Background: Glutamine metabolism is essential for Myc-induced tumors.
Results: SIRT4 regulates mitochondrial glutamine metabolism and the growth and survival of Burkitt lymphoma cells independent of Myc.
Conclusion: SIRT4 has a tumor-suppressive role in Myc-induced B cell lymphoma.
Significance: SIRT4 may be a potential target against Myc-driven and glutamine-dependent tumors.
Keywords: Cancer, Glutamine, Mitochondria, Mitochondrial metabolism, Sirtuins
Abstract
Glutamine metabolism plays an essential role for growth and proliferation of many cancer cells by providing metabolites for the maintenance of mitochondrial functions and macromolecular synthesis. Aberrant activation of the transcription factor c-Myc, e.g. caused by t(8;14) chromosomal translocation commonly found in Burkitt lymphoma, is a key driver of cellular glutamine metabolism in many tumors, highlighting the need to identify molecular mechanisms that can suppress glutamine usage in these cancers. Recently, the mitochondrial sirtuin SIRT4 has been reported to function as a tumor suppressor by regulating glutamine metabolism, suggesting that it might have therapeutic potential for treating glutamine-dependent cancers. Here, we report that SIRT4 represses Myc-induced B cell lymphomagenesis via inhibition of mitochondrial glutamine metabolism. We found that SIRT4 overexpression can dampen glutamine utilization even in Myc-driven human Burkitt lymphoma cells and inhibit glutamine-dependent proliferation of these cells. Importantly, SIRT4 overexpression sensitizes Burkitt lymphoma cells to glucose depletion and synergizes with pharmacological glycolysis inhibitors to induce cell death. Moreover, SIRT4 loss in a genetic mouse model of Myc-induced Burkitt lymphoma, Eμ-Myc transgenic mouse, greatly accelerates lymphomagenesis and mortality. Indeed, Eμ-Myc-induced B cell lymphoma cells from SIRT4 null mice exhibit increased glutamine uptake and glutamate dehydrogenase activity. Furthermore, we establish that SIRT4 regulates glutamine metabolism independent of Myc. Together, these results highlight the tumor-suppressive role of SIRT4 in Myc-induced B cell lymphoma and suggest that SIRT4 may be a potential target against Myc-induced and/or glutamine-dependent cancers.
Introduction
The MYC oncogene, which encodes a master transcription factor c-Myc (herein termed Myc), has been implicated in many human cancers by regulating cellular metabolism (1, 2). Recent studies have shown that Myc coordinates the expression of genes required for glutamine catabolism and triggers cellular dependence on glutamine as an important source for biosynthetic and bioenergetic needs of cell growth (3, 4). The efficient tumor formation driven by Myc amplification is well studied in human Burkitt lymphoma, in which Myc expression is dysregulated by the MYC/IgH chromosomal translocation (5). Previous studies have shown that increased glutamine metabolism is essential for survival and proliferation of Myc-induced Burkitt lymphoma cells (6). The Eμ-Myc transgenic mouse model, which overexpresses Myc under the control of the immunoglobulin heavy chain gene enhancer (Eμ), has constitutive Myc activation, providing an animal model to study Myc-driven lymphomas (7). These mice overexpress Myc exclusively in B cells and succumb to spontaneous pre-B and B cell lymphomas, which reach an incidence of 50% at 15–20 weeks (on a C57BL/6 background). Importantly, Myc activation/amplification-induced metabolic reprogramming triggers cellular addiction to glutamine for their growth and survival (3), highlighting the need to identify new pathways that can suppress glutamine usage even in the presence of constitutive Myc activation.
Sirtuins (SIRT1–7) are a conserved family of NAD-dependent deacetylases, deacylases, and ADP-ribosyltransferases that play essential roles in cell metabolism, stress response, and longevity (8, 9). Recently, we and others reported that the mitochondrial SIRT4 exerts tumor-suppressive activities by repressing mitochondrial glutamine metabolism, in part through modification and repression of glutamate dehydrogenase (GDH)2 (10, 11). However, little is known about how SIRT4 interacts with other oncogenic pathways that promote metabolic reprogramming in cancer cells. Because Myc supports growth and proliferation of Burkitt lymphomas, at least in part, by promoting the expression of enzymes that drive glutamine metabolism, we hypothesized that SIRT4 overexpression may be a novel mechanism for repressing Myc-induced B cell lymphomas, providing important implications for suppressing glutamine utilization in Myc-driven tumors.
In this study, we examined whether SIRT4 regulates Myc-induced B cell lymphoma. Using two human Burkitt lymphoma cell lines, we demonstrated that SIRT4 overexpression represses mitochondrial glutamine metabolism and inhibits proliferation and survival of these cells. We examined the tumor modulatory role of SIRT4 for the first time using a genetic mouse model of Myc-driven lymphoma. SIRT4 loss in Eμ-Myc transgenic mice accelerated Eμ-Myc-induced lymphomagenesis and mortality. Mechanistically, we found that SIRT4 suppresses glutamine metabolism independent of Myc. Together, these results identify a new role for SIRT4 in suppressing Myc-induced B cell lymphoma.
EXPERIMENTAL PROCEDURES
Vectors and Virus Production
The EXPANSIN7, SIRT4, and SIRT4H161Y cDNAs were recombined into pLenti CMVTRE3G Dest plasmid (Addgene). Lentiviral supernatants were generated by transient transfection of HEK293T cells and harvested 48 h after transfection.
