Background: Targeting oncogenic K-Ras for cancer therapy has remained challenging.
Results: Ubiquitination specifically occurs on the activated K-Ras orthologue in Dictyostelium via evolutionary conserved K-Ras lysines, which promotes K-Ras protein degradation.
Conclusion: Our results indicate the existence of GTP-loaded K-Ras orthologue-specific degradation system in Dictyostelium.
Significance: This work reveals a novel negative feedback regulation for the K-Ras isoform, which is critical for cytokinesis in Dictyostelium.
Keywords: Cancer Biology, Cytokinesis, Dictyostelium, Molecular Cell Biology, Ras, Signaling, Ubiquitination
Abstract
Mammalian cells encode three closely related Ras proteins, H-Ras, N-Ras, and K-Ras. Oncogenic K-Ras mutations frequently occur in human cancers, which lead to dysregulated cell proliferation and genomic instability. However, mechanistic role of the Ras isoform regulation have remained largely unknown. Furthermore, the dynamics and function of negative regulation of GTP-loaded K-Ras have not been fully investigated. Here, we demonstrate RasG, the Dictyostelium orthologue of K-Ras, is targeted for degradation by polyubiquitination. Both ubiquitination and degradation of RasG were strictly associated with RasG activity. High resolution tandem mass spectrometry (LC-MS/MS) analysis indicated that RasG ubiquitination occurs at C-terminal lysines equivalent to lysines found in human K-Ras but not in H-Ras and N-Ras homologues. Substitution of these lysine residues with arginines (4KR-RasG) diminished RasG ubiquitination and increased RasG protein stability. Cells expressing 4KR-RasG failed to undergo proper cytokinesis and resulted in multinucleated cells. Ectopically expressed human K-Ras undergoes polyubiquitin-mediated degradation in Dictyostelium, whereas human H-Ras and a Dictyostelium H-Ras homologue (RasC) are refractory to ubiquitination. Our results indicate the existence of GTP-loaded K-Ras orthologue-specific degradation system in Dictyostelium, and further identification of the responsible E3-ligase may provide a novel therapeutic approach against K-Ras-mutated cancers.
Introduction
Ras is a small GTPase that can cycle between a GTP-bound active state and a GDP-bound inactive state. GTP-loaded Ras elicits an array of downstream signaling such as Raf/MEK/ERK and PI3K/AKT pathways to promote cell survival and proliferation (1–5).
Mammalian cells encode four closely related Ras proteins, H-Ras, N-Ras, K-Ras 4A, and K-Ras 4B that are highly homologous in the first 85% of the protein and share a C-terminal CAAX box that targets them for farnesylation. The C-terminal hypervariable region (HVR)4 renders distinctive roles of Ras isoforms. The HVR of K-Ras 4B (hereafter called K-Ras), a dominant K-Ras isoform, contains a poly-basic, lysine-rich, cluster that is required for K-Ras plasma membrane localization. The HVRs of the other Ras isoforms, including K-Ras 4A, another minor K-Ras isoform, provide palmitoylation site(s) for targeting to the plasma membrane (6–10). Extensive research within the past two decades has shown that Ras isoforms differ in their ability to activate downstream effectors and promote transformation (11–13). In mouse tumor models, only the constitutively active K-Ras mutant, but not H-Ras or N-Ras mutants, promotes colorectal carcinogenesis and hyperproliferation of endoderm progenitor cells (1, 3, 14, 15). In human cancers, K-Ras is the most frequently mutated Ras isoform. Furthermore, K-Ras mutations are associated with poor clinical prognosis and resistance to chemotherapy (6, 8, 16–18). Despite numerous attempts, a small molecule inhibitor for K-Ras is yet to be developed for clinical use. Identification of KRas-specific regulatory mechanisms is likely to provide important insights into the development of novel therapeutic applications for K-Ras-mutated tumors.
To date, many of the physiological roles of Ras have been identified through studies using model organisms. However, not all model organisms have both H-Ras and K-Ras isoforms. Saccharomyces cerevisiae, a unicellular organism, has H-Ras homologue genes Ras1 and Ras2, but it does not encode for a K-Ras orthologue. Caenorhabditis elegans and Drosophila melanogaster have a single K-Ras orthologous gene but not an H-Ras orthologue. Dictyostelium discoideum, a social amoeba that has the ability to alternate between a unicellular and a multicellular form, has both K-Ras and H-Ras orthologues. Therefore, to investigate whether ubiquitination comprises an evolutionarily conserved mechanism for Ras regulation, we utilized the Dictyostelium system.
EXPERIMENTAL PROCEDURES
Materials
Anti-FLAG (M2) antibody and 3× FLAG peptide were from Sigma Aldrich; anti-Ras (Ab-3) antibody was from EMD Millipore; and anti-ubiquitin antibody (P4D1) was from Santa Cruz Biotechnology. FLAG-tagged Ras mutants were generated by the standard PCR method and subcloned into pEXP4(+). All constructs were sequenced. We produced GST-RBD in BL21 bacteria (Stratagene) and purified it on glutathione-Sepharose (Amersham Biosciences) per the manufacturer's instructions.
Cell Culture
All cell lines were cultured axenically in HL5 medium at 22 °C. Transformants were maintained in 40 μg/ml G418.
