Background: The endoplasmic reticulum aminopeptidase (ERAP) 1 is associated with HLA-B27+-ankylosing spondylitis.
Results: ERAP1 polymorphism at residues 528/575 affects the processing of HLA-B27 ligands in variant- and peptide-dependent and mutually dependent ways.
Conclusion: ERAP1 shapes HLA-B27 peptidomes through combined effects of co-occurring polymorphisms.
Significance: ERAP1-induced, peptide-mediated alterations of the immunological/pathogenetic features of HLA-B27 explain the epistasis of both molecules in ankylosing spondylitis.
Keywords: Aminopeptidase, Antigen Processing, Arthritis, Major Histocompatibility Complex (MHC), Pathogenesis, Peptides
Abstract
ERAP1 polymorphism involving residues 528 and 575/725 is associated with ankylosing spondylitis among HLA-B27-positive individuals. We used four recombinant variants to address the combined effects of the K528R and D575N polymorphism on the processing of HLA-B27 ligands. The hydrolysis of a fluorogenic substrate, Arg-528/Asp-575 < Lys-528/Asp-575 < Arg-528/Asn-575 < Lys-528/Asn-575, indicated that the relative activity of variants carrying Arg-528 or Lys-528 depends on residue 575. Asp-575 conferred lower activity than Asn-575, but the difference depended on residue 528. The same hierarchy was observed with synthetic precursors of HLA-B27 ligands, but the effects were peptide-dependent. Sometimes the epitope yields were variant-specific at all times. For other peptides, concomitant generation and destruction led to similar epitope amounts with all the variants at long, but not at short, digestion times. The generation/destruction balance of two related HLA-B27 ligands was analyzed in vitro and in live cells. Their relative yields at long digestion times were comparable with those from HLA-B27-positive cells, suggesting that ERAP1 was a major determinant of the abundance of these peptides in vivo. The hydrolysis of fluorogenic and peptide substrates by an HLA-B27 ligand or a shorter peptide, respectively, was increasingly inhibited as a function of ERAP1 activity, indicating that residues 528 and 575 affect substrate inhibition of ERAP1 trimming. The significant and complex effects of co-occurring ERAP1 polymorphisms on multiple HLA-B27 ligands, and their potential to alter the immunological and pathogenetic features of HLA-B27 as a function of the ERAP1 context, explain the epistatic association of both molecules in ankylosing spondylitis.
Introduction
ERAP1 is a multifunctional aminopeptidase of the endoplasmic reticulum involved in the final processing steps of major histocompatibility complex class I (MHC-I) ligands. Its role in this pathway is to trim peptides to the optimal size for MHC-I binding, which is 9–10 amino acid residues (1–3). Various nonsynonymous polymorphisms, involving amino acid changes in the protein (4, 5), are associated with ankylosing spondylitis (AS),4 a chronic form of arthritis typically affecting the axial skeleton and very strongly associated with HLA-B27 (6, 7). Because of strong linkage disequilibrium between polymorphisms within the same gene, it is often difficult to discern the contribution of each mutation to AS susceptibility or protection. Yet, it has been determined that this association fits a two-mutation model with contributions of both the single nucleotide polymorphisms rs301087 (coding for K528R) and rs10050860/rs17482078 (coding for D575N/R725Q), the two latter changes being indistinguishable due to tight linkage disequilibrium (8). The fact that the association of ERAP1 with AS concerns only the HLA-B27-positive disease (8, 9) strongly suggests that it is based on the functional interaction between ERAP1 and HLA-B27. In a recent study (10), we showed that the AS-associated natural polymorphism of ERAP1 variants altered the expression level of many HLA-B27-bound peptides in live cells. On the basis of that study, we proposed that the mechanism of functional interaction between these two proteins, and presumably the basis for their joint association with AS, is the alteration in the balance between epitope generation and destruction induced by ERAP1 polymorphism on the HLA-B27 peptidome.
Several in vitro studies using recombinant ERAP1 mutants and synthetic peptides have shown that the AS-protective changes Arg-528, Gln-725, and Glu-730 result in decreased enzymatic activity, whereas no effect on trimming was reported for R127P or D575N using single-residue mutants (8, 11, 12). In these studies, few peptides were tested. Because ERAP1 activity is influenced by the N-terminal residue of the substrate (13) and its sequence downstream from the N terminus (14), an incomplete and potentially misleading picture of the effect of a mutation might be obtained. In addition, the use of single mutants precluded analyzing possible combined effects among co-occurring mutations. This is particularly relevant because natural ERAP1 variants are complex allotypes that differ among each other by multiple amino acid changes (15–17), whose individual contribution to disease is not always distinguished by genetic analyses and whose concurrent influence on ERAP1 activity needs to be directly tested. Indeed, significant specificity differences, including some mediated by residue 575, were reported among natural ERAP1 variants with diverse combinations of amino acid changes, reflecting complex interactive effects in ERAP1 haplotypes on enzymatic activity (18).
In this study, four recombinant ERAP1 variants and a panel of synthetic peptides were used to analyze the context-dependent contribution of the AS-associated changes K528R and D575N to the processing of natural HLA-B27 ligands. We focused on these two residues for the following reasons: 1) as mentioned above, both K528R and D575N and/or R725Q are critical in the two-mutation model that explains the genetics of ERAP1 association with AS (8); 2) contrary to residue 725, which is known to affect ERAP1 activity (8), there are conflicting reports concerning D575N, although two studies (8, 11) found no effect of this mutation, a recent report (18) found a positive effect; and 3) both residues 528 and 575 are located outside the catalytic and peptide-binding sites of the enzyme and are thought to influence the domain rearrangements associated with the acquisition of an active ERAP1 conformation (19, 20). These three features are not fulfilled for other AS-associated polymorphisms. Two issues were specifically addressed as follows: 1) the influence of each mutation on the generation and destruction of natural HLA-B27 ligands; and 2) how the effect of a given residue in one position on ERAP1 activity and in the generation/destruction balance of HLA-B27 ligands was influenced by the residue at the other position.
EXPERIMENTAL PROCEDURES
ERAP1 Variants
The starting ERAP1 construct (a kind gift of Dr. S. C. Chang, National Taiwan University) was a pFastBac/myc-His-ERAP1 plasmid containing a full-length cDNA coding for an ERAP1 variant that, upon re-sequencing in our laboratory, showed four nonsynonymous substitutions (coding for Asp-346, Arg-514, Arg-528, and Glu-730) relative to the GenBankTM reference sequence NP-057526. Three additional ERAP1 variants (Table 1) were generated from the initial construct, hereby termed wild type, by PCR-mediated site-directed mutagenesis of codons 528 and 575. The primers used were as follows: R528K, 5′-cacttggacactgcagaagggttttcccctaataacc-3′ (sense) and 5′-ggttattaggggaaaacccttctgcagtgtccaagtg-3′ (antisense); D575N, 5′-gacattcatcaccagcaaatccaacatggtccatcgatttttgc-3′ (sense) and 5′-gcaaaaatcgatggaccatgttggatttgctggtgatgaatgtc-3′ (antisense).
TABLE 1.
ERAP1 variants used in this study
Residues associated with increased risk to AS are underlined. Residues in italics are altered relative to the reference sequence (NCBI accession number NP-057526), but have not been reported to influence AS susceptibility. Arg-528/Asp-575 is termed wild type because this was the starting variant from which the other three variants were generated by site-directed mutagenesis. SNP indicates single nucleotide polymorphism.
| SNP | Polymorphisma | R528/D575 (wild type) | Lys-528/Asp-575 | Arg-528/Asn-575 | Lys-528/Asn575 | C1Rb | WE-Ib |
|---|---|---|---|---|---|---|---|
| rs26653 | R127P | Arg | Arg | Arg | Arg | Pro | Pro |
| rs27895 | G346D | Asp | Asp | Asp | Asp | Gly | Gly |
| rs2287987 | M349V | Met | Met | Met | Met | Met | Val |
| rs78649652 | G514R | Arg | Arg | Arg | Arg | Gly | Gly |
| rs30187 | K528R | Arg | Lys | Arg | Lys | Arg | Arg |
| rs10050860 | D575N | Asp | Asp | Asn | Asn | Asp | Asn |
| rs17482078 | R725Q | Arg | Arg | Arg | Arg | Arg | Gln |
| rs27044 | Q730E | Glu | Glu | Glu | Glu | Glu | Glu |
a Naturally occurring amino acid changes are relative to the reference sequence.
b The sequence of ERAP1 from these cell lines was reported previously (10).
To our knowledge the variants in this study are not identical to any known natural allotypes. However, all the individual amino acid changes encoded in the four constructs are naturally occurring polymorphisms, and it is known from reported natural variants (10, 18) that individual polymorphisms occur in very diverse combinations. Thus, it is likely that the variants analyzed here may correspond to as yet undetected natural ones.
Generation of Recombinant Baculovirus
The correct products were confirmed by sequencing and subsequently used to transform competent DH10Bac Escherichia coli. Recombinant bacmids were isolated by standard DNA preparation methods and used to transfect Hi-5 adherent insect cells with Cellfectin II (both from Invitrogen) to produce the recombinant baculoviruses. These were harvested from the cell supernatant after 72 h, and its titer was determined by an immunofluorescence plaque assay with a mouse anti-His antibody (Qiagen, Hilden, Germany) and an Alexa 488 donkey anti-mouse antibody (Invitrogen), at 1:400 and 1:500 dilutions, respectively. Larger amounts of viruses were produced by infecting Hi-5 adherent cell cultures at a high multiplicity of infection and collecting the supernatant after 72 h.
