Background: Type IV pili are non-covalently assembled appendages, characterized now in both Gram-negative and Gram-positive bacteria.
Results: Clostridium difficile produces Type IV pili containing PilJ, a pilin with a novel dual-pilin fold.
Conclusion: Models suggest that the C-terminal pilin domain is exposed in pili, providing a unique interaction surface.
Significance: The novel fold of PilJ suggests a new mode for Type IV pilus function.
Keywords: Bacterial Adhesion, Microbiology, Microfilaments, Protein Assembly, Protein Structure
Abstract
Type IV pili are produced by many pathogenic Gram-negative bacteria and are important for processes as diverse as twitching motility, cellular adhesion, and colonization. Recently, there has been an increased appreciation of the ability of Gram-positive species, including Clostridium difficile, to produce Type IV pili. Here we report the first three-dimensional structure of a Gram-positive Type IV pilin, PilJ, demonstrate its incorporation into Type IV pili, and offer insights into how the Type IV pili of C. difficile may assemble and function. PilJ has several unique structural features, including a dual-pilin fold and the incorporation of a structural zinc ion. We show that PilJ is incorporated into Type IV pili in C. difficile and present a model in which the incorporation of PilJ into pili exposes the C-terminal domain of PilJ to create a novel interaction surface.
Introduction
Type IV pili are produced by many pathogenic Gram-negative bacteria, including Pseudomonas aeruginosa, Vibrio cholerae, Neisseria gonorrhoeae, Neisseria meningitidis, Salmonella enterica serovar Typhi, Legionella pneumophila, and enteropathogenic and enterotoxigenic Escherichia coli (1–6) and are important for diverse processes such as cellular adhesion (7), colonization (8, 9), twitching motility (10–13), biofilm formation (14), horizontal gene transfer (15, 16), and virulence (8, 9, 17). These fimbrial appendages are typically composed of many copies of a single pilin protein tightly packed in a helix to bury the hydrophobic N terminus of each subunit in the pilus core (18).
Gram-negative Type IV pilin proteins exhibit four conserved structural features: an ∼60 residue N-terminal α-helix, a loop that frequently shows helical character and leads into a central β-sheet, and a disulfide bond that bounds a C-terminal “D-region.” Type IV pilins are divided into two classes based on the length of the pre-pilin leader peptide and sequence similarity of the N-terminal α-helix. Type IVa pilins occur in a wide range of Gram-negative bacteria, whereas Type IVb pilins have been found only in bacteria capable of colonizing the human intestine and are larger, particularly in the D-region (18).
However, it has been discovered comparatively recently that Type IV pili are not exclusive to Gram-negative bacteria (19). Clostridium perfringens has the genes for and produces pili required for gliding motility (20). The genomes of all sequenced Clostridium difficile strains include one gene cluster encoding a complete set of Type IV pilin biogenesis components, a second cluster with three to six genes, and a variable number of additional pilin genes2 (19). Fimbrial appendages have been observed extending from C. difficile in vitro (21) and in a hamster model of C. difficile colonization of colonic crypts (22). Given the high burden of C. difficile infection on human disease and mortality (23), we sought to characterize the Type IV pili from this Gram-positive bacterium.
To gain insight into the diversity of Type IV pilin assemblies, we sought to characterize the Gram-positive Type IV pilin protein most likely to differ from other known family members. At 283 residues, PilJ is significantly larger than previously characterized Type IV pilins. The first 10 residues form the pre-pilin leader peptide, intermediate to the lengths of Type IVa (5–6 residues) and Type IVb (15–30 residues) pilins (18). The N-terminal residue of processed PilJ is phenylalanine, typical of Type IVa pilins. Although the sequence of the PilJ α1-N region is similar to other pilins, no significant sequence similarity to any other pilins is found in the remainder of the sequence. PilJ also contains three cysteine residues rather than the two typically found in Type IV pilins that invariably form a disulfide bond. Here, we present the crystal structure of PilJ, a C. difficile Type IV pilin protein that we found incorporated into pili on their surfaces. PilJ represents the first reported three-dimensional structure of a Gram-positive Type IV pilin protein.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
DNA sequences of PilA1 and PilJ lacking the signal peptide and codons for the N-terminal hydrophobic domain were codon-optimized for expression in E. coli, commercially synthesized (Genscript), cloned into the pET30b vector downstream of the hexahistidine tag sequence, and transformed into E. coli BL21(DE3) cells (Invitrogen). After inoculation of 1L Luria broth plus kanamycin with 20 ml of turbid overnight culture, cells were grown to A600 = 0.5 at 30 °C and induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside. After an induction period of 18 h, cultures were pelleted by centrifugation at 6000 × g for 10 min at 4 °C (Beckman Coulter); pellets were stored at −20 °C. Cell pellets were resuspended in 50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8.0, with protease inhibitors (Roche Applied Science) and lysed in a French press at 1200 p.s.i. (Sim Aminco); lysates were centrifuged at 35,000 × g for 30 min. Supernatants containing each fusion protein were applied to nickel-nitrilotriacetic acid-agarose (Qiagen) and incubated with rotation at 4 °C for 1 h. After washing, protein was eluted from the resin with increasing concentrations of imidazole in 50 mm NaH2PO4, 300 mm NaCl, pH 8.0. When necessary, column fractions were further purified by size-exclusion chromatography using a Sephacryl S-100 column. For immunoblotting and immunoabsorption, the N-terminal purification tag was cleaved from each purified pilin protein with recombinant enterokinase (Novagen) and removed by incubation with nickel-nitrilotriacetic acid resin.
Antibody Generation
Polyclonal antibodies to untagged PilA1 were raised in guinea pigs, and antibodies to untagged PilJ were raised in rabbits (Rockland Immunochemicals). Briefly, animals were immunized intradermally with purified untagged protein and complete Freund's adjuvant and boosted on days 7, 14, and 28 with protein and incomplete Freund's adjuvant. Test bleeds were taken on day 38. Terminal bleeds for rabbits were taken on day 59. The guinea pigs were boosted again, and terminal bleeds were taken on day 80.