Tandem Mass Spectrometry Analysis
FLAG-RasG expressing RasG-null cells were grown exponentially in HL5 media. The a billion of FLAG-RasG/RasG cells were then collected and lysed with radioimmune precipitation assay buffer and subjected to anti-FLAG (M2)-agarose immunoprecipitation. The beads were washed with radioimmune precipitation assay buffer and FLAG-RasG, and its ubiquitinated forms were eluted with 3×FLAG peptide. Samples were prepared from SDS-PAGE gels, stained with Coomassie Blue, and excised for mass spectrometry sequencing and ubiquitin site mapping. Gel pieces were reduced with DTT, and Cys residues were alkylated with iodoacetamide and digested overnight using modified trypsin (Promega) at 37 °C at pH 8.3. Peptides were extracted and analyzed by data-dependent microcapillary tandem mass spectrometry using both linear ion trap (Thermo Scientific, San Jose, CA) and a high resolution/mass accuracy hybrid linear ion trap LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) coupled to an EASY-nLC nanoflow HPLC (Proxeon Biosystems). MS/MS spectra acquired via collision-induced dissociation were searched against the reversed and concatenated Swiss-Prot protein database (version 55.8, UniProt) with a fixed modification for carbamidomethylation (+57.0293) and the variable modifications for oxidation (+15.9949) and ubiquitination (+114.10, GG tag) using the Sequest algorithm in Proteomics Browser software (Thermo Scientific). Peptides from RasG were identified by database scoring, and peptides modified by ubiquitin were validated manually to be sure that all b- and y- series ions were consistent with the modified residue and additional validation was performed using GraphMod software in Proteomics Browser software. The peptide false discovery rate was <1.5% based on reversed database hits.
RESULTS AND DISCUSSION
Dictyostelium RasG, K-Ras Homologue, Undergoes Ubiquitination
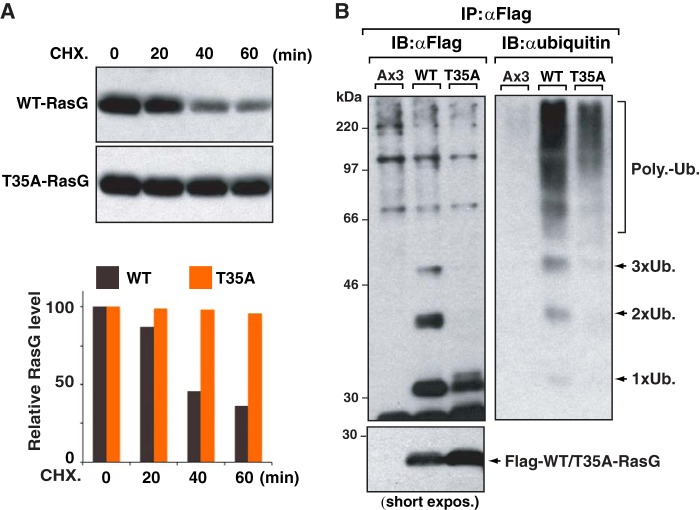
Dictyostelium has five Ras isoforms (RasB, RasC, RasD, RasG, and RasS), among which RasG is the closest homologue to human K-Ras 4B (K-Ras). RasG contains the characteristic lysine stretch of K-Ras in its C-terminal HVR, and the positions of the lysine residues in RasG are well aligned with the lysines in the HVR of human K-Ras (Fig. 1A). In contrast, the HVR of RasG has poor sequence homology to that of human H-Ras and human N-Ras. To investigate regulation of RasG, we used RasG knock-out Dictyostelium cells stably expressing FLAG-RasG (WT-RasG/ΔRasG) (11, 19) and measured protein stability of RasG in response to chemoattractant (cAMP) stimulation. The FLAG-RasG/ΔRasG cells were placed in buffer containing only Na/KPO4 and treated with the protein synthesis inhibitor, cycloheximide (CHX). The result showed that CHX led to an ∼50% decrease in RasG protein levels in ∼30 min (Fig. 1B). The half-life of the RasG protein was further shortened by chemoattractant (cAMP) stimulation. These results suggest that RasG protein stability can be dynamically decreased by extracellular stimuli.
FIGURE 1.
RasG, Dictyostelium K-Ras homologue, undergoes polyubiquitination. A, schematic diagram of G-domain of Ras (upper panel). The C-terminal HVRs of RasG and human Ras isoforms are aligned (lower panel). B, the FLAG-tagged WT-RasG/ΔRasG cells were treated with 100 μg/ml CHX for indicated times, in the presence (+) or absence (−) of 10 μm of cAMP, and cell extracts were analyzed with anti-FLAG antibody. Relative protein levels of RasG were quantified and are shown on the right. These blots are representative of two separate experiments. C, RasG undergoes polyubiquitination (Poly-Ub.RasG). FLAG-tagged RasG was immunoprecipitated (IP) from total cell lysate (TCL) of FLAG-tagged RasG stably expressing RasG knock-out cells (FLAG-RasG/ΔRasG), and immunoblotted with the indicated antibodies. Wild-type cells, Ax3 cell line, were used as control. Ubiquitinated Ras is indicated with arrowheads. These blots are representative of three separate experiments.
To investigate whether ubiquitin-mediated degradation system could account for RasG destabilization, RasG was immunoprecipitated with anti-FLAG resin from WT-RasG/ΔRasG cells. Immunoprecipitated RasG was detected with anti-FLAG antibody as an expected band (∼25 kDa). The anti-FLAG antibody also detected slower migrating bands above the 30-kDa molecular mass marker, which were not detected in the control wild-type cells (Ax3). A similar band ladder pattern was also detected with anti-Ras antibody, indicating that ladder bands were modified forms of RasG protein. The increment of RasG ladder bands was ∼9 kDa, which is equivalent to the size of ubiquitin. Consistent with this hypothesis, the slow-migrating RasG bands were recognized with anti-ubiquitin antibody (Fig. 1C). These results indicated that RasG has a ubiquitinated form and that an evolutionarily conserved mechanism of K-Ras regulation by ubiquitination may exist in Dictyostelium.