Protein Expression and Purification
Recombinant ERAP1 proteins were produced in nonadherent Hi-5 insect cells grown in Express Five serum-free medium (Invitrogen). After infection with baculovirus carrying the ERAP1 gene construct, the culture medium containing the secreted enzyme was harvested by centrifugation (3000 × g, 30 min, 4 °C). The supernatant was concentrated in a Stirred Ultrafiltration Cell (Amicon, Millipore), adjusted to 50 mm phosphate, 300 mm NaCl, 10 mm imidazole, pH 8.0, and loaded onto a Poly-prep chromatography column (Bio-Rad) pre-loaded with nickel-nitrilotriacetic acid-agarose (Qiagen). The column was washed with the same buffer, containing 20 mm imidazole, and the protein was eluted with a 40–150 mm imidazole gradient. Protein elution was checked by SDS-PAGE. Protein-containing fractions were dialyzed in Vivaspin 500 (Sartorius Stedim Biotech, Goettingen, Germany) against 50 mm Tris/HCl buffer, 1 mm DTT, pH 7.4, aliquoted, and stored at −70 °C.
Fluorogenic Substrate Hydrolysis Assay
Three μg of Leu-7-amido-4-methylcoumarin (L-AMC) (Bachem Distribution, Wei an Rhein, Germany) in 50 μl of 1 m Tris/HCl buffer, pH 8.0, was mixed with 100 ng of ERAP1 in an equal volume of 50 mm Tris/HCl, 1 mm DTT, pH 7.4 (E/S ratio, 1:30), and incubated at 37 °C up to 1 h. The substrate hydrolysis was assessed by measuring its fluorescence, at 14-s intervals, at 380 and 460 nm excitation and emission wavelengths, respectively, in a Fluostar Optima Multiwave Plate Reader (BMG Labtec, Ortenberg, Germany).
Synthetic Peptides
A total of 10 peptides, including eight precursors of prominent natural HLA-B27 ligands and two fully processed ones (Table 2), were obtained using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and purified by HPLC (purity >80%). The correct molecular weight and sequence of the synthetic peptides were confirmed by MALDI-TOF mass spectrometry (MS) using a 4800 Proteomics Analyzer (Applied Biosystems).
TABLE 2.
Digestion of peptide precursors by ERAP1 variants
The data represent the maximal combined digestion (%) of peptide species longer than the natural HLA-B27 ligands (sequences highlighted in boldface) resulting from digestion of the synthetic precursor by the indicated ERAP1 variant. It was calculated as follows: 100 − % combined yield of the corresponding species. The minimal and maximal values observed with the peptides tested with each variant are highlighted in boldface. The fully processed natural ligands RRYQKSTEL (peptide 9) and RRYQKSTELLIR (peptide 10) are not included. The data are means ± S.D. of at least three experiments.
| Peptide | Arg-528/Asp-575, % maximum | Lys-528/Asp-575, % maximum | Arg-528/Asn-575, % maximum | Lys-528/Asn-575, % maximum | Trimming scorea |
||
|---|---|---|---|---|---|---|---|
| Mean of F.R. | P1 | ||||||
| 1 | GRHHEAS.IRLPSQYNF | 15.0 ± 3.0 | 15.9 ± 2.5 | 34.7 ± 1.1 | 79.8 ± 9.2 | 1.6 | 2 |
| 2 | LGVFRKF.SRFPEALRL | 6.8 ± 1.2 | 36.5 ± 3.4 | 76.7 ± 6.9 | 95.1 ± 6.7 | 1.7 | 2 |
| 3 | NLKARNS.FRYNGLIHR | 48.4 ± 5.4 | 51.3 ± 2.4 | 63.7 ± 1.8 | 84.9 ± 1.7 | 2.0 | 2 |
| 4 | IMYKKRT.KRLVVFDAR | 29.6 ± 2.5 | 32.8 ± 0.1 | 29.9 ± 3.0 | 77 ± 2.7 | 2.0 | 1 |
| 5 | DVVYAL.KRQGRTLYGF | 10.5 ± 0.7 | 3.6 ± 0.5 | 24.6 ± 2.1 | 99.9 ± 0.5 | 2.5 | 1 |
| 6 | LYSESL.ARYGKSPYLY | 79.8 ± 0.3 | 72.8 ± 2.3 | 79.2 ± 4.1 | 77.9 ± 2.6 | 2.8 | 4 |
| 7 | LREI.RRYQKSTEL | 48 ± 1.2 | 59.3 ± 3.4 | 73.7 ± 1.8 | 74.5 ± 5.3 | 2.0 | 1 |
| 8 | LREI.RRYQKSTELLIR | 50.9 ± 5.8 | 61.2 ± 2.1 | 67.7 ± 0.8 | 85.5 ± 1.3 | 2.0 | 1 |
| Mean | 36.1 | 41.7 | 56.3 | 84.3 | |||
a Trimming susceptibility scores for each residue are based on relative cleavage efficiencies as determined in a previous study (13): score 1, 0–25% (T, H, Q, G, N, E, W, D, K, V, R, P); 2, 25–50% (F, I, S); 3, 50–75% (C); 4, 75–100% (A, L, M, Y). For each substrate the mean score of all the flanking residues (mean of F.R.) and the score of the N-terminal residue of the natural ligand (P1) are shown.
Peptide Trimming Assays
One μg of each peptide was incubated with 100 ng of ERAP1 (E/S ratio 1:10) in 50 μl of 50 mm Tris/HCl buffer, 1 mm DTT, pH 7.4, at 37 °C at various times from for 5 min to 4 h. After incubation, the reaction was stopped with 5 μl of 5% trifluoroacetic acid. The peptides in the reaction mixtures were purified with OMIX C18 pipette tips (Varian Inc. Palo Alto, CA) following the instructions of the manufacturer. The samples were dried down in a SpeedVac, dissolved in 1 μl of 30% acetonitrile, 15% isopropyl alcohol, and 0.1% trifluoroacetic acid, and sonicated for 3 min. The digestion mixtures were analyzed by MALDI-TOF MS in positive ion reflector mode at 25 kV in the mass-to-charge (m/z) range of 400–2200 as described previously (21). Peptide yields were estimated on the basis of the relative intensity of the respective ion peaks.
MS was used instead of the more conventional HPLC due to the significantly higher sensitivity of the former method and to the difficulties of separating similar and co-eluting peptide species in the relatively complex digestion mixtures generated from the long precursors used in our study. Yet to confirm the reliability of MS-based estimations, we carried out pilot experiments to assess the correspondence between both detection methods. In those experiments, the precursor peptide 7 (LREI.RRYQKSTEL) was digested with Arg-528/Asp-575 or Lys-528/Asn-575 and analyzed at every time point by MS and HPLC.
Identification of HLA-B27 Ligands in Live Cells
This was performed exactly as described previously (10). Briefly, HLA-B27-bound peptides were isolated from C1R-B*27:04 or -B*27:05 transfectant cells and from the B*27:04+ lymphoblastoid cell line Wewak I by immunopurification of HLA-B27 and acid extraction and fractionated by HPLC. Each chromatographic fraction was analyzed by MALDI-TOF MS, and individual peptides were identified by MS/MS sequencing.
RESULTS
Context-dependent Effects of AS-associated ERAP1 Polymorphism on L-AMC Hydrolysis
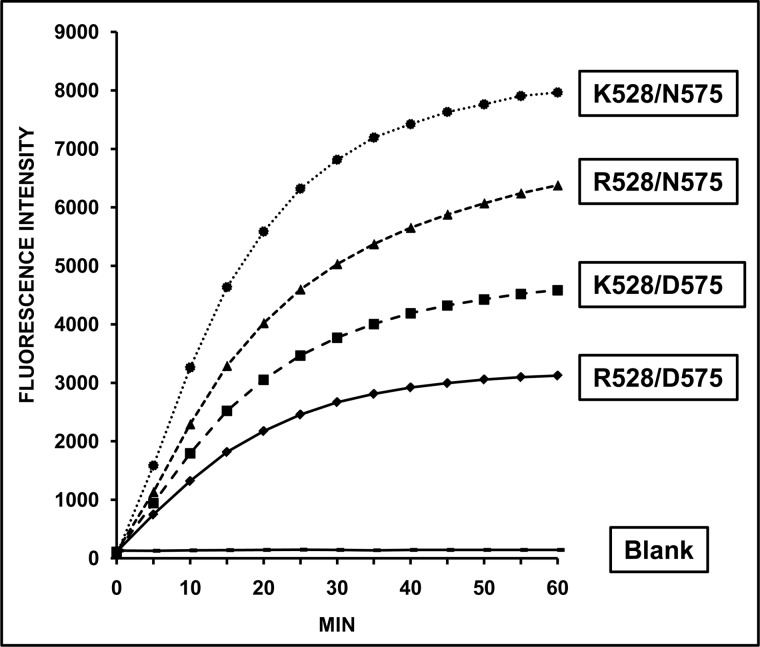
The relative activity of four ERAP1 variants differing at residue 528 (Lys/Arg), 575 (Asp/Asn), or both (Table 1) was assessed by measuring the hydrolysis of the fluorogenic L-AMC substrate as a function of time (Fig. 1). Substantial differences were observed with the following activity ranking: Lys-528/Asn-575 > Arg-528/Asn-575 > Lys-528/Asp-575 > Arg-528/Asp-575. These results indicate the following: 1) Lys-528 confers higher activity than Arg-528 if residue 575 is the same; 2) Asn-575 confers higher activity than Asp-575 regardless of residue 528; 3) the relative activity of variants differing by the K528R change depends on residue 575: Lys-528/Asn-575 > Arg-528/Asn-575 and Lys-528/Asp-575 > Arg-528/Asp-575, but Arg-528/Asn-575 > Lys-528/Asp-575; and 4) this is due to the fact that the D575N mutation increased ERAP1 activity more than R528K in this system.
FIGURE 1.
Hydrolytic activity of ERAP1 variants toward L-AMC. The indicated ERAP1 variants were incubated with the fluorogenic substrate at an E/S ratio of 1:30 (w/w), and the fluorescence intensity of the generated fluorophore was measured as a function of time. The data are means of 10–13 experiments.