Immunogold Labeling
To maximize pilus expression, the bacteria were grown anaerobically on Columbia Agar plates at 37 °C for 24 h and then incubated anaerobically at room temperature for 3 additional days. Several C. difficile colonies were suspended in 50 μl of fixative solution (2% formaldehyde, 0.5% glutaraldehyde in 50 mm cacodylate buffer, pH7.4), and 30 μl of the suspension was placed on a Formvar/carbon nickel grid (Electron Microscopy Science) for 15 min. The grids were then washed for 20 min in 0.05 m glycine in phosphate-buffered, pH 7.2, physiological saline (PBS) and blocked for 30 min with 3% skim milk plus 0.01% Tween in PBS. After 3 washes (5 min each) with incubation buffer (0.01% BSA-cTM (Aurion) in PBS), the grids were incubated for 60 min with a 100-fold dilution of either rabbit polyclonal PilJ- or guinea pig polyclonal PilA1-specific sera or of pre-immune sera. For the double-labeling experiments the grids were incubated with a 1:1 mixture of the rabbit polyclonal PilJ- and guinea pig polyclonal PilA1-specific sera or with pre-immune sera. The grids were then washed 6 times (5 min each) with incubation buffer followed by a 45-min incubation with the secondary antibody (goat anti-rabbit and/or goat-anti guinea pig IgG conjugated to 10- or 15-nm gold nanoparticles, respectively (Electron Microscopy Science), diluted 25-fold). The grids were washed again 6 times (5 min each) with incubation buffer and 3 times (5 min each) with double-distilled water, stained with 1% phosphotungstic acid, and observed using an Hitachi H-7650 Transmission Electron Microscope. One hundred random images were recorded, and the number and size of pili- or background-associated gold nanoparticles was recorded in each image by three observers who were unaware of the experimental conditions.
C. difficile Lysate Preparation
Cultures of C. difficile strain R20291 were grown on Columbia agar plates as above. Colonies were scraped off plates, resuspended in PBS, and brought to an A600 of 20. Bacteria were lysed as described previously (24); briefly, resuspended cells were frozen at −20 °C overnight, then incubated at 37 °C for 40 min. Total protein present in each lysate was quantified with a commercial bicinchoninic acid (BCA) assay kit (Thermo Scientific Pierce) according to the manufacturer's protocol.
In preparation for immunoblotting, lysates were diluted 1:1 in Laemmli buffer and heated in a boiling water bath for 10 min. Lysates and a standard curve of untagged PilJ were separated on precast 4–15% gradient Mini-PROTEAN TGX polyacrylamide gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes. Blots were blocked for 1 h with 5% nonfat dry milk, then incubated at 4 °C overnight with polyclonal rabbit anti-PilJ at a 1:10,000 dilution. Blots were washed in PBST and incubated with IRDye 800CW donkey anti-rabbit IgG H+L (Li-Cor Biosciences) at a 1:20,000 dilution for 1 h. Infrared signals were detected and quantified using the Odyssey imaging system (Li-Cor Biosciences).
Structure Determination and Refinement
PilJ, concentrated to 10 mg/ml in 10 mm MES, 100 mm NaCl, pH 6.0, was crystallized in 0.2 m ammonium sulfate, 0.1 m MES, pH 6.5, and 30% polyethylene glycol 4000 by hanging drop vapor diffusion. Crystals grew within 3 days at room temperature, forming rod-like single crystals (0.02 × 0.03 × 0.9 mm) as well as multicrystal spindles. The crystals were flash-cooled in liquid nitrogen in mother liquor supplemented with 25% (v/v) glycerol as a cryoprotectant. Diffraction data were collected at the National Light Source (NLS), Brookhaven National Laboratory at Beamline X25 with crystals diffracting to 1.98Å resolution in the space-group P212121 with a unit cell of 53 × 78 × 134 Å.
Additionally, multiwavelength anomalous dispersion data were collected at the Advanced Photon Source (APS), GM/CA-CAT, Argonne National Laboratories at Beamline 23-ID-D after an x-ray fluorescence scan revealed the presence of zinc in crystals of PilJ. Peak, edge, and remote data were collected at 1.2830, 1.2833, and 1.2320 Å, respectively. These anomalous data were used for phasing with two sites being found in each asymmetric unit. Two PilJ molecules were found in the asymmetric unit, each with a single zinc ion.
All datasets were scaled and processed using HKL2000 (25). Phenix and Coot were used for phasing, building and refinement (26–29). The phasing and initial chain building used the multiwavelength anomalous dispersion datasets. This model was subsequently refined using the higher resolution dataset, applying the phases from the anomalous data. Multiple iterative rounds of model building and refinement resulted in an Rwork factor of 18.3% and an Rfree of 23.0%. The crystallographic parameters of the refined data are summarized in Table 1.
TABLE 1.
Crystallographic parameters
Values in parentheses are for the highest resolution shell.
