Background: miR-155 is strongly induced by LPS, a response inhibited by IL-10.
Results: The Ets2 transcription factor is required for induction of miR-155 by LPS. IL-10 can subsequently decrease miR-155 via suppression of Ets2.
Conclusion: Ets2 is an important transcription factor for regulation of miR-155.
Significance: This study reports a detailed mechanism of induction of miR-155 and provides a new means of inhibition for IL-10 via suppression of Ets2.
Keywords: Ets Family Transcription Factor, Interleukin, Lipopolysaccharide (LPS), MicroRNA, Toll-like Receptors (TLR), Ets2, Interleukin 10, SHIP1, TLR4, miR-155
Abstract
MicroRNA-155 (miR-155) is highly expressed in many cancers such as B cell lymphomas and myeloid leukemia and inflammatory disorders such as rheumatoid arthritis, atopic dermatitis, and multiple sclerosis. The role of miR-155 as both a promoter of inflammation and an oncogenic agent provides a clear need for miR-155 itself to be stringently regulated. We therefore investigated the transcriptional regulation of miR-155 in response to the respective pro- and anti-inflammatory mediators LPS and IL-10. Bioinformatic analysis revealed Ets binding sites on the miR-155 promoter, and we found that Ets2 is critical for miR-155 induction by LPS. Truncation and mutational analysis of the miR-155 promoter confirmed the role of the Ets2 binding site proximal to the transcription start site for LPS responsiveness. We observed increased binding of Ets2 to the miR-155 promoter and Ets2 deficient mice displayed decreased induction of miR-155 in response to LPS. IL-10 inhibited the induction of Ets2 mRNA and protein by LPS, thereby decreasing Ets2 function on the pri-155 promoter. We have thus identified Ets2 as a key novel regulator in both the positive and negative control of miR-155 in the inflammatory response.
Introduction
The immune response to infection involves a carefully orchestrated and timed signaling cascade resulting in up-regulation of an abundance of immune and inflammatory genes, followed by a stringently regulated anti-inflammatory response to ensure a timely return to homeostasis and prevent excess inflammation (1). Toll-like receptors (TLRs)3 play a central role in this system, recognizing conserved microbial patterns and launching an efficacious and controlled immune response. These responses induced by TLRs must be meticulously regulated, and one such class of regulators is microRNAs (miRNAs). miRNAs are a class of non-coding molecules that bind to the 3′-UTR of target mRNAs, regulating their translation (2). They are highly efficient fine tuners of gene expression, exerting subtle yet essential effects throughout the genome.
miRNAs have been shown to dampen down immune responses, fine tune expression of inflammatory mediators, and control the differentiation and development of immune cell lineages. Certain miRNA, for example miR-146a, target key immune regulators to prevent excess inflammation (3). Others, such as miR-155, are proinflammatory. miR-155 is processed from a primary noncoding transcript B cell integration cluster (or pri-155), originally identified in lymphomas (4). miR-155 is potently induced in response to various TLR ligands (5) and targets a number of key players in the immune response, such as inhibiting the negative TLR4 regulator SHIP1 (6, 7) and SOCS1 (8). The targets of miR-155 in immunity and inflammation have been well characterized (9), but the detailed mechanism by which this miRNA is potently induced upon ligation of TLR receptors has not been extensively studied. We have previously shown that the anti-inflammatory cytokine IL-10 inhibits miR-155, but not the anti-inflammatory miRNAs miR-21 or miR-146a (10). This observation revealed a new axis of regulation for this anti-inflammatory cytokine on the TLR pathway. IL-10 is crucial for stifling the inflammatory response after infection and is vital to protect the host from excess inflammation (11). The exact mechanism of this inhibition, similar to much of the functions of IL-10, has not yet been unraveled in detail.
In this study, we have examined in detail the mechanism of induction of miR-155 by LPS. We have found a key role for the transcription factor Ets2 in this induction. Furthermore, IL-10 suppresses Ets2 expression and thereby inhibits the induction of miR-155 by LPS. We have therefore identified a novel transcriptional inducer of miR-155, revealed a new downstream target of IL-10 in the form of Ets2 and an important process through which IL-10 acts to exert its anti-inflammatory properties.
EXPERIMENTAL PROCEDURES
Reagents
LPS from Escherichia coli, serotype 0111:B4, was from Alexis. Recombinant mouse and human IL-10 were from R&D Biosystems. Oligonucleotides and SYBR primers were from Eurofins, and TaqMan probes were from Applied Biosystems. Ets2 antibody was from Santa Cruz Biotechnology (sc-351), and β-actin antibody was from Sigma (AC-74). STAT3 inhibitor was from Merck Millipore (catalog no. 573099).
Cell Culture and Isolation
Raw264.7 and HEK293T cell lines, obtained from the European Cell Culture Collection, and immortalized WT, myD88−/−, and TIR-domain-containing adapter-inducing interferon-β (TRIF)−/− bone marrow-derived macrophages, obtained from Dr. Elaine Kenny were maintained in Dulbecco's modified Eagle's medium. Ets2fl/fl/LysMCre+/− and matched wild type mice were obtained from Paul Hertzog (Monash Institute of Medical Research, Monash University, Victoria, Australia). Bone marrow was isolated from the tibias and femurs of C57BL/6 mice, and cells were grown in macrophage colony stimulating factor-conditioned DMEM. Peritoneal macrophages were isolated from Ets2fl/fl/LysMCre+/− or C57BL/6 mice. Purity of macrophages was confirmed by Facs analysis. Mice were euthanized by CO2, and warm DMEM was injected into the peritoneal cavity and massaged for 5 min prior to removal of lavage, which was placed on ice. The lavage was centrifuged and replated in warm DMEM medium to allow for macrophage attachment. After 1 h, nonadherent cells were removed by washing, and attached cells were harvested immediately for RNA measurement or treated with LPS (100 ng/ml) for the specified times prior to harvesting. In all cases, DMEM medium were supplemented with 10% fetal calf serum and 1% penicillin/streptomycin solution (v/v).