ERAP1 Polymorphism at Residues 528 and 575 Influences the Generation and Destruction of HLA-B27 Ligands in a Variant and Peptide-dependent Way
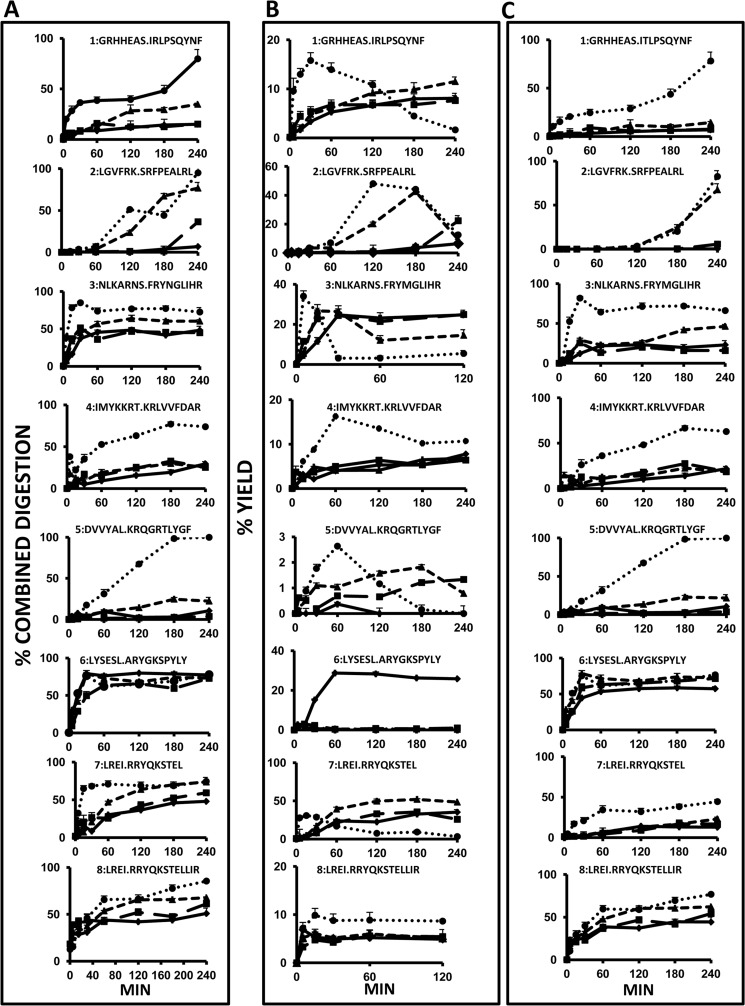
The effect of AS-associated polymorphism at each or both positions on the generation and destruction of natural HLA-B27 ligands was analyzed with 10 peptide substrates (Table 2). Peptides 1–6 were 16-mer precursors of natural ligands, including four nonamers and two decamers. In addition, two natural ligands, a 9-mer and a C-terminally extended 12-mer variant (peptides 9 and 10), as well as their N-terminally extended precursors with four flanking residues (peptides 7 and 8) were used. This panel was chosen to favor diversity of N- and C-terminal residues, as well as internal sequences, to assess peptide-dependent differences in ERAP1 trimming. The flanking and P1 residues were assigned a score of 1 to 4 based on the susceptibility of each residue to ERAP1 (Table 2). The peptides were digested at various times up to 4 h, because longer incubation times usually yielded little further digestion.
In the following analyses (Fig. 2), we computed the combined digestion of all the species longer than the natural ligand excluding the original substrate, designated as epitope precursors, the yield of the ligand, and the yield of shorter species, resulting from destructive cleavages, as a function time. The cumulative digestion of epitope precursors was chosen because the digestion rate of the enzyme is strongly dependent on the N-terminal residue of the substrate, so that the generation of the natural epitope will depend on the combined digestion rate of all the precursor species. The yield of the ligand at each time point is the epitope amount generated minus that destroyed at that time point.
FIGURE 2.
Digestion rate of synthetic peptide precursors of HLA-B27 ligands by ERAP1 variants. A, combined digestion (%) of peptide species longer than the natural ligand, excluding the original substrate. It was calculated as: 100% − the combined yield of the corresponding species; B, yield of the natural ligand; C, yield of peptides shorter than the natural ligand, resulting from destructive cleavages. ♦, Arg-528/Asp-575; ■, Lys-528/Asp-575; ▴, Arg-528/Asn-575; ●, Lys-528/Asn-575. Yields are relative to the total amount of peptide, which was estimated as the added intensity of the ion peaks corresponding to each peptide species in the MALDI-TOF MS spectrum of the digestion mixture. The data are mean ± S.D. of 3–5 experiments. Note that for any given substrate and time point, the value in A is essentially accounted for by the added yield of the corresponding ligand and its digestion product(s) in B and C, respectively.
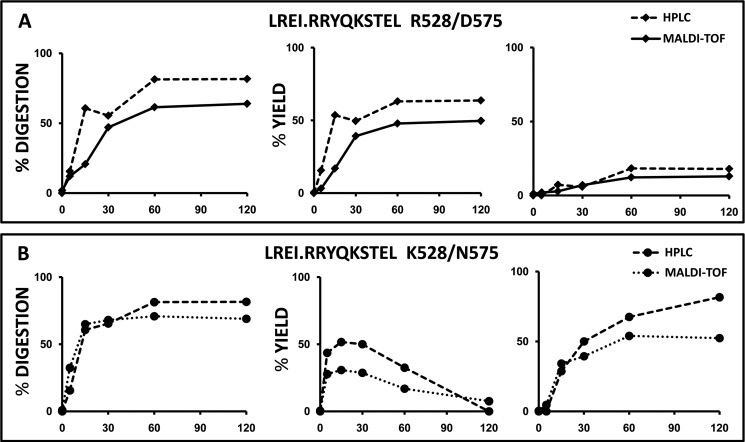
To validate the reliability of the MS-based assessment of yields of digestion products, we performed two comparative experiments in which one of the substrates in Fig. 2 (peptide 7) was digested with Arg-528/Asp-575 or Lys-528/Asn-575 and analyzed by MALDI-TOF MS and HPLC (Fig. 3). The results show that digestion rates and relative yields are similar with both techniques. The small differences observed are due to the higher sensitivity of MS that allows detection of peptide species present in very low amounts that go undetected by HPLC.
FIGURE 3.
Comparison of the digestion of a synthetic peptide substrate analyzed by MALDI-TOF MS and HPLC. A, peptide 7 was digested with Arg-528/Asp-575 at the indicated times in parallel experiments, and the digestion mixtures were analyzed by MALDI-TOF and HPLC, respectively. Conventions are as in Fig. 2. The left graph shows the combined digestion of peptide species longer than the natural ligand, excluding peptide 7 itself. Digestion was calculated as 100% − the combined yield of the corresponding species. The middle and right graphs show the % yield of the natural ligand and of shorter peptide(s), respectively. B, same substrate was digested with Lys-528/Asn-575 and analyzed as in A. The data in each panel are from a single experiment, but it was carried out separately for samples to be analyzed by either MS or HPLC.
Digestion of Epitope Precursors
The four ERAP1 variants showed distinct patterns of digestion that were both peptide- and variant-dependent (Fig. 2A). Distinct peptides were digested with very different rates and efficiencies by any given variant. The relative efficiency with which the four variants digested a given substrate also differed widely among peptides. Yet the average values of the maximal digestion of epitope precursors from peptides 1 to 8 increased in the following order: Arg-528/Asp-575 (36.1%) < Lys-528/Asp-575 (41.7%) < Arg-528/Asn-575 (56.3%) < Lys-528/Asn-575 (84.3%). Thus, the relative activity of the four ERAP1 variants toward peptidic substrates is the same as established with L-AMC. Peptide-dependent differences in the digestion of precursors correlated with the susceptibility of the flanking sequences to ERAP1 trimming (Table 2). These differences were much larger with the less active variants Arg-528/Asp-575 and Lys-528/Asp-575 (6.8–79.8 and 3.6–72.8%, respectively) than with the most active variant Lys-528/Asn-575 (74.5–99.9%). Thus, resistance of the flanking sequences to ERAP1 has a larger influence on the digestion of epitope precursors by less active variants. This is clear, for instance, when comparing peptides 1 and 2 with peptide 6 (Fig. 2A and Table 2).
Destructive Cleavages
The rate and efficiency of epitope destruction were estimated by measuring the combined yield of species shorter than the natural epitope as a function of time. In general, the patterns of epitope destruction (Fig. 2C) paralleled those of precursor digestions (Fig. 2A), showing similar variant and peptide-dependent differences. In addition, the mean maximal yield of species shorter than the natural HLA-B27 ligands for peptides 1–8 paralleled that of the digestion of precursors for each ERAP1 variant, revealing the same activity ranking (Table 3) as follows: Arg-528/Asp-575 (22.5%) < Lys-528/Asp-575 (26.8%) < Arg-528/Asn-575 (42.1%) < Lys-528/Asn-575 (75.9%). These results indicate that, in general, there is a concomitant and similar effect on both epitope generation and destruction resulting from the alterations in enzymatic activity induced by ERAP1 polymorphism.
TABLE 3.