| Native | Peak | Inflection | Remote | |
|---|---|---|---|---|
| Data Collection | ||||
| Wavelength (Å) | 1.071 | 1.2830 | 1.2833 | 1.2320 |
| Resolution range (Å) | 43.71–1.98 (2.05–1.98) | 38.93–2.70 (2.80–2.70) | 38.93–2.70 (2.83–2.70) | 38.96–2.77 (2.90–2.77) |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell (Å) | 52.83 77.86 133.66 | 52.82 77.86 133.66 | 52.83, 77.88, 133.70 | 52.87, 77.91, 133.78 |
| 90 90 90 | 90 90 90 | 90 90 90 | 90 90 90 | |
| Unique reflections | 38,121 (3,014) | 15,419 (1,482) | 15,747 (2,056) | 15,754 (2,010) |
| Completeness (%) | 97.70 (77.96) | 100.00 (96.05) | 100 (98.2) | 100 (98.1) |
| Mean I/δ(I) | 7.94 (1.93) | 12.19 (4.57) | 15.63 (7.69) | 18.52 (7.63) |
| Wilson B-factor | 24.54 | 24.65 | 29.6 | 28.6 |
| R-merge | 0.128 (0.644) | 0.118 (0.453) | 0.122 (0.472) | 0.096 (.336) |
| Refinement statistics | ||||
| Molecules in asymmetric unit | 2 | |||
| Rfactor | 0.1832 (0.2505) | |||
| Rfree | 0.2301 (0.3218) | |||
| Number of atoms | 3922 | |||
| Root mean square (bonds) | 0.009 | |||
| Root mean square (angles) | 1.15 | |||
| Ramachandran favored (%) | 98 | |||
| Ramachandran outliers (%) | 0 | |||
| Clashscore | 9.30 | |||
| Average B-factor | 28.30 | |||
| Macromolecules | 27.90 | |||
| Solvent | 31.60 | |||
| Metal ions | 18.73 | |||
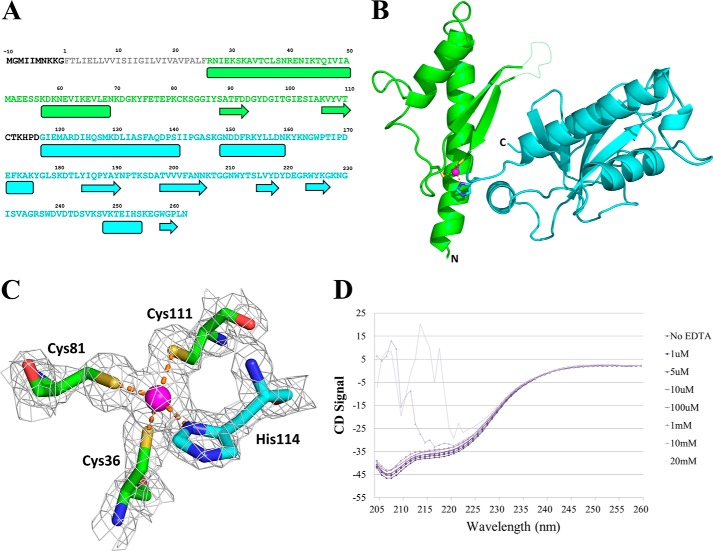
Circular Dichroism of PilJ in the Presence of EDTA
CD spectra were collected on a 10 μm solution of PilJ in 20 mm MES, 100 mm NaCl on a JASCO J-720 circular dichroism spectrophotometer. EDTA was titrated into a fixed concentration of PilJ. Each wavelength scan was referenced against a scan of a solution containing no PilJ but an identical concentration of EDTA. As the concentration of EDTA increased, PilJ could be observed slowly falling out of solution until a critical point was reached at ∼10 mm EDTA, at which point the protein precipitated leading to particles visible with the naked eye and scattering as observed in the 10 mm EDTA and 20 mm EDTA curves.
Small Angle X-ray Scattering
Small angle x-ray scattering data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL), Beamline 4-2. Scattering was measured at 2, 5, and 10 mg/ml of PilJ in 20 mm MES, 100 mm NaCl, pH 6.0. Two-dimensional diffraction images and one-dimensional intensity plots were processed using ATSAS 2.5.0 from EMBL-Hamburg. Radial distance-distribution plots were calculated with GNOM (30), and those data were used by SASTBX (31), GASBOR (32), and DAMMIF (33) to model molecular envelopes of the solution structure. The calculated intensity plot for the structure of the PilJ monomer was generated using FoXS (34) through UCSF Chimera (35) and verified using CRYSOL (36). Superimposition of the PilJ monomer from the x-ray crystal structure onto the small angle x-ray scattering (SAXS)3 envelopes was performed by collage in Situs 2.7.2 (37). Parameters are summarized in Table 3.
TABLE 3.
Small angle x-ray scattering parameters
| PilJ concentration | Rg (Gunier) | Rg (GNOM) | Dmax | Mr (Porod) | Mr (GASBOR) | Mr (DAMMIF) |
|---|---|---|---|---|---|---|
| 2 mg/ml (63 μm) | 21.45 ± 0.17 | 21.75 | 72.49 | 25173 | 29284 | 30482 |
| 5 mg/ml (158 μm) | 21.79 ± 0.06 | 22.02 | 76.52 | 25356 | 30114 | 28976 |
| 10 mg/ml (316 μm) | 21.38 ± 0.24 | 22.79 | 89.37 | 26324 | 33542 | 31024 |
Modeling the PilJ Pilus Fragment
To model a homopolymer of PilJ, we used cryo-EM data from the toxin coregulated pilus (TCP) of V. cholerae (EMD-1954) (38). We modeled the α1-N region of PilJ based on the structure of the full-length P. aeruginosa PAK pilin (39). Twenty copies of a model of PilJ, including the modeled α1-N region and the N-terminal domain but including only residues 179–193 of the C-terminal domain, were fit into the cryo-EM electron density using rigid body refinement in UCSF Chimera (35) and Coot (29) with a final correlation coefficient of 0.78 determined by Situs 2.7.2 (37). After additional rounds of refinement to remove any clashes between subunits, the missing portion of the C-terminal domain was reintroduced, completing the model.
PilJ Sequence Comparison
Sequences of C. difficile genomes were taken from the NCBI database. Sequence comparisons were made using NCBI Blast and ClustalX (40). The phylogenic tree was constructed by NJplot (41). Solvent-accessible surface area calculations were performed using CCP4 (42), and the resulting figure was made using UCSF Chimera (35).
Surface Plasmon Resonance
Using a Biacore T-100, the affinities of PilJ for both PilA1 and itself were individually measured by surface plasmon resonance. PilJ, PilA1, bovine serum albumin (BSA) and an irrelevant antibody were coupled to a CM5 chip surface by amine coupling. Using the BSA and antibody surfaces as controls, PilJ was titrated over all four surfaces using kinetic injections of concentrations ranging from 1.5 to 800 μm. Affinities were measured by steady-state equilibrium analysis.
RESULTS
Incorporation of PilJ into C. difficile Pili
To investigate the incorporation of PilJ and other pilins into C. difficile Type IV pili in vivo, we first used primary polyclonal antibodies specific to PilJ, purified to remove any immunoglobulins that cross-reacted with other C. difficile pilins, to stain C. difficile lysates in a Western blot. The quantification of those data shows that total PilJ expression under conditions conducive to the formation of pili is ∼18.25 ± 2.5 ng PilJ/μg of total protein (data not shown). The expression is lessened somewhat in a liquid culture, which disfavors the formation of Type IV pili.
