Background: Mechanisms that control expression of the splicing factor SRSF1 in human T cells are unknown.
Results: Ubiquitination and proteasome degradation of SRSF1 occur during T cell activation and in T cells from patients with systemic lupus erythematosus (SLE).
Conclusion: Ubiquitin-proteasome degradation regulates SRSF1 expression in human T cells.
Significance: Understanding how SRSF1 expression is regulated in SLE may enable new therapeutic approaches.
Keywords: Autoimmunity, Immunology, Proteasome, Protein Turnover, T Cell, T Cell Receptor, Ubiquitin
Abstract
T cells from patients with systemic lupus erythematosus (SLE) exhibit reduced expression of the critical T cell receptor (TCR)-associated CD3ζ signaling chain and are poor producers of the vital cytokine IL-2. By oligonucleotide pulldown and mass spectrometry discovery approaches, we identified the splicing regulator serine/arginine-rich splicing factor (SRSF) 1 or splicing factor 2/alternative splicing factor (SF2/ASF) to be important in the expression of CD3ζ chain. Importantly, increases in the expression of SRSF1 rescued IL-2 production in T cells from patients with SLE. In this study, we investigated the regulation of SRSF1 expression in resting and activated human T cells. We found that T cell stimulation induced a rapid and significant increase in mRNA expression of SRSF1; however, protein expression levels did not correlate with this increase. Co-engagement of CD28 induced a similar mRNA induction and reduction in protein levels. Proteasomal but not lysosomal degradation was involved in this down-regulation as evidenced by blocking with specific inhibitors MG132 and bafilomycin, respectively. Immunoprecipitation studies showed increased ubiquitination of SRSF1 in activated T cells. Interestingly, T cells from patients with SLE showed increased ubiquitination of SRSF1 when compared with those from healthy individuals. Our results demonstrate a novel mechanism of regulation of the splicing factor SRSF1 in human T cells and a potential molecular mechanism that controls its expression in SLE.
Introduction
Systemic lupus erythematosus (SLE)2 is a complex autoimmune disease of unknown etiology, which predominantly afflicts women in the reproductive years. Multi-organ damage due to deposition of pathogenic autoantibodies and immune complexes characterizes the disease. Dysregulated T cells are an important contributor to pathogenesis of this complicated disease (1). T cells from patients with SLE are poor producers of the vital cytokine interleukin (IL)-2 (2). We and others have shown that T cells from patients with SLE exhibit numerous signaling defects (3, 4), including the reduced expression of the critical T cell receptor (TCR)/CD3ζ signaling chain (5, 6). Using an mRNA oligonucleotide pulldown and mass spectrometry approach (7), we identified the serine/arginine splicing factor (SRSF) 1 or splicing factor 2/alternative splicing factor (SF2/ASF) binding to the CD3ζ 3′-untranslated region (UTR) (8). We showed that SRSF1 regulates alternative splicing of the CD3ζ 3′-UTR to promote the generation of a full-length version over a defective truncated splice variant and thus enhances CD3ζ expression in human T cells. Interestingly, we found altered expression of SRSF1 in T cells from SLE patients. Average Srsf1 mRNA expression was lower in T cells from SLE patients when compared with those from healthy individuals. SRSF1 protein expression was decreased in SLE T cells, more so in patients with worse disease. Importantly, increasing SRSF1 expression by transient transfection into SLE T cells rescued IL-2 production (9). The mechanisms of SRSF1 regulation in human T cells are not known, and understanding these would help identify the processes involved in its altered expression in SLE T cells.
SRSF1 or SF2/ASF is a prototype member of the serine/arginine-rich (SR) family of splicing proteins. The N-terminal RNA binding domain of this protein contains two RNA recognition motifs, whereas the C-terminal domain has SR dipeptide repeats and is critical for protein-protein interactions. Not only does SRSF1 regulate constitutive splicing of pre-mRNA, but also, it is an important determinant of alternative splicing (10). Besides alternative splicing, SRSF1 has been shown to regulate diverse aspects of gene regulation, including mRNA stability (11, 12), translation (13), and also transcription (9, 14). Very little is known regarding its role and regulation in immune cells and specifically in T cells. Antigen activation of T cells has been described to influence numerous alternative splicing events (15), including those of the adhesion molecule CD44 (16) and signaling proteins such as CD45, which was shown to be regulated by SRSF1 (17). However, not much is known about the control of this splicing regulator during T cell activation.
T cell activation not only triggers the activation and increased expression of downstream effectors, but also interestingly down-regulates certain molecules simultaneously. For example, TCR/CD3 triggering induces a rapid and sustained down-regulation of the CD3ζ chain, which is mediated by ligand-induced endocytosis, ubiquitination, and lysosomal degradation (18). The IκBα inhibitory component is targeted for ubiquitin-proteasome degradation, which is essential for nuclear translocation of NFκB and activation of downstream targets (19, 20). The ubiquitin-proteasome system is an important cellular mechanism of protein degradation, which allows for the removal of aberrant, misfolded, aged, or excess proteins and generates peptides and amino acids that can be recycled. Ubiquitin is a small, 76-amino acid (∼8-kDa) protein and is ubiquitously expressed. It is conjugated through the glycine residue at the C-terminal end with the side chain of a lysine residue on the target protein. A series of enzymes, activating (E1), carrier (E2), and ligase (E3), are involved in the activation of ubiquitin, recognition of substrate, and conjugation of ubiquitin to the substrate. Polyubiquitin chain-tagged proteins are ultimately degraded by a large protease called the 26 S proteasome. A recent study showed that T cell stimulation drives the proteasomal degradation of Argonaute 2, a core effector protein of the microRNA-induced silencing complex (21). Another study showed that the splicing factor SRSF5 is down-regulated by proteasome degradation and that this occurs simultaneously with increase in mRNA expression during late erythroid differentiation (22). Whether SRSF1 undergoes similar regulation at the protein level during T cell activation is not known.
In this study, we show for the first time that T cell activation induces a rapid and significant increase in the mRNA expression of SRSF1, whereas protein expression does not mimic this increase. We further show that bypassing the proximal TCR signaling with phorbol myristic acid (PMA) and ionomycin induces a similar phenotype with even stronger down-regulation of protein expression. This discrepancy between the mRNA and protein expression is not due to excessive mRNA decay, but rather due to active protein degradation mediated by the proteasome. Immunoprecipitation of SRSF1 revealed increased ubiquitination in activated T cells. Finally, we show that T cells from patients with SLE exhibit increased ubiquitinated forms of SRSF1 when compared with healthy individuals.
EXPERIMENTAL PROCEDURES
T Cells
Peripheral blood from healthy donors was obtained from the Kraft Family Blood Donor Center (Dana Farber Cancer Institute, Boston, MA) and the Blood Donor Center (Boston Children's Hospital, Boston, MA). Peripheral blood from SLE patients, rheumatoid arthritis (RA) patients, and age-, race-, and gender-matched healthy individuals was drawn at the Rheumatology Clinic at the Beth Israel Deaconess Medical Center (Boston, MA). All patients fulfilled at least four of the American College of Rheumatology (ACR) classification criteria for SLE or RA, respectively. Informed consent was obtained from all study participants. All studies were approved by the institutional review board. Total T cells were purified by negative selection using the Rosette Sep T cell isolation kit (Stem Cell Technologies, Vancouver, Canada).