Bioinformatic Analysis of pri-155 Promoter
Nucleotide sequences for Homo sapiens and eight orthologous sequences were obtained for the upstream region of the pri-155 promoter from the Ensembl database (assembly GRCH37.p8). Upstream regions were taken as 2500 bases upstream and 500 downstream from the transcription start site. The identification of evolutionarily conserved transcription factor binding sites was performed using PhyloGibbs; a Gibbs sampling technique that utilizes phylogenetic footprinting. PhyloGibbs identifies both evolutionarily conserved and over-represented binding sites utilizing only sequence data and without the use of binding profile/experimental date. PhyloGibbs was used to analyze the orthologous upstream sequences using all possible binding sizes between 4 and 20, fixing all other parameters at the default settings. Secondary identification of transcription factor binding sites was performed using JASPAR and ConSite. JASPAR is a transcription factor binding profile database, which performs a single sequence comparison to all high-quality transcription factor models. ConSite is a web-based tool that performs both transcription factor model comparisons in combination with phylogenetic footprinting leading to results of greater significance. A default binding threshold of 0.8 was used for the both the JASPER and ConSite analysis.
RNA Isolation and Real Time PCR
Cells (primary bone marrow-derived macrophages (BMDMs), Raw264.7, immortalized BMDM, or primary peritoneal macrophages) were plated 1 day prior to stimulation. Cells were stimulated with LPS ± IL-10 as indicated in the figure legends. Total RNA was extracted using the RNeasy kit (Qiagen), modified to obtain small RNA species. cDNA for miRNA and mRNA analysis was prepared from 5–100 ng/ml total RNA using the high-capacity cDNA archive kit (Applied Biosystems) according to the manufacturer's instructions and incorporating TaqMan primers for miR-155 and RNU6B for miRNA analysis. miRNA expression was measured by Taqman analysis using specific Taqman Assays for miR-155 or RNU6B (Applied Biosystems) according to the manufacturer's instructions. mRNA expression was measured using SYBR Green-based chemistry (KAPA-Sybr) using the following primers: Pri-mmu-155, 5′-aaa cca gga agg gga agt gt-3′ (forward) and 5′-caa gag tca ccc tgc tgg at-3′ (reverse); Ets2, 5′-aat gca ggc acc aaa cta cc-3′ (forward) and 5′-gtc ctg gct gat gga aca gt- 3′ (reverse); Ets1, 5′-tcc aga cag aca cct tgc ag-3′ (forward) and 5′-ggt gag gcg gtc aca act at-3′ (reverse); GAPDH, 5′-ttc acc acc atg gag aag gc-3′ (forward) and 5′-ggc atg gac tgt ggt cat ga-3′ (reverse); SHIP1, 5′-ggt ggt acg gtt tgg aga ga-3′ (forward) and 5′-atg ctg agc ctc tgt ggt ct-3′ (reverse). miRNA and mRNA expression were measured on the 7900 RT-PCR system (Applied Biosystems), and fold changes in expression were calculated by the ΔΔCT method using RNU6B as an endogenous control for miRNA analysis and GAPDH as an endogenous control for mRNA expression. All fold changes are expressed normalized to non-stimulated control for each cell type.
Enzyme-linked Immunosorbent Assay
Murine TNF-α expression was measured from the supernatants of stimulated cells using an enzyme-linked immunosorbent assay DuoSet kit (R&D Biosystems) according to the manufacturer's instructions.
Protein Expression
Differentiated BMDM or Raw264.7 cells were seeded at 4 × 105 in six-well plates and stimulated with LPS ± IL-10 as indicated in the figure legends. Cells were lysed in low stringency lysis buffer complete with protease inhibitors. Protein concentration was then determined using the Coomassie Bradford reagent (Pierce). Lysates were resolved on 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membrane. Membranes were blocked in 5% (w/v) dried milk in TBS-T (50 mm Tris/HCl, pH 7.6, 150 mm NaCl, and 0.1% (v/v) Tween 20) before being immunoblotted with anti-Ets2 (Santa Cruz Biotechnology) or anti-β-actin (AC-74, Sigma) antibodies (1:1000 or 1:10,000, respectively) in 5% (w/v) dried milk in TBS-T at 4 °C overnight, or at room temperature for at least 2 h. Membranes were then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody diluted 1:2000 in 5% (w/v) dried milk in TBS-T for 1 h. Blots were developed by enhanced chemiluminescence according to the manufacturer's instructions (Cell Signaling Technology).
Luciferase Assays
Pri-155 luciferase plasmid along with the Ets1 mutant (M_pri-155) were a kind gift from Eric Flemington (Tulane University, New Orleans, LA). Truncated pri-155 plasmids were generated by PCR amplification using specific primers to create plasmids of varying lengths that were recloned in to the luciferase plasmid at the NheI and HinDIII sites. The reverse primer, 5′-tca ccg ccg ggt taa ctg-3′, was used for each truncation along with specific forward primers as follows: T1_pri-155, 5′-ctg aga ccc atg aat gag-3′; T2_pri-155, 5′-ctc tta ggg acc tgc tgg-3′; T3_pri-155, 5′-caa agg ttg gag ccc aag-3′; T4_pri-155, 5′-cag aaa agg cgc ctg gtc-3′. Raw264.7 cells seeded at 2 × 105/ml in 24-well plates were transfected using 6% JetPEI macrophage transfection reagent as per the manufacturer's protocol, with each plasmid and TK-Renilla. Cells were rested for 24–48 h prior to stimulation with LPS. In all cases, cells were lysed in passive lysis buffer before being analyzed for both luciferase and TK-Renilla activity as described previously (23). Data were normalized to TK-Renilla activity and represented as mean ± S.D. for triplicate determinations where fold changes are expressed normalized to non-stimulated control.
Chromatin Immunoprecipitation
Raw264.7 cells were set up at 4 × 105cell/ml in DMEM, treated with LPS ± IL-10 for 24 h and fixed by adding a final concentration of 1% formaldehyde to each culture dish. Flasks were incubated for 10 min at room temperature. A 1/20 volume of 2.5 m glycine was then added to each flask and allowed to set at room temperature for 5 min prior to washing in PBS and resuspension in 6 ml of ChIP lysis buffer (SDS lysis buffer with leupeptin, aprotinin, and PMSF) and immediately snap frozen in liquid nitrogen. The samples were thawed and resuspended in SDS:Triton buffer and then sonicated at 22% intensity, 10 × 30 s per sample, placing on ice in between pulses. Preblocked protein A beads were incubated with samples prior to overnight incubation with primary antibodies; anti-Ets2 (Santa Cruz Biotechnology, sc22803) and anti-HA (Sigma, H6908). Quantitative RT-PCR was carried out using primers for either the pri-155 promoter consensus Ets binding site (0/+9), a non-Ets binding site (−1003/−995). Data are presented as percent of input.
