of Drg1 requires ATP loading and results in inhibition of ATP hydrolysis in this site. As a consequence the physiological activity of Drg1, i.e. the release of Rlp24 from pre-60S particles, is blocked, and further progression of cytoplasmic preribosome maturation is prevented. Our results identify the first target of an inhibitor of ribosome biogenesis and provide the mechanism of inhibition of a key step in large ribosomal subunit formation.
Introduction
Ribosomes translate the genetic information into the amino acid sequence of proteins and are composed of one large and one small subunit containing ribosomal RNAs (rRNA) and ribosomal proteins. In eukaryotic cells, the formation of ribosomes involves >200 trans-acting factors (nonribosomal proteins), most of which are essential (for a recent introduction to ribosome biogenesis see Ref. 1). Ribosome biogenesis starts with the transcription of precursors of ribosomal RNA (pre-rRNA) in the nucleolus. Joining of ribosomal and nonribosomal proteins with pre-rRNA results in formation of precursor particles for the large and small subunits. A complex maturation pathway driven by transiently joining nonribosomal proteins leads to export-competent particles that are transported through the nuclear pore complex into the cytoplasm where final maturation steps take place. These include release and recycling of shuttling maturation and export factors and incorporation of late joining ribosomal proteins (for review, see Ref. 2). We and others demonstrated recently that the AAA2 protein Drg1 is a key factor in cytoplasmic pre-60S maturation in yeast (3–6). Drg1 is composed of an N domain and the two ATPase domains D1 and D2 and forms hexamers in the presence of ATP (7–9). The protein is recruited to cytoplasmic pre-60S particles by direct interaction with the shuttling protein Rlp24 (3). The C-terminal domain of Rlp24 then stimulates ATP hydrolysis in both AAA domains of Drg1. ATP hydrolysis in D2 is essential and triggers the release of Rlp24 from pre-60S particles whereas ATP hydrolysis in D1 is required for the subsequent dissociation from Rlp24. The release of Rlp24 by Drg1 represents a critical event in pre-60S maturation and is a prerequisite for downstream maturation steps like the release of other shuttling factors and association of late joining cytoplasmic proteins (3).
We showed previously that the drug diazaborine blocks ribosome biogenesis in yeast (10). Diazaborine is a heterocyclic boron-containing compound with strong antimicrobial activity (11). In Gram-negative bacteria the drug blocks fatty acid biosynthesis by inhibiting the enoyl-ACP reductase FabI (12, 13). In yeast, fatty acid biosynthesis is not affected by diazaborine (9). Instead, the drug specifically interferes with large ribosomal subunit formation (10). Resistance to diazaborine in yeast is mediated by certain allelic forms of DRG1 (8). However, the exact relationship between Drg1 and the drug remained elusive, because the basal ATPase activity of Drg1 was not inhibited by diazaborine in vitro, and direct binding of radiolabeled inhibitor could not be demonstrated using equilibrium dialysis (9).
We show here that diazaborine binds to Drg1 and specifically blocks ATP hydrolysis in the D2 domain, thereby preventing Rlp24 release from pre-60S particles. Thus, our results identify the target and describe the mechanism of action of the first specific inhibitor of the ribosome biogenesis pathway.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
The yeast and bacterial strains used in the present study are listed in Table 1. All plasmids used are listed in Table 2. Chromosomal deletions or gene fusions were generated by homologous recombination using PCR products to transform the respective yeast strain as described (14). Strains were grown either in yeast extract peptone dextrose (YPD) complex medium or for plasmid maintenance, in synthetic dextrose complete medium supplemented with the appropriate amino acids.
TABLE 1.
Yeast and bacterial strains used in this study
| Genotype | Source | |
|---|---|---|
| Yeast strains | ||
| Arx1-TAP | MATα leu2 ura3 his3 trp1 ARX1-TAP::TRP1MX6 | (5) |
| GZYX18 | MATα ura3 his3 leu2 trp1 drg1–18 ARX1-TAP::TRP1MX6 | (5) |
| FWY111 | MATa ura3 leu2 his3 ade2 trp1 afg2–18 | (9) |
| Arx1-TAP drg1-1 | MATα leu2 ura3 his3 trp1 drg1–1 ARX1-TAP::TRP1MX6 | This study |
| W303 | MATa ade2 ura3 his3 leu2 trp1 | S. D. Kohlwein |
| HKY51 | MATa ade2 ura3 his3 leu2 trp1 drg1-1 | This study |
| Arx1-YFP | See W303, ARX1-YFP::HISMX6 | (6) |
| Arx1-YFP drg1-1 | See HKY51, ARX1-YFP::HISMX6 | This study |
| Arx1-YFP drg1-18 | See FWY111, ARX1-YFP::HISMX6 | (6) |
| Nog1-GFP | See W303, NOG1-GFP::HISMX6 | (5) |
| Nog1-GFP drg1-1 | See HKY51, NOG1-GFP::HISMX6 | This study |
| Nog1-GFP drg1-18 | See FWY111, NOG1-GFP::HISMX6 | (6) |
| Tif6-GFP | See W303, TIF6-GFP::HISMX6 | (5) |
| Tif6-GFP drg1-1 | See HKY51, TIF6-GFP::HISMX6 | This study |
| Tif6-GFP drg1-18 | See FWY111, TIF6-GFP::HISMX6 | (6) |
| Bud20-YFP | See W303, BUD20-YFP::HISMX6 | (6) |
| Bud20-YFP drg1-1 | See HKY51, BUD20-YFP::HISMX6 | This study |
| Bud20-YFP drg1-18 | See FWY111, BUD20-YFP::HISMX6 | (6) |
| Nop7-YFP | See W303; NOP7-YFP::HISMX6 | (6) |
| Nop7-YFP drg1-1 | See HKY51, NOP7-YFP::HISMX6 | This study |
| Nop7-YFP drg1-18 | See FWY111, NOP7-YFP::HISMX6 | (6) |
| Rlp24-TAP | MATα ade2 ura3 leu2 trp1 RLP24-TAP::TRP1MX6 | (5) |
| Rlp24-TAP drg1-1 | See Rlp24-TAP, drg1–1 | This study |
| BY4743 Δafg2/AFG2 | MATa/MATα his3/his3 leu2/leu2 met15/MET15 LYS2/lys2 ura3/ura3 YLR397c::kanMX4/YLR397c | Euroscarf |
| GZAFG2 | MAT α, ura3, leu2, his3, lys2, trp1, drg1::kanMX4 [pRS316-Drg1] | This study |
| E. coli strain | ||
| BL21 Codon Plus (DE3)-RIPL | B F− ompT hsdS (rB− mB−) dcm+ Tetr gal λ (DE3) endA Hte [argU ileY leuWproL Camr] | Stratagene |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant marker | Source |
|---|---|---|
| pAZ7 | GST-DRG1 | (9) |
| pAZ8 | GST-drg1-1 | (9) |
| pDRG1-EQ1 | GST-drg1E346Q | (3) |
| pDRG1-EQ2 | GST-drg1E617Q | (3) |
| pDRG1-KA1 | GST-drg1K292A | This study |
| pDRG1-KA2 | GST-drg1K563A | This study |
| pRS315 | CEN, 2μ, LEU2 | (32) |
| pRS315-DRG1 | DRG1 | (3) |
| pRS315-drg1-1 | drg1-1 | This study |