Antibodies and Reagents
Anti-SRSF1 (clone 96), anti-Ubiquitin (clone Ubi-1) and control mouse IgG antibodies were purchased from Life Technologies. Anti-CD3ζ antibody and HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CD3 (OKT3 clone) antibody was purchased from Bio X Cell (West Lebanon, NH), and anti-CD28 antibody was from BioLegend (San Diego, CA). Goat anti-mouse cross linker, MG132, and bafilomycin A1 were purchased from EMD Millipore (Billerica, MA). Actinomycin D, PMA, ionomycin, and cycloheximide were purchased from Sigma-Aldrich. Polyubiquitinated conjugates (FK1) mAb, caspase-3 inhibitor Z-Asp-Glu-Val-Asp-FMK (DEVD), and pan caspase inhibitor Z-Val-Ala-Asp-FMK (VAD) were from Enzo Life Sciences (Farmingdale, NY).
T Cell Activation
T cells were cultured in complete RPMI medium 5 × 106 cells/ml in 24-well plates and stimulated with soluble anti-CD3 (5 μg/ml), anti-CD28 (2.5 μg/ml), and cross linker (2.5 μg/ml) antibodies or with PMA (25 ng/ml) and ionomycin (0.5 μg/ml) for the time points indicated. In some experiments, cells were pretreated with the proteasome inhibitor MG132 (5–10 μm) or the lysosomal inhibitor bafilomycin A1 (10 μm) for 2 h before activation. Inhibitors were kept in cultures for the duration of the activation.
RT-PCR
Total RNA was isolated using the RNeasy plus extraction kit (Qiagen, Valencia, CA). 200 ng of RNA was reverse-transcribed using the RNA to cDNA EcoDry premix (Clontech). Real time PCR amplification was carried out with SYBR Green I master mix on a LightCycler 480 (Roche Applied Science) using the program: initial denaturation at 95 °C for 5 min, 40 cycles of amplification; denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; 1 cycle of melting curves, 95 °C for 15 s, 65 °C for 2 min, and 97 °C (continuous); and a final cooling at 37 °C. All PCR reactions were performed in triplicate. Threshold cycle (Ct) values were used to calculate relative mRNA expression by the ΔCt relative quantification method. Primer sequences are: SRSF1: forward 5′-TCT CTG GAC TGC CTC CAA GT-3′ and reverse 5′-GGC TTC TGC TAC GAC TAC GG-3′; IL-2: forward 5′-CAC ACT CAC AGT AAC CTC AAC TCC T-3′ and reverse 5′-GTG GGA AGC ACT TAA TTA TCA AGT CAG TG-3′; housekeeping gene cyclophilin A: forward 5′-TTC ATC TGC ACT GCC AAG AC-3′ and reverse 5′-TCG AGT TGT CCA CAG TCA GC-3′.
Protein Gel Electrophoresis and Immunoblotting
Cells were washed with PBS, pelleted, and lysed in radioimmunoprecipitation assay buffer (Boston Bioproducts, Ashland, MA) supplemented with complete mini protease inhibitor mixture (Roche Applied Science). Lysates were mixed with SDS sample buffer and reducing agent, heated at 70 °C for 10 min, resolved on 4–12% Bis-Tris SDS-PAGE gels, and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% nonfat milk in Tris-buffered saline with 0.05% Tween 20 (TBS-T) for 1 h, incubated with primary antibody (1:1000; or 1:4000 β-actin) overnight at 4 °C, washed three times with TBS-T, incubated with HRP-conjugated secondary antibody (1:2000) for 1 h at room temperature, washed three times with TBS-T, developed using chemiluminescence detection reagents, and imaged with the Fujifilm LAS-4000 imager. Densitometry was performed with the Quantity One software (Bio-Rad).
Immunoprecipitation Experiments
Immunoprecipitations were performed using the Direct IP kit (Pierce) according to the manufacturer's instructions. Briefly, cells were lysed in immunoprecipitation/lysis buffer (15–20 μl/106 cells) with protease inhibitor mixture and the deubiquitinating enzyme inhibitor, iodoacetamide (10 mm). Lysates were precleared with control resin beads for 1 h on a rotator at 4 °C. SRSF1 antibody (3 μg) or a control mouse IgG was coupled to protein A/G resin beads on a rotator for 2 h at room temperature. After several washes, antibody-coupled resin was incubated with precleared lysates overnight at 4 °C. Several washes were performed to remove unbound proteins, and bound proteins were eluted with elution buffer. Elutes were mixed with sample buffer and reducing agent and heated at 70 °C for 10 min. Gel electrophoresis and immunoblotting were performed as described in the above section.
Statistical Analyses
The Student's t test was used for statistical analysis, and a p value < 0.05 was considered statistically significant.
RESULTS
T Cell Activation Increases mRNA but Not Protein Expression of the Splicing Factor SRSF1
We asked whether and how the mRNA and protein expression of the splicing factor SRSF1 is modulated during initial T cell activation. T cells purified from peripheral blood of healthy individuals were stimulated with anti-CD3 plus anti-CD28 antibodies over a period of 24 h. Total mRNA and protein were isolated from the cells. Srsf1 mRNA expression was assessed by reverse transcription and quantitative real time PCR. We previously showed that over a period of 72 h of T cell activation, there is a gradual increase in SRSF1 expression notably at 48 and 72 h (8). However, a careful examination of the initial 24 h of stimulation revealed that although Srsf1 mRNA expression rapidly increased 4–5-fold (Fig. 1A), protein expression did not change significantly during the activation period, or even decreased to some extent (Fig. 1, B and C). We further tested whether triggering the TCR alone with anti-CD3 or with CD28 co-stimulation would affect the expression of SRSF1. We found that in both conditions, a similar increase in Srsf1 mRNA occurred (Fig. 2A), although protein decrease was slightly more pronounced in the co-stimulated cells especially at 24 h (Fig. 2B). CD28 co-stimulation activates the JNK pathway and is critical for the stabilization of IL-2 mRNA, and results in the exponential increase in IL-2 production. Therefore, as an internal experimental control, we measured IL-2 mRNA expression levels. As expected we observed significantly higher levels of IL-2 mRNA in the CD28 co-stimulated cells (Fig. 2C). These results indicate that the SRSF1 mRNA and protein expression undergo differential regulation in human T cells upon TCR stimulation.
FIGURE 1.
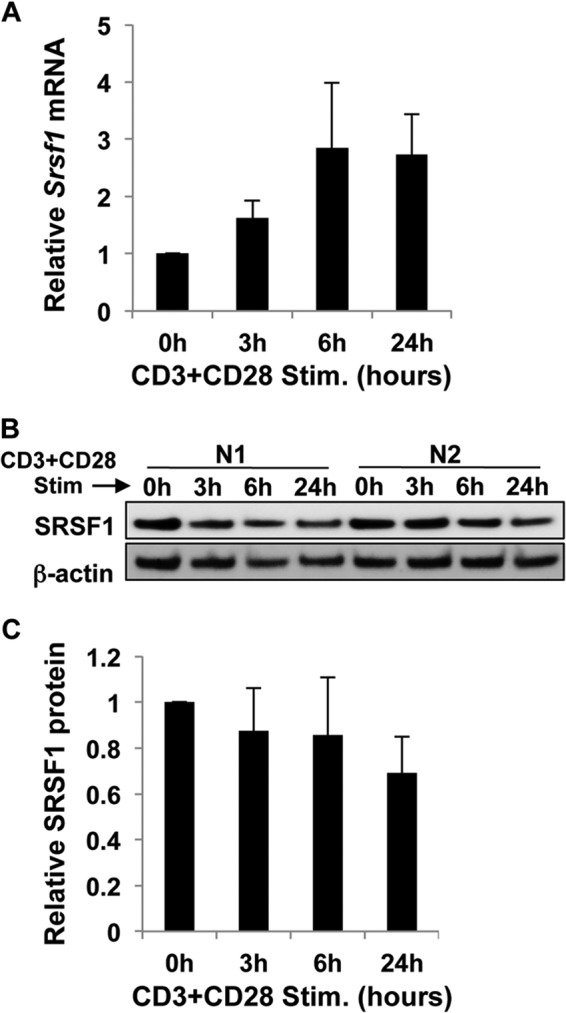
T cell activation induces mRNA but not protein expression of SRSF1. T cells were activated with anti-CD3 and anti-CD28 for 0, 3, 6, and 24 h. A, cells were collected, and total RNA was isolated and reverse-transcribed. SRSF1 expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graph shows average values from n = 6 donors, and error bars represent S.D. Stim., stimulated. B, total protein was extracted from T cells and immunoblotted for SRSF1 and β-actin. Representative blots from two donors (N1 and N2) are shown. C, densitometric quantitation of SRSF1 from immunoblots was performed and normalized to β-actin. Graph shows average values from n = 6 donors, and error bars represent S.D.