Affinity Purification with Biotinylated Oligonucleotides
Oligonucleotides for the terminal Ets binding site on the pri-155 promoter were annealed 90–95 °C for 3–5 min and then heat block was allowed to cool to room temperature (forward, 5′BIO-gcgccggcttcctgtacgc-3′; reverse, 5′-cgcgtacaggaagccggcg-3′). Raw264.7 cells were seeded at 4 × 105 cell/ml and treated with LPS ± IL-10. 24 h later, cells were lysed in 100 μl of oligonucleotide buffer (25 mm Tris, 5% glycerol, 50 mm EDTA, 5 mm NaF, Nonidet P-40 1%, 1 mm DTT, 150 mm NaCl, and protease and phosphatase inhibitors), and snap-frozen. Samples were then thawed on ice and diluted with a further 900 μl of oligonucleotide buffer containing no NaCl. 10 μl sample of lysate was kept to which 40 μl of 5× SDS buffer was added. Remaining lysates were then precleared with 20 μl of prewashed streptavidin-agarose beads, rotating at 4 °C for 15 min before centrifuging at 2500 rpm for 5 min at 4 °C. Supernatants were removed to a fresh tube with 30 μl of prewashed streptavidin-agarose beads and 30 μg of 5′-biotinylated oligonucleotide. Binding was performed for 2 h at 4 °C, rotating. Samples were centrifuged for 5 min at 4 °C to pellet the beads, which were washed three times before 50 μl of 5× SDS sample buffer was added to the beads. Samples were loaded on a 10% gel and Ets2 binding detected by Western blotting as described previously.
RESULTS
The pri-155 Promoter Is Controlled by Ets Transcription Factors
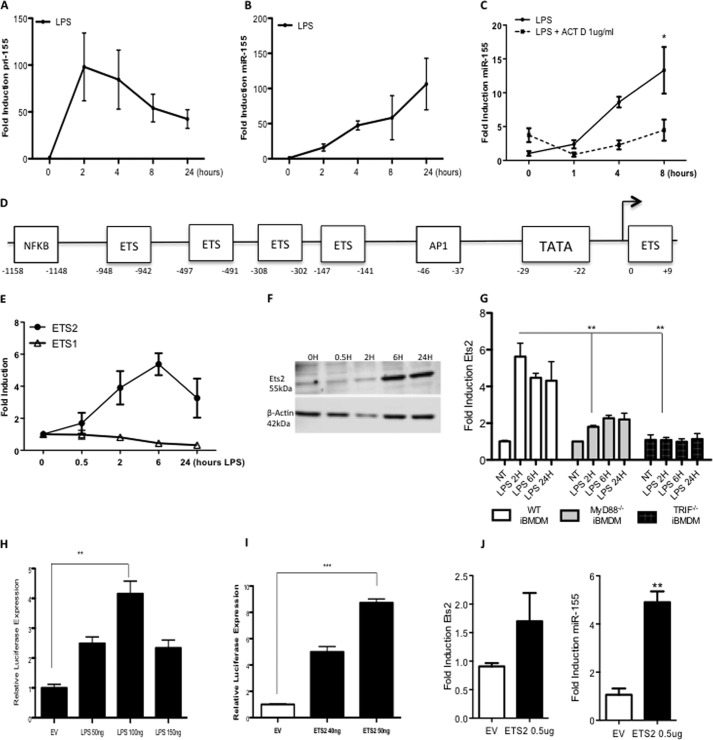
As miR-155 has previously been shown to be potently induced in response to the TLR4 ligand LPS (5), we wished to investigate the mechanism of this induction. The strong induction over time of pri-155 and miR-155 by LPS in BMDM was first verified (Fig. 1, A and B). IL-6 was also measured as a positive control for LPS treatment (data not shown). The potent transcriptional inhibitor, actinomycin D, blocked the induction of miR-155 by LPS as shown in Fig. 1C. The miR-155 promoter gene pri-155 was next investigated bioinformatically to determine key transcription factor binding sites. Regions 2500 bases upstream and 500 downstream from the transcription start site were analyzed in the miR-155 gene. The identification of evolutionarily conserved transcription factor binding sites was performed using PhyloGibbs across eight different, species including human and mouse. Combining the most significant results from PhyloGibbs and JASPAR identified ETS family binding sites as stand-out candidates. Further verification was carried out using ConSite, which verified all sites in the previous tests. The analysis indicated the miR-155 promoter was heavily controlled by the Ets family of transcription factors. Five of the putative Ets binding sites are identified in the schematic shown in Fig. 1D. Positions are indicated relative to the transcription initiation site as described previously (12).
FIGURE 1.
The pri-155 promoter is controlled by Ets transcription factors. A–C, primary BMDMs from C57BL/6 mice (n = 3) were treated with LPS (100 ng/ml) for 0–24 h (A and B) or pretreated with actinomycin D (ACT D; 1 μg) prior to LPS stimulation for 24 h (C). RT-PCR analysis of RNA was carried out with primers specific for pri-155 or mature miR-155 as indicated. Expression is normalized to that of GAPDH (for pri-155 expression) or RNU6B (for miR-155 expression) and is presented relative to that of untreated controls. Data are presented as the mean ± S.E. of one representative experiment shown of three independent experiments (n = 3 per experiment). D, bioinformatic analysis of the pri-155 promoter revealed highly conserved Ets transcription factor binding sites. E and F, primary BMDM from C57BL/6 mice (n = 3) were treated with LPS (100 ng/ml) for 0–24 h. RT-PCR analysis of RNA was carried out with primers specific for Ets2 or Ets1 mRNA (E), and protein levels were measured by Western blot with antibodies specific for Ets2 or β-actin (F). Data are representative of three separate experiments, with each point assayed in triplicate, with error bars representing S.D. G, wild type, MyD88−/−, or TRIF−/− immortalized BMDMs (iBMDM) were plated at 5 × 105/ml and stimulated with LPS for 0–24 h prior to lysis for RNA extraction. Ets2 or GAPDH control was measured by real-time PCR. Data are the mean ± S.D. of triplicate determinations from three independent experiments. H and I, pri-155 promoter activation was assayed in Raw264.7 cells using a pri-155 luciferase reporter construct and increasing doses of LPS (50–100 ng) (H), or plasmid encoding Ets2 (40–50 ng) or empty vector (EV) control (I). Luciferase activity was determined 24 h after transfection and normalized to TK Renilla control. Results are presented as the mean ± S.D. for triplicate determinations and are representative of three separate experiments. J, Raw264.7 cells were transfected with 0.5 μg of Ets2 plasmids or empty vector control and lysed for RNA. miR-155 expression was measured by real-time PCR. Statistical analysis was carried out using Student's t test; *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
As we were interested in the induction of miR-155 in an inflammatory context, we next examined the induction of Ets family members in response to the TLR4 ligand LPS. Fig. 1E shows that Ets2 is induced over time is response to LPS, whereas the closely related Ets family member Ets1 is unaffected by LPS stimulation. Further investigation of Ets2 revealed it is also induced strongly at the protein level (Fig. 1F) and that induction by LPS is MyD88- and TRIF-dependant (Fig. 1G), confirming it as a key modulator of downstream TLR4 responses.