Cell Culture
Ramos and Raji human Burkitt lymphoma cells (kindly provided by Dr. Anthony G. Letai, Dana-Farber Cancer Institute, Boston, MA) were maintained in RPMI (Invitrogen) supplemented with 10% fetal bovine serum (Clontech) and penicillin/streptomycin (Invitrogen). Stable cell lines expressing indicated cDNAs were generated by lentiviral transduction in the presence of 8 μg/ml Polybrene followed by selection with appropriate antibiotic resistance markers. Cells were induced by incubation with doxycycline (1 μg/ml) (Clontech). To establish B cell lymphoma lines, lymph nodes from diseased mice were propagated in RPMI supplemented with 10% FBS, penicillin/streptomycin, and 2-mercaptoethanol (Invitrogen).
Animal Studies
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee, the standing committee on animals at Harvard. Eμ-Myc transgenic mice (catalogue name, C57BL/6J-Tg(IghMyc)22Bri/J) were purchased from The Jackson Laboratory. Eμ-Myc males were crossed with Sirt4−/− females with the C57BL/6 background (12), and then Eμ-Myc/Sirt4+/− male offspring were crossed with Sirt4+/− females to generate Eμ-Myc/Sirt4+/+ mice and Eμ-Myc/Sirt4−/− mice. The expected genotypes were obtained in Mendelian ratios. The transgene was detected by PCR amplification according to the manufacturer's instructions. Animals were monitored twice weekly after birth for palpable tumors and signs of illness, and only mice showing enlarged lymphoid organs at necropsy were designated as having lymphomas.
Glutamine and Glucose Measurements
Glutamine, ammonia, glucose, and lactate levels in culture media were measured using the BioProfile FLEX analyzer (Nova Biomedical), as described previously (10). Briefly, fresh media were added to a six-well plate of cells, and metabolite levels in the media were measured at the indicated times and normalized to the number of cells in each well.
Cell Proliferation Assay
Cells were grown in complete culture media or in the absence of glucose or glutamine in 6-well plates at 105 cells per well for the indicated days and counted. Cell number was immediately analyzed on a Z1 Coulter Counter.
Flow Cytometric Measurement of Cell Death
Cells were harvested, pelleted by centrifugation, and resuspended in PBS containing 3% FBS. The measurement of cell death was performed by flow cytometry using propidium iodide staining, as described previously (10).
Western Blotting
Cells were lysed with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, and 0.5% Nonidet P-40) supplemented with protease inhibitor cocktail (Roche Applied Science). Cell lysates were separated by SDS-PAGE and immunoblotting. Antibodies for human and mouse SIRT4 were described previously (12). Antibodies used were: anti-actin (Sigma), anti-Myc (Santa Cruz Biotechnology), anti-GLS (Abcam), and anti-GDH (US Biological).
Quantitative RT-PCR
Total RNA was prepared with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 1 μg of total RNA was reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad). Diluted cDNAs were analyzed by real-time PCR using SYBR Green I master mix on a LightCycler 480 (Roche Applied Science). The level of gene expression was normalized to β-actin. The primer sequences were: CCTTTCGCTCATACTCTACCAC and AAACACTACCAAGCCCAGG for human ASCT2; TCGGAAAGCTGTACTGGTTG and TCTGTTCCCCACAATCCAAG for human actin; CATCTACTTCCTCTTCACCCG and CCACACCATTCTTCTCCTCTAC for mouse Asct2; ATGGCTTTTGCGTTTGTCTG and GAGGTATCCAAAGGTCGCTG for mouse Sn2; and AGCCATGTACGTAGCCATCC and CTCTCAGCTGTGGTGGTGAA for mouse actin.
GDH Activity Measurement
GDH activity was measured with the GDH activity assay kit (Biovision) according to the manufacturer's instructions. Briefly, cells were counted and homogenized. The cell extracts were used for the enzyme assays, in which the NADH produced by GDH was measured with a microplate reader (Agilent Technologies).
Statistical Analysis
Unpaired two-tailed Student's t test was performed unless otherwise noted. All experiments were performed at least two or three times. For the mice survival study, the log rank (Mantel-Cox) test was performed.
RESULTS
SIRT4 Suppresses Mitochondrial Glutamine Metabolism in Human Burkitt Lymphoma Cells
Recent studies by our laboratory and others have shown that SIRT4 limits glutamine anaplerosis and acts as a tumor suppressor in vitro and in vivo (10, 11). The Myc oncogene promotes the expression of genes involved in metabolic reprogramming of cells toward glutaminolysis and triggers cellular dependence on glutamine for their growth and survival (4, 13). However, the interaction between Myc and SIRT4 has never been investigated. Thus, we sought to probe whether SIRT4 can repress glutamine metabolism and tumorigenesis in Myc-driven tumors.
First, we examined whether elevated SIRT4 expression represses cellular glutamine metabolism in Myc-induced B cell lymphomas. As tumor cells may readily adapt their fuel utilization for growth and survival, we generated a novel doxycycline (Dox)-inducible system to acutely increase SIRT4 expression in Ramos or Raji human Burkitt lymphoma cell lines. These cells contained Dox-inducible EXPANSIN7 plant protein (pEXP7; control), human SIRT4 (SIRT4), or a catalytic mutant of SIRT4 (SIRT4H161Y) (10) constructs, such that Dox treatment resulted in a rapid induction of each protein (Fig. 1, A and B). The resulting cells provide an important new tool to assess the immediate effects of SIRT4 induction on cellular metabolism.