HLA-B27 epitope destruction by ERAP1 variants
The data represent the maximal % yield of peptide species shorter than the natural HLA-B27 ligands (sequences highlighted in boldface) by the indicated ERAP1 variant. For each variant, the minimal and maximal (Max)values observed with the peptides tested are highlighted in boldface. Peptides 9 (RRYQKSTEL) and 10 (RRYQKSTELLIR) are not included. The data are means ± S.D. of at least three experiments.
| Peptide | Arg-528/Asp-575, % Max | Lys-528/Asp-575, % Max | Arg-528/Asn-575, % Max | Lys-528/Asn-575, % Max | |
|---|---|---|---|---|---|
| 1 | GRHHEAS.IRLPSQYNF | 6.9 ± 0.4 | 9.1 ± 0.9 | 14.4 ± 1.1 | 78.1 ± 9.2 |
| 2 | LGVFRKF.SRFPEALRL | 0.3 ± 0.1 | 5.6 ± 1.2 | 67.5 ± 6.9 | 82.6 ± 6.7 |
| 3 | NLKARNS.FRYNGLIHR | 23.1 ± 2.7 | 25.7 ± 0.8 | 46.4 ± 1.8 | 81.7 ± 1.7 |
| 4 | IMYKKRT.KRLVVFDAR | 21.8 ± 2.5 | 27.5 ± 0.1 | 22.4 ± 3.0 | 66.6 ± 2.7 |
| 5 | DVVYAL.KRQGRTLYGF | 10.5 ± 0.7 | 3.6 ± 0.5 | 22.8 ± 2.1 | 99.9 ± 0.5 |
| 6 | LYSESL.ARYGKSPYLY | 58.5 ± 2.6 | 71.5 ± 2.3 | 78.4 ± 4.1 | 76.6 ± 2.6 |
| 7 | LREI.RRYQKSTEL | 14.1 ± 0.3 | 17 ± 3.5 | 23 ± 2.2 | 44.5 ± 2.6 |
| 8 | LREI.RRYQKSTELLIR | 44.5 ± 3.1 | 54.2 ± 2.1 | 62.2 ± 0.8 | 76.8 ± 1.3 |
| Mean | 22.5 | 26.8 | 42.1 | 75.9 | |
Generation of HLA-B27 Ligands
All the natural ligands were generated with the four ERAP1 variants, albeit with widely different efficiencies (Table 4). Because of the concomitant increase of cleavages leading to generation and destruction of the epitopes as a function of ERAP1 activity, the mean maximal yield of the eight ligands was similar for Arg-528/Asp-575 (14.0%) and Lys-528/Asp-575 (12.5%) and only slightly higher with the most active ones Arg-528/Asn-575 (17.9%) and Lys-528/Asn-575 (19.2%). The average time at which the maximal yield of epitopes was obtained was longest with the less active variant Arg-528/Asp-575 (101.9 min) and shortest with the most active variant Lys-528/Asn-575 (35.6 min). A similar activity ranking was reflected at the mean earliest time at which the natural ligands were detected in the digestion mixtures (Table 4).
TABLE 4.
HLA-B27 epitope generation by ERAP1 variants
Detection indicates the time (in min) and % yield where the HLA-B27 ligand was detected at ≥1% of the total digestion mixture with the indicated ERAP1 variant. Because peptide 5 did not reach this value with Arg-528/Asp-575, the longest digestion time was assigned. Maximum indicates the earliest time (in min), and the % yield where the HLA-B27 ligand was detected at ≥80% of the maximal value observed with the indicated ERAP1 variant. The data are means ± S.D. of at least three experiments.
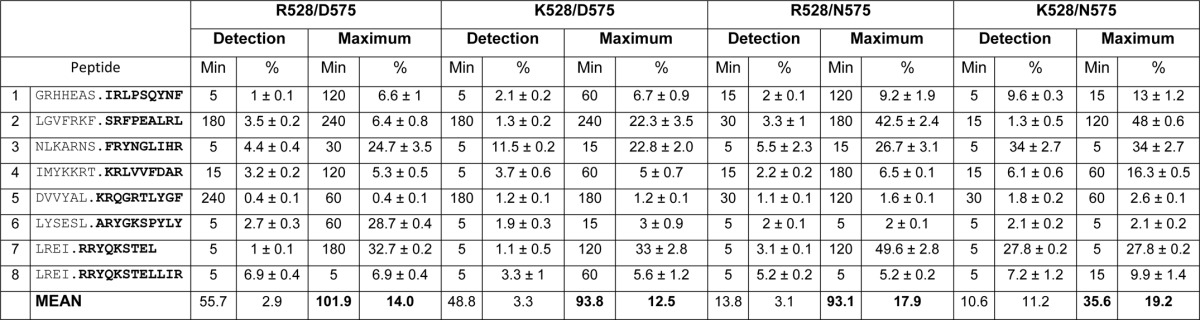
Significant peptide-to-peptide and variant-dependent differences were observed in the maximal amount of epitope and, more importantly, in the time at which this was obtained (Fig. 2B). Four situations were distinguished. For peptides 1, 3, 5, and 7, the highest epitope yields were obtained with the most active enzyme, Lys-528/Asn-575, at shorter times, and the tendency was reversed at longer times due to more efficient destruction of the epitope by this enzyme. A second pattern was observed with peptides 4 and 8. For these two substrates, the epitope yields were similar with Arg-528/Asp-575, Lys-528/Asp-575, and Arg-528/Asn-575 and were consistently higher with Lys-528/Asn-575 at all times. A third pattern concerned substrate 2. Again, the highest epitope yield was obtained at the shortest time with Lys-528/Asn-575, and the rate of production was slower than for all other peptides. Significant epitope amounts were also produced with Arg-528/Asn-575 but at a slower rate. The less active enzymes produced the epitope only at the longest digestion times. Finally, substrate 6 generated significant amounts of epitope only with the less active enzyme, Arg-528/Asp-575. With the three other variants, the epitope was detected in very low amounts and only at short times. This pattern was due to the very efficient digestion of substrate 6 by all four variants. However, although the digestion of the epitope precursors was slightly higher with Arg-528/Asp-575 (Fig. 2A), the destruction of the natural ligand by this variant was less efficient (Fig. 2C). Thus, the generation/destruction balance favored epitope production by the less active variant in this case. Significantly, peptide 6 had, among those in this study, the flanking and P1 residues most susceptible to ERAP1 trimming (Table 2). The earliest times at which the natural ligands were detected and their yields at these times also showed significant peptide-to-peptide and variant-dependent differences and reflected the same activity ranking among the four variants (Table 4).
In summary, these results indicate the following. 1) The four ERAP1 variants differ in their enzymatic activity toward peptide substrates with the same ranking as determined with L-AMC. 2) Globally, both the cleavages leading to generation and to destruction of the natural ligands increase with the enzymatic activity. 3) As a result, the mean maximal yield of the HLA-B27 ligands was rather similar for all four variants. 4) Significant differences were observed in the processing of individual substrates, both in the way (rate and yield) in which the same variant processed different substrates and in the way in which different variants processed the same substrate. 5) With only one exception epitope production was significantly higher at shorter times with the most active enzyme, while at longer times the differential epitope destruction by the different variants tended to decrease or even reverse relative epitope yields. 6) In one out of eight substrates analyzed, the generation/destruction balance favored the preferential production of the natural ligand by the less active ERAP1 variant.
Generation of Peptide Length Variants in Vitro
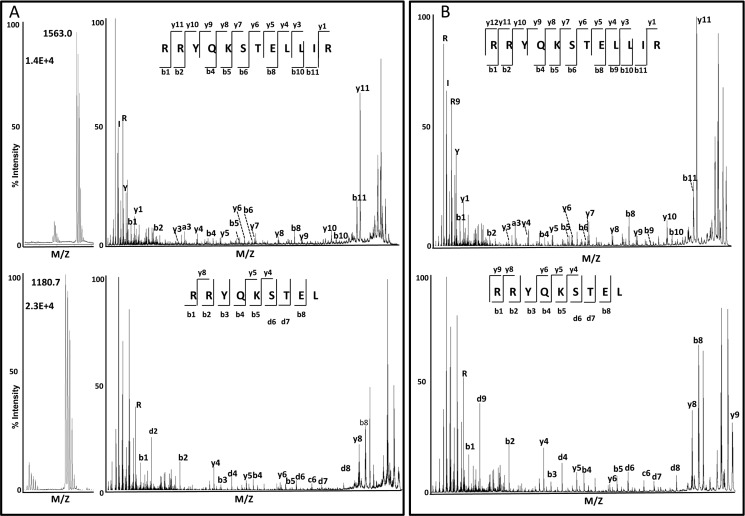
Related natural ligands differing in length by C-terminal extensions are frequent in the HLA-B27 and other MHC-I peptidomes. An example is RRYQKSTEL, a prominent ligand of B*27:05 and other HLA-B27 subtypes (22), and its C-terminally extended variant RRYQKSTELLIR, found in B*27:04 (10) and B*27:05 (Fig. 4). To examine the influence of ERAP1 polymorphism in the generation of these peptides, we digested their N-terminally extended precursors, peptides 7 and 8, and the natural ligands themselves (peptides 9 and 10) with the four ERAP1 variants.
FIGURE 4.
Identification of RRYQKSTELLIR and RRYQKSTEL from C1R-B*27:05 cells. A, MALDI-TOF MS spectra (left panel) from the HPLC fractions 150 and 121, respectively, corresponding to the maximum of the elution profile of each peptide during the fractionation of the B*27:05-bound peptide pool. Only the relevant ion peaks are shown. The sequence of both ligands was determined by MALDI-TOF/TOF MS/MS (right panel); B, MALDI-TOF/TOF MS/MS spectra of the corresponding synthetic peptides.
The cumulative digestion of epitope precursors from substrates 7 and 8 was very efficient with all four variants, especially with Arg-528/Asn-575 and Lys-528/Asn-575 (Fig. 2A). However, although for substrate 7 digestion of the natural ligand was very inefficient with all but the most active enzyme, destructive cleavages were more prominent on peptide 8 (Fig. 2C), as expected from the larger length of RRYQKSTELLIR. Thus, the yield of the shorter epitope RRYQKSTEL was significantly higher than that of RRYQKSTELLIR with all four enzymes (Fig. 2B). With the most active variant, the maximal yield of epitope 7 was obtained at short times and decreased later due to further digestion. With the three other variants, the longer epitope was generated at a faster rate, so that at the shortest times (up to 15 min) the yield of the longer ligand (about 3–7%, depending on time and enzyme variant) was comparable or even higher than the yield of the shorter one (about 1–5%).
The direct digestion of the natural ligands, peptides 9 and 10, showed dramatic differences (Fig. 5). RRYQKSTEL showed a significant resistance to ERAP1. At the longest digestion time only the most active variant fully digested the nonamer, whereas with the three other enzymes, about 35–55% of the peptide remained undigested. At shorter times, such as 60 min, as much as 40–88% of the ligand remained undigested with any given variant. In contrast, RRYQKSTELLIR was completely degraded by all four variants at 180 min of digestion. It was already fully digested after 15 min by Lys-528/Asp-575 and Arg-528/Asn-575 and about 85% by Lys-528/Asn-575. Only the less active variant, Arg-528/Asp-575, showed a significantly lower degradation rate with this peptide, which nevertheless was much faster than for RRYQKSTEL.