| pRS315-drg1-3 | drg1-3 | This study |
| pRS315-drg1-4 | drg1-4 | This study |
| pRS315-drg1-11 | drg1-11 | This study |
| pGEX6P-1 | ori ampR lacIq PreScission Cleavage site GST tag | GE Healthcare |
| pGEX-Rlp24C | GST-RLP24C | (3) |
| pET32a | ori ampR lacIq Trx tag-S tag HIS6 tag | Novagen |
| pET32-Rlp24C | HIS6-RLP24C (441–599 bp) | (3) |
| pET28 | ori ampR lacIq HIS6 tag | Novagen |
| pET28-N172 | HIS6-NUP116 (1–516 bp) | (3) |
Tandem Affinity Purification (TAP)
Preribosomal particles were purified according to the standard TAP protocol (15, 16) from 4 liters of yeast culture. Strains were treated for 1 h with 100 μg/ml diazaborine 2b18 (11), equivalent to 370 μm. The drug was a kind gift from Gregor Högenauer, a retired professor of the University of Graz. Extracts were prepared in buffer A (20 mm HEPES-NaOH, pH 7.5, 10 mm KCl, 2.5 mm MgCl2, 1 mm EGTA, 1 mm DTT, 0.5 mm PMSF, and Complete® protease inhibitor mixture (Roche Applied Science)) and incubated with 300 μl of settled IgG beads (GE Healthcare) for 90 min at 4 °C. Beads were washed four times with 2 ml of buffer A and once with 2 ml of buffer B (buffer A plus 100 mm NaCl without the addition of protease inhibitors). Elution of the preribosomal particles from the IgG beads was performed by digestion with the tobacco etch virus protease, which cleaves between the protein A tag and the calmodulin binding moiety of the TAP tag. Subsequent purification on calmodulin beads was performed as described (5).
Fluorescence Microscopy
Strains were grown in YPD to early log phase and treated with 370 μm diazaborine for 1 h. Cells were then inspected for GFP/YFP fluorescence with a Zeiss Axioskop fluorescence microscope using a suitable band pass filter for GFP or YFP, respectively. For immune fluorescence microscopy, an Rlp24-TAP-tagged strain was used. After diazaborine treatment, cells were fixed using buffered formaldehyde. After washing, cells were spheroplasted with zymolyase (VWR) in potassium phosphate buffer, pH 7.0, containing 1.2 m sorbitol and 30 mm β-mercaptoethanol. Spheroplasts were spotted on polylysine-treated slides, washed with PBS containing 0.1% Triton X-100, and incubated with BSA to block nonspecific adsorption. As primary antibody a polyclonal anti-protein A antibody (Sigma) and as secondary antibody a goat anti-rabbit-AB conjugated with rhodamine (Amersham Biosciences) were used. Chromatin was stained with DAPI (Invitrogen).
Expression and Purification of Proteins
Drg1 and mutant variants thereof were expressed in yeast cells as described previously (9). Cell disruption was carried out in a bead mill (Merckenschlager) in the presence of 0.6-mm glass beads for 4 min with CO2 cooling. Fusion proteins were expressed in Escherichia coli BL21 codon plus (Table 1). GST-Rlp24C was purified via the GST tag using GSH-agarose beads (Sigma) as described in Ref. 3. His6 versions of Rlp24C and the Nup116 fragment (codons 1–172) were purified by Ni2+ chelating chromatography on nickel-nitrilotriacetic acid columns (Qiagen) as described previously (17). All expression plasmids are listed in Table 2.
Protein-Protein Interaction
Interaction assays of Drg1 and Rlp24C in the absence or presence of 370 μm diazaborine were carried out as described in Ref. 3. The eluates were then analyzed by SDS-PAGE and Coomassie staining.
ATPase Activity Assay
ATPase activity was determined using the Malachite Green phosphate assay from BioAssaySystems through measurement of the formation of free inorganic phosphate (Pi) as described previously (3). If not otherwise stated, a concentration of 1 mm ATP was used. All samples were measured in triplicate of at least two independent experiments. The enzymatic parameters were calculated with GraphPad Prism.
In Vitro Release Assay
Pre-60S particles, stalled at an early cytoplasmic maturation step by incubation of the Arx1-TAP drg1-ts strain at 37 °C for 1 h, were purified from 12 liters of late log phase cultures using the TAP protocol described above. Subsequent release reactions were performed as described previously (3) in the presence of Drg1 variants, His6Nup116 fragment, and ATP. When indicated, 370 μm diazaborine was added, or ATP was replaced by AMP-PNP. After incubation for 45 min at room temperature, the supernatants containing released proteins were collected by centrifugation and analyzed by SDS-PAGE and Western blotting using polyclonal antibodies.
Differential Scanning Fluorometry (DSF)
Differential scanning fluorometry was performed in a Corbett Rotor-Gene 6000 device with a constant fluorescence measurement at each stage (excitation 470 nm/emission 555 nm) based on the method described in Ref. 18. Samples contained 1.8 μm Drg1 and different concentrations of ATP and/or diazaborine in DSF buffer (20 mm HEPES-KOH, pH 7.0, 150 KOAc, 5 mm Mg(OAc)2, 1 mm DTT). SYPRO® Orange (Sigma) was added to a final dilution of 1:1000, and the reaction mixtures were incubated on ice for 30 min. 40-μl samples were subjected in duplicate to thermal unfolding using a gradient from 25 to 99 °C. The temperature was raised 1 °C per stage with a delay of 5 s after each step. The results were analyzed using the Rotor-Gene 6000 Series software 1.7.
Generation of a Homology Model of Drg1
The model of Drg1 was built with the automatic homology modeling module (19) of the YASARA program (20). Initially, 10 models were built using the two closest templates in the Protein Data Bank (1R7R and 3CF3, which are both crystal structures of murine p97 protein) and five slightly different alignments per template (21). The modeling procedure involved the SCWRL algorithm (22) for side chain rotamer prediction and hydrogen bonding network optimization (23), as well as an energy minimization with explicit solvent shell (19) to generate the final models. Surprisingly, the models based on template 3CF3 (24) scored consistently better, even though 1R7R was solved at higher resolution (3.6 versus 4.25 Å). The best model obtained a structure validation Z-score (24) of −2.2 (average of dihedral and packing Z-scores), which is good considering the low resolution template. A Protein Data Bank file of the model is available from the authors upon request.