To determine whether PilJ was incorporated into pili, we stained C. difficile with primary polyclonal antibodies against PilJ and another C. difficile pilin, PilA1, which is encoded by the first gene in the pil operon, and secondary antibodies labeled with 10-nm (PilA1) and 15-nm (PilJ) gold nanoparticles. These micrographs clearly show the presence of both PilA1 and PilJ in C. difficile pili (Fig. 1A); tallies of pili-associated nanoparticles per field are shown in Fig. 1B. Importantly, several individual pili are labeled with both particles, suggesting that both pilins can be found within the same pilus. At the resolution of immunogold staining, however, it is impossible to determine whether PilA1 and PilJ are incorporated into such heteropolymeric pili in any regular pattern.
FIGURE 1.
PilJ expression and localization. A, electron micrographs stained with anti-PilA1 and anti-PilJ antibodies and immunogold-labeled secondary antibodies with particle sizes 10 nm (PilA1) and 15 nm (PilJ). Nanoparticles associated with pili are labeled with either gray triangles (10 nm, PilA1) or black triangles (15 nm, PilJ). B, quantification of immunogold labeling. The average number of particles per field is shown as a horizontal line. The error bars show one standard deviation.
High Resolution Structure of PilJ
We determined the x-ray crystal structure of soluble PilJ to 1.98Å resolution (Table 1). The overall fold of PilJ is unique among Type IV pilins and contains two distinct domains (Fig. 2, A and B). The two PilJ domains share certain structural features of Gram-negative Type IV pilins. PilJ is the only Type IV pilin with two consecutive domains, although some pseudopilins exhibit insertion domains (43, 44).
FIGURE 2.
PilJ three-dimensional structure. A, the sequence of PilJ with the secondary structure outlined below. The prepilin leader sequence is shown in black, and the α1-n region is in gray. The N-terminal domain is green, and the C-terminal domain is blue. Helices are shown as boxes, and strands are shown as arrows. B, schematic representation of the structure of PilJ. The N-terminal domain is green, the C-terminal domain is blue, and the zinc atom is magenta. The disordered loop spanning residues 94–102 is indicated with a dotted line. C, the zinc-binding site of PilJ. Cysteines 36, 81. and 111 from the N-terminal domain are in green, histidine 114 from the C-terminal domain is in cyan, and the zinc atom is in magenta. The gray mesh shows the bounds of a 2Fo − Fc electron density map. D, CD spectra of PilJ are shown in increasing concentrations of EDTA.
The N-terminal helix of PilJ, starting with residue 26, shows a typical Type IV pilin protein fold as does the α-β loop, which exhibits a large degree of α-helical character. The β sheet of the N-terminal domain is unique in that it consists of only two β strands, whereas all other Type IV pilin protein β sheets contain at least three strands. Nine residues in the loop between these two strands are disordered in the crystal structure.
Joining the two domains of PilJ is a zinc atom that is ligated by all three cysteine residues in PilJ, Cys-36, Cys-81, and Cys-111 as well as by His-114 (Fig. 2C). Notably, we solved the structure by multiwavelength anomalous dispersion phasing from a single zinc atom per PilJ monomer. Only these three cysteine residues exist in PilJ, and this metal binding site is a unique characteristic of the PilJ fold. Its position and the paucity of interdomain buried surface area (∼200 Å2) suggest that the ligated zinc atom stabilizes the interaction between the two domains, as supported by chelation-induced unfolding (Fig. 2D).
After the unusual two-strand β sheet and a short loop, another long (25-residue) α helix, oriented at ∼70° to the N-terminal helix, forms the start of the C-terminal domain of PilJ. This helix is followed by another α-β loop that includes an additional incomplete α helix. The C-terminal PilJ domain also includes a five-stranded β sheet with additional shorter helices surrounding it and, thus, forms a more complete Type IV pilin-like fold than does the N-terminal domain (Fig. 3A).
FIGURE 3.

Structural comparison of the PilJ N-terminal and C-terminal domains. A, the N-terminal and C-terminal domains of PilJ. The initial α helices are in magenta, the αβ loops are in green, and the first two strands of each β-sheet are in blue. B, the N-terminal domain is depicted in green, and the C-terminal domain is in cyan.
Each of the two individual domains of PilJ are structurally homologous (45) to numerous major and minor Type IV pilins as well as Type II secretion pseudo-pilins (Table 2). A pairwise comparison of the two domains in PilJ also indicates significant structural similarity (Z-score = 3.7) (Fig. 3B). Despite this commonality of fold between the individual PilJ domains and known pilins, no portion of the soluble PilJ protein is similar in sequence to any other known protein.
TABLE 2.
Structural similarity to PilJ domains
rmsd, root mean square deviation; lali, length of alignment; nres, number of aligned residues; %id, percentage of sequence identity.
| Number | Chain | Z | rmsd | lali | nres | %id | PDB Description |
|---|---|---|---|---|---|---|---|
| PilJ N-terminal domain | |||||||
| 1 | 1T92-B | 6.2 | 2.7 | 72 | 108 | 15 | General secretion pathway protein G (GspG) |
| 5 | 3GN9-A | 5.8 | 2.7 | 70 | 112 | 13 | Type II secretory pathway, pseudopilin EpsG |
| 8 | 2OPD-B | 5.6 | 2.2 | 68 | 121 | 9 | PilX |
| 12 | 3G20-A | 5.6 | 2.6 | 70 | 114 | 11 | Type II secretion protein |
| 24 | 3HG9-A | 5.0 | 2.4 | 61 | 119 | 5 | PilM |
| 26 | 1OQV-A | 4.9 | 2.8 | 70 | 171 | 6 | Toxin-coregulated pilus subunit |
| 80 | 2RET-G | 4.0 | 2.6 | 55 | 81 | 7 | Pseudopilin EpsI |
| PilJ C-terminal domain | |||||||
| 1 | 2OPD-B | 4.5 | 4.2 | 91 | 121 | 8 | PilX |
| 59 | 1T92-B | 3.1 | 5.6 | 72 | 108 | 10 | General secretion pathway protein G (GspG) |
| 79 | 3GN9-C | 3.0 | 2.8 | 67 | 111 | 13 | Type II secretory pathway, pseudopilin EpsG |
| 91 | 3G20-A | 2.9 | 2.8 | 69 | 114 | 9 | Type II secretion protein |
| 96 | 3CI0-I | 2.9 | 3.1 | 60 | 83 | 8 | pseudopilin GspI |
| 97 | 3CFI-G | 2.9 | 2.9 | 56 | 74 | 9 | Type II secretory pathway, pseudopilin EpsI |
Self-association of PilJ Headgroups
As interactions between pilin head-groups are an important component of Type IV pilus formation (38), we used SAXS and surface plasmon resonance to show that the soluble PilJ protein that we crystallized is able to associate with itself and other pilin proteins.