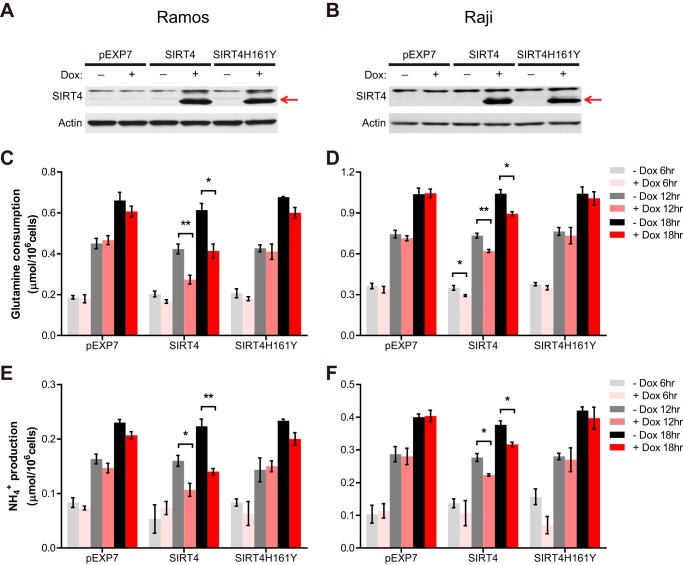
FIGURE 1.
SIRT4 represses glutamine anaplerosis in human Burkitt lymphoma cells. A and B, SIRT4 protein in whole cell lysates from Ramos (A) and Raji (B) cells treated with or without Dox (1 μg/ml) for 12 h was detected by immunoblotting. β-Actin served as a loading control. C and D, glutamine uptake in pEXP7, SIRT4, and SIRT4H161Y Ramos (C) and Raji (D) cells incubated with or without doxycycline for the indicated times (n = 3). E and F, ammonia production in pEXP7, SIRT4, and SIRT4H161Y Ramos (E) and Raji (F) cells incubated with or without doxycycline for the indicated times (n = 3). Data are mean ± S.E. *, p < 0.05 and **, p < 0.01.
We observed that Dox-induced SIRT4 overexpression resulted in a pronounced reduction in glutamine consumption in both Burkitt lymphoma cell lines, whereas overexpression of pEXP7 or SIRT4H161Y did not affect fuel utilization (Fig. 1, C and D). In the mitochondria, glutamine is metabolized via glutaminase (GLS) to glutamate and ammonia (NH4+) and further converted to the tricarboxylic acid (TCA) cycle intermediate α-ketoglutarate via GDH or aminotransferases. It has been reported that in glioblastoma cells, most of the ammonia (>90%) in the media was derived from this glutamine anaplerosis (14). Indeed, we found that SIRT4-induced reduction of glutamine consumption was accompanied by a reduction in ammonia production from cells (Fig. 1, E and F). However, we observed no obvious change in glucose uptake and lactate secretion (data not shown), consistent with our previous study showing that SIRT4 directly regulates mitochondrial glutamine metabolism but not glucose consumption (10). Thus, these data demonstrate that SIRT4 overexpression represses mitochondrial glutamine metabolism in Myc-driven human Burkitt lymphoma cells.
SIRT4 Suppresses the Growth of Human Burkitt Lymphoma Cells
Refilling the mitochondrial carbon pool by glutamine anaplerosis is essential for the biosynthetic roles of mitochondria and the rapid proliferation of cancer cells (3, 15). As our results showed that glutamine anaplerosis is repressed by SIRT4 overexpression in Burkitt lymphoma cells (Fig. 1), we next tested the idea that SIRT4-mediated repression of mitochondrial glutamine metabolism would inhibit the proliferation of Burkitt lymphoma cells. Indeed, we found that Dox-induced SIRT4 overexpression significantly repressed proliferation of both Burkitt lymphoma cell lines, whereas overexpression of pEXP7 or SIRT4H161Y did not (Fig. 2, A and B). GLS is the first enzyme for mitochondrial glutamine metabolism (16), and its inhibition limits glutamine entry into the TCA cycle (6, 17). It has been shown that SIRT4 overexpression no longer inhibits glutamine uptake when GLS1 is reduced (10). To examine whether SIRT4 represses cell proliferation through the repression of mitochondrial glutamine metabolism, we cultured SIRT4-inducible lymphoma cells with BPTES (18), an inhibitor of GLS, and measured proliferation with or without Dox treatment. Importantly, control and SIRT4-induced cells proliferated at similar rates under these conditions (Fig. 2C). Remarkably, BPTES treatment inhibits cell proliferation more profoundly than SIRT4 overexpression, suggesting that SIRT4 functions downstream of GLS and represses proliferation of Burkitt lymphoma cells by limiting mitochondrial glutamine metabolism. Taken together, in culture, SIRT4 overexpression can repress the growth of Myc-driven tumor cells.