RESULTS
Diazaborine Blocks Cytoplasmic Pre-60S Maturation
We reported previously that certain drg1 mutants are defective in the release of shuttling proteins from cytoplasmic pre-60S particles (3, 5, 6). Most shuttling proteins are released and recycled into the nucleus soon after nuclear export of pre-60S particles into the cytoplasm. Therefore, these proteins show low abundance in purifications of late preribosomal particles and a mainly nuclear steady-state localization. In contrast, inhibition of shuttling factor release by Drg1 inactivation results in their increased occurrence in preparations of late pre-60S particles and a mislocalization into the cytoplasm (3, 5, 6) (Fig. 1B). Furthermore, the failure to release these proteins prevents the binding of later joining factors to these pre-60S particles (3, 5).
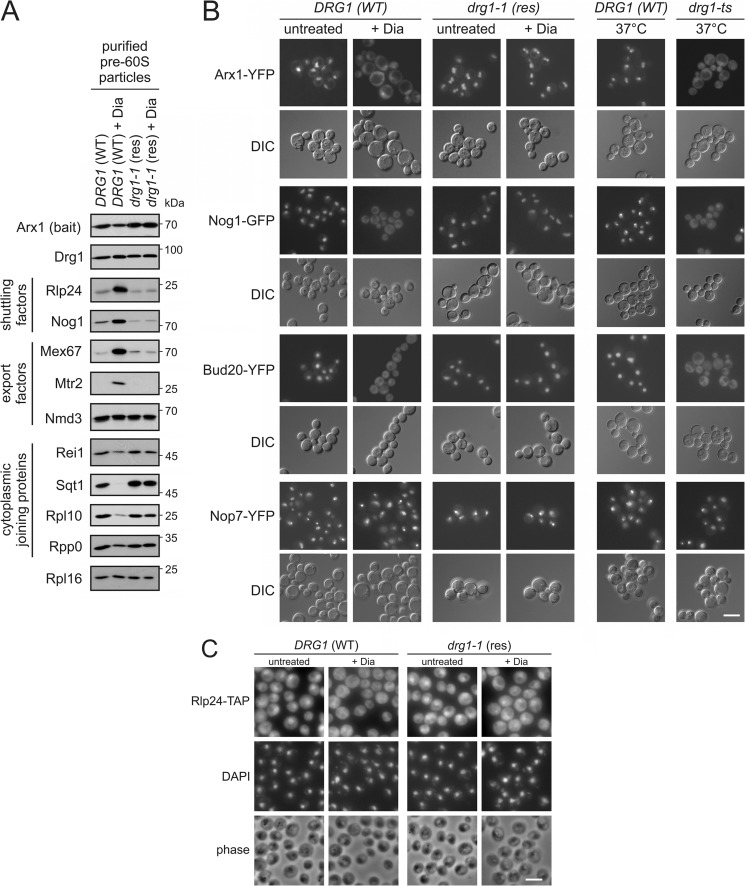
FIGURE 1.
Diazaborine blocks maturation of late pre-60S particles. A, diazaborine treatment leads to the enrichment of shuttling and export factors in purifications of late pre-60S particles. The DRG1 wild-type strain (DRG1 (WT)) and the diazaborine resistant drg1-1 mutant (drg1-1 (res)) were treated with 370 μm diazaborine for 1 h. Afterward pre-60S particles were isolated with Arx1-TAP as bait protein and analyzed by Western blotting with antibodies directed against shuttling proteins, export factors, and late joining cytoplasmic factors. B, diazaborine (Dia) triggers the cytoplasmic accumulation of shuttling factors. Yeast strains expressing chromosomal fusions of shuttling pre-60S maturation factors Arx1, Nog1, or Bud20 with YFP or GFP in the DRG1 wild-type (DRG1 (WT)) and the diazaborine resistant drg1-1 mutant (drg1-1 (res)) were treated with diazaborine. After 1 h, samples were inspected by fluorescence microscopy. A strain expressing a chromosomal fusion of the strictly nuclear pre-60S maturation factor Nop7 with YFP was used as a control. As a control for cytoplasmic accumulation, the localization of the respective fluorescent proteins was also inspected in the drg1-ts background. DIC, differential interference contrast picture. C, the shuttling factor Rlp24 accumulates in the cytoplasm after treatment with diazaborine. Immune fluorescence with TAP-tagged Rlp24 is shown. The DRG1 wild-type strain (DRG1 (WT)) or the diazaborine-resistant drg1-1 mutant (drg1-1 (res)) expressing Rlp24-TAP fusion proteins were treated with diazaborine for 1 h. Thereafter cells were fixed, and Rlp24-TAP was detected by immunofluorescence. DAPI, DAPI staining of the DNA in the nucleus. Phase, phase contrast pictures to monitor morphological integrity of the cells. White scale bars in B and C, 5 μm.
The resistant phenotype of certain drg1 alleles prompted us to investigate whether diazaborine triggers phenotypes similar to those caused by functional inactivation of Drg1. To investigate whether diazaborine also affects shuttling factor release, we purified late pre-60S particles by TAP using Arx1 as bait protein. As shown in Fig. 1A, drug treatment of the wild-type strain resulted in increased levels of the shuttling proteins Rlp24 and Nog1 and of the export factors Mex67 and Mtr2 in these pre-60S purifications. As expected, due to its near stoichiometric presence on Arx1 particles, the levels of Nmd3 showed only a minor increase relative to the bait protein. Concomitantly, decreased levels of the late joining cytoplasmic proteins Sqt1 and Rpl10, and to a lesser extent Rei1, were detected in pre-60S preparations upon diazaborine treatment. In contrast, no alterations in the composition of late pre-60S particles were observed upon diazaborine treatment of the resistant drg1-1 mutant, which carries a V725E exchange in the polypeptide chain. These results suggest a block of shuttling protein release from the pre-60S particles in the presence of diazaborine.
We next asked whether this blockage takes place in the cytoplasm as is the case in drg1-ts mutants (5, 6). For this purpose GFP or YFP fusions of representative shuttling factors were inspected by fluorescence microscopy after diazaborine treatment. The shuttling proteins Arx1, Nog1, Bud20, and Rlp24, which showed a predominantly nuclear localization in untreated cells, accumulated in the cytoplasm after treatment with diazaborine (Fig. 1, B and C). In contrast, no cytoplasmic mislocalization was observed in the resistant drg1-1 strain or after diazaborine treatment of strains expressing a YFP fusion of the strictly nuclear pre-60S maturation factor Nop7. The phenotype of diazaborine-treated cells strongly resembled that observed in a drg1-ts mutant shifted to the restrictive temperature (Fig. 1B and Ref. 6).
Together, these results demonstrate that the drug causes a failure to release shuttling factors from pre-60S particles, which leads to their accumulation in the cytoplasm in pre-60S bound form. Notably, as Drg1 was still associated with pre-60S particles after treatment with diazaborine, the drug does not prevent association of the AAA-ATPase with pre-60S particles in vivo.
Diazaborine Inhibits the Release of Rlp24 from Pre-60S Particles
Targeting of Drg1 to preribosomal particles as well as activation of ATP hydrolysis are mediated by the C-terminal domain of the shuttling factor Rlp24 (3). To investigate whether diazaborine affects binding of Drg1 to Rlp24C, we analyzed their interaction using GST pulldown assays. As shown in Fig. 2A, diazaborine had no effect on the in vitro interaction between Drg1 and Rlp24. This is consistent with the presence of Drg1 on pre-60S particles from the diazaborine-treated strain (Fig. 1A).