RasG Undergoes Activation-dependent Ubiquitination
Next, we investigated whether RasG protein stability might be modulated by RasG activation and/or its downstream signaling. We introduced a point mutation (Thr-35 to Ala) in RasG that interferes with the GTP-mediated switch transition and therefore abrogates effector binding of Ras (14, 15, 20, 21). To study the mutant RasG stability, we used vegetative Dictyostelium cells rather than developmentally competent cells because mutation of RasG can affect Dictyostelium development (16–18, 22, 23). The WT-RasG/ΔRasG cells or T35A-RasG/ΔRasG cells were grown to exponential phase and then treated with cycloheximide. Consistent with the result seen in developed cells, CHX treatment led to a 50% decrease in wild-type RasG protein levels within 30 min (Fig. 2A) in the vegetative growth condition. In contrast, the half-life of T35A-RasG protein was significantly extended.
FIGURE 2.
Polyubiquitination and degradation of RasG requires RasG activation and its downstream effector. A, T35A-RasG mutant increased protein stability. The WT- and T35A-RasG stably expressing cells were treated with 100 μg/ml CHX for indicated times, and cell extracts were analyzed with anti-FLAG antibody. Relative protein levels of RasG were quantified by ImageJ software and shown in the bottom panel. These blots are representative of two separate experiments. B, T35A-RasG mutant decreased ubiquitination level. FLAG-tagged WT- and T35A- RasG were immunoprecipitated (IP) from total cell lysate (TCL) of FLAG-tagged RasG stably expressing RasG knock-out cells (FLAG-RasG/ΔRasG) and immunoblotted (IB) with the indicated antibodies. Wild-type cells, Ax3 cell line, were used as control. Ubiquitinated Ras is indicated with arrowheads. short expos., short exposure; Poly-Ub., polyubiquitination.
To investigate whether RasG ubiquitination is triggered by the downstream effector pathway of RasG, ubiquitination levels of wild-type RasG and T35A-RasG were assessed. Non-ubiquitinated T35A-RasG expression levels were ∼2 times more than WT-RasG. However, the ubiquitination level of T35A-RasG was ∼3 times less than that of WT-RasG. Taking into consideration the increased protein stability of T35A-RasG, these results suggest that RasG induces its own ubiquitination via downstream effector pathway for its degradation. Notably, mono-ubiquitinated T35A-RasG was still detectable, albeit at lower levels compared with wild-type RasG. This suggests the existence of yet another RasG ubiquitination pathway that is independent of RasG activity. However, such a pathway may not be relevant for RasG degradation, as T35A-RasG is more stable compared with wild-type RasG (Fig. 2B).
Ubiquitination Sites of RasG Are Located in the Guanine Nucleotide-binding Domain and C-terminal Hypervariable Region
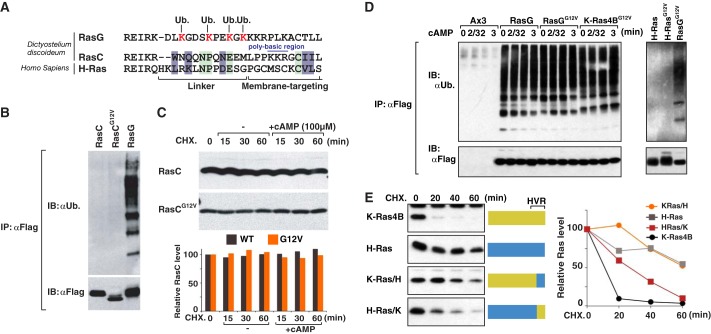
To gain further insight into the role of RasG ubiquitination, we determined ubiquitination sites in RasG. FLAG-RasG protein and its ubiquitinated forms were purified with anti-FLAG resin followed by FLAG peptide elution. The bands migrating at the positions for mono- and diubiquitinated form of RasG were digested by trypsin and subjected to LC-MS/MS analysis. Major ubiquitination sites were found at Lys residues 169, 173, 176, and Lys-178, and a minor fraction of Lys-117 ubiquitination was found (Fig. 3 A–C)5. The four major ubiquitination sites are located in the C-terminal HVR. HVR can be subdivided into two regions, the linker region and membrane-targeting region (19, 20, 24, 25). Interestingly, all four ubiquitinated lysine residues are evolutionarily conserved from Dictyostelium RasG to human K-Ras4A and K-Ras4B and are all located in the linker region (Fig. 3A).
FIGURE 3.
Ubiquitination of RasG occurred on K-Ras specific lysine residues. A, ubiquitination (Ub.) sites in RasG was identified by tandem mass spectrometry and shown in the red boxed lysine residues. The sequence of the RasG ubiquitination sites and corresponding region of human Ras isoforms are aligned (lower panel). The identified ubiquitination sites were mutated to arginine residues in 4KR and 7KR RasG mutants as shown in the lower panel. B–D, diagram of LC/MS/MS identified in vivo ubiquitination sites in RasG and ubiquitin. FLAG-tagged RasG was stable expressed and immunoprecipitated with anti-FLAG antibody. The IP was run out on SDS-PAGE, stained with Coomassie, and the band corresponding to monoubiquitinated RasG (B), diubiquitinated (di-Ub.) RasG (C and D) was cut out, and isolated and subjected to tryptic digest and LC/MS/MS analysis. Amino acid coverage map showing the tryptic peptides sequenced by LC-MS/MS are in dark green and the detected ubiquitination sites are highlighted in magenta. Light green and blue highlight oxidations of Asn/Gln and Met, which are most likely an in vitro processing artifact.