First, we employed SAXS analysis to probe the behavior of PilJ in solution. The SAXS profile of PilJ is typical of a folded globular protein and agrees well at lower concentrations with the logarithmic scale curve (log(I) versus q) predicted based on the structure of the monomer from our x-ray crystal structure using CRYSOL (Fig. 4A) (36). We also used the radial distance distribution function calculated from the SAXS data to derive a molecular envelope using spherical harmonics in SASTBX, DAMMIF, and GASBOR (31–33). The envelopes created by each of the three programs are similar, and there is excellent agreement with the PilJ monomer structure observed in the crystal. The superimposition of the crystal structure monomer into the SASTBX envelope has a correlation coefficient of 0.94 and is shown in Fig. 4B. No aggregation was observed even at the highest concentration (10 mg/ml) as evidenced by the flat slope of the low q region of the intensity plot in Fig. 4A and the Kratky plot (Fig. 4C). The radius of gyration (Rg) values calculated from the Guinier plot (Fig. 4D) and from the Fourier method used by GNOM (30) are in good agreement with the values being nearly constant across the experimental concentration range (21–23 Å) and are similar to the value calculated from the crystal structure (19 Å). The larger value of Rg in solution most likely stems from the absent electron density corresponding to the expression tag in the crystal structure.
FIGURE 4.
Self-association of Pilin headgroups. A, log-scale intensity plot of SAXS profiles at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). B, superimposition of PilJ x-ray crystal structure (cyan ribbon) into envelope calculated for 2 mg/ml PilJ by SASTBX (gray mesh). C, Kratky Plot (I × q2 versus q) at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). D, radial distribution function calculated by GNOM at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). Error bars are shown as hash marks. The inset shows a Gunier plot (ln(I(q)) versus q2) of the region used to calculate the radius of gyration.
The SAXS data clearly show that the soluble PilJ construct is primarily monomeric over the entire experimental concentration range. The intensity curves were compared with a theoretical scattering curve predicted from the structure of the monomer by FoXS (Fig. 4A). Although this produced generally good agreement, the χ2 values were unexpectedly high. We attribute some of this discrepancy to molecular flexibility either of the purification tag or between the two domains. However at higher concentrations, the profile of the experimental SAXS data diverges further from the predicted profile for the monomer. The increasing deviation of the SAXS profile from the curve predicted for the monomeric protein does not stem from aggregation, and this shift can also be seen in the radial distribution functions calculated for each of the three concentrations (Fig. 4C) by an increase in the value of Dmax and by an increasing molecular weight predicted by Porod's law (Table 3). These trends suggest that PilJ might be able to self-associate to some degree even in the absence of the hydrophobic α1-N helix.
To determine the ability of PilJ head groups to self-associate, we measured their binding by surface plasmon resonance, which indicates that soluble PilJ can bind to a surface coated with PilJ. These binding studies show an affinity of ∼300 μm for the self-association of PilJ proteins missing the α1-N helical region (Fig. 5). Additionally, as immunogold staining also revealed the presence of PilA1 in C. difficile Type IV pili, we measured the association of the PilJ and PilA1 pilin headgroups. The affinity of this heterotypic interaction was ∼4-fold stronger (Kd = 70 μm).
FIGURE 5.
Surface plasmon resonance binding analysis of PilJ and PilA1. Surface plasmon resonance binding of soluble PilJ to PilJ (A) and PilA1 (B) surfaces. The binding titrations are shown as insets, with the steady-state values at each concentration depicted as black squares. The equilibrium fits are shown as gray curves.
Model of the Incorporation of PilJ into Type IV Pili
To understand how PilJ might be positioned in the formation of a C. difficile Type IV pilus, we used the crystal structure of PilJ to build a molecular model of a hypothetical fragment of a Type IV pilus composed of repeating units of PilJ protein. We first modeled the structure of the hydrophobic α1-N domain of PilJ using the crystal structure of the full-length P. aeruginosa PAK pilin (39) as has been done with other truncated pilin constructs (46, 47). The model of the full-length PilJ monomer is shown in Fig. 6A.
FIGURE 6.
Model of PilJ pilus formation. A, model of full-length PilJ. The α1-N helix is shown in gray, the N-terminal domain is in green, and the C-terminal domain is in cyan. B, superimposition of selected regions of PilJ onto TcpA. The PilJ N-terminal domain is shown in green, and the selected portion of the C-terminal domain is in cyan. TcpA is shown in gray. C, space-fill model of truncated PilJ chains modeled into a pilus fragment and superimposed onto electron density from an electron micrograph of the V. cholera TCP (gray mesh). Each chain of the truncated PilJ model is colored individually. D, space-fill and ribbon model of a PilJ pilus formed from full-length PilJ; each chain is colored individually.