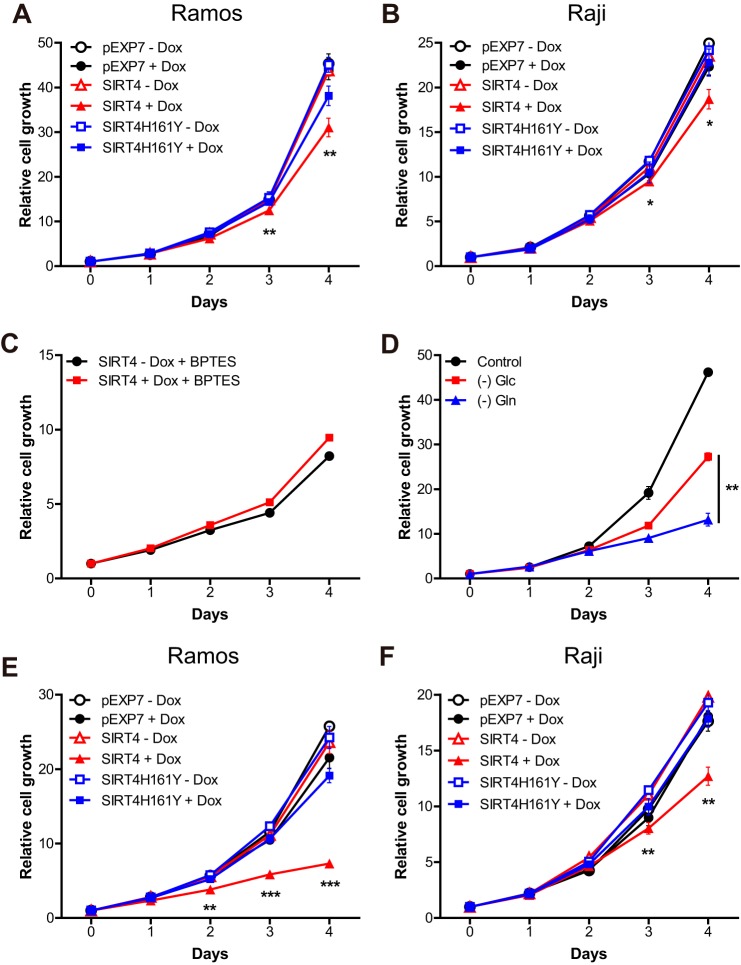
FIGURE 2.
SIRT4 represses proliferation of Burkitt lymphoma cells through mitochondrial glutamine metabolism. A and B, growth curves of pEXP7, SIRT4, and SIRT4H161Y Ramos (A) and Raji (B) cells cultured in 6-well plates with or without doxycycline (1 μg/ml). Cell number was measured every 24 h for 4 consecutive days (n = 3). C, growth curves of control or doxycycline-treated SIRT4 Ramos cells cultured in standard media or media supplemented with BPTES (10 μm) (n = 3). D, growth curves of Ramos cells were cultured in standard media or media without glucose ((−) Glc) or glutamine ((−) Gln) (n = 3). E and F, growth curves of pEXP7, SIRT4, and SIRT4H161Y Ramos (E) and Raji (F) cells cultured in glucose-depleted media with or without doxycycline (n = 3). Data are mean ± S.D. *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
Because previous studies showed that the overexpression of Myc sensitizes cells to glutamine withdrawal (17), we measured the proliferation of Ramos and Raji cells under glutamine deprivation. Although Burkitt lymphoma cells sustained their growth in glucose-depleted media, the growth rates of these cells were significantly diminished with glutamine deprivation (Fig. 2D and data not shown), indicating that glutamine metabolism is important for cell proliferation of human Burkitt lymphoma cells. Mitochondrial glutamine metabolism supports the refilling of the mitochondrial TCA cycle in the absence of glucose, and glutamine metabolism alone can sustain cell growth in Myc-induced Burkitt lymphoma cells (6). To further examine whether SIRT4 overexpression suppressed cell proliferation through glutamine metabolism, we grew these cells in glucose-deprived media, thereby stalling glycolytic flux and forcing the cells to rely on mitochondrial glutamine metabolism for proliferation. Under these conditions, pEXP7 and SIRT4H161Y cell lines grew at the same rate with or without Dox treatment (Fig. 2, E and F). Strikingly, we observed that Dox-induced SIRT4 overexpression dramatically repressed proliferation of both Burkitt lymphoma cells (Fig. 2, E and F). Notably, the magnitude of growth inhibition caused by SIRT4 overexpression was highly increased in these conditions when compared with the inhibition under glucose-repleted media (Fig. 2, A and B). Together, these data demonstrate that SIRT4 inhibits proliferation of Myc-induced Burkitt lymphoma cells by limiting glutamine entry into the mitochondrial metabolism.
SIRT4 Synergizes with Glycolytic Inhibition to Induce the Cell Death of Burkitt Lymphoma Cells
Mitochondrial glutamine catabolism is essential for sustaining cell viability in the absence of glucose (14, 19), and ectopic Myc expression drives cells to be dependent on glutamine for survival (17). To assess the functional relevance of decreased mitochondrial glutamine metabolism by SIRT4 overexpression on the survival of Myc-driven B cell lymphoma cells, we deprived Burkitt lymphoma cells of glucose, thereby shifting cellular dependence to glutamine to maintain viability. Both Burkitt lymphoma cells were able to survive and proliferate in glucose-deficient media (Fig. 2D and data not shown). The addition of Dox did not affect the cell survival of pEXP7- and SIRT4H161Y-overexpressing Ramos and Raji cell lines (Fig. 3, A and B). However, Dox-induced SIRT4 overexpression in both cell lines significantly induced cell death in glucose-deprived media (Fig. 3, A and B), suggesting that SIRT4 overexpression reduced the ability of Burkitt lymphoma cells to utilize glutamine for mitochondrial energy production, which is essential for cell survival.