FIGURE 5.
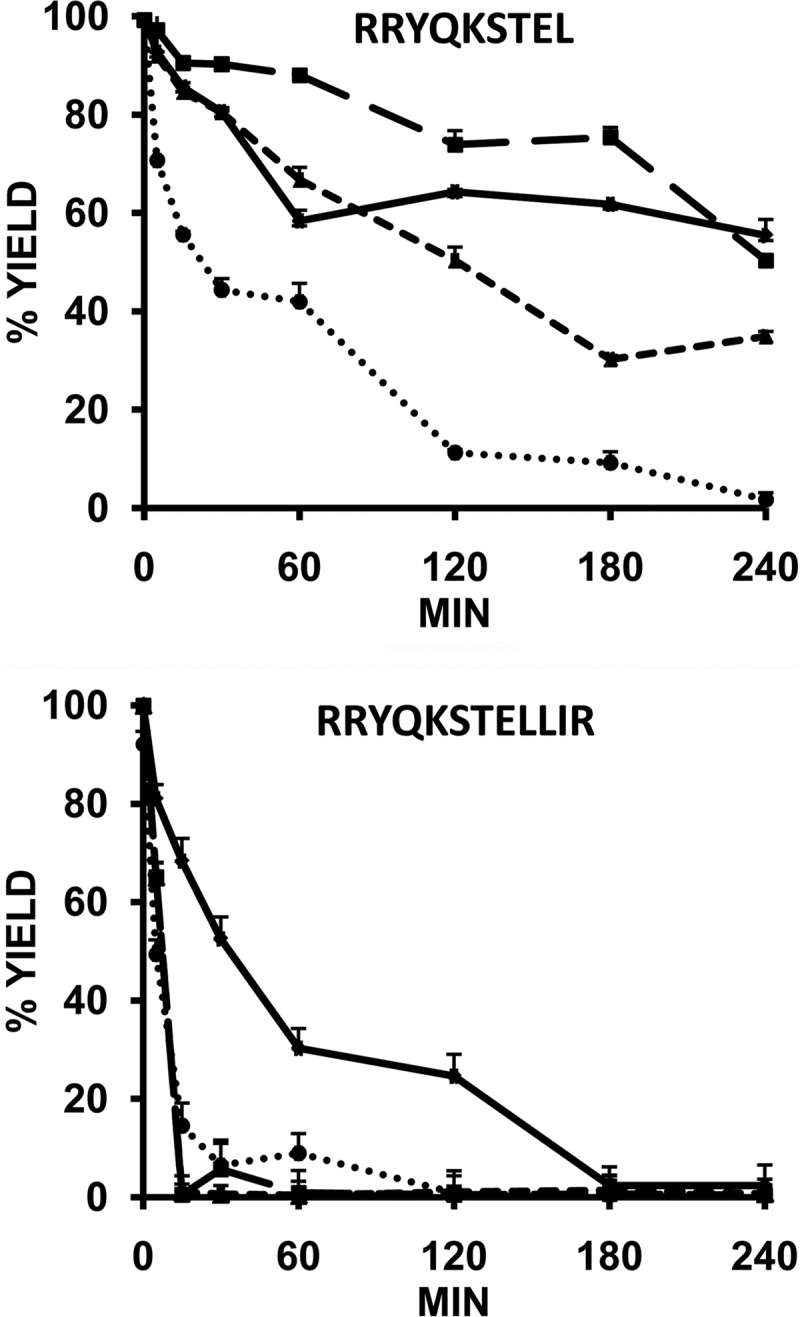
Digestion of two related HLA-B27 ligands of distinct length by ERAP1 variants. The indicated peptides were digested at an E/S ratio 1:10 (w/w) at various times with the following ERAP1 variants: Arg-528/Asp-575 (♦); Lys-528/Asp-575 (■); Arg-528/Asn-575 (▴); and Lys-528/Asn-575 (●).The data are means ± S.D. of three experiments.
Generation of Peptide Length Variants in Live Cells
We asked to what extent the more efficient generation of RRYQKSTEL, relative to RRYQKSTELLIR, observed in vitro might hold in live cells, where other variables can influence the endogenous processing of these peptides. Thus, we examined their recovery in the HLA-B*27:04 and B*27:05 peptidomes isolated from the lymphoid cell lines Wewak I (B*27:04+), C1R-B*27:04, and C1R-B*27:05. The endogenous ERAP1 variants of these cell lines (10) are not identical to any of those used in vitro, but ERAP1 in C1R has Arg-528 + Asp-575 and in Wewak I has Arg-528 + Asn-575 (Table 1). The HLA-B27 peptidome isolated from each cell line was fractionated by HPLC, and each fraction was analyzed by MALDI-TOF MS. Each of the peptides was identified by MS/MS sequencing of the corresponding ion peak in the MALDI-TOF spectrum (Fig. 4). The added intensity of the corresponding ion peak in the consecutive HPLC fractions in which the eluted peptide was detected was taken as an estimation of its abundance. Because of the nonquantitative nature of MALDI-TOF MS, the measurements were carried out in eight independent preparations from C1R-B*27:04, four from Wewak I and three from C1R-B*27:05, and the average intensity of the ion peak corresponding to each of the peptides in all the experiments was calculated for each cell line. The ratio between the average ion peak intensity obtained for the 9- and 12-mer was taken as an estimation of the relative recovery of both peptides in each cell line, and this was compared with the yield of the corresponding peptides upon in vitro digestion of their precursors by Arg-528/Asp-575 and Arg-528/Asn-575. The 9-mer was recovered with higher yield than the 12-mer in ratios of about 4–8-fold from the three cell lines. These ratios were very similar to those obtained in vitro at 60 min or larger digestion times (Table 5). These results suggest that, despite the many variables that can influence the amount of MHC-I ligands in vivo, the generation of RRYQKSTEL and RRYQKSTELLIR by ERAP1 may be a major determinant of their abundance in live cells.
TABLE 5.
Generation of length variants of HLA-B27 ligands in vitro and in live cells
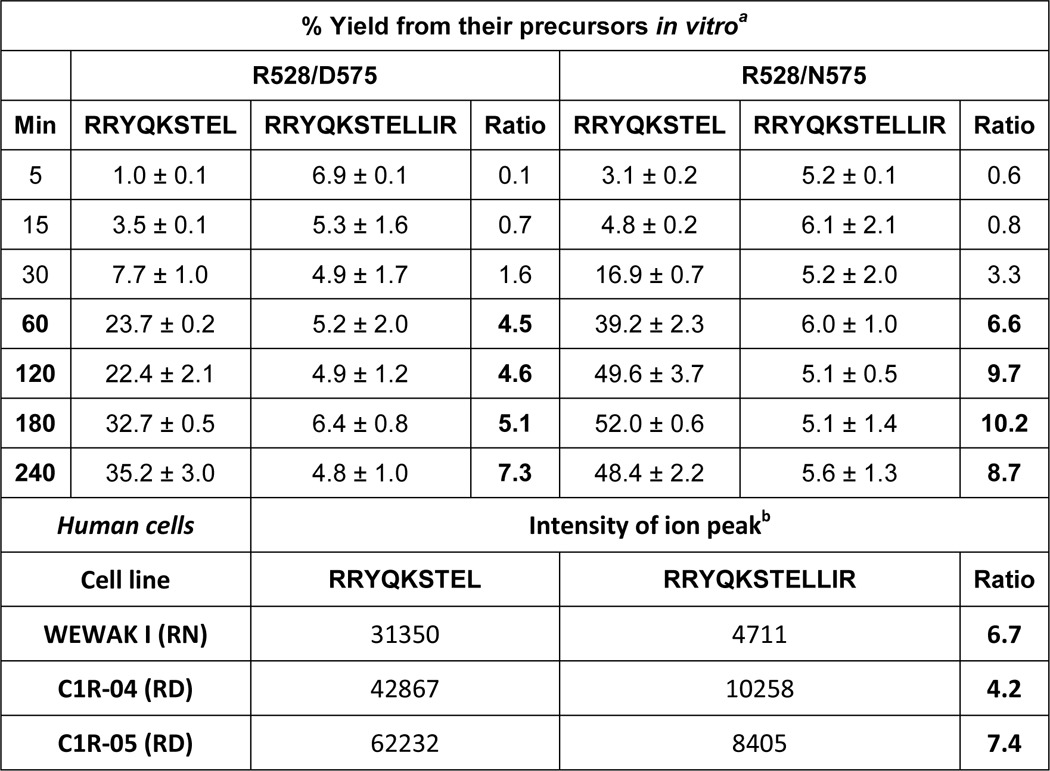
a The data are from the digestions of peptides 7 and 8 at the indicated times (Fig. 2B) and are means ± S.D. from three (RRYQKSTELLIR) or five (RRYQKSTEL) independent experiments.
b The data are means from four (Wewak I), eight (C1R-04), and three (C1R-05) independent preparations, respectively.