RasG Receives Lys-48-linked Polyubiquitination
Linkage-specific ubiquitination often determine the fate of the ubiquitinated proteins. Potentially seven different types of ubiquitin linkages can be formed, depending on whether the ubiquitins are attached to Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, or Lys-63 on the ubiquitin (26–28). To determine the ubiquitin linkage, we undertook the LC-MS/MS analysis. LC-MS/MS detected 90% of entire ubiquitin protein from the diubiquitinated RasG. Strikingly, we found that diubiquitinated RasG contained Lys-48-linked ubiquitination in the diubiquitinated form of RasG (Fig. 3D), whereas there was no ubiquitinated fragment of other regions detected, including Lys-63.
To further verify the result, we immunoprecipitated RasG for Western blot analysis and then blotted the membrane with linkage specific ubiquitin antibodies. We observed a strong signal by Lys-48-linkage-specific ubiquitin antibody. Next we tested whether polyubiquitinated form of RasG contain Lys-63-linked ubiquitin. The anti-Lys-63-linked ubiquitin antibody displayed robust signal in the total cell lysate fraction, which are distinctive pattern compared with the anti-Lys-48-linked ubiquitin antibody, consistent with previous reports (26–28). However we could not detect Lys-63-linked ubiquitin signal in the RasG-immunoprecipitated sample (Fig. 4A). These data are consistent with the LC-MS/MS result (Fig. 3D).
FIGURE 4.
Lys-48-linked ubiquitination of RasG regulates RasG stability and cytokinesis. A, WT-RasG receives Lys-48-linked polyubiquitination. FLAG-tagged WT- and 7KR-RasG mutants cells were treated with the indicated proteasome inhibitors for 40 min. WT- and 7KR-RasG were immunoprecipitated (IP) from total cell lysate (TCL) and immunoblotted (IB) with the indicated antibodies. Only MG132 displayed inhibition of proteasome inhibition, leading accumulation of polyubiquitinated proteins. B, 4KR- and 7KR-RasG mutants decreased ubiquitination level. Ubiquitination level of the indicated RasG mutants was assessed as in Fig. 1A. These blots are representative of two separate experiments. Relative polyubiquitinated RasG levels were assessed by quantification of poly- and non ubiquitinated (Ub.) RasG signal intensity by ImageJ software. Polyubiquitination signal was divided by non-ubiquitinated RasG signal to estimate the relative ubiquitination level of WT- and RasG mutants. C, 4KR- and 7KR-RasG mutants increased protein stability. Protein stability of the indicated RasG mutants was assessed as in Fig. 2. These blots are representative of two separate experiments. D, 4KR-RasG mutant increased number of nuclei in suspension culture. WT and 4KR mutant expressing RasG -null cells were shaken for 3 days, plated, and stained with DAPI. Random fields were photographed, and fractions of cells containing the designated number of nuclei were determined. Averages of two experiments comprising >200 cells for each line are shown. These blots are representative of two separate experiments. DMSO, dimethyl sulfoxide.
To further verify the finding, we examined the effect of proteasome inhibitors on ubiquitination of RasG. We tested four proteasome inhibitors, which have been widely used, and increased concentration to 5∼10-fold the commonly used dose. Unfortunately, lactacystin, epoxomicin, and ALLN did not induce any detectable changes of ubiquitination signal of cellular proteins. This is in agreement with the previous studies that Dictyostelium cellular proteins were refractory to MG132, MG262, and lactacystin, presumably due to the restricted permeability to and high rate of drug exclusion from the Dictyostelium or both (29, 30). At 100 μm concentration, the treatment of MG132 slightly induced accumulation of the polyubiquitinated cellular proteins (Fig. 4A). Under this condition, there was further accumulation of the Lys-48-ubiquitinated RasG. This suggests that Lys-48-linked ubiquitination of RasG is degraded, if not all, via the ubiquitin-proteasome system.
C-terminal Lysine Cluster Ubiquitination Leads to RasG Destabilization
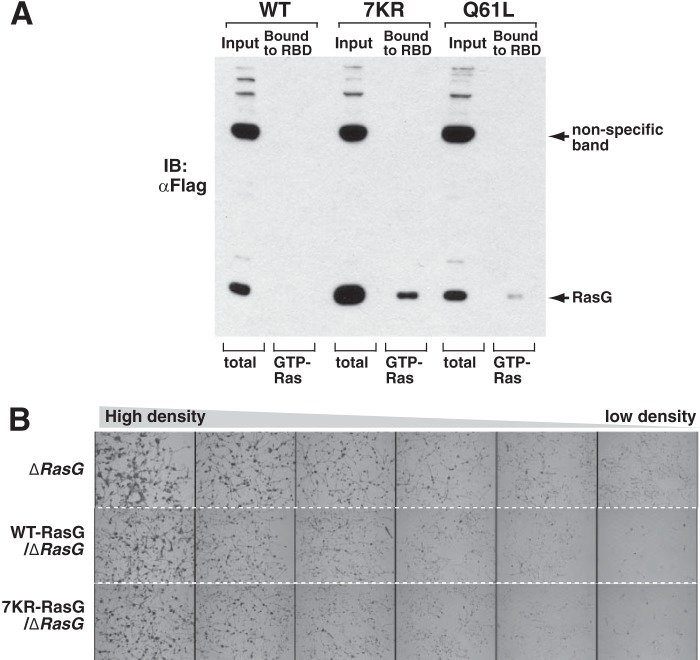
To further investigate the function of ubiquitination in the HVR of RasG, we made a RasG mutant where all four lysines found to be ubiquitinated are substituted with arginines (4KR-RasG). We also made a mutant that has the seven lysine residues in the membrane-targeting region substituted for arginine residues (7KR-RasG) (Fig. 4, A and B). FLAG-tagged 4KR-RasG and 7KR-RasG mutants were stably expressed into RasG-null cells, and their ubiquitination levels were assessed. Fig. 4B shows that the ubiquitination level of 7KR-RaG was 9 times lower than that of wild-type RasG. The results of Figs. 1B and 2A suggest that activated-RasG receives polyubiquitination. Importantly, we found that polyubiquitination levels were increased ∼2 times in G12V-RasG and Q61L-RasG constitutively active mutants, which are locked in GTP-bound state, compared with that of wild-type RasG. Consistent with the results of Figs. 1 and 2, this suggests the activated form of RasG receives polyubiquitination. To further verify this, we introduce mutation of 4KR into Q61L-RasG rather than WT-RasG because 4KR mutation may reduce RasG activation by non-ubiquitin-related pathway. Strikingly, 4KR-Q61L-RasG reduced ubiquitination level, 17 times less than Q61L-RasG (Fig. 4B). The results indicated that these four lysine residues found by mass spectrometry analysis were bona fide ubiquitination sites. However, both 7KR- and 4KR-Q61L mutants still receive a small fraction of polyubiquitination, suggesting that there is another site(s) for polyubiquitination. It is also possible that the mutation of the C-terminal lysine may lead to ubiquitination of another site(s), which is otherwise not ubiquitinated.