Having confirmed that Ets2 is induced by LPS and that the miR-155 promoter is controlled by many Ets binding sites, we next investigated the effect of Ets2 on the pri-155 promoter. First, we confirmed our previous finding that LPS can drive the pri-155 promoter. Pri-155 promoter constructs fused to a luciferase reporter were transfected into Raw264.7 cells and treated with LPS (50–150 ng), which drove the pri-155 promoter 5-fold at 100 ng (Fig. 1H), confirming that miR-155 is regulated by LPS at the transcriptional level. Subsequently, pri-155 promoter constructs fused to a luciferase reporter were cotransfected with plasmids encoding Ets2. Fig. 1I shows that overexpression of Ets2 could drive the pri-155 promoter in a dose-dependent manner. To investigate the effect of Ets2 on mature miR-155, gain of function studies were utilized, where Ets2 was overexpressed and mature miR-155 was measured by real-time PCR. As revealed in Fig. 1J, 0.5 μg Ets2 is sufficient to induce a 5-fold increase in mature miR-155, indicating Ets2 alone is sufficient for the transcription of mature miR-155, thus focusing our study on Ets2 regulation of miR-155 in response to LPS.
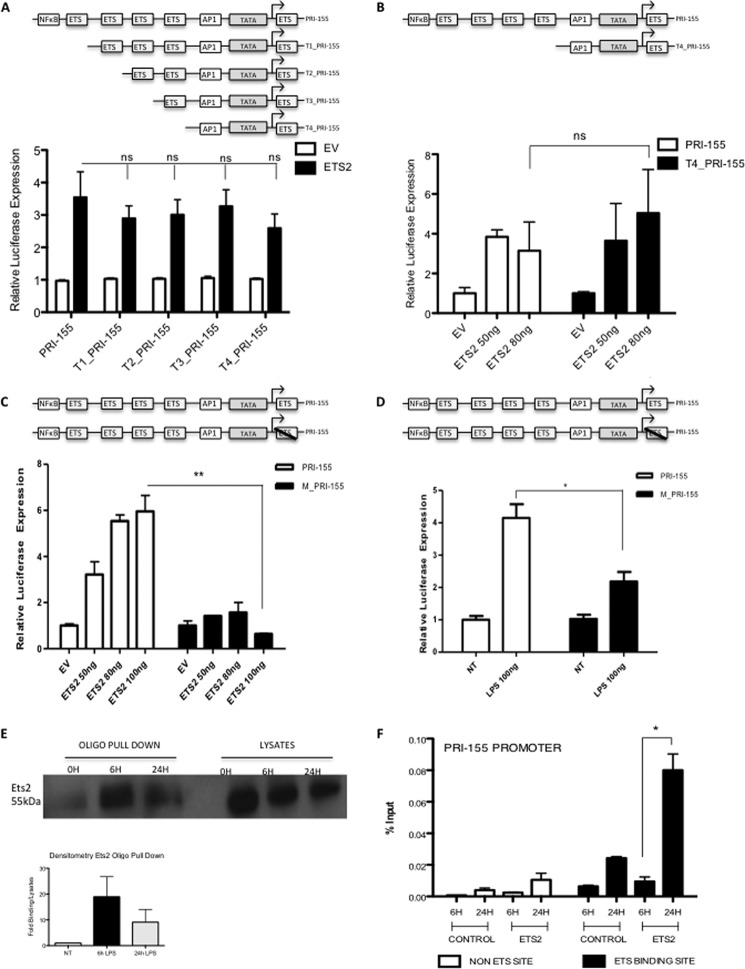
Ets2 Binds to a Highly Conserved Site on the pri-155 Promoter
Having confirmed that Ets2 and LPS can drive the pri-155 promoter and that LPS could induce Ets2, the regions of the promoter that were most important for this interaction were next examined. Four additional pri-155 luciferase constructs were generated, each truncated to contain one less Ets binding site until only the terminal Ets binding site and the TATA box remained (T1-T4_PRI-155). A schematic of these promoter constructs is shown in Fig. 2A, top panel. Each construct was co-transfected with an Ets2 expression plasmid, and interestingly, it was evident that Ets2 could continue to drive the truncated pri-155 promoters (T1-T4_PRI-155) to the same extent as the full-length pri-155 promoter (Fig. 2A, lower panel). This was examined in more detail by transfecting higher amounts of plasmid encoding Ets2 with the pri-155 luciferase construct with only the terminal Ets binding site remaining. Again Ets2 could be seen to drive the T4_PRI-155 construct to an equal extent as the full-length pri-155 (Fig. 2B). This finding implied that the terminal Ets binding site might be key for Ets2 to drive transcription of the pri-155 promoter. We therefore examined the effect of overexpression of Ets2 on a pri-155 construct in which the terminal Ets binding site is mutated (12) because we could not create a truncation as this site occurs 3′ to the TATA box. Fig. 2C reveals that Ets2 can no longer drive induction of the pri-155 promoter when this terminal Ets binding site is mutated (M_PRI-155). Additionally, induction of pri-155 by LPS was strongly decreased when the terminal Ets binding site was mutated as shown in Fig. 2D. Together, these findings indicate that Ets2 requires the terminal Ets binding site on the pri-155 promoter for induction by LPS.
FIGURE 2.