FIGURE 2.
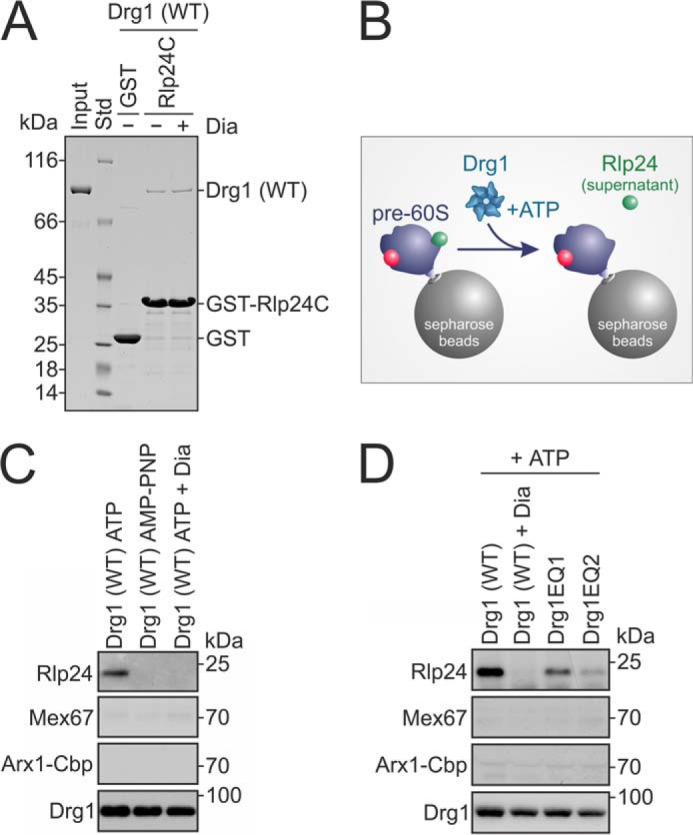
Diazaborine blocks in vitro release of Rlp24. A, diazaborine (Dia) does not prevent the interaction between Drg1 (WT) and Rlp24. GST-Rlp24C was immobilized on glutathione-agarose and incubated with Drg1 and 1 mm ATP in the presence or absence of 370 μm diazaborine. After washing and elution, eluates were analyzed by SDS-PAGE and Coomassie staining. Input, purified Drg1 used in the assay; Std, protein standard. GST served as control. B, experimental schema shows in vitro release assay of C and D. Pre-60S particles isolated from a drg1-ts mutant after incubation at the restrictive temperature using Arx1-TAP as bait protein are immobilized on Sepharose beads. After incubation with Drg1 (WT) and ATP, released proteins (i.e. Rlp24) are found in the supernatant. Nup116, which was present in all samples, was omitted from the schema for simplicity reasons. C, diazaborine blocks Rlp24 release. The nonhydrolyzable ATP analog AMP-PNP or ATP and 370 μm diazaborine were added to the release assay described in B. The supernatants were analyzed by SDS-PAGE and Western blotting with antibodies directed against Drg1 and Rlp24. Mex67 and Arx1-TAP, which are not released by Drg1, were analyzed as negative controls. D, ATP hydrolysis in the D2 domain of Drg1 is essential for the in vitro release of Rlp24. The release assay was performed as in C, using either wild-type Drg1, wild-type Drg1 and diazaborine, or the D1 or D2 ATP hydrolysis-defective Drg1 variants Drg1EQ1 and Drg1EQ2, all in the presence of ATP. Whereas Drg1EQ1 showed only slightly reduced release activity, Drg1EQ2 was almost completely unable to release Rlp24, resembling diazaborine-treated wild-type Drg1.
Because the physiological function of Drg1 is to release Rlp24 from pre-60S particles, we investigated whether diazaborine interferes with this activity using our recently developed in vitro release assay (3) (Fig. 2B). Briefly, pre-60S particles purified from a drg1-ts mutant were bound to calmodulin beads and incubated with ATP and Drg1 in the absence or presence of diazaborine. As our previous work showed that the nucleoporin Nup116 is required for the in vitro release of Rlp24 from pre-60S particles by Drg1 (3) it was also added to the reaction mixture. The amounts of released Rlp24 in the supernatants were then analyzed by Western blotting. As shown in Fig. 2C, addition of diazaborine prevented the release of Rlp24 almost completely (95.0 ± 3.0% inhibition compared with the untreated control). A similar result was obtained with the nonhydrolyzable nucleotide analog AMP-PNP (96.7 ± 1.9% inhibition), which could be an indication that diazaborine interferes with ATP hydrolysis of Drg1. To compare the effects of diazaborine treatment with those caused by inactivation of ATP hydrolysis in the two individual AAA domains, we investigated mutant variants defective for ATP hydrolysis in the first (Drg1EQ1) or the second (Drg1EQ2) AAA domain. As shown in Fig. 2D, Drg1EQ1 was still able to release Rlp24 from purified pre-60S particles, although to a lesser extent than the wild-type protein (inhibition of 57.2 ± 10.8%). In contrast, a much stronger inhibition of release was observed when Drg1EQ2 was used (89.0 ± 4.5%). Thus, diazaborine blocks Rlp24 release to a similar extent as inactivation of ATP hydrolysis in the D2 domain of Drg1. These results are consistent with in vivo data showing that only ATP hydrolysis in D2 is essential for growth and for the release of Rlp24 (3).
Binding of Diazaborine to Drg1 Requires Loading of ATP in D2
The inhibition of the physiological activity of Drg1 by diazaborine suggests direct interaction of the drug with Drg1. We therefore measured binding of diazaborine to Drg1 by DSF (18). This method makes use of the stain SYPRO® Orange, which emits fluorescence only when bound to hydrophobic regions of proteins that become exposed upon denaturation. This property enables the recording of melting curves of proteins. The melting temperature can then be deduced from the first derivative of this curve. Binding of low molecular weight compounds to proteins frequently results in an increase of their melting temperature (25). Therefore, plotting the increase in melting temperature against the compound concentration allows estimation of quantitative binding parameters.
When diazaborine was added in the absence of ATP, no alteration of the melting curve was detected (Fig. 3A). However, when 5 mm ATP and 740 μm diazaborine were simultaneously present in the sample, a shift of the melting curve to higher temperatures was observed compared with the sample with ATP alone (Fig. 3B). We conclude that diazaborine binds directly to Drg1 and that this interaction requires ATP. The order of drug and nucleotide addition did not influence the melting temperature. In addition, increasing ATP concentrations did not inhibit binding of the drug to Drg1, suggesting that diazaborine does not compete with the nucleotide for binding to Drg1. We next asked whether ATP binding to the D1 or D2 domain is required to allow interaction with the inhibitor. For this purpose, we made use of the Drg1KA1 and Drg1KA2 variants, which are unable to bind ATP in the first and second AAA domain, respectively. In the presence of ATP, the Drg1KA1 variant showed an additional peak in the melting curve when diazaborine was present in the sample (Fig. 3C). Thus, nucleotide binding to the D1 domain is not required for diazaborine binding. In contrast, the melting curve of the Drg1KA2 variant was not influenced by diazaborine, demonstrating that ATP binding into the D2 domain of Drg1 is a prerequisite for interaction with the inhibitor (Fig. 3D).