We have further investigated the role of the HVR ubiquitination on RasG protein stability. As shown in Fig. 4C, wild-type RasG protein level was quickly decreased after CHX treatment, while 4KR- and 7KR-RasG mutants significantly increased protein stability. Consistent with the elevated Lys-48-linked polyubiquitination level (Fig. 4, A and B), the degradation of G12V- and Q61L-RasG was faster than that of wild-type RasG. Importantly, Q61L-RasG destabilization was completely reversed by the four-lysine residue mutation. These results indicate that ubiquitination of HVR in RasG regulates RasG protein stability.
RasG Ubiquitination Pathway Is Critical for Cytokinesis
RasG is critical for cytokinesis (20, 21, 31). Recently, it has been shown that Cdc42, a Rho family GTPase essential for cytokinesis, is down-regulated during mitotic exit, which is required for proper cytokinesis (22, 23, 32). To assess the functional significance of RasG ubiquitination, we assessed cytokinesis of wild-type- and 4KR-RasG-expressing RasG-null cells using suspension culture where cytokinesis-deficient mutants become multinucleated (20, 24, 25, 33, 34). Consistent with previous reports, RasG-null cells became multinucleated after 3 days of shaking culture in suspension. The expression of wild-type RasG prevented the emergence of multinucleated cells (Fig. 4D). However, the expression of 4KR-RasG mutant failed to reverse the cytokinesis defect of RasG-null cells.
Next, to test whether KR-mutant retains activity, we first performed a biochemical assay. The GST-Ras-binding domain pulldown assay reveals that 7KR mutant has high basal activity, which is more than Q61L mutant. Thus, the 7KR mutant undergoes GTP loading and binds to its effector molecules (Fig. 5A). However, the 7KR mutant may no longer be the same as WT-RasG because the mutant protein level is higher than the WT, similar to its basal activation levels.
FIGURE 5.
Effect of ubiquitination on the basal RasG activity and developmental activity. A, 7KR-RasG increases basal activity. RasG wild-type (WT) or mutants were stably expressed in ΔRasG cells, and their activation level was analyzed by a GST-RBD pulldown assay of Ras as described in Ref. 51. Q61L-RasG has much higher basal activity than WT-RasG. 7KR mutant increases basal activity, presumably because the 7KR-RasG could evade from the activation-dependent RasG degradation. B, 7KR-RasG cells retain developmental activity, similar to WT-RasG cells. RasG wild-type (WT) or 7KR mutant were stably expressed in ΔRasG cells and plated on non-nutrient agar to induce the developmental process as described previously (52, 53). Although the impeding mutation of GTPase activity of RasG has shown to attenuate the developmental process (35), the 7KR-RasG cells displayed highly comparable developmental progression to WT-RasG cells. IB, immunoblot.
Therefore, we next examined whether 7KR-RasG has a role in developmental process of Dictyostelium, which requires proper regulation of chemotaxis and gene expression. The cells were seeded on the development plate at various cell densities, and their developmental processes were monitored for the developmental process for 48 h. Interestingly, although constitutive RasG activation has been shown to lead to developmental defects in Dictyostelium (35), the 7KR-RasG mutant cells have normal developmental functioning, indistinguishable to that of WT-RasG cells (Fig. 5B). The biochemical and developmental analyses suggest that ubiquitin-mediated regulation of RasG is not involved in the normal developmental process even though the 7KR-mutant has higher basal Ras activity. It should be noted that cells under the developmental assay condition, which does not have nutrients, do not proliferate. Furthermore, cytokinesis defect of RasG cells are only prominent when they are cultured in suspension (20, 24, 25). Thus, the cytokinesis defect of RasG mutant cells was suppressed in the developmental assay where cells were plated on agar.
Cytokinesis failure is one of the well described events that induce aneuploidy and genomic instability (36–38). Furthermore, it has been shown that Ras is activated at the cellular poles during cytokinesis in Dictyostelium (14, 31, 39, 40) and that K-Ras is shown to have an important role in cytokinesis and genome stability in mammalian cells (32, 41, 42). The findings in Dictyostelium RasG potentially reflect the circumstance of tumorigenesis driven by hyper-activation of K-Ras: the expression of mutated K-Ras not only activates downstream signaling but also induces aneuploidy and promotes tumorigenesis (33, 34, 43, 44).