FIGURE 2.
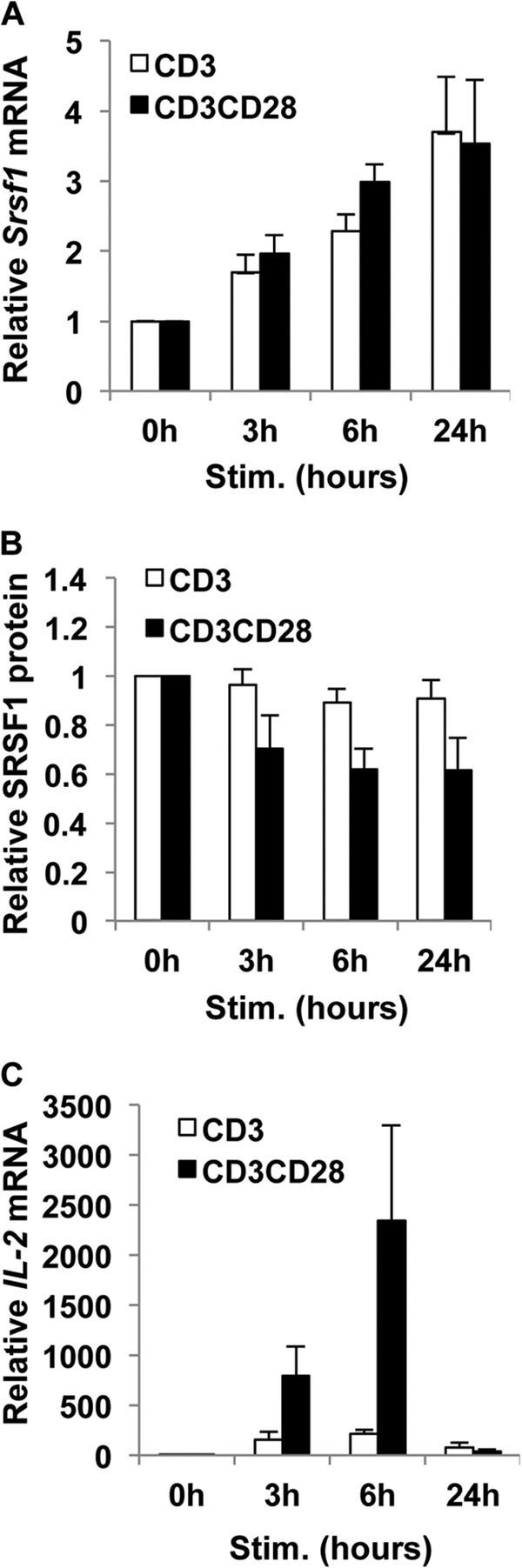
T cell activation with or without co-stimulation induces the discrepant mRNA and protein expression of SRSF1. T cells were activated with anti-CD3 alone or anti-CD3 plus anti-CD28 for 0, 3, 6, and 24 h. A, cells were collected, and total RNA was isolated and reverse-transcribed. SRSF1 expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graph shows average values from n = 5 donors, and error bars represent S.E. Stim., stimulated. B, total protein was extracted from T cells and immunoblotted for SRSF1 and β-actin. Representative blots are shown. Densitometric quantitation of SRSF1 from immunoblots was performed and normalized to β-actin. Graph shows average values from n = 4 donors, and error bars represent S.E. C, IL-2 expression was measured by quantitative PCR and normalized to cyclophilin A housekeeping gene. Densitometric quantitation of SRSF1 from immunoblots was performed and normalized to β-actin. Graph shows average values from n = 5 donors, and error bars represent S.E.
Bypassing the TCR Induces Differential mRNA and Protein Expression of SRSF1
TCR activation induces tyrosine phosphorylation of signaling intermediates and intracellular signaling pathways to activate diacylglycerol, inositol triphosphate, and the Ras mitogen-activated protein kinase (MAPK) pathways. Diacylglycerol induces NFκB activation, and inositol triphosphate triggers calcium flux to activate nuclear factor of activated T-cells (NFAT), whereas the MAPK pathway activates c-Fos, a component of the c-Fos/c-Jun AP1 transcription factor. Nuclear translocation of these transcriptional activators leads to cellular activation, proliferation, and differentiation. To assess whether TCR proximal or distal signaling is required for the differential mRNA and protein regulation of SRSF1, we used PMA and the calcium ionophore ionomycin to activate T cells. PMA activates diacylglycerol, and ionomycin activates calcium channels, thus activating the same intracellular signaling cascade, although bypassing the proximal TCR signaling entirely. As seen in Fig. 3A, we observed a similar increase in mRNA expression of SRSF1. Interestingly, the down-regulation of SRSF1 protein was more evident using this mode of stimulation (Fig. 3, B and C). These results show that the mRNA induction and protein down-regulation are independent of the proximal TCR signaling.
FIGURE 3.