As no three-dimensional structures exist at any resolution for Gram-positive Type IV pili, we modeled a homopolymer of PilJ based on the electron density of the TCP of Gram-negative V. cholerae (39). A superposition of PilJ with TcpA, the TCP major pilin, revealed structural similarity in regions of the PilJ N-terminal domain, especially in the N-terminal α-helix and α-β loop as well as in a region of the C terminus analogous to the α3-region of TcpA. Residues in the TcpA α3-region and α-β loop had been found previously to mediate contacts between head groups of individual TcpA pilin subunits in the TCP (47). Because the C-terminal domain of PilJ is unique among all Type IV pilin proteins and has no equivalent in the structure of TcpA, our initial model for fitting the pilin subunit into the electron micrograph of TcpA was restricted to the N-terminal domain and residues 169–183 near the C terminus, which were preserved due to their similarity in structure and position to the region of the α3-region of TcpA. We superimposed the truncated PilJ with TcpA (Fig. 6B) and fit multiple copies of it into the electron density of the TCP with a correlation coefficient of 0.78. This fit is illustrated in Fig. 6C, which shows the model of the truncated PilJ forming a similar left-handed helix to the one modeled for TcpA. Contacts between subunits occurred between the α1-N domains and between the α-β loop of subunit i-1 and the C-terminal region of subunit i. Although there is no obvious “glue” between the subunits, the interface between the head groups in the model included two sets of acidic and basic residues that might potentially form salt bridges. They are residues Asp-71 and Lys-180 as well as Lys-73 and Asp-181.
After rigid-body refinement of this truncated PilJ filament, we replaced the truncated PilJ subunits with full-length PilJ structures. The reintroduction of the C-terminal domain produced no steric clashes. As shown in Fig. 6D, the protrusion of the C-terminal domain is such that regions of a pilus composed primarily of PilJ would be both wider than any known pilus, ∼120 Å, and more striated. The surface-exposed regions of PilJ in our model lie nearly entirely within the C-terminal domain with the interface being formed by residues 69–77 of the α-β loop and 178–183 in the C-terminal domain.
Surface Polymorphisms in PilJ
The sequences of 20 publically available C. difficile genomes exhibit 14 polymorphic sites differing in the number of variations from the reference sequence (R20291) from 1 to 12 as shown in Table 4. PilJ is well conserved, with even the most divergent strains exhibiting a sequence identity of 96%. The largest cluster of identical sequences contains our reference and seven other strains, all of which belong to the 027 ribotype (NAP1). The 12 sequences that diverge from that of R20291 contain two distinct clusters, 1 of 4 identical sequences, 3 of which stem from ribotype 078 (NAP7/8) strains, and another of 3 identical sequences including 2 017 ribotypes. This same clustering pattern, a large NAP1 cluster, a smaller NAP7/8 cluster, and a small number of sequences falling between these two consensus sequences, was observed previously in a genome-wide comparison of 14 C. difficile strains (48). The lone exception to this rule is the sequence of CIP 107932, which clusters with those of the NAP7/8 strains despite the overall genetic similarity between CIP 107932 and NAP1 strains. A phylogenic tree showing strains clustered by their PilJ sequence is shown in Fig. 7A.
TABLE 4.
Polymorphisms in PilJ
Unk, unknown.
| Strain | Ribotype | Variations from R20291 |
|---|---|---|
| R20291 | 027 | Unk |
| QCD-66C23 | 027 | Unk |
| CD196 | 027 | Unk |
| 2007855 | 027 | Unk |
| QCD-76w55 | 027 | Unk |
| QCD-37 × 79 | 027 | Unk |
| BI-1 | 027 | Unk |
| QCD-97b34 | 027 | Unk |
| BI-9 | 001 | I118V, I124V |
| QCD-63q42 | 001 | I118V, I124V |
| ATCC 43255 | 087 | I101R, I118V, I124V |
| 70-100-2010 | D58G, I63M, I118V, N191S | |
| 630 | 012 | V21I, D58G, I63M, I118V, A136S, N191S |
| CF5 | 017 | S55Q, E68Q, I118V, I124V, V245A |
| M68 | 017 | S55Q, E68Q, I118V, I124V, V245A |
| 002-P50-2011 | Unk | S55Q, E68Q, I118V, I124V, V245A |
| CIP 107932 | Unk | S55P, E61K, E65D, I101G, K113E, I118V, I124V, G148S, N163K, S195P |
| M120 | 078 | S55P, E61K, E65D, I101G, K113E, I118V, I124V, G148S, N163K, S195P |
| QCD-23m63 | 078 | S55P, E61K, E65D, I101G, K113E, I118V, I124V, G148S, N163K, S195P |
| NAP08 | 078 | S55P, E61K, E65D, I101G, K113E, I118V, I124V, G148S, N163K, S195P |
FIGURE 7.
Sequence variation in PilJ. A, unrooted phylogenic tree of C. difficile strains by PilJ sequence. B, model of full-length PilJ. Polymorphic residues in PilJ are represented with side chains rendered as spheres, colored by the number of variations from the reference sequence within the set of 20 C. difficile strains compared here. The blue surface shows the area of each pilin occluded from solvent by the formation of a pilus fragment and hence potentially part of a pilin-pilin interface.
Only 2 of the 14 polymorphisms are found in the occluded volume of our modeled PilJ pilus (Fig. 7B). The variable sites of PilJ are, conversely, largely excluded from those regions that form the putative pilin-pilin interface, suggesting that those regions have been conserved evolutionarily to form structural contacts in pili. However, as the composition of C. difficile Type IV pili is still under investigation, it remains unclear to what extent those contacts are made between PilJ subunits as opposed to other C. difficile pilins. The variable sites are also restricted to one face of PilJ, primarily in the C-terminal domain and the α-β loop (this is the left side of PilJ as it is depicted in Fig. 6). Accordingly, this exposed surface may mediate a PilJ pilus-specific function, such as adhesion.
DISCUSSION
It has been perhaps three billion years since Gram-positive bacteria, Gram-negative bacteria, and archaea last shared a common ancestor, and yet Type IV pili are found in each of these diverse life forms, suggesting that extant systems diverged from a primordial form at least that old. The first high resolution three-dimensional structure of a Gram-positive type IV pilin, PilJ, which we present here, substantially broadens our understanding of these proteins and the formation of pili. Numerous structural features of PilJ that do not exist in the previously characterized Type IV pilin proteins of Gram-negative bacteria contribute to this knowledge, including the following. (i) PilJ contains no disulfide bonds, and hence the C terminus has no D-region. (ii) The three cysteine residues present in PilJ form a zinc-binding site to incorporate a structural metal. (iii) The initial β-sheet of PilJ is smaller than previously characterized pilins, containing only two β-strands. (iv) Finally and perhaps most importantly, PilJ contains two Type IV pilin protein-like domains anchored at the joint by the structural metal ion. This division into multiple domains implies that each has a separate function, and because the position of the C-terminal domain is unprecedented, it follows that PilJ, in the context of the pili in which it is found, may have functions that are distinct from other pilin proteins.