H-Ras Homologue, RasC, Does Not Undergo Ubiquitination
Next, we investigated whether other Ras isoforms in Dictyostelium are ubiquitinated similarly to RasG. To test this possibility, we focused on Dictyostelium RasC, a functional orthologue of human H-Ras, which is a critical mediator of chemoattractant-induced PI3K and AKT activation (36, 38). RasC does not have lysine residues in its linker region, and RasC shares several residues in the linker region with mammalian H-Ras (Fig. 6A). FLAG-tagged wild-type RasC was stably expressed into RasC-null mutant cells (WT-RasC/ΔRasC). The expression of RasC complemented the developmental defect of RasC-null cells indicating that the exogenously expressed RasC protein was functional. Unlike RasG, we did not detect ubiquitination of RasC (Fig. 6B). We also expressed G12V-RasC, a mutant that is constitutively bound to GTP and found that G12V-RasC did not undergo ubiquitination either. Consistent with this finding, both wild-type and G12V-RasC proteins are very stable (Fig. 6C). These results reveal that RasC is refractory to ubiquitination-mediated degradation.
FIGURE 6.
Ubiquitination specifically occurs on K-Ras, but not H-Ras. A, the sequence of the RasG ubiquitination (Ub.) sites and corresponding region of Dictyostelium RasC and human H-Ras are aligned. B, Dictyostelium RasC does not undergo ubiquitination. FLAG-tagged WT- and G12V-RasC were stably expressed in RasC-null cells. Ubiquitination level of RasC and RasG was assessed as in Fig. 1. These blots are representative of two separate experiments. C, RasC is a highly stable protein. Protein stability of RasC was assessed as in Fig. 2. These blots are representative of two separate experiments. D, human K-Ras undergo polyubiquitination, whereas human H-Ras is refractory to ubiquitination in Dictyostelium. FLAG-tagged K-Ras and H-Ras were stably expressed in ΔRasG cells. The ubiquitination level of K-Ras and H-Ras were assessed as in Fig. 1. These blots are representative of two separate experiments. E, C-terminal HVR determined the protein stability of K-Ras and H-Ras in Dictyostelium. A schematic diagram of K-Ras and H-Ras chimeric mutants (left). Human K-Ras, H-Ras, and chimeric mutants were stably expressed in ΔRasG cells, and their protein stability was assessed as in Fig. 2. These blots are representative of two separate experiments. IB, immunoblot.
Ubiquitination Specifically Occurs on K-Ras Orthologue, but Not on H-Ras Orthologue, in Dictyostelium
Sequence alignment revealed that the identified ubiquitination sites of RasG were conserved in mammalian K-Ras 4A and 4B, but not in H-Ras (Figs. 1A and 6A). Therefore, we next examined whether human K-Ras protein could be recognized for ubiquitination in Dictyostelium cells. Human G12V-K-Ras4B was stably expressed into RasG-null cells and immunoprecipitated with anti-FLAG agarose. Western blotting with anti-ubiquitin antibody revealed that G12V-K-Ras was highly ubiquitinated in Dictyostelium at levels similar to RasG ubiquitination (Fig. 6D). To further examine whether human Ras proteins undergo ubiquitination in Dictyostelium in an isoform-specific manner, human wild-type and G12V-H-Ras were stably expressed in Dictyostelium. Fig. 6D, right panel, shows that in Dictyostelium, ubiquitination of wild-type H-Ras and G12V-H-Ras was not observed in contrast to the ubiquitination of K-Ras. The most significant difference of K-Ras and H-Ras is their C-terminal HVR region. To further examine the importance of K-Ras HVR for ubiquitination, we have generated a chimeric H-Ras protein that harbors the HVR of K-Ras (H-Ras/K) and a chimeric K-Ras that harbors the HVR of H-Ras (K-Ras/H). Fig. 6E shows that a half-life of K-Ras/H protein prolonged about six times more than that of wild-type K-Ras, a half-life of H-Ras/K mutant was about three times less than wild-type H-Ras.
Using biochemical and genetic approaches, we have found that RasG, a Dictyostelium K-Ras orthologue, undergoes ubiquitination and that the extent of ubiquitination is dependent on downstream effector signaling. Ubiquitination specifically occurred on lysine residues that are conserved in K-Ras HVR linker domain. RasG ubiquitination regulates RasG protein levels and is required for proper cytokinesis. Ectopically expressed human K-Ras can be polyubiquitinated in Dictyostelium. However, human H-Ras and Dictyostelium RasC, a H-Ras orthologue, are refractory to ubiquitination. Thus, Dictyostelium has a ubiquitination system that is specific for activated RasG that can also target human K-Ras.
Recently, three types of ubiquitin-mediated Ras regulatory pathways have been discovered: 1) ubiquitination of Ras isoforms at Lys-117 or Lys-147 leads to RasGEF-independent Ras activation (14, 39, 40, 45); 2) ubiquitination of H-Ras by the E3-ubiquitin ligase RabEX5 results in H-Ras endomembrane translocation (41, 42); 3) ubiquitination of phosphorylated Ras isoforms by βTrCP, another E3 ligase, results in Ras proteasome-mediated degradation (43, 44). So far, K-Ras-specific degradation has not been described in any system. Thus, this is the first report that demonstrates a negative feedback regulation of a K-Ras orthologue. Furthermore, it is conceivable that ubiquitination at the K-Ras-specific lysines may alternate subcellular localization of RasG because of its proximity to the HVR.
There are >500 E3 ligases in the human genome, whereas the Dictyostelium genome has <100 potential E3-ligases. In our database search, there was no putative E3 ligase that harbors a Ras binding domain. Because E3 ligase often consists of several subunits, we assume that the activated RasG/K-Ras-specific E3 ligase may form a protein complex that contains subunit binding to activated-Ras. We also speculate that RasG/K-Ras E3 ligase may recognize GTP-RasG/KRas, which is in the binding status with its effector, to specifically regulate effector pathway. This could be analogous mechanism to the ubiquitin-mediate effector selection of Ras, which we have reported recently (14, 39, 45). We expect that active RasG-specific E3 ligase has a role in cytokinesis, based on our observation. Thus, Dictyostelium mutants that have a cytokinesis defect may contain the RasG E3 ligase. Future biochemical purification and/or genetic analysis should reveal the responsible machinery for RasG ubiquitination.