ERAP1 Polymorphism Influences the Competition between Peptidic and Nonpeptidic Substrates
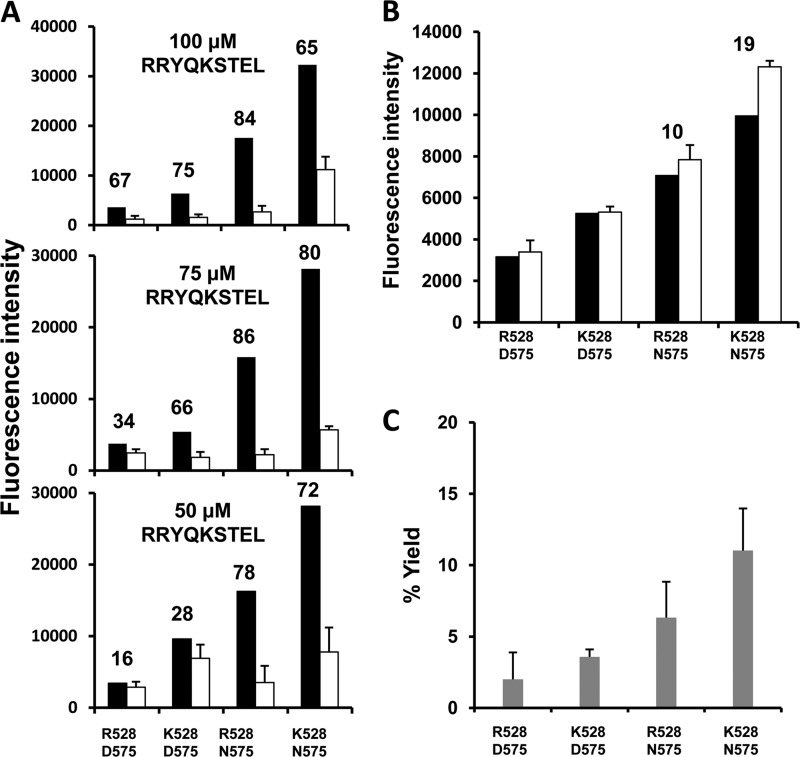
In the following experiments, we examined to what extent the polymorphism at residues 528 and 575 influenced the inhibition of L-AMC hydrolysis by a peptidic substrate. Each ERAP1 variant was incubated with L-AMC in the presence of various concentrations of the natural HLA-B27 ligand RRYQKSTEL (Fig. 6A). The peptide inhibited L-AMC hydrolysis with an efficiency that increased with the activity of the variant: the inhibition was lowest for Arg-528/Asp-575, increased for Lys-528/Asp-575, and was highest for Arg-528/Asn-575 and Lys-528/Asn-575. With the latter variant, a decreased inhibition was observed at the highest peptide concentration. Because short peptides that are not further trimmed by ERAP1 allosterically activate the hydrolysis of small nonpeptidic substrates (12, 19, 23), we tested the possibility that the diminished inhibition with Lys-528/Asn-575 was due to an activating effect of the RYQKSTEL octamer, arising from the digestion of the 9-mer. In the presence of 100 μm octamer, L-AMC hydrolysis was particularly activated with this variant (Fig. 6B). Accordingly, the highest production of the octamer from 100 μm nonamer in the same conditions as in Fig. 6A was obtained with Lys-528/Asn-575 (Fig. 6C). These results indicate that, although ERAP1 polymorphism affects the hydrolysis of peptidic and nonpeptidic substrates, peptides compete advantageously in a more active context.
FIGURE 6.
Influence of ERAP1 polymorphism on the inhibition of L-AMC hydrolysis by a natural HLA-B27 ligand. A, L-AMC was incubated at 100 μm concentration, with the indicated ERAP1 variants at an E/S ratio of 1:30 (w/w), in the presence of 100, 75, or 50 μm of RRYQKSTEL for 1 h. The fluorescence intensity of the processed fluorogenic substrate in the absence (black) or in the presence of peptide (white) is indicated. The figures above the histograms indicate the % inhibition of L-AMC hydrolysis. B, L-AMC was incubated for 1 h at 100 μm concentration, with the indicated ERAP1 variants, at an E/S ratio of 1:30 (w/w), in the absence (black) or in the presence (white) of 100 μm of the RYQKSTEL octamer, resulting from the digestion of the nonamer used in A. Figures above the histogram bars of the most active variants indicate the % activation of L-AMC hydrolysis. C, percent yield of RYQKSTEL after incubation of 100 μm RRYQKSTEL with the indicated ERAP1 variants in exactly the same conditions as in A. The data are means ± S.D. of three experiments.
ERAP1 Polymorphism Influences the Inhibition of Peptide Processing by Short Peptides
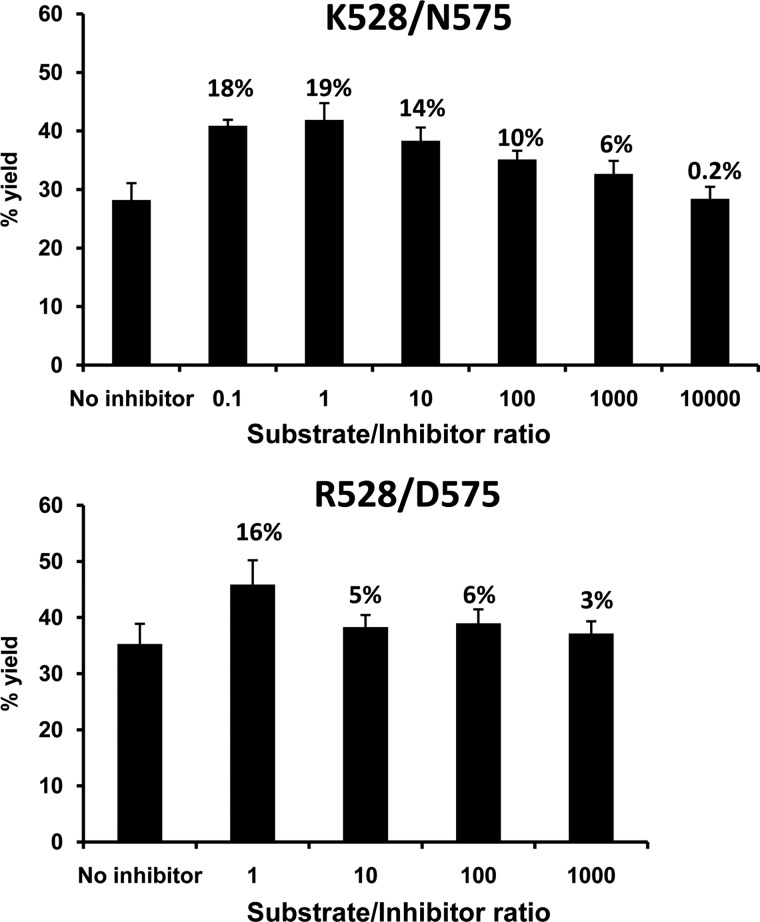
Because short peptides inhibit the processing of longer substrates (19), we examined whether ERAP1 polymorphism influenced the extent of this inhibition. Peptide 6, a 16-mer that was efficiently processed by all ERAP1 variants (Fig. 2), was tested for the inhibition of its processing in the presence of the RYQKSTEL octamer. The recovery of the precursor substrate after 4 h of digestion by the two ERAP1 variants with the largest activity differences, Arg-528/Asp-575 and Lys-528/Asn-575, was analyzed as a function of the octamer concentration (Fig. 7). At high inhibitor concentration (weight ratio 1:1 or higher), the inhibition was similar (19 and 16%) for both enzymes. However, whereas the inhibition fell almost to background levels (6%), at 10:1 substrate/inhibitor ratio for Arg-528/Asp-575, a 1000:1 ratio was required to reach this level with Lys-528/Asn-575. Thus, as with L-AMC, the octamer inhibited peptide trimming by the most active variant more efficiently than for the less active variant.
FIGURE 7.
Influence of ERAP1 polymorphism on the inhibition of peptide trimming by a short peptide. Peptide 6 (LYSELARYGKSPYLY) was incubated with the indicated ERAP1 variants in the presence of various amounts of RYQKSTEL for 4 h at an E/S ratio of 1:10. In each panel, the % yield of the precursor substrate, relative to the total digestion mixture, was plotted for the indicated substrate/inhibitor ratios. The figures above the histograms indicate the % inhibition of substrate hydrolysis. The data are means ± S.D. of three experiments.
DISCUSSION
To assess the contribution of this study to our understanding of the functional ERAP1/B27 interaction in AS, the following three issues must be considered: 1) the influence of ERAP1 polymorphism on the mechanism of peptide trimming; 2) the complexity of natural ERAP1 variants, which usually differ among each other by multiple amino acid residues; and 3) the correspondence of in vitro assays with the situation in live cells.
X-ray diffraction studies (19, 20) have virtually confirmed the molecular ruler mechanism of ERAP1 trimming (23), which is dependent on the affinity and length of the substrate and requires a peptide-binding site that is topologically distinct from the catalytic site. A conformational transition, involving global domain movements, is required for optimal enzymatic activity. Thus, ERAP1 polymorphism can influence peptide trimming by affecting the following: 1) the catalytic site or its immediate environment (i.e. residue 349); 2) the peptide-binding site and substrate/enzyme interactions (i.e. residue 730); and 3) the domain rearrangement. Residue 528 is located at the interdomain II to III region, which is the hinge for the domain rotation taking place during the conformational transition. This probably explains the functional effects of K528R. In contrast, residue 575 is located in a loop of eight residues connecting two β-strands in the middle of domain III, away from interdomain junctions or other functional sites. There are no obvious effects of the D575N mutation on ERAP1 activity that one can infer from just considering its topology. They are not due to changes in the glycosylation pattern, because Asn-575 is not in a consensus glycosylation sequence. Presumably, they are also not due to just an influence on the peptide-binding site, because the mutation also affected the hydrolysis of L-AMC, which binds close to the catalytic site. Thus, it is conceivable that D575N may affect the conformation and/or flexibility of domain III in a way that could alter the conformational transition required for increased activity. This transition induces critical changes in the active site (19, 20), which could explain the observed influence of residue 575 on the hydrolysis of the fluorogenic substrate.
Given the potential of single-residue polymorphisms to alter ERAP1 activity, combined effects among co-occurring mutations in natural variants are very likely. Thus, to understand the role of natural ERAP1 polymorphism in vivo, the effect of a mutation must be assessed as a function of its structural context. Only recently has the effect of natural ERAP1 haplotypes on peptide trimming been analyzed in live cells (10, 18, 24). These studies showed that ERAP1 activity depends on the precise combination of amino acid changes in a given variant.