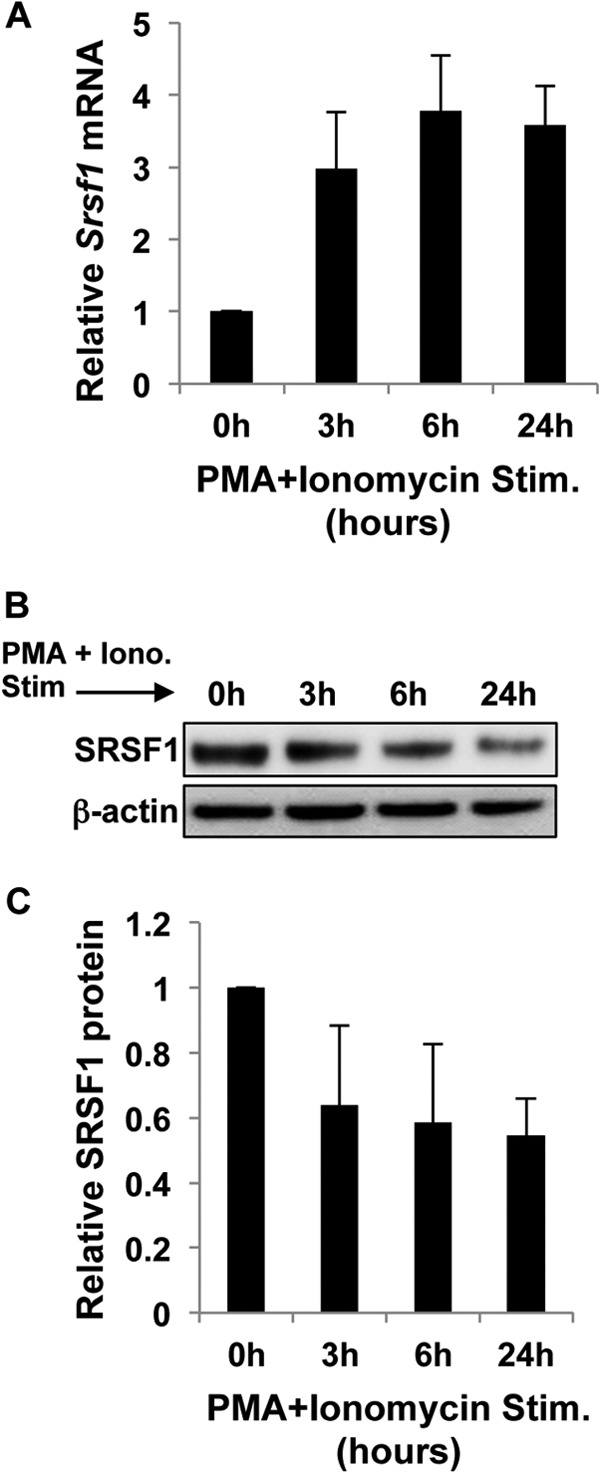
Increased mRNA and decreased protein expression of SRSF1 occurs independently of the proximal TCR signaling. T cells were activated with PMA and ionomycin for 0, 3, 6, and 24 h. A, cells were collected, and total RNA was isolated and reverse-transcribed. SRSF1 expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graph shows average values from n = 3 donors, and error bars represent S.D. Stim., stimulated. B, total protein was extracted from T cells and immunoblotted for SRSF1 and β-actin. Representative blots are shown. Iono., ionomycin. C, densitometric quantitation of SRSF1 from immunoblots was performed and normalized to β-actin. Graph shows average values from n = 3 donors, and error bars represent S.D.
Srsf1 mRNA Stability Is Not Altered during T cell Activation
We asked whether despite the increase in SRSF1 mRNA expression, post-transcriptional half-life of the transcripts was compromised. Accordingly, we tested whether T cell stimulation affected the mRNA stability of SRSF1. T cells were stimulated with anti-CD3 and anti-CD28 or left unstimulated, and actinomycin D was added to block transcription. SRSF1 mRNA expression was measured over a period of 2 h. We found no significant differences in mRNA decay of SRSF1 in resting versus stimulated T cells and observed a normal half-life of about 1.5 h in both conditions (Fig. 4A). T cell activation induces IL-2 transcription and enhanced stabilization of IL-2 mRNA, which leads to the exponential increase in IL-2 production. Therefore, as an internal experimental control, we measured the half-life of IL-2 mRNA, which is well known to increase upon T cell activation. We observed significantly higher mRNA stability of IL-2 mRNA in stimulated cells when compared with unstimulated cells (Fig. 4B). These results indicate that the mRNA stability of SRSF1 is not affected by T cell activation.
FIGURE 4.
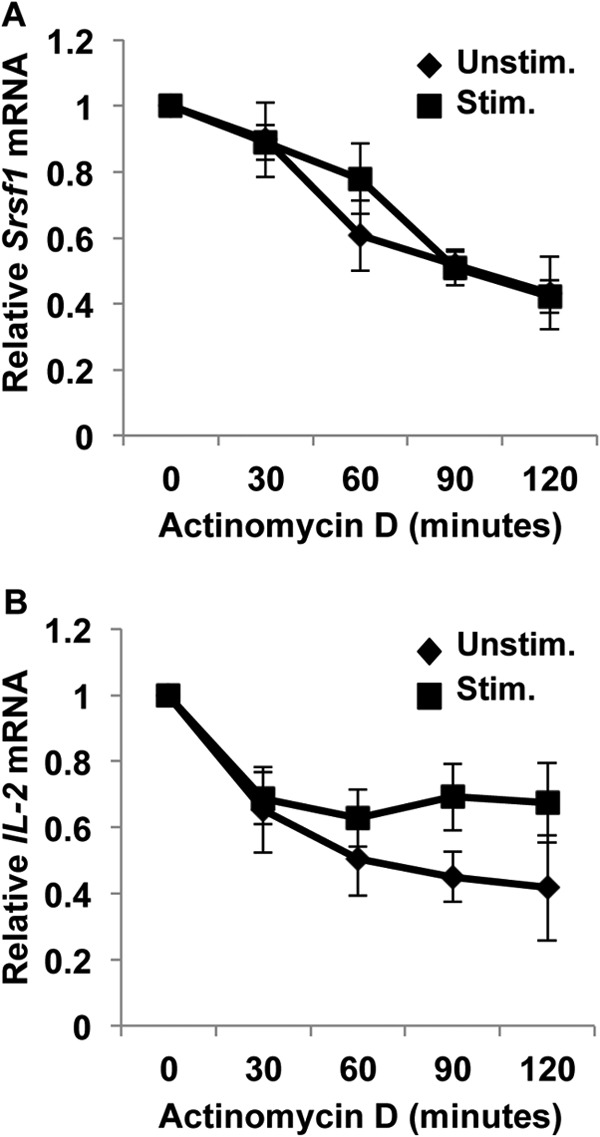
T cell activation does not induce mRNA degradation of SRSF1. T cells were activated with anti-CD3 plus anti-CD28 (Stim.) or left unstimulated (Unstim.) for 3 h. Actinomycin D (10 μg/ml) was added to cultures, and cells were collected at 0, 30, 60, 90, and 120 min later. Total RNA was isolated and reverse-transcribed. A and B, SRSF1 (A) and IL-2 (B) expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graphs show average values from n = 5 donors, and error bars represent S.E.
Proteasome but Not Lysosome Degradation Regulates Protein Expression of SRSF1 during T cell Activation
T cell activation is shown to induce degradation of molecules such as CD3ζ, IκB, and Bcl-10 via lysosomal and proteasomal degradation pathways (18, 20, 23). To test whether similar mechanisms play a role in SRSF1 protein regulation, we added inhibitors of the proteasome or the lysosome during T cell activation. As seen in Fig. 5, A and B, the addition of the proteasome inhibitor MG132 blocked the decrease in protein expression at 5 h after activation. This effect was reversible at 24 h of activation, as evidenced by the near complete loss of protein at this time point, although this may additionally represent cellular toxicity that can occur after prolonged incubation with MG132. The addition of lysosomal inhibitor bafilomycin did not show this restoration, indicating that the proteasome rather than the lysosome is involved in the degradation of SRSF1. The addition of MG132 and bafilomycin together did not restore SRSF1 expression at the 24-h time point (Fig. 5A, lower panel). It is known that upon T cell stimulation, the CD3ζ chain along with the entire TCR complex is internalized by endocytosis followed by endosome-lysosomal fusion and proteolysis by lysosomal enzymes. CD3ζ chain is also degraded by the proteasome. Therefore, as control, we assessed CD3ζ expression and found as expected that blocking either the lysosome or the proteasome limited its degradation (Fig. 5, A and C). These results indicate that T cell stimulation actively induces the degradation of SRSF1, and this contributes to the observed discrepancy between mRNA and protein expression of SRSF1.
FIGURE 5.