Currently, it is unclear whether mammalian cells have a similar mechanism of K-Ras regulation. Several studies in human cancers revealed that increased Ras protein levels in tumors can be attributed to suppressing microRNAs against Ras and/or genomic amplification of the Ras locus. It is possible that a K-Ras-specific degradation system may act as tumor suppressor and thus suppressed in cancers. Consistent with this notion, it has been shown that expression of proapoptotic Ras effectors such as Nore1 and RASSFs (Ras association domain family) are frequently down-regulated by promoter methylation or chromosomal rearrangement in various cancers (46–50). These observations support the possibility that K-Ras degradation pathway might be conserved from Dictyostelium to human, which could be down-regulated in cancer cells. Future studies to identify an E3 ligase specific to RasG and K-Ras are required and may provide a novel approach to target K-Ras-mutated cancers.
Acknowledgments
We gratefully acknowledge the members of the University of Cincinnati Cancer Institute, the UC Brain Tumor Center and the Izayoi Meeting for stimulating discussions and helpful suggestions, Dr. Melanie Cushion, Dr. Antonis Koromilas, Mary Kemper, Dillon Chen, and Janice Connelly for help in critical reading and preparing this manuscript. We sincerely thank Dr. Lewis Cantley for the critical comments, support, and encouragement. We also thank Drs. Ronald Warnick, Sarah Kozma, Carol Mercer, and George Thomas for generous support and feedback. We thank Xuemei Yang and Min Yuan for help with mass spectrometry experiments.
This work was supported in part by National Institutes of Health Grants 2P01CA120964 (to J. M. A.) and 5P30CA006516 (to J. M. A.) and the University of Cincinnati (to A. T. S.).
J. M. Asara, unpublished observations.
- HVR
- hypervariable region
- CHX
- cycloheximide
- 7KR-RasG
- seven lysine residues in the membrane-targeting region substituted for arginine residues
- 4KR-RasG
- four lysines found to be ubiquitinated are substituted with arginines.
REFERENCES
- 1. Quinlan M. P., Quatela S. E., Philips M. R., Settleman J. (2008) Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol. Cell Biol. 28, 2659–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karnoub A. E., Weinberg R. A. (2008) Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haigis K. M., Kendall K. R., Wang Y., Cheung A., Haigis M. C., Glickman J. N., Niwa-Kawakita M., Sweet-Cordero A., Sebolt-Leopold J., Shannon K. M., Settleman J., Giovannini M., Jacks T. (2008) Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wennerberg K., Rossman K. L., Der C. J. (2005) The Ras superfamily at a glance. J. Cell. Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 5. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 6. Downward J. (2003) Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 7. Hancock J. F. (2003) Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 8. Malumbres M., Barbacid M. (2003) RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 9. Prior I. A., Hancock J. F. (2012) Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 23, 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mor A., Philips M. R. (2006) Compartmentalized ras/mapk signaling. Annu. Rev. Immunol. 24, 771–800 [DOI] [PubMed] [Google Scholar]
- 11. Sasaki A. T., Chun C., Takeda K., Firtel R. A. (2004) Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keller J. W., Haigis K. M., Franklin J. L., Whitehead R. H., Jacks T., Coffey R. J. (2007) Oncogenic K-RAS subverts the antiapoptotic role of N-RAS and alters modulation of the N-RAS: gelsolin complex. Oncogene 26, 3051–3059 [DOI] [PubMed] [Google Scholar]
- 13. Voice J. K., Klemke R. L., Le A., Jackson J. H. (1999) Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 274, 17164–17170 [DOI] [PubMed] [Google Scholar]
- 14. Sasaki A. T., Carracedo A., Locasale J. W., Anastasiou D., Takeuchi K., Kahoud E. R., Haviv S., Asara J. M., Pandolfi P. P., Cantley L. C. (2011) Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 4, ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A. (2001) Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl. Acad. Sci. U.S.A. 98, 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khosla M., Spiegelman G. B., Weeks G. (1996) Overexpression of an activated rasG gene during growth blocks the initiation of Dictyostelium development. Mol. Cell Biol. 16, 4156–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thiery R., Robbins S., Khosla M., Spiegelman G. B., Weeks G. (1992) The effects of expression of an activated rasG mutation on the differentiation of Dictyostelium. Biochem. Cell Biol. 70, 1193–1199 [DOI] [PubMed] [Google Scholar]
- 18. Jaffer Z. M., Khosla M., Spiegelman G. B., Weeks G. (2001) Expression of activated Ras during Dictyostelium development alters cell localization and changes cell fate. Development 128, 907–916 [DOI] [PubMed] [Google Scholar]
- 19. Gorfe A. A., Hanzal-Bayer M., Abankwa D., Hancock J. F., McCammon J. A. (2007) Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 50, 674–684 [DOI] [PubMed] [Google Scholar]
- 20. Tuxworth R. I., Cheetham J. L., Machesky L. M., Spiegelmann G. B., Weeks G., Insall R. H. (1997) Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolourani P., Spiegelman G., Weeks G. (2010) Ras proteins have multiple functions in vegetative cells of Dictyostelium. Eukaryot. Cell 9, 1728–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Onishi M., Ko N., Nishihama R., Pringle J. R. (2013) Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol. 202, 311–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Atkins B. D., Yoshida S., Saito K., Wu C. F., Lew D. J., Pellman D. (2013) Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 202, 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Lozanne A., Spudich J. A. (1987) Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091 [DOI] [PubMed] [Google Scholar]