Besides obviating context-dependent effects, previous in vitro studies using single mutants (8, 11, 12) cannot be directly generalized to HLA-B27 and to the situation in live cells due to some limitations, which we tried to overcome in our study. Because ERAP1 trimming is strongly dependent on the structure of the substrate (13, 14, 23), extrapolating the effects of ERAP1 polymorphism on other peptides to HLA-B27 ligands is unreliable. Thus, we examined the effect of AS-associated changes on peptide precursors of natural HLA-B27 ligands that were selected for structural diversity, to better assess peptide-dependent effects. Moreover, in vitro conditions, using recombinant enzymes and single substrates, differ significantly from those in human cells, where ERAP1 acts in the presence of ERAP2 (25) and of complex peptide pools that may influence ERAP1 activity (12). These considerations do not invalidate in vitro approaches but impose a cautious extrapolation and, ideally, a correlation with data from human cells, as done in this study.
The hydrolysis of L-AMC confirmed the lower activity of Arg-528 relative to Lys-528 in a given context, as reported in previous studies (8, 11, 12) and revealed that Asn-575 conferred higher activity compared with Asp-575. Although the magnitude of this difference was dependent on residue 528, Asn-575 was more active than Asp-575 in both the Arg-528 and Lys-528 contexts. In contrast, the relative activity of Arg-528 and Lys-528 variants was dependent on residue 575, demonstrating the context-dependent effect of these AS-associated mutations in complex allotypes. The induction of higher activity by D575N in the Arg-528 context was independently observed with natural ERAP1 variants and N-terminally extended precursors of an H-2Kb-restricted epitope (18). D575N is so far unique among the AS-protective mutations in that it enhanced ERAP1 activity, because K528R, R725Q, and Q730E have the opposite effect.
The relative activity of the four ERAP1 variants toward peptide substrates paralleled that with L-AMC when their joint effects over multiple substrates were considered. However, significant peptide-dependent differences were observed, as expected from the strong dependence of ERAP1 on the structure of the substrate.
We previously reported that AS-associated ERAP1 polymorphism induced quantitative differences in the HLA-B27 peptidome from human cells, affecting many peptides expressed in distinct ERAP1 contexts (10). Now we found that epitope amounts, which result from the balance between their generation and destruction, were strongly dependent on both ERAP1 activity and digestion time. Frequently, faster generation of the epitope by the most active variant led to higher yield at shorter times, whereas destructive cleavages tended to equal or decrease epitope yields, relative to less active variants, at long reaction times. This may be relevant to the mechanism of peptide transfer from ERAP1 to the MHC molecule. If ERAP1 would act in close spatial connection with the peptide-loading complex, this could allow a fast transfer of the natural ligand to the MHC, which would protect it from further degradation. This mechanism would be compatible with a protective effect of MHC-I molecules from destructive ERAP-mediated trimming (26), and it would enable the most active variants with an advantage to generate many MHC ligands. Alternatively, if ERAP1 generates a peptide pool in the endoplasmic reticulum that is uncoupled to the peptide-loading mechanism of MHC-I molecules, allowing for a more extensive iteration of substrate trimming, the most active ERAP1 variants would not be necessarily advantageous over less active ones in generating the MHC ligands, as observed in our experiments at long digestion times. This latter alternative explains why a more active ERAP1 variant, with AS-predisposing polymorphisms, reduced the presentation of multiple HLA-B27-restricted ligands, relative to a less active variant expressing multiple AS-protective polymorphisms in a recent study (24). Furthermore, to our knowledge, there is no consistent evidence for any coupling between ERAP1 and the peptide-loading complex of MHC-I that could allow fast peptide transfer to MHC and prevent further epitope degradation.
Given the significant differences between peptide digestion in vitro and in live cells, where many different variables may condition the expression level of HLA-B27 ligands, extrapolating our results to the situation in vivo must be done with great caution. Yet the close correlation found between the relative yields of RRYQKSTEL and RRYQKSTELLIR, both in vitro and in live cells in two distinct ERAP1 contexts, strongly suggests that ERAP1 processing may be a major determinant of the relative amounts of these two ligands in vivo. Of note, that this correlation held only at relatively long digestion times further supports that there is no coordinated peptide transfer from ERAP1 to the MHC-I molecule. Instead, ERAP1 presumably generates peptide pools that become available for MHC-I binding in the ER.
That the inhibition of L-AMC hydrolysis by a natural HLA-B27 ligand increased with ERAP1 activity suggests a dominant effect of ERAP1 polymorphism on peptide substrates, relative to small nonpeptidic ones. A likely explanation may be that the conformational transition induced by the peptide substrate to the active state of the enzyme favors peptide hydrolysis over that of a small one. Alternatively, the peptide-induced transition to the closed/active conformation may limit access of L-AMC to the catalytic site to an extent that would depend on the efficiency with which the enzyme is activated upon peptide binding.
It has been suggested that small peptides may unproductively bind in the catalytic site, without inducing the transition to the active state, and block the binding of longer, productive substrates (19). Our study showed that the inhibition of the trimming of a peptide substrate by the octamer product of a natural HLA-B27 ligand depended on the activity of the ERAP1 variant. Thus, ERAP1 polymorphism influences the inhibition of peptide processing by short peptides resulting from the destructive cleavage of natural ligands. Although the competition among peptide substrates, as presumably occurs in vivo (12), was not addressed here, our observations show that the influence of ERAP1 polymorphism on peptide trimming has a regulatory component affecting substrate competition, including that mediated by small digestion products.
In conclusion, this study demonstrates that the AS-associated residues 528 and 575 influence ERAP1 function in a mutual context-dependent way, so that the relative activity of variants carrying a given change at one of these positions depends on the polymorphism at the other position. Although for most of the AS-associated polymorphisms the protective alleles (i.e. Arg-528, Gln-725, and Glu-730) diminish ERAP1 activity, the protective Asn-575 change had the opposite effect. The various residue combinations at both positions affected the generation and destruction of HLA-B27 ligands in a variant and peptide-dependent way. The resulting effect is both extensive and complex. In some cases, both epitope generation and destruction increased similarly with ERAP1 activity, resulting in little alteration in the final yield of the natural ligand at long digestion times. In these cases the ligand was obtained with highest yield by the most active variant at short digestion times, when generation dominated over destruction. In other cases, relative epitope yields among variants were maintained at all times. Our results support that the mechanism of functional interaction between ERAP1 and HLA-B27 involves a widespread alteration in the balance between generation and destruction of HLA-B27 ligands induced by AS-associated ERAP1 polymorphism. This results in large variant- and peptide-dependent effects on epitope levels and, presumably, in the generation or not of particular ligands in a given ERAP1 context, consistent with observations in live cells (10).
Thus, the involvement of ERAP1 in the pathogenesis of AS can be envisaged as a far-reaching influence in the shaping of the HLA-B27 peptidome, affecting peptide binding and presentation in quantitative and qualitative ways, depending on the particular combination of polymorphic residues. As shown here and also noted recently (24) some HLA-B27 epitopes are more extensively destroyed by a more active ERAP1 variant, resulting in their higher production in a less active context. However, other epitopes are predominantly generated in a more active one, as shown here (i.e. peptides 4 and 8) and in live cells (10).
Besides the obvious effects on antigen presentation, the influence of ERAP1 polymorphism on the peptidome may affect folding and stability of HLA-B27, which also depend on the bound peptides (27). Through its effect on these features, ERAP1 could influence the mechanisms by which HLA-B27 activates the IL23/IL17 axis in spondyloarthropathies, either through misfolding (28) or activation of KIR3DL2+ CD4+ T cells mediated by surface heavy chain homodimers (29), which are generated upon endosomal recycling of HLA-B27 (30). How these features are affected by AS-associated ERAP1 polymorphism is currently a major issue in HLA-B27 research.
Acknowledgments
We thank Dr. Shi-Chung Chang (National Taiwan University) for providing the ERAP1 construct and Sergio Ciordia and Juan P. Albar (Proteomics Facility, Centro Nacional de Biotecnología, Madrid) for help in MS. We also acknowledge our colleagues at the Centro de Biología Molecular Severo, Ochoa Carlos Alvarez-Navarro and Juan J. Cragnolini (currently at Massachusetts Institute of Technology, Cambridge, MA), for help in the generation of recombinant ERAP1 proteins, Noel García-Medel for analytical software, and Jorgina Satrústegui for help in fluorometry.
This work was supported by Grant SAF2011/25681 (Plan Nacional de I+D+i) (to J. A. L. C.) and an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
- AS
- ankylosing spondylitis
- L-AMC
- Leu-7-amido-4-methylcoumarin.