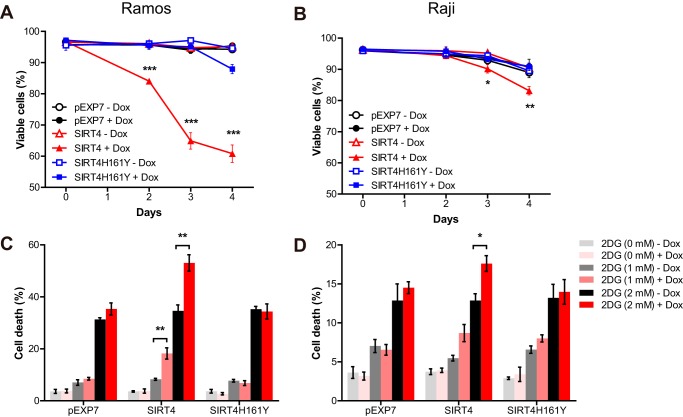
FIGURE 3.
SIRT4 sensitizes Burkitt lymphoma cells to the inhibition of glucose metabolism. A and B, pEXP7, SIRT4 and SIRT4H161Y Ramos (A) and Raji (B) cells cultured in glucose-depleted media with or without doxycycline (1 μg/ml). At the indicated time points, cell viability was measured via propidium iodide exclusion assay (n = 3). Data are mean ± S.D. C and D, pEXP7, SIRT4, and SIRT4H161Y Ramos (C) and Raji (D) cells were given the indicated doses of 2-deoxyglucose (2DG) with or without doxycycline. Cell viability was measured 72 h after treatment via propidium iodide exclusion assay (n = 3). Data are mean ± S.E. *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
To further test the effect of SIRT4 overexpression on the sensitivity of Burkitt lymphoma cells to glycolysis inhibition, we cultured lymphoma cells with 2-deoxyglucose, an inhibitor of glucose metabolism, and assessed the cell viability after Dox treatment. Consistent with our previous results, Dox-induced SIRT4 overexpression sensitized Burkitt lymphoma cells to 2-deoxyglucose-induced cell death, whereas overexpression of pEXP7 or SIRT4H161Y did not (Fig. 3, C and D). These data provide further support for the critical role of SIRT4 in glutamine metabolism and survival of Myc-induced Burkitt lymphoma cells and demonstrate that the combination treatments with SIRT4 overexpression and glycolysis inhibitors potently synergize to kill tumor cells with elevated Myc signaling.
Loss of SIRT4 Accelerates Lymphomagenesis and Mortality in Eμ-Myc Transgenic Mice
Loss of SIRT4 increased glutamine-dependent cell proliferation and tumorigenesis (10). We found that SIRT4 overexpression represses glutamine utilization, proliferation, and survival of Myc-induced Burkitt lymphoma cells. To extend these cellular findings to an in vivo model, we crossed Eμ-Myc transgenic mice with SIRT4 knock-out (KO) mice (12) to generate Eμ-Myc/SIRT4KO mice and Eμ-Myc/SIRT4 wild type (WT) littermates. This is the first investigation of the tumor-suppressive role of SIRT4 in a genetic mouse model of human cancer.
To examine whether the loss of SIRT4 affects tumorigenesis in this mouse model, the survival of Eμ-Myc/SIRT4KO mice (n = 26) was compared with the survival of Eμ-Myc/SIRT4WT mice (n = 24). Importantly, we found that the loss of SIRT4 significantly decreased the survival of Eμ-Myc mice (p = 0.0238 with the log rank test) (Fig. 4A). The median survival of Eμ-Myc/SIRT4KO mice was 139 days, which was significantly shorter than Eμ-Myc/SIRT4WT mice (195 days, p < 0.05). Although there was no significant difference between the survival of Eμ-Myc/SIRT4 heterozygous (n = 30) and Eμ-Myc/SIRT4WT mice, there was a slight trend that Eμ-Myc/SIRT4Het mice died earlier than Eμ-Myc/SIRT4WT mice, with a median survival of 150 days (Fig. 4A). At autopsy, the spleen weights of Eμ-Myc/SIRT4KO mice were heavier than those of Eμ-Myc/SIRT4WT mice (Fig. 4B), consistent with the increased mortality of Eμ-Myc/SIRT4KO mice. As we hypothesize that SIRT4 loss accelerates lymphomagenesis, we next examined lymphoma incidence in these mice at 11 and 16 weeks of age. Approximately 41% (14/34) and 71% (29/41) of Eμ-Myc/SIRT4KO animals developed B cell lymphomas at 11 and 16 weeks, respectively, whereas 23% (7/30) and 52% (22/42) of age-matched Eμ-Myc/SIRTWT mice developed lymphomas (Fig. 4C). None of the nontransgenic SIRT4 WT or KO mice developed lymphoma during a comparable period of monitoring (Fig. 4C). As reported previously (5), tumors arising in these mice expressed B220 but were heterogeneous with respect to the amount of IgM expression (Fig. 4D, left). We detected no differences in the distribution of these tumor types when comparing tumors from SIRT4 WT versus KO mice (Fig. 4D, right). Taken together, our data demonstrate that SIRT4 has tumor-suppressive activities in a genetic mouse model of human Burkitt lymphoma.