The bifurcated nature of the PilJ structure suggests a clearer differentiation of function between self-association (largely the product of the N-terminal domain) and non-self-interaction (the probable function of the C-terminal domain) than is observed in previously characterized Type IV pilin proteins. Although the interaction of the α-β loop with the C terminus likely contributes to pilus formation through favorable electrostatic interactions, the burial of hydrophobic surface area occurs almost exclusively in the N-terminal domain, primarily through the α1 helix. Our model suggests a multilayer approach to pilus formation by PilJ, where the most hydrophobic regions of the pilin are buried deep in the pilus core and interact primarily with each other or other pilins, whereas the interfaces between the more solvent-exposed head groups are less hydrophobic in nature. The commonality of fold between the two domains of PilJ suggests they share a common ancestral sequence, but the two sequences have since diverged beyond sequence recognition. This may reflect a differentiation of function between the two PilJ domains. The C-terminal domain comprises a larger portion of the pilin head-group and includes accessory α-helices in addition to the primary α-helix at its N terminus, unlike the N-terminal domain, which consists entirely of the N-terminal α-helix, the α-β loop, and a small β-sheet.
We expect the that the incorporation of a structural metal also stems from the requirements of a dual-pilin fold. Although the exact affinity of PilJ for zinc has not been determined, cysteine3histidine1 zinc binding sites have been shown to bind to zinc with subpicomolar affinity (49). The combination of high affinity and the relatively high physiological concentration of zinc (∼12–16 μm) (50) should allow C. difficile to form T4P incorporating PilJ without difficulty under physiological conditions.
Although the exact composition of C. difficile Type IV pili is only now being determined, numerous genetic factors have led us to hypothesize that PilA1 is the major pilin in the majority of C. difficile Type IV pili.2 However, our data show a robust level of PilJ expression, and immunogold electron microscopy clearly shows the presence of PilJ in C. difficile pili as well. Additionally, double immunogold staining appears to show the presence of PilJ and PilA1 in the same pilus. An interaction between the two is also supported by binding studies between the soluble PilA1 and PilJ constructs. The protrusion of the C-terminal domain of PilJ from the pilus core in our model of PilJ incorporation implies that interactions between C. difficile Type IV pili and other molecules occur largely through the C-terminal domain of PilJ rather than through PilA1 or the N-terminal domain of PilJ.
Type IV pili are known to be involved in a variety of functions, including twitching motility, biofilm formation, bacterial autoaggregation, adhesion to host cells, and horizontal gene transfer, but each of these functions is mediated by a similar binding event, be it to cell host cells, other bacteria, or in the case of horizontal transfer, DNA. The use of Type IV pili as adhesive molecules in Gram-negative strains is well documented (51), and a Type IV pilus of another Gram-positive strain, Ruminococcus albus, has been found to adhere to cellulose both in vivo and as isolated pili (52). Placing adhesive molecules on appendages can stem from a variety of selective pressures, including the use of those appendages for motility and avoiding close proximity between cell membranes (51). Because C. difficile PilJ has an entirely novel C-terminal domain, we hypothesize that this radically different exposed surface mediates a pilus-specific, rather than a structural function, most likely forming an equally unusual binding interface.
As many processes in microbial infection and colonization incorporate adhesion, the possibility of blocking those pathways through antibodies or other molecules binding to the C-terminal domain of PilJ presents an intriguing potential avenue for anti-C. difficile therapeutics. Our x-ray crystal structure of PilJ provides a structural basis for such future inhibitor development. However, further experimentation is necessary to determine whether the Type IV pili of C. difficile mediate adhesion to host cells, biofilm formation, motility, or some other process and to elucidate the role of PilJ.
In conclusion, we demonstrate that, through their incorporation of PilJ, Type IV pili of C. difficile present a novel pilin-like surface, making PilJ an attractive target for therapeutic efforts to combat the increasing prevalence of Clostridium difficile infection.
Acknowledgments
We thank the staff at Argonne National Laboratory Advanced Photon Source beam line GMCA-CAT, Brookhaven National Laboratory National Synchrotron Light Source beam line X29, and Dr. Daniel A. Bonsor for technical assistance with x-ray data collection and structure determination as well as the staff at Stanford Synchrotron Radiation Lightsource beam line 4-2 for technical assistance with SAXS data collection. We also thank Dr. Angela Wilks for the use of her CD spectrophotometer.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 AI095190 and R21 AI105881.
The atomic coordinates and structure factors (code 4IXJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Maldarelli, G., De Masi, L., von Rosenvinge, E., Carter, M., and Donnenberg, M. (2014) Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile. Pathogens Dis., in press.
- SAXS
- small angle x-ray scattering
- TCP
- toxin coregulated pilus.