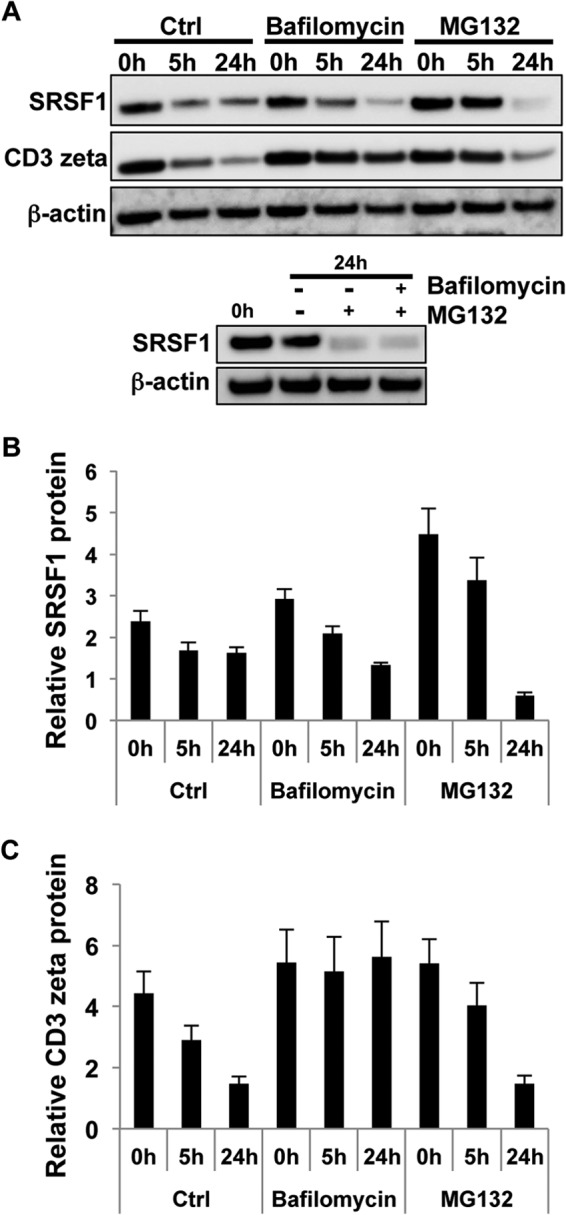
T cell activation induces proteasomal but not lysosomal degradation of SRSF1. A, T cells were pretreated with either lysosomal inhibitor bafilomycin A1 and/or proteasome inhibitor MG132 for 2 h and then activated with soluble anti-CD3 and anti-CD28 antibodies for 0, 5, and 24 h. Total protein was extracted from T cells and immunoblotted for SRSF1, CD3ζ, and β-actin. Ctrl, control. B, densitometric quantitation of SRSF1 from immunoblots was performed and normalized to β-actin. Graphs show average values from n = 3 donors, and error bars represent S.E. C, densitometric quantitation of CD3ζ from immunoblots was performed and normalized to β-actin. Graphs show average values from n = 3 donors, and error bars represent S.E.
Increased Ubiquitination of SRSF1 in Activated T Cells
Proteins destined for proteolysis are tagged with multiple ubiquitin molecules and then targeted to the proteasome for degradation. To determine the ubiquitination state of SRSF1, we immunoprecipitated SRSF1 from protein extracts of resting and activated T cells. SRSF1 immunoprecipitates were resolved on SDS-PAGE gels and immunoblotted with an anti-ubiquitin antibody. As seen in Fig. 6, lysates showed decreased expression of SRSF1 after activation, which was restored with MG132 inhibitor treatment. Increased ubiquitination was observed in activated T cells when compared with resting cells, and further accumulation of ubiquitinated complexes was seen in MG132-treated cells, more so at 5 h of activation. We included a negative control IgG antibody in the immunoprecipitation experiments and immunoblotted lysates with the anti-ubiquitin antibody as positive control (Fig. 6B). To confirm whether SRSF1 is mono- or polyubiquitinated, we reprobed the membranes with a polyubiquitin-specific (FK1) antibody and observed similarly increased ubiquitination in MG132-treated cells (Fig. 6B, lower panel). Densitometric quantitation of ubiquitinated SRSF1 normalized to total SRSF1 from the lysates showed a significant increase in ubiquitination in MG132-treated cells (Fig. 6C).
FIGURE 6.
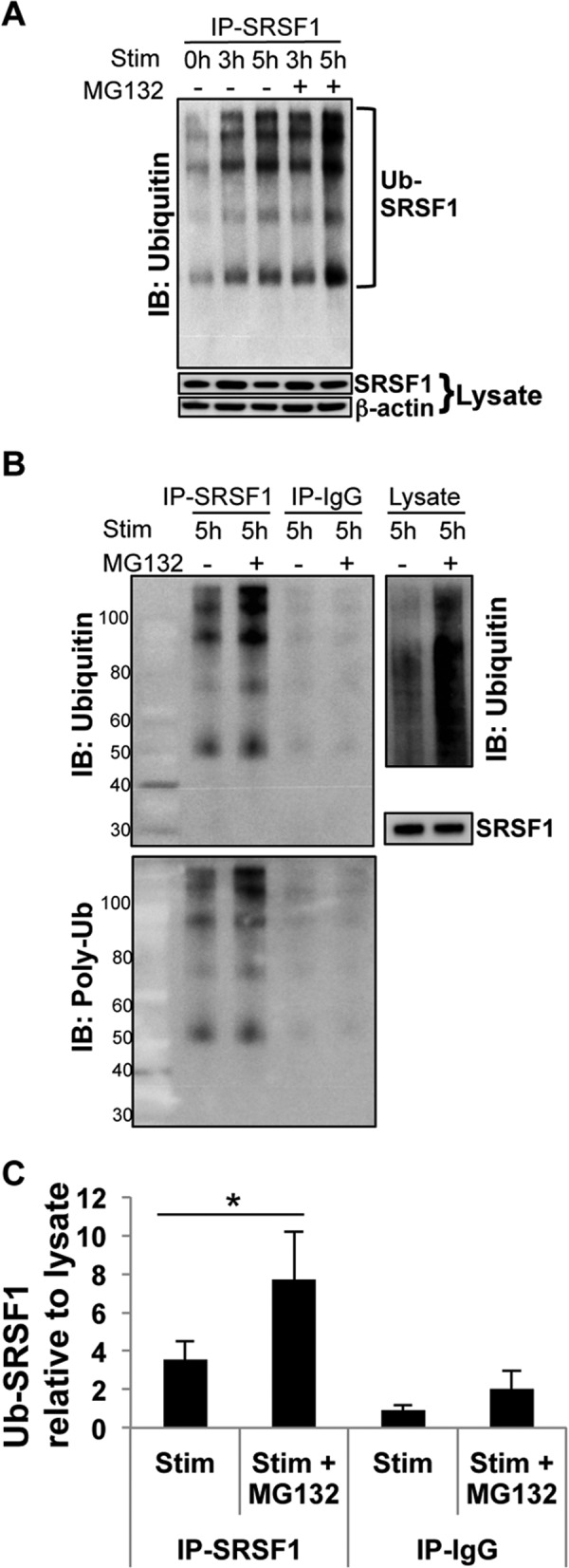
T cell activation induces increased ubiquitination of SRSF1. A, T cells were pretreated with proteasome inhibitor MG132 for 2 h and activated with PMA and ionomycin for the indicated time points. Total protein was extracted, and SRSF1 was immunoprecipitated (IP) using a monoclonal antibody. Immunoprecipitates were resolved on gels and immunoblotted (IB) for ubiquitin. Lysates were resolved on gels and immunoblotted for SRSF1 and β-actin. A representative blot is shown from a total of n = 5 donors. Stim., stimulated. Ub-SRSF1, ubiquitinated SRSF1. B, T cells were pretreated and activated as in A. Total protein was immunoprecipitated using anti-SRSF1 and control IgG antibodies. Immunoprecipitates were resolved on gels and immunoblotted with anti-ubiquitin antibody (upper left panel), and stripped and reprobed with a polyubiquitin (Poly-Ub) (FK1) antibody (lower panel). Lysates were resolved and immunoblotted with a polyubiquitin (FK1) antibody (upper right panel) and SRSF1 antibody (lower right panel). Representative blots are shown from one of n = 8 donors. C, densitometric quantitation of ubiquitinated SRSF1 (Ub-SRSF1) immunoblots from B was performed and normalized to SRSF1 from lysates. Graph shows average values from n = 8 donors, and error bars represent S.E. * indicates p value < 0.05.