REFERENCES
- 1. York I. A., Chang S. C., Saric T., Keys J. A., Favreau J. M., Goldberg A. L., Rock K. L. (2002) The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat. Immunol. 3, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 2. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. (2002) An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 3. Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. (2002) ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 [DOI] [PubMed] [Google Scholar]
- 4. Wellcome Trust Case Control Consortium, Australo-Anglo-American Spondylitis Consortium (TASC), Burton P. R., Clayton D. G., Cardon L. R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D. P., McCarthy M. I., Ouwehand W. H., Samani N. J., Todd J. A., Donnelly P., Barrett J. C., Davison D., Easton D., Evans D. M., Leung H. T., Marchini J. L., Morris A. P., Spencer C. C., Tobin M. D., Attwood A. P., Boorman J. P., Cant B., Everson U., Hussey J. M., Jolley J. D., Knight A. S., Koch K., Meech E., Nutland S., Prowse C. V., Stevens H. E., Taylor N. C., Walters G. R., Walker N. M., Watkins N. A., Winzer T., Jones R. W., McArdle W. L., Ring S. M., Strachan D. P., Pembrey M., Breen G., St Clair D., Caesar S., Gordon-Smith K., Jones L., Fraser C., Green E. K., Grozeva D., Hamshere M. L., Holmans P. A., Jones I. R., Kirov G., Moskivina V., Nikolov I., O'Donovan M. C., Owen M. J., Collier D. A., Elkin A., Farmer A., Williamson R., McGuffin P., Young A. H., Ferrier I. N., Ball S. G., Balmforth A. J., Barrett J. H., Bishop T. D., Iles M. M., Maqbool A., Yuldasheva N., Hall A. S., Braund P. S., Dixon R. J., Mangino M., Stevens S., Thompson J. R., Bredin F., Tremelling M., Parkes M., Drummond H., Lees C. W., Nimmo E. R., Satsangi J., Fisher S. A., Forbes A., Lewis C. M., Onnie C. M., Prescott N. J., Sanderson J., Matthew C. G., Barbour J., Mohiuddin M. K., Todhunter C. E., Mansfield J. C., Ahmad T., Cummings F. R., Jewell D. P., Webster J., Brown M. J., Lathrop M. G., Connell J., Dominiczak A., Marcano C. A., Burke B., Dobson R., Gungadoo J., Lee K. L., Munroe P. B., Newhouse S. J., Onipinla A., Wallace C., Xue M., Caulfield M., Farrall M., Barton A., Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee, Bruce I. N., Donovan H., Eyre S., Gilbert P. D., Hilder S. L., Hinks A. M., John S. L., Potter C., Silman A. J., Symmons D. P., Thomson W., Worthington J., Dunger D. B., Widmer B., Frayling T. M., Freathy R. M., Lango H., Perry J. R., Shields B. M., Weedon M. N., Hattersley A. T., Hitman G. A., Walker M., Elliott K. S., Groves C. J., Lindgren C. M., Rayner N. W., Timpson N. J., Zeggini E., Newport M., Sirugo G., Lyons E., Vannberg F., Hill A. V., Bradbury L. A., Farrar C., Pointon J. J., Wordsworth P., Brown M. A., Franklyn J. A., Heward J. M., Simmonds M. J., Gough S. C., Seal S.; Breast Cancer Susceptibility Collaboration (UK), Stratton M. R., Rahman N., Ban M., Goris A., Sawcer S. J., Compston A., Conway D., Jallow M., Newport M., Sirugo G., Rockett K. A., Bumpstead S. J., Chaney A., Downes K., Ghori M. J., Gwilliam R., Hunt S. E., Inouye M., Keniry A., King E., McGinnis R., Potter S., Ravindrarajah R., Whittaker P., Widden C., Withers D., Cardin N. J., Davison D., Ferreira T., Pereira-Gale J., Hallgrimsdo'ttir I. B., Howie B. N., Su Z., Teo Y. Y., Vukcevic D., Bentley D., Brown M. A., Compston A., Farrall M., Hall A. S., Hattersley A. T., Hill A. V., Parkes M., Pembrey M., Stratton M. R., Mitchell S. L., Newby P. R., Brand O. J., Carr-Smith J., Pearce S. H., McGinnis R., Keniry A., Deloukas P., Reveille J. D., Zhou X., Sims A. M., Dowling A., Taylor J., Doan T., Davis J. C., Savage L., Ward M. M., Learch T. L., Weisman M. H., Brown M. (2007) Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harvey D., Pointon J. J., Evans D. M., Karaderi T., Farrar C., Appleton L. H., Sturrock R. D., Stone M. A., Oppermann U., Brown M. A., Wordsworth B. P. (2009) Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum. Mol. Genet. 18, 4204–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brewerton D. A., Hart F. D., Nicholls A., Caffrey M., James D. C., Sturrock R. D. (1973) Ankylosing spondylitis and HL-A 27. Lancet 1, 904–907 [DOI] [PubMed] [Google Scholar]
- 7. Schlosstein L., Terasaki P. I., Bluestone R., Pearson C. M. (1973) High association of an HL-A antigen, W27, with ankylosing spondylitis. N. Engl. J. Med. 288, 704–706 [DOI] [PubMed] [Google Scholar]
- 8. Evans D. M., Spencer C. C., Pointon J. J., Su Z., Harvey D., Kochan G., Oppermann U., Opperman U., Dilthey A., Pirinen M., Stone M. A., Appleton L., Moutsianas L., Moutsianis L., Leslie S., Wordsworth T., Kenna T. J., Karaderi T., Thomas G. P., Ward M. M., Weisman M. H., Farrar C., Bradbury L. A., Danoy P., Inman R. D., Maksymowych W., Gladman D., Rahman P., Spondyloarthritis Research Consortium of Canada (SPARCC), Morgan A., Marzo-Ortega H., Bowness P., Gaffney K., Gaston J. S., Smith M., Bruges-Armas J., Couto A. R., Sorrentino R., Paladini F., Ferreira M. A., Xu H., Liu Y., Jiang L., Lopez-Larrea C., Díaz-Peña R., López-Vázquez A., Zayats T., Band G., Bellenguez C., Blackburn H., Blackwell J. M., Bramon E., Bumpstead J. S., Casas J. P., Corvin A., Craddock N., Deloukas P., Dronov S., Duncanson A., Edkins S., Freeman C., Gillman M., Gray E., Gwilliam R., Hammond N., Hunt S. E., Jankowski J., Jayakumar A., Langford C., Liddle J., Markus H. S., Mathew C. G., McCann O. T., McCarthy M. I., Palmer C. N., Peltonen L., Plomin R., Potter S. C., Rautanen A., Ravindrarajah R., Ricketts M., Samani N., Sawcer S. J., Strange A., Trembath R. C., Viswanathan A. C., Waller M., Weston P., Whittaker P., Widaa S., Wood N. W., McVean G., Reveille J. D., Wordsworth B. P., Brown M. A., Donnelly P., Australo-Anglo-American Spondyloarthritis Consortium (TASC), Wellcome Trust Case Control Consortium 2 (WTCCC2) (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C. M., Ho H. H., Chang S. W., Wu Y. J., Lin J. C., Chang P. Y., Wu J., Chen J. Y. (2012) ERAP1 genetic variations associated with HLA-B27 interaction and disease severity of syndesmophytes formation in Taiwanese ankylosing spondylitis. Arthritis Res. Ther. 14, R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-Medel N., Sanz-Bravo A., Van Nguyen D., Galocha B., Gómez-Molina P., Martín-Esteban A., Alvarez-Navarro C., de Castro J. A. (2012) Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol. Cell. Proteomics 11, 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goto Y., Hattori A., Ishii Y., Tsujimoto M. (2006) Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 12. Evnouchidou I., Kamal R. P., Seregin S. S., Goto Y., Tsujimoto M., Hattori A., Voulgari P. V., Drosos A. A., Amalfitano A., York I. A., Stratikos E. (2011) Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J. Immunol. 186, 1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hearn A., York I. A., Rock K. L. (2009)) The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 183, 5526–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evnouchidou I., Momburg F., Papakyriakou A., Chroni A., Leondiadis L., Chang S. C., Goldberg A. L., Stratikos E. (2008) The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE 3, e3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maksymowych W. P., Inman R. D., Gladman D. D., Reeve J. P., Pope A., Rahman P. (2009)) Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis Rheum. 60, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 16. Choi C. B., Kim T. H., Jun J. B., Lee H. S., Shim S. C., Lee B., Pope A., Uddin M., Rahman P., Inman R. D. (2010)) ARTS1 polymorphisms are associated with ankylosing spondylitis in Koreans. Ann. Rheum. Dis. 69, 582–584 [DOI] [PubMed] [Google Scholar]
- 17. Szczypiorska M., Sánchez A., Bartolomé N., Arteta D., Sanz J., Brito E., Fernández P., Collantes E., Martínez A., Tejedor D., Artieda M., Mulero J. (2011)) ERAP1 polymorphisms and haplotypes are associated with ankylosing spondylitis susceptibility and functional severity in a Spanish population. Rheumatology 50, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 18. Reeves E., Edwards C. J., Elliott T., James E. (2013) Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J. Immunol. 191, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen T. T., Chang S. C., Evnouchidou I., York I. A., Zikos C., Rock K. L., Goldberg A. L., Stratikos E., Stern L. J. (2011) Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 18, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von Delft F., Kavanagh K. L., Brown M. A., Bowness P., Wordsworth P., Kessler B. M., Oppermann U. (2011) Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cragnolini J. J., García-Medel N., Lopez de Castro J. A. (2009) Endogenous processing and presentation of T-cell epitopes from Chlamydia trachomatis with relevance in HLA-B27-associated reactive arthritis. Mol. Cell. Proteomics 8, 1850–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez de Castro J. A., Alvarez I., Marcilla M., Paradela A., Ramos M., Sesma L., Vázquez M. (2004) HLA-B27: a registry of constitutive peptide ligands. Tissue Antigens 63, 424–445 [DOI] [PubMed] [Google Scholar]
- 23. Chang S. C., Momburg F., Bhutani N., Goldberg A. L. (2005) The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 17107–17112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seregin S. S., Rastall D. P., Evnouchidou I., Aylsworth C. F., Quiroga D., Kamal R. P., Godbehere-Roosa S., Blum C. F., York I. A., Stratikos E., Amalfitano A. (2013) Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity 46, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., van Endert P. M. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 [DOI] [PubMed] [Google Scholar]
- 26. Kanaseki T., Blanchard N., Hammer G. E., Gonzalez F., Shastri N. (2006) ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity 25, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marcilla M., López de Castro J. A. (2008) Peptides: the cornerstone of HLA-B27 biology and pathogenetic role in spondyloarthritis. Tissue Antigens 71, 495–506 [DOI] [PubMed] [Google Scholar]
- 28. DeLay M. L., Turner M. J., Klenk E. I., Smith J. A., Sowders D. P., Colbert R. A. (2009) HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 60, 2633–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bowness P., Ridley A., Shaw J., Chan A. T., Wong-Baeza I., Fleming M., Cummings F., McMichael A., Kollnberger S. (2011) Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J. Immunol. 186, 2672–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bird L. A., Peh C. A., Kollnberger S., Elliott T., McMichael A. J., Bowness P. (2003) Lymphoblastoid cells express HLA-B27 homodimers both intracellularly and at the cell surface following endosomal recycling. Eur. J. Immunol. 33, 748–759 [DOI] [PubMed] [Google Scholar]