Ets2 binds to a highly conserved binding site on the pri-155 promoter. Pri-155 promoter constructs were truncated sequentially such that one Ets binding site was deleted at a time (T1-T4_pri-155), only the terminal Ets binding site remained (T4_pri-155) or the terminal Ets binding site was mutated (M_pri-155). A, pri-155 promoter and T1-T4_pri-155 promoter activation was assayed in HEK293T cells using an Ets2 expression plasmid. B, pri-155 promoter and T4_pri-155 promoter activation was assayed in Raw264.7 cells using increasing doses of Ets2 expression plasmid (50–80 ng). C and D, pri-155 promoter and M_pri-155 promoter activation was assayed in Raw264.7 cells with increasing doses of Ets2 expression plasmid (50–100 ng) (C) or LPS (100 ng/ml) (D). Luciferase activity was determined 24–48 h after transfection and normalized to TK Renilla controls. E, Raw264.7 cells were treated with LPS for 0, 6, and 24 h and lysed, and an oligonucleotide (oligo) pulldown assay was carried out with the terminal Ets binding site oligonucleotide sequence at position 0/+9. Samples were probed for Ets2 by Western blot. Densitometry was carried out relative to lysates and are relative to non-treated control (NT). Bar graph represents the mean of two independent experiments. F, Raw264.7 cells were treated with LPS for 6 or 24 h, after which a ChIP assay was performed. Primers specific for the terminal Ets binding site on pri-155 (black bars); and additionally, a non-Ets-binding site (white bars) were designed, and binding events were measured as percent of input by real time PCR using antibodies against HA (control antibody) or Ets2. In all cases, results are presented as the mean ± S.D. for triplicate determinations and are representative of at least three separate experiments. Statistical analysis was carried out using Student's t test. *, p < 0.05; **, p < 0.001.
We next examined binding interaction at this site using a number of novel approaches. First, oligonucleotide pulldown assays were employed to investigate binding at this terminal Ets site. Oligonucleotides specific to the Ets binding site were incubated with lysates from Raw264.7 cells treated with LPS for 6 and 24 h, and Fig. 2E reveals Ets2 binding to this specific and highly conserved site on the pri-155 promoter in response to LPS. To investigate this finding further, and in an endogenous context, ChIP at the key terminal Ets binding site was next carried out. This technique allows us to examine the endogenous binding of transcription factors to chromatin in response to LPS. As shown in Fig. 2F, Ets2 binds to the pri-155 promoter at 24 h of LPS treatment (black bars). Ets2 does not bind to a non-Ets-binding site (white bars) or the β-actin promoter (data not shown). These findings confirmed that LPS induces Ets2 binding to the Ets binding site at the initiation site on the miR-155 promoter, whose function we have confirmed by mutational analysis.
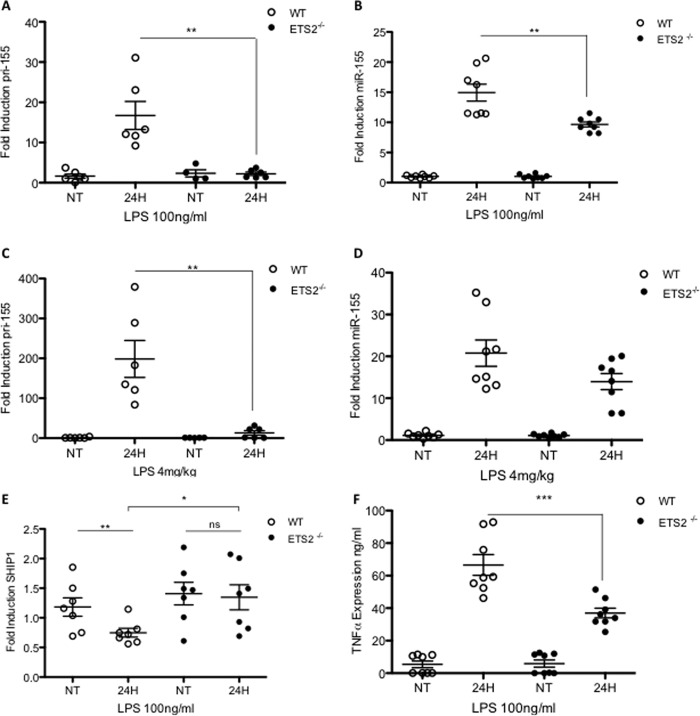
pri-155 Expression Is Ablated and miR-155 Induction Is Reduced in Response to LPS in Ets2−/− Mice
We next investigated miR-155 induction in cells from Ets2−/− conditionally generated mice. In these mice, the Ets2 gene is under the control of the lysM promoter, creating knock-outs specific to the myeloid lineage. Peritoneal macrophages were prepared from Ets2−/−, Ets2+/−, and wild type C57BL/6 mice and stimulated in vitro with LPS for 24 h. The macrophages from the Ets2−/− mice lacked induction of pri-155 in response to LPS (Fig. 3A), confirming the importance of this transcription factor in LPS-mediated induction of pri-155. The production of mature miR-155 was also significantly reduced in Ets2−/− mice in comparison with wild type mice (Fig. 3B).
FIGURE 3.
pri-155 and mature miR-155 induction are reduced in response to LPS in Ets2−/− mice. A and B, primary peritoneal macrophages from WT, Ets2+/−, or Ets2−/− mice were treated with LPS (100 ng/ml) for 0 or 24 h in vitro. C and D, WT or Ets2−/− mice were administered an intraperitoneal injection of LPS (4 mg/kg) or PBS for 24 h in vivo prior to culling and extraction of peritoneal macrophages. E and F, primary peritoneal macrophages from WT or Ets2−/− mice were treated with LPS (100 ng/ml) for 0 or 24 h in vitro. RT-PCR analysis of RNA was carried out with primers specific for pri-155, miR-155, or SHIP1, as indicated. Expression is normalized to that of GAPDH (pri-155/SHIP1) or RNU6B (miR-155) and is presented relative to that of untreated controls. F, supernatants from peritoneal macrophages stimulated in vitro were harvested, and TNF-α levels were measured by ELISA. In all cases, data are the mean of n = 6–8 mice, with each data point representing an individual mouse, and with error bars representing S.E. Statistical analysis was carried out using Student's t test. *, p < 0.05; **, p < 0.001; ***, p < 0.0001. NT, non-treated.