Increased Ubiquitination of SRSF1 in T Cells from Patients with SLE
We asked whether ubiquitination modulates SRSF1 protein expression in T cells from SLE patients when compared with healthy individuals and patients with other autoimmune diseases such as RA. We obtained T cells from 13 patients with SLE, 5 patients with RA, and age-, race-, and gender-matched healthy controls. SLE patients were all female, age range between 22 and 44 years: six Caucasian, six African American, and one mixed race. SLE disease activity index (SLEDAI) scores ranged from 0 to 7. Patients with RA were all female, age range between 37 and 73 years: two Caucasian and three African American. T cells were isolated, and total lysates were immunoprecipitated using an SRSF1-specific antibody and immunoblotted for ubiquitin (Fig. 7A, upper panels). In parallel, immunoblots for SRSF1 showed the protein in the same conjugates, confirming the presence of SRSF1 in these ubiquitinated complexes (Fig. 7A, lower panels). Total lysates were immunoblotted with anti-ubiquitin antibody as a positive control and showed a smearing pattern of ubiquitinated complexes (Fig. 7B). Densitometric quantitation of ubiquitinated SRSF1 normalized to total SRSF1 from lysates showed a significant increase in ubiquitination in SLE patients when compared with healthy individuals (p = 0.03), whereas there was no significant difference in ubiquitination in RA patients when compared with healthy individuals (p = 0.87) (Fig. 7C). These results indicate that T cell activation induces increased ubiquitination of SRSF1, and patients with SLE exhibit this feature as well.
FIGURE 7.
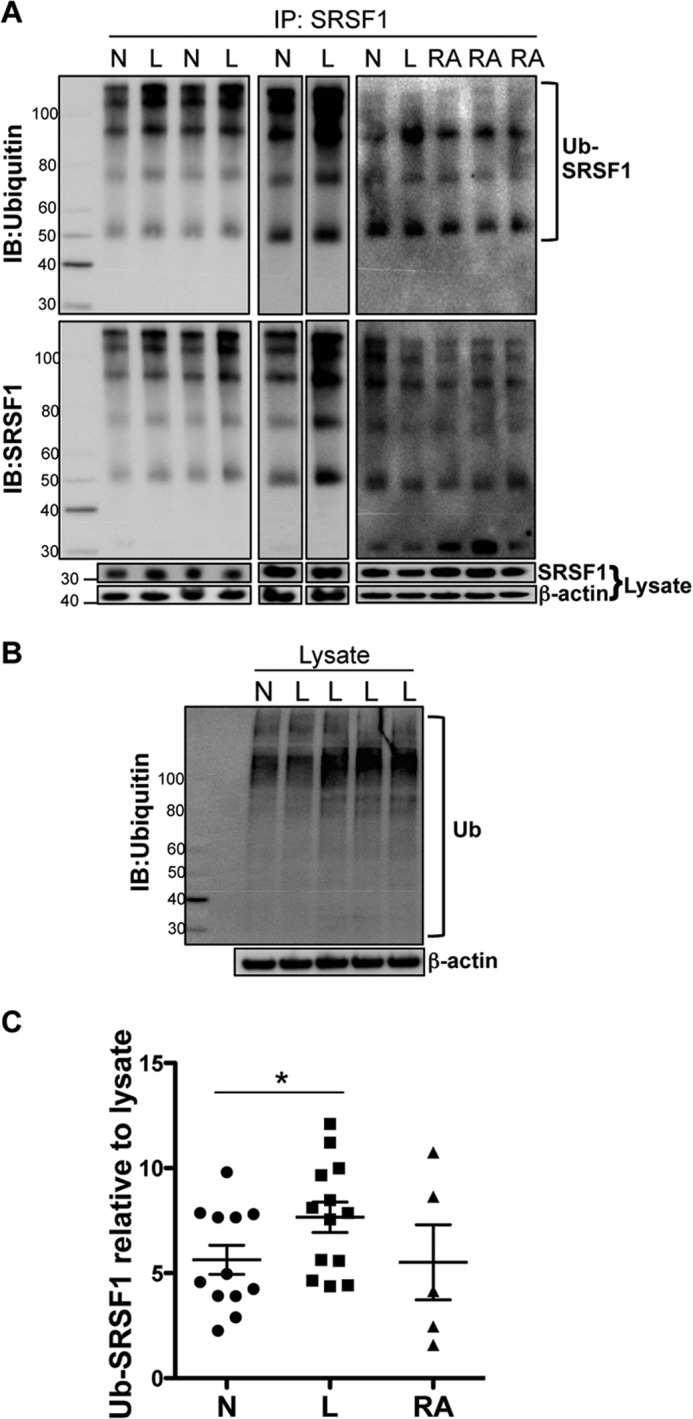
Increased ubiquitination of SRSF1 in T cells from patients with SLE. T cells were purified from peripheral blood of healthy individuals (N), patients with SLE (L), and patients with rheumatoid arthritis (RA). A, total protein was extracted, and SRSF1 was immunoprecipitated (IP) using a monoclonal antibody. Immunoprecipitates were resolved on SDS-PAGE gels and immunoblotted (IB) for ubiquitin (upper left and middle panels), a polyubiquitin antibody (upper right panel), and SRSF1 (lower panels). Total lysates were resolved on gels and immunoblotted for SRSF1 and β-actin. Ub-SRSF1, ubiquitinated SRSF1. B, total lysates were resolved on a gel and immunoblotted for ubiquitin (Ub) and β-actin. C, densitometric quantitation of ubiquitinated SRSF1 (Ub-SRSF1) immunoblots from A was performed and normalized to SRSF1 from lysates. Graph shows average values from N (n = 12), L (n = 13), and RA (n = 5) donors, and error bars represent S.E. * indicates p value < 0.05.
Caspase- but Not Calpain Protease-mediated Degradation Contributes to the Regulation of SRSF1 Protein Expression during T Cell Activation
We previously showed that increased caspase-3 expression and activity contribute to the reduced expression of CD3ζ in T cells from patients with SLE (24). Interestingly, the SRSF1 amino acid sequence bears several caspase-3 recognition DXXD motifs where D is aspartic acid and X is any amino acid. Based on these concepts, we asked whether mechanisms involving caspases or proteases play a role in SRSF1 expression during T cell activation. We added the caspase-3 inhibitor (DEVD) during T cell activation and observed a dose-dependent effect on restoration of SRSF1 protein expression (Fig. 8, A and B). Additionally, the pan-caspase inhibitor (VAD) was also able to restore protein expression in activated T cells. The calpain protease inhibitor calpeptin was added to cultures and did not have any effect on SRSF1 expression (Fig. 8, C and D). These results indicate that caspase-mediated degradation contributes to the expression of SRSF1 protein in activated T cells.
FIGURE 8.