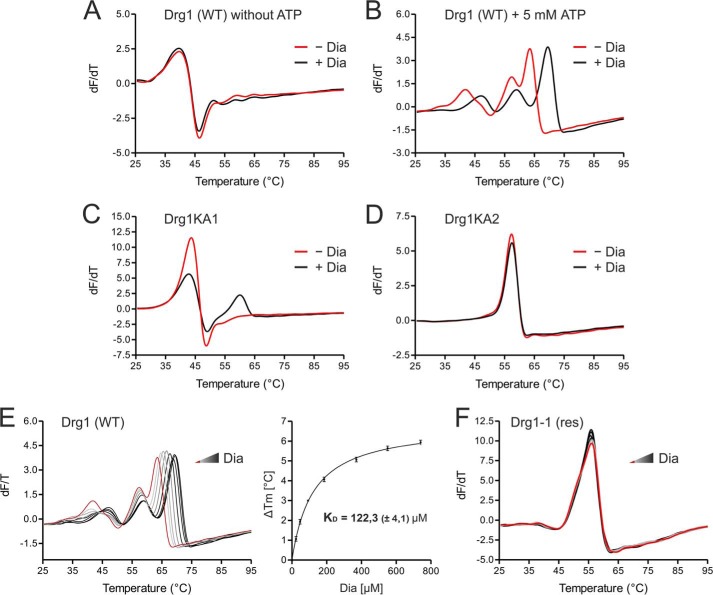
FIGURE 3.
Binding of diazaborine requires ATP loading into the D2 domain of Drg1. Binding of diazaborine (Dia) to Drg1 was investigated by DSF. 1.8 μm Drg1 (WT) or mutant variants thereof were incubated with different diazaborine concentrations in the absence or presence of ATP, and melting curves were recorded in the presence of SYPRO® Orange in a Rotor-Gene 6000. A and B, binding of diazaborine to Drg1 requires the presence of ATP. The purified Drg1 protein was incubated without (red curve) or with (black curve) 740 μm diazaborine in the absence (A) or presence (B) of 5 mm ATP. Addition of diazaborine resulted in a shift of the melting curve to higher temperatures in the presence of ATP, demonstrating binding of the inhibitor to the protein. C and D, diazaborine binding to Drg1 requires ATP loading into D2. Melting curves of Drg1 variants unable to bind ATP into the first (Drg1KA1 (C)) or the second (Drg1KA2 (D)) AAA domain were investigated in the presence of 5 mm ATP without (red curve) or with 740 μm diazaborine (black curve). Diazaborine treatment resulted in a change of the melting curve of Drg1KA1 (C) but not Drg1KA2 (D), showing that loading of ATP into D2 is necessary for diazaborine binding. E, diazaborine binding to Drg1 (WT) in the presence of 5 mm ATP was assessed quantitatively. Melting curves were recorded without diazaborine (red line) or in the presence of increasing diazaborine concentrations (0–740 μm; light gray to black gradient, left panel). Right panel, plotting of the increase in melting temperature (in relation to the untreated sample) over the drug concentration allows the estimation of a drug binding curve. The calculated KD (± S.D.) of three independent measurements is shown. F, the melting curve of the diazaborine-resistant protein Drg1-1 (res) shows no alterations even with 740 μm diazaborine, suggesting that the inhibitor does not bind to this protein.
To obtain quantitative binding parameters, we recorded melting curves of Drg1 in the presence of 5 mm ATP and increasing diazaborine concentrations. As shown in Fig. 3E (left panel), a concentration-dependent shift of the melting temperature was observed. Plotting the increase in melting temperature (relative to the sample containing ATP alone) against the drug concentration resulted in a hyperbolic drug binding curve with a KD of 122.3 ± 4.1 μm diazaborine (mean of three biological replicates, right panel). In contrast to the wild-type, the protein from the diazaborine-resistant Drg1-1 variant did not show a change in melting temperature even at high drug concentrations (Fig. 3F). We conclude that the resistance of the Drg1-1 protein to diazaborine arises from the failure of the inhibitor to bind to this variant.
Resistance Is Caused by Amino Acid Exchanges in the Nucleotide Binding Pocket of D2
The drg1-1 allele was originally identified together with several other alleles in a random mutagenesis screen for diazaborine-resistant mutants (8) (Fig. 4A). However, the mechanism of how these alleles mediate resistance was unknown. As indicated in Fig. 4B, all exchanges identified in the mutants are located in the second AAA domain of Drg1. To better understand the mechanism of resistance, we calculated a model of Drg1 based on the crystal structure of the closely related AAA-ATPase p97. p97 shows 45% sequence identity with Drg1 and is the closest relative for which a crystal structure is available. As shown in Fig. 4C, all amino acid exchanges conferring resistance to diazaborine cluster around the nucleotide binding pocket of the D2 domain, suggesting that the inhibitor binds in close proximity to the nucleotide.
FIGURE 4.