We also examined miR-155 in vivo. Ets2−/− and matched wild type C57BL/6 mice were administered an intraperitoneal injection of LPS or PBS and 24 h later were culled for harvesting of peritoneal macrophages. The induction of pri-155 and miR-155 in response to LPS was again observed strongly in the wild type mice, and this induction was reduced in the Ets2−/− mice. Pri-155 induction is completely ablated (Fig. 3C), and miR-155 is reduced (Fig. 3D) in Ets2−/− mice in vivo, further supporting the evidence that Ets2 is a necessary transcription factor for the LPS-mediated induction of pri-155 and miR-155.
miR-155 target genes in macrophages from WT and Ets2−/− mice were next investigated. As shown in Fig. 3E, LPS significantly suppresses the miR-155 target SHIP1 in WT peritoneal macrophages, consistent with published data on SHIP1 induction and repression by miR-155 in BMDMs (6). In Ets2−/− peritoneal macrophages, basal SHIP1 is elevated, and LPS can no longer decrease it. This finding implicated Ets2 for the first time having a downstream effect on a microRNA target. There is evidence to suggest that miR-155 is required for stabilizing TNF-α expression (14). As shown in Fig. 3F, Ets2−/− mice deficient in miR-155 show significantly decreased TNF-α expression. These data further confirm Ets2 as a key regulator of miR-155, with downstream impacts on inflammatory target genes, thus implicating Ets2 in the proinflammatory response to LPS and uncovering the mechanism of induction of miR-155.
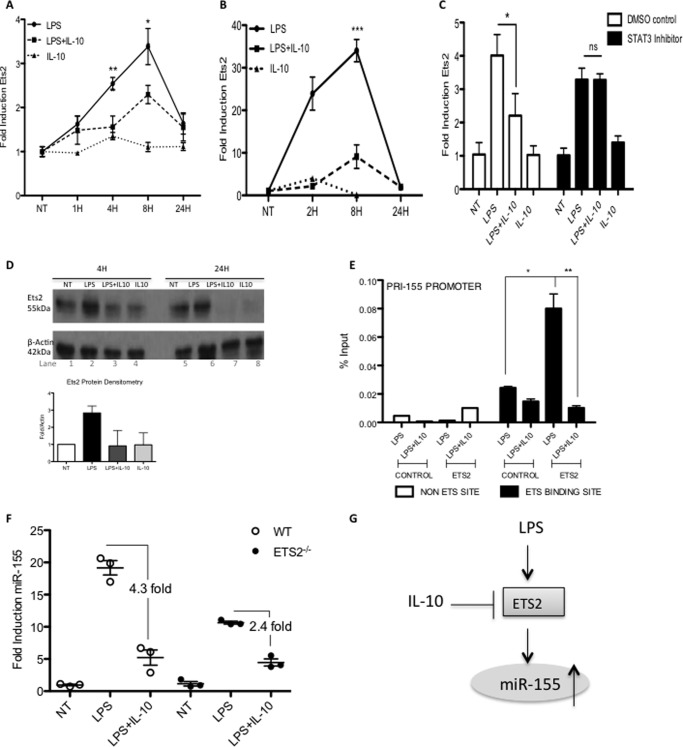
IL-10 Inhibits Ets2 Protein Expression and Binding to the miR-155 Promoter
Having determined that induction of miR-155 by LPS involves Ets2, we next examined whether the inhibitory effect of IL-10 on miR-155 might involve the targeting of Ets2. Interestingly, Ets2 mRNA induction by LPS is strongly inhibited by IL-10 in Raw264.7 cells (Fig. 4A) and primary BMDM (Fig. 4B) over time. Little is known about the mechanism through which IL-10 exactly exerts its anti-inflammatory effects on TLR signaling. It has been established that STAT3 is highly important for IL-10 to exert its effects (15); thus, to understand this further, we utilized a STAT3 inhibitor. Our data revealed that IL-10 is acting through the STAT3 pathway to inhibit Ets2 and can no longer exert its anti-inflammatory effects with just 1 μm Stat3 inhibitor (Fig. 4C, black bars). These data correlate with the original manuscript detailing the inhibition of miR-155 by IL-10, which was also found to be STAT3-dependent (10). Inhibition of Ets2 by IL-10 was also evident at the protein level. Fig. 4D shows how there is a basal expression of Ets2 in Raws (lane 1). LPS can increase expression (lane 2), and IL-10 can inhibit this effect (lane 3). IL-10 alone can also reduce the basal expression (lane 4). This effect is also evident at 24 h (lanes 5–8). This is the first time IL-10 has been implicated in the control of Ets2, and due to the novel finding that IL-10 can inhibit Ets2 mRNA and protein, we furthered our investigation by examining whether this finding impacted directly on the pri-155 promoter. Chromatin immunoprecipitation assay in Fig. 4E reveals that IL-10 can reduce LPS induced binding of Ets2 to the pri-155 promoter (black bars). This novel result reveals IL-10 may also be dependent on Ets2 to regulate miR-155.
FIGURE 4.
IL-10 inhibits Ets2 induction by LPS. Raw264.7 cells (A) or primary BMDMs from C57BL/6 mice (B) were treated with LPS (100 ng/ml) and IL-10 (20 ng/ml) for 0–24 h. C, Raw264.7 cells were treated with STAT3 inhibitor (0.5–1 μm) or dimethyl sulfoxide (DMSO) control 1 h prior to stimulation with LPS (100 ng/ml) and IL-10 (20 ng/ml) for 4 h. Real time PCR results are presented as the mean ± S.E. of three independent experiments, each carried out in triplicate. D, Raw264.7 cells were treated with LPS (100 ng/ml) and IL-10 (20 ng/ml) for 0–24 h. Protein levels were measured by Western blot with antibodies specific for Ets2 or β-actin. Densitometry was carried out relative to β-actin controls and expressed as fold over non-treated samples. Densitometry is the mean of three independent experiments ± S.D. E, Raw264.7 cells were treated with LPS (100 ng/ml) ± IL-10 (20 ng/ml) for 24 h after which a ChIP assay was performed. Primers specific for the terminal Ets binding site on the pri-155 promoter at position 0/+9 (black bars) were designed, in addition to a non Ets-binding site at position −995/−1003 (white bars). Binding events were measured as percent of input by real time PCR using antibodies against HA (control antibody) or Ets2. Results are presented as the mean ± S.D. for triplicate determinations and are representative of two independent experiments. F, primary peritoneal macrophages from WT or Ets2−/− mice (n = 3) were treated with LPS (100 ng/ml) ± IL-10 (20 ng/ml) for 24 h in vitro prior to lysis for RNA extraction. miR-155 was measured by real time PCR. Data are the mean of n = 3 mice ± S.E. Statistical analysis was carried out using Student's t test. *, p < 0.05; **, p < 0.001. G, schematic illustrating possible mechanism of control of miR-155 via Ets2.
Finally, we investigated the ability of IL-10 to inhibit miR-155 in wild type and Ets2-deficient peritoneal macrophages. Our data, shown in Fig. 4F, revealed that IL-10 could not inhibit miR-155 to the same extent in Ets2−/− mice (2.4-fold decrease) compared with wild type mice (4.3-fold decrease). This finding indicates Ets2 plays a role in both the pro- and anti-inflammatory control of miR-155 and reveals for the first time that IL-10 may act through Ets family members and thereby impacts on the inflammatory process.