REFERENCES
- 1. Stone B. J., Abu Kwaik Y. (1998) Expression of multiple pili by Legionella pneumophila. Identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66, 1768–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strom M. S., Lory S. (1993) Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol 47, 565–596 [DOI] [PubMed] [Google Scholar]
- 3. Zhang X. L., Tsui I. S., Yip C. M., Fung A. W., Wong D. K., Dai X., Yang Y., Hackett J., Morris C. (2000) Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Girón J. A., Ho A. S., Schoolnik G. K. (1991) An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710–713 [DOI] [PubMed] [Google Scholar]
- 5. Lee K. K., Sheth H. B., Wong W. Y., Sherburne R., Paranchych W., Hodges R. S., Lingwood C. A., Krivan H., Irvin R. T. (1994) The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit. Mol. Microbiol. 11, 705–713 [DOI] [PubMed] [Google Scholar]
- 6. Taniguchi T., Fujino Y., Yamamoto K., Miwatani T., Honda T. (1995) Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect. Immun. 63, 724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudel T., Scheurerpflug I., Meyer T. F. (1995) Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373, 357–359 [DOI] [PubMed] [Google Scholar]
- 8. Herrington D. A., Hall R. H., Losonsky G., Mekalanos J. J., Taylor R. K., Levine M. M. (1988) Toxin, toxin-coregulated pili, and the Toxr regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tacket C. O., Taylor R. K., Losonsky G., Lim Y., Nataro J. P., Kaper J. B., Levine M. M. (1998) Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect. Immun. 66, 692–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bradley D. E. (1980) A function of Pseudomonas aeruginosa PAO polar pili. Twitching motility. Can. J. Microbiol. 26, 146–154 [DOI] [PubMed] [Google Scholar]
- 11. Henrichsen J. (1983) Twitching motility. Annu. Rev. Microbiol. 37, 81–93 [DOI] [PubMed] [Google Scholar]
- 12. Merz A. J., So M., Sheetz M. P. (2000) Pilus retraction powers bacterial twitching motility. Nature 407, 98–102 [DOI] [PubMed] [Google Scholar]
- 13. Wall D., Kaiser D. (1999) Type IV pili and cell motility. Mol. Microbiol. 32, 1–10 [DOI] [PubMed] [Google Scholar]
- 14. O'Toole G. A., Kolter R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304 [DOI] [PubMed] [Google Scholar]
- 15. Seifert H. S., Ajioka R. S., Marchal C., Sparling P. F., So M. (1988) DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature 336, 392–395 [DOI] [PubMed] [Google Scholar]
- 16. Yoshida T., Kim S. R., Komano T. (1999) Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181, 2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bieber D., Ramer S. W., Wu C. Y., Murray W. J., Tobe T., Fernandez R., Schoolnik G. K. (1998) Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118 [DOI] [PubMed] [Google Scholar]
- 18. Craig L., Pique M. E., Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 19. Melville S., Craig L. (2013) Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 77, 323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varga J. J., Nguyen V., O'Brien D. K., Rodgers K., Walker R. A., Melville S. B. (2006) Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other clostridia. Mol. Microbiol. 62, 680–694 [DOI] [PubMed] [Google Scholar]
- 21. Borriello S. P., Davies H. A., Barclay F. E. (1988) Detection of fimbriae amongst strains of Clostridium difficile. FEMS Microbiol. Lett. 49, 65–67 [Google Scholar]
- 22. Goulding D., Thompson H., Emerson J., Fairweather N. F., Dougan G., Douce G. R. (2009) Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1. Infect. Immun. 77, 5478–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhuptani P., Natavio A., Forsyth L. (2012) Evaluation of clinical outcomes with monotherapy versus combination therapy in the treatment of Clostridium difficile infection. Crit. Care Med. 40, U105–U105 [Google Scholar]
- 24. Lawley T. D., Croucher N. J., Yu L., Clare S., Sebaihia M., Goulding D., Pickard D. J., Parkhill J., Choudhary J., Dougan G. (2009) Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191, 5377–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 31. Liu H., Morris R. J., Hexemer A., Grandison S., Zwart P. H. (2012) Computation of small angle scattering profiles with three-dimensional Zernike polynomials. Acta Crystallogr. A 68, 278–285 [DOI] [PubMed] [Google Scholar]
- 32. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys J 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lammie D., Osborne J., Aeschlimann D., Wess T. J. (2007) Rapid shape determination of tissue transglutaminase using high-throughput computing. Acta Crystallogr. D Biol. Crystallogr. 63, 1022–1024 [DOI] [PubMed] [Google Scholar]
- 34. Schneidman-Duhovny D., Hammel M., Sali A. (2010) FoXS. A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 38, W540–W544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 36. Svergun D., Barberato C., Koch M. H. J. (1995) CRYSOL. A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 37. Wriggers W., Birmanns S. (2001) Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133, 193–202 [DOI] [PubMed] [Google Scholar]
- 38. Li J., Egelman E. H., Craig L. (2012) Structure of the Vibrio cholerae type IVb pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J. Mol. Biol. 418, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., Yeager M., Forest K. T., Tainer J. A. (2003) Type IV pilin structure and assembly. X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 40. Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405 [DOI] [PubMed] [Google Scholar]
- 41. Perrière G., Gouy M. (1996) WWW-query. An on-line retrieval system for biological sequence banks. Biochimie 78, 364–369 [DOI] [PubMed] [Google Scholar]
- 42. Collaborative Computational Project, Number 4 (1994) Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 43. Korotkov K. V., Hol W. G. (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 15, 462–468 [DOI] [PubMed] [Google Scholar]
- 44. Giltner C. L., Nguyen Y., Burrows L. L. (2012) Type IV pilin proteins. Versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography. Implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 47. Li J., Lim M. S., Li S., Brock M., Pique M. E., Woods V. L., Jr., Craig L. (2008) Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 16, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Forgetta V., Oughton M. T., Marquis P., Brukner I., Blanchette R., Haub K., Magrini V., Mardis E. R., Gerding D. N., Loo V. G., Miller M. A., Mulvey M. R., Rupnik M., Dascal A., Dewar K. (2011) Fourteen-genome comparison identifies DNA markers for severe-disease-associated strains of Clostridium difficile. J. Clin. Microbiol. 49, 2230–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramboarina S., Morellet N., Fournié-Zaluski M. C., Roques B. P., Moreller N. (1999) Structural investigation on the requirement of CCHH zinc finger type in nucleocapsid protein of human immunodeficiency virus 1. Biochemistry 38, 9600–9607 [DOI] [PubMed] [Google Scholar]
- 50. Lu J., Stewart A. J., Sadler P. J., Pinheiro T. J., Blindauer C. A. (2008) Albumin as a zinc carrier. Properties of its high-affinity zinc-binding site. Biochem. Soc. Trans. 36, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 51. Kline K. A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 [DOI] [PubMed] [Google Scholar]
- 52. Rakotoarivonina H., Jubelin G., Hebraud M., Gaillard-Martinie B., Forano E., Mosoni P. (2002) Adhesion to cellulose of the Gram-positive bacterium R. albus involves type IV pili. Microbiology 148, 1871–1880 [DOI] [PubMed] [Google Scholar]