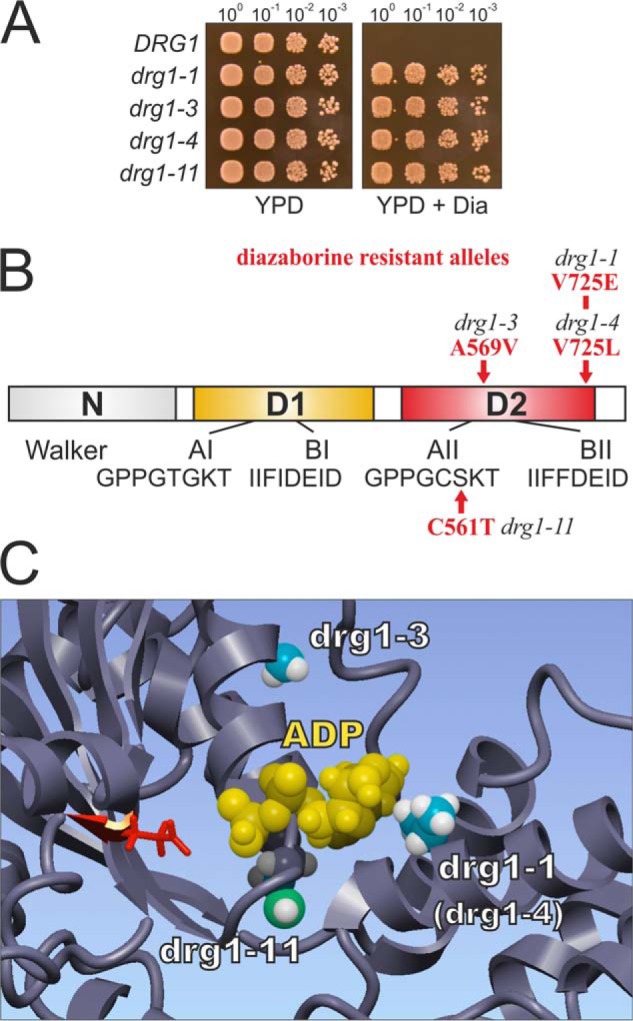
Mutations leading to diazaborine resistance cluster around the ATP binding pocket of the D2 domain of Drg1. A, the drg1-1, drg1-3, drg1-4, and drg1-11 alleles confer resistance to diazaborine (Dia). Serial dilutions of a drg1 deletion strain complemented by centromeric plasmids with the indicated drg1 alleles were spotted onto YPD plates or YPD plates containing 370 μm diazaborine. Plates were incubated at 30 °C for 3 days. B, schematic shows Drg1 with the amino acid exchanges causing diazaborine resistance highlighted in red. The two AAA-ATPase domains D1 (yellow) and D2 (red) and the N domain (gray), as well as the Walker A and Walker B motifs are indicated. C, structural model of the region around the nucleotide binding pocket in the D2 domain of Drg1 with bound ADP is shown. Amino acid residues which are exchanged in the diazaborine-resistant drg1-1, drg1-3, and drg1-11 alleles are shown in the ball mode. The conserved glutamate residue (Glu-617) of the Walker B motif involved in ATP hydrolysis is depicted in red. The drg1-4 allele contains a V725L exchange and affects the same residue as the Drg1-1 exchange.
Diazaborine Specifically Inhibits ATP Hydrolysis in the D2 Domain of Drg1
The data shown above demonstrate that diazaborine binding to Drg1 requires the presence of nucleotide. Next we tested whether the drug blocks ATP hydrolysis by Drg1. However, consistent with previous data, the basal ATPase activity of Drg1 was largely unaffected by the inhibitor (Ref. 9 and Fig. 5A). Because the ATPase activity of Drg1 is stimulated by the C-terminal domain of Rlp24 (3) (Rlp24C), we also tested the effect of diazaborine on the stimulated ATPase activity. Indeed, in the presence of saturating Rlp24C concentrations, the wild-type protein was inhibited by 370 μm diazaborine by ∼25%. Higher drug concentrations (i.e. 740 μm) did not further decrease the measured activity. No inhibition of the resistant Drg1-1 protein was found even with the highest tested drug concentration (Fig. 5A). Notably, when the wild-type Drg1 protein was stimulated with lower concentrations of Rlp24C in the range of the KD (28 nm (3)), we observed a more pronounced inhibition (∼60%) in the presence of diazaborine (Fig. 5B). In contrast, no inhibition of the resistant Drg1-1 variant was observed throughout all tested Rlp24 concentrations (Fig. 5C).
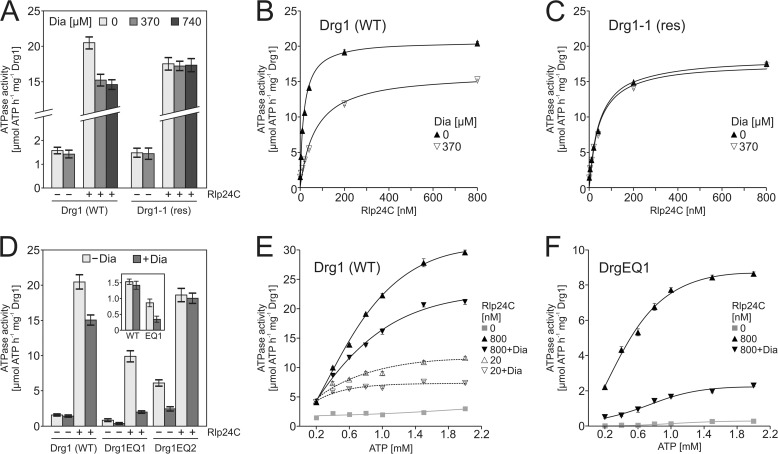
FIGURE 5.
Diazaborine (Dia) only inhibits ATP hydrolysis in the D2 domain of Drg1. A, the ATPase activity of Drg1 is inhibited by diazaborine. ATPase activity of 1200 nm (corresponding to 200 nm hexamer) purified wild-type Drg1 (WT) or the Drg1-1 (res) protein was measured in the absence or presence of Rlp24C (800 nm) and diazaborine (370 μm). In addition, the inhibition of the stimulated ATPase activity was also tested in the presence of 740 μm diazaborine. Error bars, S.D. of three biological replicates. B and C, Drg1 wild-type protein (B) or the diazaborine-resistant variant Drg1-1 (C) were incubated in the presence of different Rlp24C concentrations (0–800 nm) and 370 μm diazaborine. Error bars, S.E. of three biological replicates. D, only the second AAA domain of Drg1 is inhibited by diazaborine. Effect of diazaborine (370 μm) on the ATPase activity of mutant variants of Drg1 defective for ATP hydrolysis in the first (Drg1EQ1) or second (Drg1EQ2) AAA domain is shown. ATPase activity was measured in the absence or presence of 800 nm Rlp24C. Inset, enlarged display of the basal activity of the Drg1 wild-type (WT) protein and the Drg1EQ1 variant (EQ1) in the presence and absence of diazaborine. Error bars, S.D. of two biological replicates. E and F, ATP dependence of the inhibition of Drg1 and Drg1EQ1 by diazaborine. The ATPase activity of Drg1 in the presence of 20 nm and 800 nm Rlp24C (E) and of the Drg1EQ1 variant in the presence of 800 nm Rlp24 (F) was tested using increasing ATP concentrations without and with 370 μm diazaborine. The ATPase activity in the absence of Rlp24 (basal activity) is shown as a gray line. Error bars, S.E. of two biological replicates.
The fact that ATP hydrolysis was not completely abolished by the inhibitor suggests that diazaborine only targets one of the two AAA domains of Drg1 and that in the presence of saturating Rlp24 concentrations the activity of the insensitive domain is prevailing. Because we have previously shown that Rlp24 stimulates ATP hydrolysis in both AAA domains of Drg1 (3), we aimed to identify which of the two domains is targeted by the drug. For this purpose we purified the Drg1EQ1 and Drg1EQ2 variants. The fact that these proteins can hydrolyze ATP only in either the first (EQ2) or the second (EQ1) ATP binding site allowed us to dissect the influence of the drug on the individual AAA domains. In the presence of saturating Rlp24 concentrations, diazaborine inhibited the ATPase activity of Drg1EQ1 by 80%. Additionally, the basal activity of Drg1EQ1 was also reduced by the drug by 60 ± 11%, showing that diazaborine specifically blocks ATP hydrolysis in the second AAA domain (Fig. 5D, inset). In contrast, the ATPase activity of the first AAA domain was not inhibited in the presence of Rlp24, as the Drg1EQ2 variant was insensitive to the inhibitor. Notably, the basal ATPase activity observed with this variant, which is 4-fold higher than that of the wild-type (Fig. 5D and Ref. 3), was reduced by diazaborine. However, our data clearly demonstrate that the inhibitor specifically targets the second AAA domain of Drg1 (Figs. 3 and 4). We therefore suggest that in Drg1EQ2, the observed inhibition of ATP hydrolysis in D1 in the absence of Rlp24 is likely an indirect consequence of diazaborine binding into D2. Together with the elevated basal activity of Drg1EQ2 compared with the wild-type protein, this result indicates a pronounced communication from the D2 to the D1 domain.