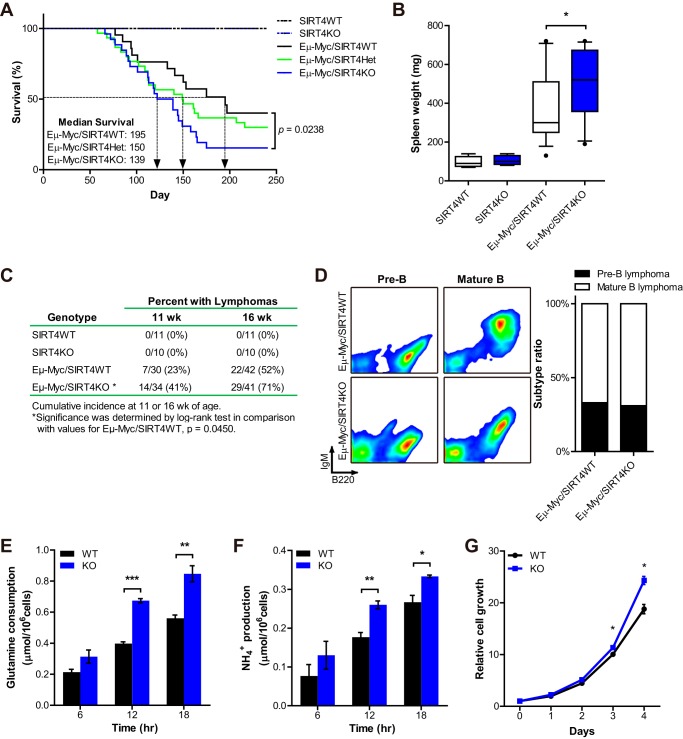
FIGURE 4.
SIRT4 loss accelerates Myc-driven B cell lymphoma. A, analysis of overall survival of Eμ-Myc/SIRT4WT (n = 24), Eμ-Myc/SIRT4Het (n = 30), and Eμ-Myc/SIRT4KO (n = 26) mice. p = 0.0238 by the log rank test. B, spleen weight of WT (n = 5), KO (n = 5), Eμ-Myc/SIRT4WT (n = 15), and Eμ-Myc/SIRT4KO (n = 14) mice at autopsy. The boxes represent the interquartile range; whiskers represent the 10th–90th percentile range; and bars represent the median. C, cumulative lymphoma incidences at 11 and 16 weeks of age. Significance between Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice was determined by log rank test. p = 0.0450. D, left, analysis of lymph node cell suspensions isolated from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice. Dot plots show B220 and IgM expression on the cell population from independent mice that were examined after the onset of illness by using flow cytometry. Right, relative frequencies of lymphoma types of Eμ-Myc/SIRT4WT (n = 12) and Eμ-Myc/SIRT4KO (n = 13) mice: pre-B lymphoma (B220+IgM−) and mature B lymphoma (B220+IgM+). E and F, glutamine uptake (E) and ammonia production (F) from B cell lymphoma cells from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice incubated for the indicated times (n = 3). Data are mean ± S.E. G, growth curves of B cell lymphoma cells from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice. Cell number was measured every 24 h for 4 consecutive days (n = 3). Data are mean ± S.D. *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
Next, to assess whether the mechanistic functions of SIRT4 were important in these Eμ-Myc B cell lymphomas, we measured the usage of glutamine and glucose in the cells derived from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO B cell lymphomas. Consistent with our previous data, SIRT4 loss increased glutamine uptake and ammonia production of these cells (Fig. 4, E and F), but not glucose uptake and lactate production (data not shown). These data support the model that SIRT4 loss enhances glutamine metabolism, contributing to the growth of B cell lymphomas. Indeed, SIRT4 KO lymphoma cells grew faster than WT cells (Fig. 4G). Thus, these results surprisingly show that even the accelerated glutamine usage in a Myc-driven tumor cell can be further induced by SIRT4 loss.
SIRT4 Regulates Cellular Glutamine Metabolism in a Myc-independent Manner
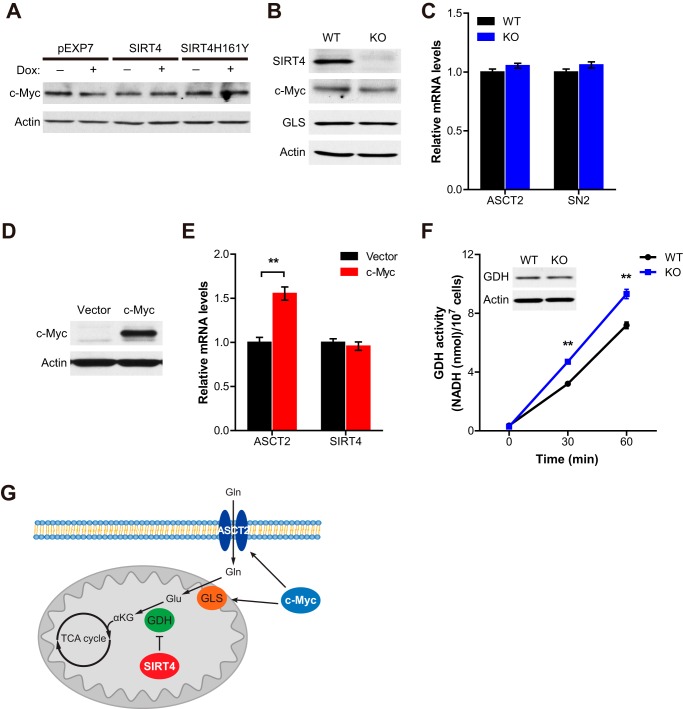
Myc regulates the transcription of diverse target genes, and it stimulates mitochondrial glutaminolysis, which is essential for its proliferative and oncogenic functions (4). It has been shown that the modulation of Myc transcriptional activity, through a dominant-interfering Myc mutant, exerts profound effects on tumor growth (20). Because SIRT4 expression levels regulate cellular glutamine metabolism even in tumor cells with elevated Myc expression, we hypothesize that SIRT4 regulates glutamine metabolism independent of Myc. To test this idea, we first examined whether Myc expression is changed by SIRT4 expression levels. We found that Myc protein levels were not affected by SIRT4 overexpression or SIRT4 loss (Fig. 5, A and B). We next assessed whether SIRT4 could repress glutamine metabolism in Myc-driven tumors by repressing the activity of Myc. It has been shown that Myc regulates glutamine metabolism by enhancing the transcription of glutamine transporters, ASCT2 and SN2 (4), and by elevating GLS expression through posttranscriptional regulation (21). When we examined the expression levels of these Myc target genes in SIRT4 WT and KO lymphoma cells, we observed no obvious change (Fig. 5, B and C), suggesting that SIRT4 does not regulate glutamine metabolism by inhibiting Myc itself. Conversely, we also assessed whether SIRT4 expression was regulated by Myc. As reported, Myc overexpression significantly induced the transcription of its target gene ASCT2, whereas SIRT4 expression was not changed (Fig. 5, D and E), demonstrating that SIRT4 is not a target of Myc in this system.