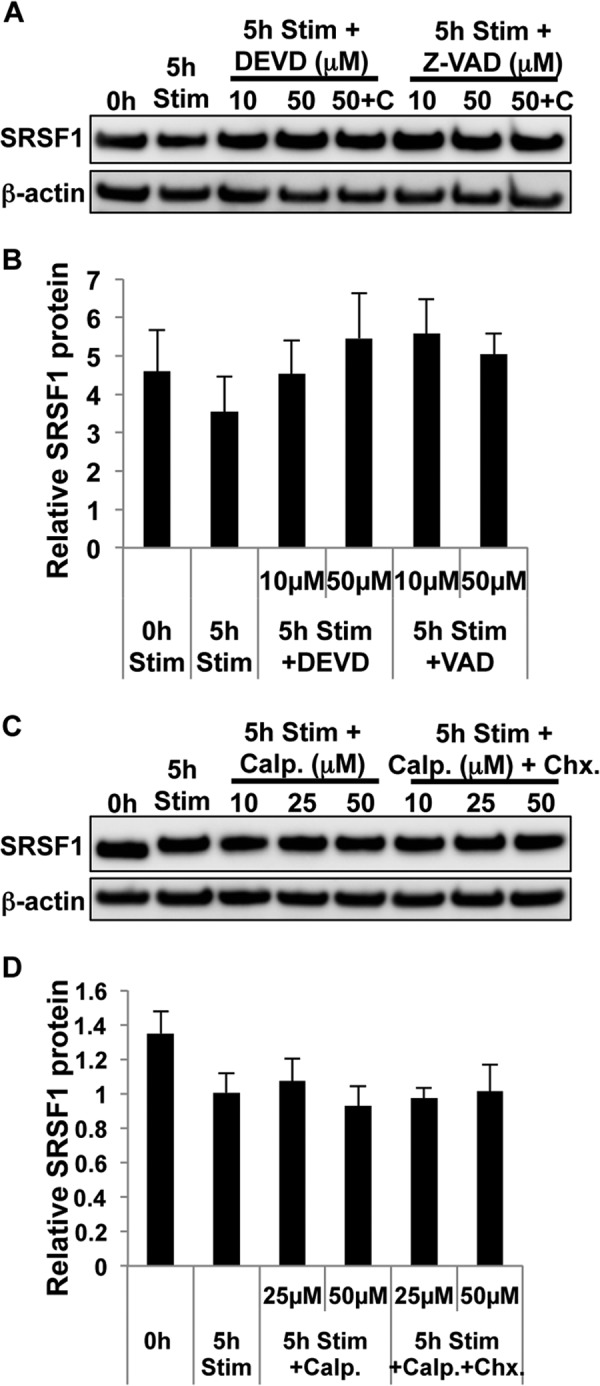
T cell activation induces caspase-mediated degradation of SRSF1. A, T cells were pretreated with the protein translation inhibitor cycloheximide (C, 10 μg/ml) for 10 min followed by either caspase-3 inhibitor DEVD or pan-caspase inhibitor VAD for 1 h and then activated with soluble anti-CD3 and anti-CD28 antibodies for 5 h. Total protein was extracted from T cells and immunoblotted for SRSF1 and β-actin. 5h Stim, 5-h stimulation. B, densitometric quantitation of SRSF1 immunoblots from A was performed and normalized to β-actin. C, T cells were pretreated with cycloheximide (Chx, 10 μg/ml) for 10 min followed by protease inhibitor calpeptin (Calp.) for 1 h, and then activated with soluble anti-CD3 and anti-CD28 antibodies for 5 h. Total protein was extracted from T cells and immunoblotted for SRSF1 and β-actin. D, densitometric quantitation of SRSF1 immunoblots from C was performed and normalized to β-actin. Graphs show average values from n = 4 donors, and error bars represent S.E.
DISCUSSION
In this study, we present several novel findings. First, we show that Srsf1 mRNA expression is significantly induced upon T cell activation; however, protein expression does not change or even decreases. Second, we show that triggering the TCR alone or with co-stimulation induces the discrepant SRSF1 mRNA and protein expression. Third, bypassing the TCR with PMA plus ionomycin stimulation induces a more pronounced down-regulation of protein expression. Fourth, we show that proteasome- and caspase-mediated degradation but not lysosome- or protease-mediated degradation are responsible for the SRSF1 protein decrease during activation. Finally, we show increased ubiquitination of SRSF1 in T cells from some patients with SLE when compared with healthy individuals.
It appears that the levels of cellular SRSF1 expression are tightly controlled. SRSF1 regulates alternative splicing of tumor suppressor genes such as BIN1 (25), and genes involved in apoptosis such as caspase-9 and Bcl-x (26). Accordingly, high levels of SRSF1 can lead to oncogenic transformation (25), whereas depletion of SRSF1 can lead to genomic instability and cell cycle arrest (27). T cells down-regulate several cellular proteins after stimulation, either to propagate signaling or to dampen and prevent excessive signaling or activation-induced cell death. For example, TCR stimulation induces lysosomal degradation of the CD3ζ chain (18) and of Bcl-10, a negative regulator of TCR-induced NF-κB signaling activity (23). Also, TCR signaling induces proteasome-mediated degradation of the NF-κB inhibitor IκBα (20). Src-like adapter proteins (SLAP) are required for the targeted ubiquitination and degradation of the TCR signaling components (28). Although we find that T cell stimulation induces a rapid and significant increase in mRNA levels, a tight control of SRSF1 protein in T cells during the early activation phase may prevent premature expansion of T cells in response to antigenic stimulation. Thus ubiquitination and proteasome degradation may serve as homeostatic regulation of the levels of SRSF1.
Interestingly, CD28 co-stimulation led to slightly more pronounced decline of SRSF1 protein levels (Fig. 2B). It has been shown that CD28 and CTLA4 mediate their effect on the threshold of T cell activation via an E3 ubiquitin ligase Cbl-b (29), a molecule critical for maintenance of the balance between tolerance and autoimmunity (30). It would be interesting to examine whether SRSF1 mediates a similar effect on fine-tuning the T cell response.
Although we observed no significant differences in the rate of mRNA decay in activated T cells, it is possible that SRSF1 may also be regulated at the level of mRNA translation. The long 3′-UTR contains ultraconserved elements involved in post-transcriptional regulation, and SRSF1 was shown to negatively regulate its own expression (31). Translational repression is also mediated by microRNA, and this may be another mechanism of SRSF1 regulation. Several microRNAs are known to be modulated in T cells during activation and differentiation (32). A computational (TargetScan) search of the 3′-UTR of SRSF1 revealed numerous predicted microRNAs targeting putative sites including miR-214 and miR-155, which are known to increase during CD4 T cell activation (33). It would be interesting to examine their specific role in SRSF1 regulation.
Besides proteasomal degradation, ubiquitin modification determines other aspects of protein metabolism to regulate vital cellular processes. Substrate proteins may be modified by monoubiquitin, multiple monoubiquitin, or polyubiquitin chains. Monoubiquitination can regulate proteasome-independent cellular processes such as protein localization, transcriptional activation, and chromatin structure. Multiple monoubiquitination has effects on localization of proteins such as p53 and receptor tyrosine kinases, whereas short ubiquitin chains are intermediate in protein degradation. Many proteins are heterogeneously polyubiquitinated as evidenced by ubiquitin smears above 100 kDa of molecular mass in SDS-PAGE gels (30, 34). In our studies, ubiquitinated SRSF1 was seen mainly in distinct bands even with a polyubiquitin-specific FK1 antibody. It is likely that ubiquitin modification of SRSF1 also regulates other aspects of SRSF1 function such as cellular localization and activity.