DISCUSSION
In the study of miRNA, the majority of research to date focuses on the interesting target genes and pathways on which miRNAs impact, as opposed to the detailed mechanisms by which miRNAs themselves are controlled and induced. In the context of inflammation, miRNAs have been shown to be crucial for the development, differentiation, and orchestration of a smooth and timely immune response and subsequent resolution. Therefore, it is extremely important to understand how these miRNA are induced, inhibited, and controlled to maintain control of their downstream target genes. In this study, we identify Ets2 as a critical transcription factor in the induction of miR-155 by LPS and, furthermore, demonstrate inhibition of Ets2 as a mechanism for suppression of miR-155 by IL-10. This finding not only uncovers a mechanism of induction of miR-155 at the transcriptional level, it also reveals a new downstream target gene of IL-10 and a possible mechanism through which IL-10 may act to exert its anti-inflammatory properties.
miR-155 is highly important in the inflammatory response to infection, with miR-155−/− mice suffering extremely stunted immune responses, including defective lymphocyte and antigen presenting function and impaired immunological memory (16). Additionally, the mice are highly resistant to autoimmune diseases such as experimental autoimmune encephalomyelitis, again highlighting the extent of the decrease in immune activation of these miR-155−/− mice (17). Numerous studies and review articles have investigated and collated miR-155 target genes and their effects in various inflammatory phenotypes (18–20), but studies investigating the exact induction of this miRNA from the promoter level are sparse.
To understand the potent induction of miR-155 by LPS, detailed bioinformatic analysis was carried out and revealed a number of conserved Ets binding sites on the pri-155 promoter, and luciferase reporter assays indicated the terminal Ets binding site adjacent to the TATA box was highly important. Ets transcription factors are one of the largest families of transcription factors known and have wide-ranging roles from cell cycle regulation, cell differentiation, and a more recently discovered role in regulation of the immune response (21). For example, Ets binding sites have been discovered in the promoter regions of a number of immune genes, including those encoding the T cell receptor α and β subunits, interleukin 5, interleukin 12 (22, 23), and additionally, many viruses such as moloney sarcoma virus. Ets2 is induced in response to LPS, others have shown the protein kinase C pathway may be involved in this induction (24). It may also be phosphorylated on threonine 72 by LPS, but its exact mechanism of induction has not been fully elucidated (25). Moreover, our finding that Ets2 is MyD88- and TRIF-dependent has further confirmed it as a TLR-regulated gene, likely important in the immune response. Up-regulation of Ets2 has been found in 30% of rheumatoid arthritis tissue samples (26), and our study furthers the novel evidence for an immunological role for Ets2. Our investigation of induction of miR-155 by Ets2 led us to examine immune responses in the Ets2−/− mice. The mice were found to be deficient in induction of pri-155 and miR-155 and, as a result, had reciprocally altered levels of miR-155 inflammatory target genes SHIP1 and TNF-α. This interesting finding emphasizes a role for the Ets2 transcription factor in innate immunity and indicates Ets2 may control many downstream target genes through its induction and control of miR-155. Although there is a significant reduction of miR-155 induction in the Ets2−/− mice some miR-155 expression remains, most likely due to separate LPS-mediated post-transcriptional mechanism of induction of miR-155 from pri-155 by maturation factors such as KH-type splicing regulatory protein (KSRP, also known as KHSRP) (27). KSRP binds with high affinity to the terminal loop of the target miRNA precursors and promotes their maturation (28) and indicates that miR-155 can be post-transcriptionally regulated further after induction by LPS.
Conflicting studies have recently reported that regulation of miR-155 does not occur at the transcriptional level (29); however, many of these studies were based heavily on the inability of LPS to drive a pri-155 luciferase reporter construct. Conversely, we and others (30, 31) have repeatedly shown LPS can drive induction of the pri-155 promoter in a dose-dependent manner. Our data are additionally supported by chromatin immunoprecipitation and oligonucleotide pulldown assays displaying increased binding of Ets2 to the pri-155 promoter in the presence of LPS, Ets2−/− mice displaying ablated induction of pri-155 in response to LPS, and finally, the addition of the transcriptional inhibitor actinomycin D strongly reducing mature miR-155 induction. Other studies have additionally shown miR-155 is transcriptionally regulated in other cells types such as by AP1 in B cells (12) and STAT3 in T cells (32), supporting our evidence of transcriptional regulation of miR-155.
miR-155 is a potently proinflammatory microRNA and thus requires stringent control to prevent excess inflammation. Eμ-transgenic miR-155 mice exhibit excessive inflammation and uncontrolled responses to inflammatory stimuli, including excessive TNFα and hypersensitivity to septic shock (33). In the context of macrophage activation by LPS, our study has shown that Ets2 is a key transcription factor for miR-155 induction. Due to this excessive inflammation association with aberrant expression of miR-155, it is of utmost importance that miR-155 be carefully controlled and down-regulated. One such mechanism of this inhibition is through the anti-inflammatory cytokine IL-10 (10). IL-10 has an irreplaceable role in negatively regulating inflammation, and any additional insight as to how this occurs is of high value in this field. IL-10 acts primarily by blocking the genes which code for those molecules central to propagating the inflammatory response (8), as illustrated for example by profound colitis in IL-10−/− mice (13, 34, 35). However, much of the mechanism of action of IL-10 is still unknown, and our study sought to investigate how IL-10 is inhibiting miR-155. Our data established the novel finding that IL-10 is acting to inhibit Ets2 mRNA and protein, both basally and in response to LPS stimulation. This interesting finding was relevant for control of miR-155, as this potent decrease in Ets2 expression resulted in decreased Ets2 recruitment to the pri-155 promoter in the presence of LPS and decreased mature miR-155. Additionally, Ets2 knock-out mice could not decrease miR-155 to the same extent after IL-10 treatment, further supporting the importance of Ets2 for control and regulation of miR-155. IL-10 still decreased miR-155 expression to some extent in Ets2-deficient cells, although not to the same extent as in wild type cells. We are currently exploring the Ets2-independent component being targeted by IL-10.