FIGURE 5.
SIRT4 regulates mitochondrial glutamine metabolism of Myc-driven B cell lymphoma cells independent of Myc. A, Myc protein in whole cell lysates from Ramos cells treated with or without doxycycline (1 μg/ml) for 12 h was detected by immunoblotting. β-Actin served as a loading control. B, SIRT4, GLS, and Myc proteins in B cell lymphoma cells from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice were detected by immunoblotting. β-Actin served as a loading control. C, relative mRNA expression levels of indicated genes in B cell lymphoma cells from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice (n = 3). β-Actin was used as an endogenous control for quantitative RT-PCR. D, Myc protein in HEK293T cells expressing empty vector or c-Myc was detected by immunoblotting. β-Actin served as a loading control. E, relative mRNA expression levels of indicated genes in HEK293T cells expressing empty vector or c-Myc (n = 3). β-Actin was used as an endogenous control for quantitative RT-PCR. F, GDH activity in B cell lymphomas from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO mice (n = 3). NADH produced by GDH was measured from extracts of each cell line (data are mean ± S.D.). G, a proposed model illustrating the regulation of Myc-induced B cell lymphoma by SIRT4. Data are mean ± S.E. **, p < 0.01.
SIRT4 is a negative regulator of GDH activity (12), and GDH contributes to the inhibitory role of SIRT4 in glutamine metabolism (10, 11). To test whether SIRT4 regulates GDH activity in Myc-driven lymphoma cells, we measured GDH activity in cells derived from Eμ-Myc/SIRT4WT and Eμ-Myc/SIRT4KO B cell lymphomas. We found that GDH activity was significantly increased in SIRT4 KO lymphoma cells when compared with WT cells (Fig. 5F). Taken together, these results provide critical evidence that SIRT4 regulates mitochondrial glutamine metabolism and proliferation of B cell lymphoma cells in a Myc-independent manner and that the loss of this important regulatory node accelerates Myc-induced lymphomagenesis (Fig. 5G).
DISCUSSION
In this study, we reveal the profound impact of SIRT4 on mitochondrial glutamine metabolism and proliferation using in vitro and in vivo Myc-driven tumor models. Here we report that SIRT4 suppresses glutamine metabolism even in Myc-activated Burkitt lymphoma cells and inhibits glutamine-dependent proliferation of these cells. Importantly, SIRT4 overexpression sensitizes Burkitt lymphoma cells to glucose-free media and synergizes with glycolysis inhibitors to induce cell death. Moreover, SIRT4 loss can further activate glutamine usage and accelerate Myc-induced lymphomagenesis and mortality of Eμ-Myc transgenic mice. These data demonstrate that SIRT4 can modulate glutamine metabolism in addition to the regulation by Myc.
Our findings are consistent with previous studies showing that SIRT4 functions as a tumor suppressor by regulating mitochondrial glutamine metabolism (10, 11). Moreover, we found that SIRT4 KO mice spontaneously develop several types of tumors, including lung tumor, liver tumor, mammary tumor, and lymphoma, and that SIRT4 expression is decreased in many human cancers (10).
Here, we show that the loss of SIRT4 further accelerates Myc-induced B cell lymphomas. It is a common feature of proto-oncogenes that their oncogenic potential is accompanied with cooperating mutations. For example, the expression of miR-17∼92 cluster, frequently amplified in human cancer, is required for Myc-induced B cell lymphoma by counterbalancing the pro-apoptotic activity of Myc (22). Thus, SIRT4 loss may create a tumor-permissive environment to provide additional metabolic advantages, supporting tumor cell survival and proliferation.
Many therapeutic strategies have been developed to target metabolic pathways of cancer cells (23). For example, targeting glucose metabolism through glycolysis inhibitors has been used to treat tumors (24, 25). However, cancer cells retain the capacity for metabolic adaptation to survive periods of diminished glucose metabolism by activating alternative metabolic pathways (23), and some cancer cells depend on a high rate of glutaminolysis for their continued growth and survival. For instance, Myc-driven tumor cells display addiction to glutamine metabolism (3) and can sustain their viability and proliferation under glucose-depleted conditions (6, 21). We also showed that Myc-induced human Burkitt lymphoma cells are able to survive and proliferate in glucose-deficient media. Importantly, SIRT4 overexpression sensitizes these cells to glucose withdrawal-induced cell death and significantly inhibits glucose-independent proliferation. Moreover, the combination of SIRT4 overexpression and glycolysis inhibitors significantly increased the death of lymphoma cells. Thus, our current work highlights the therapeutic potential of SIRT4 for targeting metabolic flexibility and adaptation of Myc-driven tumors.