The ubiquitin-proteasome system has been shown to be important in autoimmune disease and specifically in SLE. We previously reported that T cells from SLE patients exhibit increased ubiquitinated forms of the CD3ζ signaling chain (35). The TRIM21 (Ro52/SSA) autoantigen prevalent in SLE was identified as a ubiquitin ligase and shown to target the polyubiquitin-mediated degradation of IRF3 to turn off the type I IFN response (36). Sera from patients with SLE were shown to have autoantibodies against the proteasome α-type subunit C9 (37). Our findings of increased ubiquitinated forms of SRSF1 in T cells from patients with SLE, similar to that seen during activation of normal T cells, suggest that this maybe due to the ongoing TCR stimulation by autoantigens.
In conclusion, our study reveals a novel mechanism of regulation of the splicing factor SRSF1 in human T cells, and a potential mechanism for its altered expression in SLE.
Acknowledgments
We thank Dr. Caroline Jefferies and Dr. Elisa Lazzari (Department of Molecular and Cellular Therapeutics at the Royal College of Surgeons in Ireland) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI42269 (to G. C. T.) and K01 AR060781 (to V. R. M.).
- SLE
- systemic lupus erythematosus
- TCR
- T cell receptor
- SRSF1
- serine/arginine splicing factor 1
- PMA
- phorbol myristic acid
- RA
- rheumatoid arthritis
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- DEVD
- Z-Asp-Glu-Val-Asp-FMK
- VAD
- Z-Val-Ala-Asp-FMK
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone.
REFERENCES
- 1. Tsokos G. C. (2011) Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 2. Linker-Israeli M., Bakke A. C., Kitridou R. C., Gendler S., Gillis S., Horwitz D. A. (1983) Defective production of interleukin 1 and interleukin 2 in patients with systemic lupus erythematosus (SLE). J. Immunol. 130, 2651–2655 [PubMed] [Google Scholar]
- 3. Moulton V. R., Tsokos G. C. (2011) Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res. Ther. 13, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perl A. (2010) Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity 43, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brundula V., Rivas L. J., Blasini A. M., París M., Salazar S., Stekman I. L., Rodríguez M. A. (1999) Diminished levels of T cell receptor ζ chains in peripheral blood T lymphocytes from patients with systemic lupus erythematosus. Arthritis Rheum. 42, 1908–1916 [DOI] [PubMed] [Google Scholar]
- 6. Nambiar M. P., Mitchell J. P., Ceruti R. P., Malloy M. A., Tsokos G. C. (2003) Prevalence of T cell receptor ζ chain deficiency in systemic lupus erythematosus. Lupus 12, 46–51 [DOI] [PubMed] [Google Scholar]
- 7. Moulton V. R., Kyttaris V. C., Juang Y. T., Chowdhury B., Tsokos G. C. (2008) The RNA-stabilizing protein HuR regulates the expression of ζ chain of the human T cell receptor-associated CD3 complex. J. Biol. Chem. 283, 20037–20044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moulton V. R., Tsokos G. C. (2010) Alternative splicing factor/splicing factor 2 regulates the expression of the ζ subunit of the human T cell receptor-associated CD3 complex. J. Biol. Chem. 285, 12490–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moulton V. R., Grammatikos A. P., Fitzgerald L. M., Tsokos G. C. (2013) Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 1845–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Black D. L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 11. Lemaire R., Prasad J., Kashima T., Gustafson J., Manley J. L., Lafyatis R. (2002) Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 16, 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun D., Novotny M., Bulek K., Liu C., Li X., Hamilton T. (2011) Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat. Immunol. 12, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michlewski G., Sanford J. R., Cáceres J. F. (2008) The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30, 179–189 [DOI] [PubMed] [Google Scholar]
- 14. Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X. D. (2008) The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15, 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinez N. M., Lynch K. W. (2013) Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol. Rev. 253, 216–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matter N., Herrlich P., König H. (2002) Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691–695 [DOI] [PubMed] [Google Scholar]
- 17. Motta-Mena L. B., Heyd F., Lynch K. W. (2010) Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol. Cell 37, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valitutti S., Müller S., Salio M., Lanzavecchia A. (1997) Degradation of T cell receptor (TCR)-CD3-ζ complexes after antigenic stimulation. J. Exp. Med. 185, 1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baeuerle P. A., Baltimore D. (1988) IκB: a specific inhibitor of the NF-κB transcription factor. Science 242, 540–546 [DOI] [PubMed] [Google Scholar]
- 20. Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 21. Bronevetsky Y., Villarino A. V., Eisley C. J., Barbeau R., Barczak A. J., Heinz G. A., Kremmer E., Heissmeyer V., McManus M. T., Erle D. J., Rao A., Ansel K. M. (2013) T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210, 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Breig O., Baklouti F. (2013) Proteasome-mediated proteolysis of SRSF5 splicing factor intriguingly co-occurs with SRSF5 mRNA upregulation during late erythroid differentiation. PLoS One 8, e59137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scharschmidt E., Wegener E., Heissmeyer V., Rao A., Krappmann D. (2004) Degradation of Bcl10 induced by T-cell activation negatively regulates NF-κB signaling. Mol. Cell. Biol. 24, 3860–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnan S., Kiang J. G., Fisher C. U., Nambiar M. P., Nguyen H. T., Kyttaris V. C., Chowdhury B., Rus V., Tsokos G. C. (2005) Increased caspase-3 expression and activity contribute to reduced CD3ζ expression in systemic lupus erythematosus T cells. J. Immunol. 175, 3417–3423 [DOI] [PubMed] [Google Scholar]
- 25. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chalfant C. E., Rathman K., Pinkerman R. L., Wood R. E., Obeid L. M., Ogretmen B., Hannun Y. A. (2002) De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J. Biol. Chem. 277, 12587–12595 [DOI] [PubMed] [Google Scholar]
- 27. Li X., Wang J., Manley J. L. (2005) Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 19, 2705–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dragone L. L., Shaw L. A., Myers M. D., Weiss A. (2009) SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol. Rev. 232, 218–228 [DOI] [PubMed] [Google Scholar]
- 29. Li D., Gál I., Vermes C., Alegre M. L., Chong A. S., Chen L., Shao Q., Adarichev V., Xu X., Koreny T., Mikecz K., Finnegan A., Glant T. T., Zhang J. (2004) Cutting edge: Cbl-b: one of the key molecules tuning CD28- and CTLA-4-mediated T cell costimulation. J. Immunol. 173, 7135–7139 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J. (2004) Ubiquitin ligases in T cell activation and autoimmunity. Clin. Immunol. 111, 234–240 [DOI] [PubMed] [Google Scholar]
- 31. Sun S., Zhang Z., Sinha R., Karni R., Krainer A. R. (2010) SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bronevetsky Y., Ansel K. M. (2013) Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 253, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sonkoly E., Ståhle M., Pivarcsi A. (2008) MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18, 131–140 [DOI] [PubMed] [Google Scholar]
- 34. Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nambiar M. P., Enyedy E. J., Fisher C. U., Krishnan S., Warke V. G., Gilliland W. R., Oglesby R. J., Tsokos G. C. (2002) Abnormal expression of various molecular forms and distribution of T cell receptor ζ chain in patients with systemic lupus erythematosus. Arthritis Rheum. 46, 163–174 [DOI] [PubMed] [Google Scholar]
- 36. Higgs R., Ní Gabhann J., Ben Larbi N., Breen E. P., Fitzgerald K. A., Jefferies C. A. (2008) The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181, 1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feist E., Dörner T., Kuckelkorn U., Schmidtke G., Micheel B., Hiepe F., Burmester G. R., Kloetzel P. M. (1996) Proteasome α-type subunit C9 is a primary target of autoantibodies in sera of patients with myositis and systemic lupus erythematosus. J. Exp. Med. 184, 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]