We have demonstrated previously that the two AAA domains of Drg1 exhibit different affinities for the nucleotide (9). To test the influence of ATP loading into D2 on the inhibition of Drg1, we measured the enzymatic activity of wild-type Drg1 and the Drg1EQ1 variant in the presence of Rlp24 and different ATP concentrations (Fig. 5, E and F). At the lowest tested nucleotide concentration of 0.2 mm ATP, similar ATP hydrolysis was measured with the wild-type protein in the presence and in the absence of the drug, suggesting that under these conditions only the diazaborine-insensitive D1 domain is active (Fig. 5E). In the presence of higher nucleotide concentrations, the inhibitor caused a pronounced reduction of ATP hydrolysis. In contrast to the wild-type protein, the Drg1EQ1 variant displayed reduced ATP hydrolysis rates in the presence of diazaborine at all tested nucleotide concentrations, even at 0.2 mm (Fig. 5F; compare first data points in Fig. 5, E and F). Taken together, these results suggest that the D2 domain represents the low affinity ATP binding site which is specifically targeted by diazaborine.
Blockage of ATP Hydrolysis in D2 Correlates with Growth Inhibition
To evaluate how the inhibition of ATP hydrolysis in D2 by diazaborine affects growth, we measured the ATPase activity as well as the growth of yeast cells in the presence of increasing drug concentrations. We focused on the Drg1EQ1 variant as in the wild-type protein, both AAA domains are activated under saturating Rlp24C concentrations, but only D2 is inhibited by diazaborine (Fig. 5D). As shown in Fig. 6A, in the presence of saturating Rlp24C concentrations, the ATPase activity of both, the Drg1 wild-type and the EQ1 variant was inhibited by increasing diazaborine concentrations. The Ki of diazaborine for the D2 domain was estimated to approximately 26 μm. A significant decrease in cell density of both the wild-type and the drg1EQ1 mutant was observed at a drug concentration corresponding to the Ki of diazaborine for the D2 domain (Fig. 6B). These results demonstrate a clear correlation between the inhibition of ATP hydrolysis in D2 and the decrease in proliferation of yeast cells in vivo.
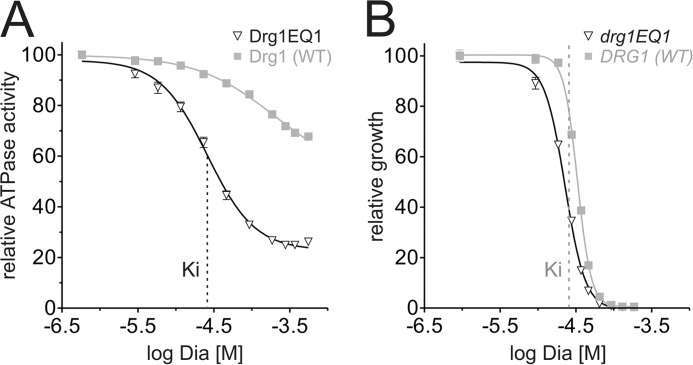
FIGURE 6.
Blockage of ATP hydrolysis in D2 parallels growth inhibition. A, the ATPase activity of Drg1EQ1 was measured in the presence of 800 nm Rlp24C using increasing concentrations of diazaborine (Dia). The relative activity values were plotted against the logarithmic inhibitor concentration. The Ki determined with Drg1EQ1 is indicated by a dashed line. The activity of wild-type Drg1 (WT) is shown as a control. B, growth inhibition of the drg1EQ1 mutant is shown upon diazaborine treatment. Cells were grown in the presence of increasing diazaborine concentrations for 24 h, the A600 was measured, and the relative A600 values were plotted against the logarithmic inhibitor concentration. The inhibitor concentration corresponding to the Ki determined in A is indicated by a dashed line.
DISCUSSION
Ribosome biogenesis in eukaryotic cells involves the coordinated activity of >200 trans-acting factors. Because most of these factors are essential, ribosome biogenesis provides a promising target for antimicrobial and anti-tumor chemotherapy. In this study, we investigated the mechanism of action of the first specific inhibitor of this pathway, the heterocyclic boron-containing compound diazaborine. Our data demonstrate that the AAA-ATPase Drg1 is the direct target of diazaborine. Drg1 is only found in eukaryotic cells and was previously shown to be a key factor of pre-60S ribosome maturation in yeast. Accordingly, treatment with the drug leads to the same phenotypic consequences as those caused by the functional inactivation of Drg1, i.e. a failure to release Rlp24 from pre-60S particles and a block in downstream maturation (Fig. 1, compare with Ref. 3).
What is the mechanism of inhibition of the drug? Our work shows that diazaborine specifically binds into the second AAA domain of Drg1 and thereby inhibits ATP hydrolysis in this site. However, the drug does not act as a competitive inhibitor for ATP. Instead, ATP binding to D2 is a prerequisite for diazaborine to recognize Drg1. Nucleotide binding is also required for inhibitor interaction with the enoyl-ACP reductase (FabI), which is the target of diazaborine in Gram-negative bacteria. For FabI, it was shown that the NAD+ cofactor has to bind to the enzyme to enable interaction with the inhibitor (12, 13). Structural studies showed that in the active site of FabI, the boron atom of diazaborine forms a covalent bond with the 2′-OH of the nicotinamide ribose moiety of the NAD+ cofactor (26, 27). It is however currently unknown whether or not binding of diazaborine to Drg1 also involves the formation of a covalent bond between ATP and the inhibitor. Our homology model of Drg1 shows that all up to now identified amino acid exchanges causing resistance to diazaborine are located in close proximity to the nucleotide in the D2 domain of the protein (Fig. 4C). This observation supports the possibility of a close contact between ATP and diazaborine within the D2 domain of Drg1. Therefore, formation of covalent intermediates or at least tight binding between a nucleotide cofactor and the inhibitor could be a general principle for the mode of action of diazaborine. Our structural model also offers an explanation for the resistance mechanism: The mutations resulting in the resistant phenotype in the drg1-1, drg1-3, and drg1-4 alleles introduce amino acid residues with bulkier side chains than those present in the wild-type and therefore may reduce the accessibility of the binding pocket for the drug. As diazaborine is unable to bind to the Drg1-1 protein, the resistance mechanism may involve the occlusion of diazaborine from its binding site near the D2 nucleotide binding pocket of Drg1.