Previous studies have shed light on the importance of mitochondrial glutamine metabolism for oncogene-induced tumorigenesis, and the significance of SIRT4 in this node is just beginning to emerge. It has been shown that glutamine catabolism by the TCA cycle is essential for Kras-mediated tumorigenicity (26). Also, the inhibition of mitochondrial glutamine metabolism by targeting GLS represses Rho GTPases-induced transformation (27). Recently, Csibi et al. (11) found that mammalian target of rapamycin complex 1 (mTORC1) promotes mitochondrial glutamine metabolism by transcriptionally repressing SIRT4 expression and that SIRT4 overexpression reduces proliferation and transformation of tuberous sclerosis 2 mutant cells. In this study, we uncover the profound impact of SIRT4 on mitochondrial glutamine metabolism and tumor survival and proliferation in glutamine-dependent Myc-driven tumor models. Thus, these findings suggest that the tumor-suppressive activity of SIRT4, by regulating mitochondrial glutamine metabolism, could apply to other oncogene-induced tumors and provide a new avenue for tumor therapy.
Acknowledgments
We thank Ditte Lee for technical assistance and Natalie German and Yoshinori Ishikawa for helpful discussions. We are also grateful to Anthony G. Letai for providing cell lines.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AG032375 (to M. C. H.).
- GDH
- glutamate dehydrogenase
- Dox
- doxycycline
- pEXP7
- expansin7
- GLS
- glutaminase
- BPTES
- bis-2-(5-phenylacetoamido-1,2,4-thiadiazol-2-yl)ethyl sulfide
- TCA
- tricarboxylic acid.
REFERENCES
- 1. Dang C. V., Le A., Gao P. (2009) MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dang C. V. (2010) Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 70, 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wise D. R., Thompson C. B. (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. (1985) The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 [DOI] [PubMed] [Google Scholar]
- 6. Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., Zimmerman L. J., Liebler D. C., Slebos R. J., Lorkiewicz P. K., Higashi R. M., Fan T. W., Dang C. V. (2012) Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michalak E. M., Jansen E. S., Happo L., Cragg M. S., Tai L., Smyth G. K., Strasser A., Adams J. M., Scott C. L. (2009) Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 16, 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haigis M. C., Guarente L. P. (2006) Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 9. Finkel T., Deng C. X., Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeong S. M., Xiao C., Finley L. W., Lahusen T., Souza A. L., Pierce K., Li Y. H., Wang X., Laurent G., German N. J., Xu X., Li C., Wang R. H., Lee J., Csibi A., Cerione R., Blenis J., Clish C. B., Kimmelman A., Deng C. X., Haigis M. C. (2013) SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Csibi A., Fendt S. M., Li C., Poulogiannis G., Choo A. Y., Chapski D. J., Jeong S. M., Dempsey J. M., Parkhitko A., Morrison T., Henske E. P., Haigis M. C., Cantley L. C., Stephanopoulos G., Yu J., Blenis J. (2013) The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 13. Dang C. V. (2011) Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb. Symp. Quant. Biol. 76, 369–374 [DOI] [PubMed] [Google Scholar]
- 14. Yang C., Sudderth J., Dang T., Bachoo R. M., McDonald J. G., DeBerardinis R. J. (2009) Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 69, 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Curthoys N. P., Watford M. (1995) Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutrition 15, 133–159 [DOI] [PubMed] [Google Scholar]
- 17. Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. (2007) Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 178, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson M. M., McBryant S. J., Tsukamoto T., Rojas C., Ferraris D. V., Hamilton S. K., Hansen J. C., Curthoys N. P. (2007) Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 406, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choo A. Y., Kim S. G., Vander Heiden M. G., Mahoney S. J., Vu H., Yoon S. O., Cantley L. C., Blenis J. (2010) Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell 38, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., Evan G. I. (2008) Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., Dang C. V. (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mu P., Han Y. C., Betel D., Yao E., Squatrito M., Ogrodowski P., de Stanchina E., D'Andrea A., Sander C., Ventura A. (2009) Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23, 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tennant D. A., Durán R. V., Gottlieb E. (2010) Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 [DOI] [PubMed] [Google Scholar]
- 24. Maschek G., Savaraj N., Priebe W., Braunschweiger P., Hamilton K., Tidmarsh G. F., De Young L. R., Lampidis T. J. (2004) 2-deoxy-d-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res. 64, 31–34 [DOI] [PubMed] [Google Scholar]
- 25. Papaldo P., Lopez M., Cortesi E., Cammilluzzi E., Antimi M., Terzoli E., Lepidini G., Vici P., Barone C., Ferretti G., Di Cosimo S., Nistico C., Carlini P., Conti F., Di Lauro L., Botti C., Vitucci C., Fabi A., Giannarelli D., Marolla P. (2003) Addition of either lonidamine or granulocyte colony-stimulating factor does not improve survival in early breast cancer patients treated with high-dose epirubicin and cyclophosphamide. J. Clin. Oncol. 21, 3462–3468 [DOI] [PubMed] [Google Scholar]
- 26. Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., Chandel N. S. (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 107, 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J. B., Erickson J. W., Fuji R., Ramachandran S., Gao P., Dinavahi R., Wilson K. F., Ambrosio A. L., Dias S. M., Dang C. V., Cerione R. A. (2010) Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]