- 25. Fukui Y. (1990) Structure and function of the cytoskeleton of a Dictyostelium myosin- defective mutant. J. Cell Biol. 110, 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim H. T., Kim K. P., Uchiki T., Gygi S. P., Goldberg A. L. (2009) S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 28, 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nathan J. A., Kim H. T., Ting L., Gygi S. P., Goldberg A. L. (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohanty S., Lee S., Yadava N., Dealy M. J., Johnson R. S., Firtel R. A. (2001) Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langenick J., Araki T., Yamada Y., Williams J. G. (2008) A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 121, 3524–3530 [DOI] [PubMed] [Google Scholar]
- 31. Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N., Firtel R. A. (2007) G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178, 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang G., Mercado-Uribe I., Multani A. S., Sen S., Shih I. e. M., Wong K. K., Gershenson D. M., Liu J. (2013) RAS promotes tumorigenesis through genomic instability induced by imbalanced expression of Aurora-A and BRCA2 in midbody during cytokinesis. Int. J. Cancer 133, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo J., Emanuele M. J., Li D., Creighton C. J., Schlabach M. R., Westbrook T. F., Wong K. K., Elledge S. J. (2009) A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker D. J., Dawlaty M. M., Wijshake T., Jeganathan K. B., Malureanu L., van Ree J. H., Crespo-Diaz R., Reyes S., Seaburg L., Shapiro V., Behfar A., Terzic A., van de Sluis B., van Deursen J. M. (2013) Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat. Cell Biol. 15, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reymond C. D., Gomer R. H., Nellen W., Theibert A., Devreotes P., Firtel R. A. (1986) Phenotypic changes induced by a mutated ras gene during the development of Dictyostelium transformants. Nature 323, 340–343 [DOI] [PubMed] [Google Scholar]
- 36. Charest P. G., Shen Z., Lakoduk A., Sasaki A. T., Briggs S. P., Firtel R. A. (2010) A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell 18, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujiwara T., Bandi M., Nitta M., Ivanova E. V., Bronson R. T., Pellman D. (2005) Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437, 1043–1047 [DOI] [PubMed] [Google Scholar]
- 38. Bolourani P., Spiegelman G. B., Weeks G. (2006) Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17, 4543–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker R., Lewis S. M., Sasaki A. T., Wilkerson E. M., Locasale J. W., Cantley L. C., Kuhlman B., Dohlman H. G., Campbell S. L. (2013) Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 20, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X. S., Shankar S., Dhanasekaran S. M., Ateeq B., Sasaki A. T., Jing X., Robinson D., Cao Q., Prensner J. R., Yocum A. K., Wang R., Fries D. F., Han B., Asangani I. A., Cao X., Li Y., Omenn G. S., Pflueger D., Gopalan A., Reuter V. E., Kahoud E. R., Cantley L. C., Rubin M. A., Palanisamy N., Varambally S., Chinnaiyan A. M. (2011) Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Disc. 1, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jura N., Scotto-Lavino E., Sobczyk A., Bar-Sagi D. (2006) Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679–687 [DOI] [PubMed] [Google Scholar]
- 42. Xu L., Lubkov V., Taylor L. J., Bar-Sagi D. (2010) Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr. Biol. 20, 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim S. E., Yoon J. Y., Jeong W. J., Jeon S. H., Park Y., Yoon J. B., Park Y. N., Kim H., Choi K. Y. (2009) H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitylation. J. Cell Sci. 122, 842–848 [DOI] [PubMed] [Google Scholar]
- 44. Jeong W. J., Yoon J., Park J. C., Lee S. H., Lee S. H., Kaduwal S., Kim H., Yoon J. B., Choi K. Y. (2012) Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci. Signal. 5, ra30. [DOI] [PubMed] [Google Scholar]
- 45. Baker R., Wilkerson E. M., Sumita K., Isom D. G., Sasaki A. T., Dohlman H. G., Campbell S. L. (2013) Differences in the regulation of k-ras and h-ras isoforms by monoubiquitination. J. Biol. Chem. 288, 36856–36862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vavvas D., Li X., Avruch J., Zhang X. F. (1998) Identification of Nore1 as a potential Ras effector. J. Biol. Chem. 273, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 47. van Engeland M., Roemen G. M., Brink M., Pachen M. M., Weijenberg M. P., de Bruïne A. P., Arends J. W., van den Brandt P. A., de Goeij A. F., Herman J. G. (2002) K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene 21, 3792–3795 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y. J., Ahsan H., Chen Y., Lunn R. M., Wang L. Y., Chen S. Y., Lee P. H., Chen C. J., Santella R. M. (2002) High frequency of promoter hypermethylation ofRASSF1A andp16 and its relationship to aflatoxin B1-DNA adduct levels in human hepatocellular carcinoma. Mol. Carcinog. 35, 85–92 [DOI] [PubMed] [Google Scholar]
- 49. Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M. (2002) Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12, 253–265 [DOI] [PubMed] [Google Scholar]
- 50. Pfeifer G. P., Dammann R., Tommasi S. (2010) RASSF proteins. Curr. Biol. 20, R344–5 [DOI] [PubMed] [Google Scholar]
- 51. Sasaki A. T., Firtel R. A. (2009) Spatiotemporal regulation of Ras-GTPases during chemotaxis. Methods Mol. Biol. 571, 333–348 [DOI] [PubMed] [Google Scholar]
- 52. Takeda K., Sasaki A. T., Ha H., Seung H. A., Firtel R. A. (2007) Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 282, 11874–11884 [DOI] [PubMed] [Google Scholar]
- 53. Lee S., Comer F. I., Sasaki A., McLeod I. X., Duong Y., Okumura K., Yates J. R., 3rd, Parent C. A., Firtel R. A. (2005) TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16, 4572–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]