Taken together, our work shows that ribosome biogenesis in general and particularly energy-consuming enzymes within this pathway may represent promising targets for the development of novel inhibitors. These enzymes include additional AAA-ATPases, like Rix7 and Rea1, but also a number of RNA helicases and GTPases (1). Furthermore, AAA-ATPases have emerged as interesting new targets for low molecular weight inhibitors in recent years. For example, significant efforts were undertaken to identify inhibitors for p97/VCP (28–31), which is closely related to Drg1 but involved in different cellular pathways. The mechanistic insights gained from our study will therefore be helpful for the rational design of novel inhibitors of other AAA-ATPases.
Acknowledgments
We thank Kerstin Klein and Anke Loregger for expert help during early stages of this work; Micheline Fromont-Racine, Bernard L. Trumpower, Juan P. Ballesta, Arlen W. Johnson, Ed Hurt, and Sabine Rospert for generously supplying reagents; and Tamsyn Stanborough for critically reading the manuscript.
This work was supported by grants from the European Union Seventh Framework Programme (FP7/2007–2013) under Grant 289350 (to E. K.) and Austrian Science Fund (FWF) Grants P21991 and P26136 (to H. B.).
- AAA
- ATPases associated with diverse cellular activities
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- DSF
- differential scanning fluorometry
- res
- diazaborine-resistant
- TAP
- tandem affinity purification
- YPD
- yeast extract peptone dextrose.
REFERENCES
- 1. Kressler D., Hurt E., Bassler J. (2010) Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 2. Panse V. G., Johnson A. W. (2010) Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., Unterweger S., Rechberger G., Pertschy B., Bergler H. (2012) Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 199, 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lo K. Y., Li Z., Bussiere C., Bresson S., Marcotte E. M., Johnson A. W. (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39, 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., Högenauer G., Fromont-Racine M., Bergler H. (2007) Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell Biol. 27, 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bassler J., Klein I., Schmidt C., Kallas M., Thomson E., Wagner M. A., Bradatsch B., Rechberger G., Strohmaier H., Hurt E., Bergler H. (2012) The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 32, 4898–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thorsness P. E., White K. H., Ong W. C. (1993) AFG2, an essential gene in yeast, encodes a new member of the Sec18p, Pas1p, Cdc48p, TBP-1 family of putative ATPases. Yeast 9, 1267–1271 [DOI] [PubMed] [Google Scholar]
- 8. Wendler F., Bergler H., Prutej K., Jungwirth H., Zisser G., Kuchler K., Högenauer G. (1997) Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 272, 27091–27098 [DOI] [PubMed] [Google Scholar]
- 9. Zakalskiy A., Högenauer G., Ishikawa T., Wehrschütz-Sigl E., Wendler F., Teis D., Zisser G., Steven A. C., Bergler H. (2002) Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae: decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 277, 26788–26795 [DOI] [PubMed] [Google Scholar]
- 10. Pertschy B., Zisser G., Schein H., Köffel R., Rauch G., Grillitsch K., Morgenstern C., Durchschlag M., Högenauer G., Bergler H. (2004) Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 24, 6476–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grassberger M. A., Turnowsky F., Hildebrandt J. (1984) Preparation and antibacterial activities of new 1,2,3-diazaborine derivatives and analogues. J. Med. Chem. 27, 947–953 [DOI] [PubMed] [Google Scholar]
- 12. Bergler H., Wallner P., Ebeling A., Leitinger B., Fuchsbichler S., Aschauer H., Kollenz G., Högenauer G., Turnowsky F. (1994) Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J. Biol. Chem. 269, 5493–5496 [PubMed] [Google Scholar]
- 13. Kater M. M., Koningstein G. M., Nijkamp H. J., Stuitje A. R. (1994) The use of a hybrid genetic system to study the functional relationship between prokaryotic and plant multi-enzyme fatty acid synthetase complexes. Plant Mol. Biol. 25, 771–790 [DOI] [PubMed] [Google Scholar]
- 14. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 15. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 16. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 17. Schmitt J., Hess H., Stunnenberg H. G. (1993) Affinity purification of histidine-tagged proteins. Mol. Biol. Rep. 18, 223–230 [DOI] [PubMed] [Google Scholar]
- 18. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 19. Krieger E., Joo K., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krieger E., Koraimann G., Vriend G. (2002) Increasing the precision of comparative models with YASARA NOVA: a self-parameterizing force field. Proteins 47, 393–402 [DOI] [PubMed] [Google Scholar]
- 21. Mückstein U., Hofacker I. L., Stadler P. F. (2002) Stochastic pairwise alignments. Bioinformatics 18, S153–S160 [DOI] [PubMed] [Google Scholar]
- 22. Canutescu A. A., Shelenkov A. A., Dunbrack R. L., Jr. (2003) A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12, 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krieger E., Dunbrack R. L., Jr., Hooft R. W., Krieger B. (2012) Assignment of protonation states in proteins and ligands: combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 819, 405–421 [DOI] [PubMed] [Google Scholar]
- 24. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 25. Waldron T. T., Murphy K. P. (2003) Stabilization of proteins by ligand binding: application to drug screening and determination of unfolding energetics. Biochemistry 42, 5058–5064 [DOI] [PubMed] [Google Scholar]
- 26. Baldock C., Rafferty J. B., Sedelnikova S. E., Bithell S., Stuitje A. R., Slabas A. R., Rice D. W. (1996) Crystallization of Escherichia coli enoyl reductase and its complex with diazaborine. Acta Crystallogr D Biol. Crystallogr 52, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 27. Levy C. W., Baldock C., Wallace A. J., Sedelnikova S., Viner R. C., Clough J. M., Stuitje A. R., Slabas A. R., Rice D. W., Rafferty J. B. (2001) A study of the structure-activity relationship for diazaborine inhibition of Escherichia coli enoyl-ACP reductase. J. Mol. Biol. 309, 171–180 [DOI] [PubMed] [Google Scholar]
- 28. Bursavich M. G., Parker D. P., Willardsen J. A., Gao Z. H., Davis T., Ostanin K., Robinson R., Peterson A., Cimbora D. M., Zhu J. F., Richards B. (2010) 2-Anilino-4-aryl-1,3-thiazole inhibitors of valosin-containing protein (VCP or p97). Bioorg. Med. Chem. Lett. 20, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 29. Chou T. F., Deshaies R. J. (2011) Development of p97 AAA-ATPase inhibitors. Autophagy 7, 1091–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magnaghi P., D'Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., Gasparri F., Cozzi L., Cucchi U., Orrenius C., Polucci P., Ballinari D., Perrera C., Leone A., Cervi G., Casale E., Xiao Y., Wong C., Anderson D. J., Galvani A., Donati D., O'Brien T., Jackson P. K., Isacchi A. (2013) Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 9, 548–556 [DOI] [PubMed] [Google Scholar]
- 31. Wang Q., Shinkre B. A., Lee J. G., Weniger M. A., Liu Y., Chen W., Wiestner A., Trenkle W. C., Ye Y. (2010) The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One 5, e15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]