Our findings are likely to be significant in the context of inflammation, as many diseases correlating with low IL-10 also correlate with elevated miR-155, such as colitis, arthritis, and experimental autoimmune encephalomyelitis. Because IL-10 can so potently inhibit miR-155, it is possible that anti-miR-155 could have utility as an alternative to IL-10 therapy as an anti-inflammatory strategy.
Overall, the mode of regulation identified in this study have uncovered a mechanism of miR-155 induction and repression through modulation of Ets family transcription factors. This not only gives us new insight into the regulation of miR-155 but also provides important new information on the mechanism of action of IL-10.
Acknowledgment
We acknowledge Erik Flemington for providing Pri-155 promoter constructs.
This work was supported by the Science Foundation Ireland, Health Research Board/Marie Curie CoFund, and the European Research Council. The Monash Institute of Medical Research was supported by the Australian National Health and Medical Research Council, Australian Research Council, Health Research Board/Marie Curie CoFund, and the Victorian government's operational infrastructure support program.
- TLR
- Toll-like receptor
- miRNA
- microRNA
- BMDM
- bone marrow-derived macrophage
- TRIF
- TIR-domain-containing adapter-inducing interferon-β.
REFERENCES
- 1. Janeway C. A., Jr., Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 2. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Neill L. A., Sheedy F. J., McCoy C. E. (2011) MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 [DOI] [PubMed] [Google Scholar]
- 4. Clurman B. E., Hayward W. S. (1989) Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 9, 2657–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. An H., Xu H., Zhang M., Zhou J., Feng T., Qian C., Qi R., Cao X. (2005) Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS response primarily through a phosphatase activity- and PI-3K-independent mechanism. Blood 105, 4685–4692 [DOI] [PubMed] [Google Scholar]
- 8. Wang P., Hou J., Lin L., Wang C., Liu X., Li D., Ma F., Wang Z., Cao X. (2010) Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 185, 6226–6233 [DOI] [PubMed] [Google Scholar]
- 9. Quinn S. R., O'Neill L. A. (2011) A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 23, 421–425 [DOI] [PubMed] [Google Scholar]
- 10. McCoy C. E., Sheedy F. J., Qualls J. E., Doyle S. L., Quinn S. R., Murray P. J., O'Neill L. A. (2010) IL-10 inhibits miR-155 induction by toll-like receptors. J. Biol. Chem. 285, 20492–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray P. J. (2006) Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6, 379–386 [DOI] [PubMed] [Google Scholar]
- 12. Yin Q., Wang X., McBride J., Fewell C., Flemington E. (2008) B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 283, 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. (1993) Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 [DOI] [PubMed] [Google Scholar]
- 14. Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. (2011) Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 286, 1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murray P. J. (2006) STAT3-mediated anti-inflammatory signalling. Biochem. Soc. Trans. 34, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 16. Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Connell R. M., Kahn D., Gibson W. S., Round J. L., Scholz R. L., Chaudhuri A. A., Kahn M. E., Rao D. S., Baltimore D. (2010) MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., Santos M. A., Pierre P. (2009) MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 20. Lindsay M. A. (2008) microRNAs and the immune response. Trends Immunol. 29, 343–351 [DOI] [PubMed] [Google Scholar]
- 21. Gallant S., Gilkeson G. (2006) ETS transcription factors and regulation of immunity. Archivum Immunologiae et Therapiae Experimentalis 54, 149–163 [DOI] [PubMed] [Google Scholar]
- 22. Wang C. Y., Petryniak B., Ho I. C., Thompson C. B., Leiden J. M. (1992) Evolutionarily conserved Ets family members display distinct DNA binding specificities. J. Exp. Med. 175, 1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blumenthal S. G., Aichele G., Wirth T., Czernilofsky A. P., Nordheim A., Dittmer J. (1999) Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J. Biol. Chem. 274, 12910–12916 [DOI] [PubMed] [Google Scholar]
- 24. Boulukos K. E., Pognonec P., Sariban E., Bailly M., Lagrou C., Ghysdael J. (1990) Rapid and transient expression of Ets2 in mature macrophages following stimulation with cMGF, LPS, and PKC activators. Genes Dev. 4, 401–409 [DOI] [PubMed] [Google Scholar]
- 25. Sweet M. J., Stacey K. J., Ross I. L., Ostrowski M. C., Hume D. A. (1998) Involvement of Ets, rel and Sp1-like proteins in lipopolysaccharide-mediated activation of the HIV-1 LTR in macrophages. J. Inflamm. 48, 67–83 [PubMed] [Google Scholar]
- 26. Dittmer J., Nordheim A. (1998) Ets transcription factors and human disease. Biochim. Biophys. Acta 1377, F1–11 [DOI] [PubMed] [Google Scholar]
- 27. Ruggiero T., Trabucchi M., De Santa F., Zupo S., Harfe B. D., McManus M. T., Rosenfeld M. G., Briata P., Gherzi R. (2009) LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 23, 2898–2908 [DOI] [PubMed] [Google Scholar]
- 28. Trabucchi M., Briata P., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2011) KSRP Promotes the Maturation of a Group of miRNA Precuresors. Adv. Exp. Med. Biol. 700, 36–42 [DOI] [PubMed] [Google Scholar]
- 29. Cheung S. T., So E. Y., Chang D., Ming-Lum A., Mui A. L. (2013) Interleukin-10 inhibits lipopolysaccharide induced miR-155 precursor stability and maturation. PloS One 8, e71336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y., Liu W., Sun T., Huang Y., Wang Y., Deb D. K., Yoon D., Kong J., Thadhani R., Li Y. C. (2013) 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J. Immunol. 190, 3687–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai Y., Diao Z., Sun H., Li R., Qiu Z., Hu Y. (2011) MicroRNA-155 is involved in the remodelling of human-trophoblast-derived HTR-8/SVneo cells induced by lipopolysaccharides. Hum. Reprod. 26, 1882–1891 [DOI] [PubMed] [Google Scholar]
- 32. Escobar T., Yu C. R., Muljo S. A., Egwuagu C. E. (2013) STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 54, 4017–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 34. Rennick D., Davidson N., Berg D. (1995) Interleukin-10 gene knock-out mice: a model of chronic inflammation. Clin. Immunol. Immunopathol. 76, S174–178 [DOI] [PubMed] [Google Scholar]
- 35. Berg D. J., Kühn R., Rajewsky K., Müller W., Menon S., Davidson N., Grünig G., Rennick D. (1995) Interleukin-10 is a central regulator of the response to LPS in murine models of endotoxic shock and the Shwartzman reaction but not endotoxin tolerance. J. Clin. Invest. 96, 2339–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]