Abstract
In this study, a fingerprint of steroid saponins, the major bioactive constituents in the crude extracts from Dioscorea zingiberensis C. H. Wright (DZW), has been established for the first time by high-performance liquid chromatography coupled with evaporative light scattering detector (HPLC-ELSD) and the simultaneous characterization of the steroid saponins by high-performance liquid chromatography coupled with electrospray ionization-mass spectrometry and quadrupole tandem time-of-fight mass analyzers detection (HPLC-ESI-Q/TOF). These HPLC analyses were both carried out on a Welchrom C18 column (250 mm × 4.6 mm I.D., 5 μm) with a mobile phase composed of water and acetonitrile under gradient elution. There were 68 common characteristic peaks in the fingerprints, in which 12 of them were confirmed by comparing their mass spectra and retention times with those of the reference compounds. In order to identify the other unknown peaks, their fragmentation behaviors characteristic for the major groups of steroid saponins from DZW with six types of aglycone skeletons were discussed in detail, and possible MS/MS fragmentation pathways were proposed for aiding the structural identification of these components. According to the summarized fragmentation patterns, these peaks were tentatively assigned by matching their empirical molecular formula with those of the published compounds, or by elucidating their quasi-molecular ions and fragment ions referring to available literature information when the reference standards were unavailable. As a result, 22 steroid saponins were found in DZW for the first time. In addition, the quantitative analysis of the 12 known peaks was accomplished at the same time which indicated that there was a great variability in the amount of these active compounds in different batches in the crude extracts. This approach could demonstrate that the fingerprint could be considered to be a suitable tool to comprehensively improve the quality control of DZW, and the identification and structural elucidation of the peaks in the fingerprint may provide important experimental data for further pharmacological and clinical researches.
Keywords: fingerprint, steroid saponins, HPLC-ELSD, HPLC-ESI-Q/TOF
1. Introduction
It is well known that traditional Chinese medicine (TCM) is a complex mixture containing tens or even hundreds of chemically different constituents which, unlike pure synthetic drugs, often exhibit the synergic therapeutic effects [1–3]. In the standardization quality control of TCM, direct qualification and quantification of the naturally-occurring active constituents is a desirable criterion. However, it would be impossible if some of the active markers or reference compounds in the complex TCM are not commercially available. In addition, most of the natural products are so complex that the analysis during quality control of the TCM is rather tedious. So the traditional quality control methods require a number of very severe challenges [4, 5]. Therefore, it is absolutely necessary to develop more sensitive and reliable analytical methods for the quality control of TCM in order to guarantee its efficacy and safety when they are utilized in clinical practices.
The application of quadrupole time-of-flight tandem mass spectrometry (Q-TOF), which can yield empirical chemical formula based on the accurate masses of molecular ions and detailed fragmentation information, is a rapid and sensitive technique with great accuracy and precision for structural elucidation [6–8]. So, direct coupling of high-performance liquid chromatography with tandem mass spectrometry (HPLC-MS/MS) could facilitate informative and high-throughput screening of chemical constituents, and it has been proven to be a powerful tool for the rapid on-line analysis of the known compounds as well as identification of the structures of unknown compounds in complex TCM [9, 10]. Because of aforementioned characteristics, this technology will be widely employed to control the quality of TCM in the future. The chromatographic fingerprint of a plant material, as a comprehensive quantifiable identification method to show chemical information of herbal medicines, has been an efficient approach for the evaluation and quality control of complex TCM in recent years [11, 12]. This chromatographic fingerprint technique could be used to determine the identity, authenticity, batch-to-batch consistency of the herbal medicines, and it is also useful to overcome the limitations when using few marker compounds [13]. Consequently, the method would be more valid and efficient than the traditional methods in quality control of TCM.
Dioscorea zingiberensis C.H.Wright (DZW), a native plant in China, is one of the most commonly used raw materials in a traditional Chinese medicine (TCM) [14]. The steroid saponins, especially the water-soluble ones present primarily in the rhizomes, are the main bioactive components that have been used in clinic for the treatment of coronary heart disease, because of enhansing coronary blood flow (MBF), improving peripheral circulation and depressing platelet aggregation, as well as decreasing cholesterol and triglyceride in blood for many years [15]. What’s more, diosgenin, which exists in the form of steroid saponins and extracted mainly from the tubers of Dioscorea zingiberensis, is an important precursor for the synthesis of steroid hormone drugs and steroidal contraceptives such as adrenal cortex hormone, sex hormone, progestational hormone and anabolic steroid [16, 17]. In China, DZW is the preferred species for the production of diosgenin, as it has the highest contents of steroid saponins among the Dioscorea L. plants [18]. Although separation and purification of steroid saponins from DZW have been challenged by some researchers for many years, only a small fraction of them, about twenty, have been identified, because of the structural complexity, especially their sugar linkages. Many published papers have reported the determination of some steroid saponins in DZW [19], such as zingiberensis new saponin, deltonin, and dioscin, using analytical methods mainly including high performance liquid chromatography (HPLC). Obviously, only identification and quantitative determination of these few compounds alone are not sufficient for the comprehensive quality control of the DZW, since TCM is a complicated system. Because of these reasons, it is necessary to develop an effective and reliable method for qualitative and quantitative analysis of DZW, which can analyze as many steroid saponins as possible in DZW to ensure its safety and efficacy.
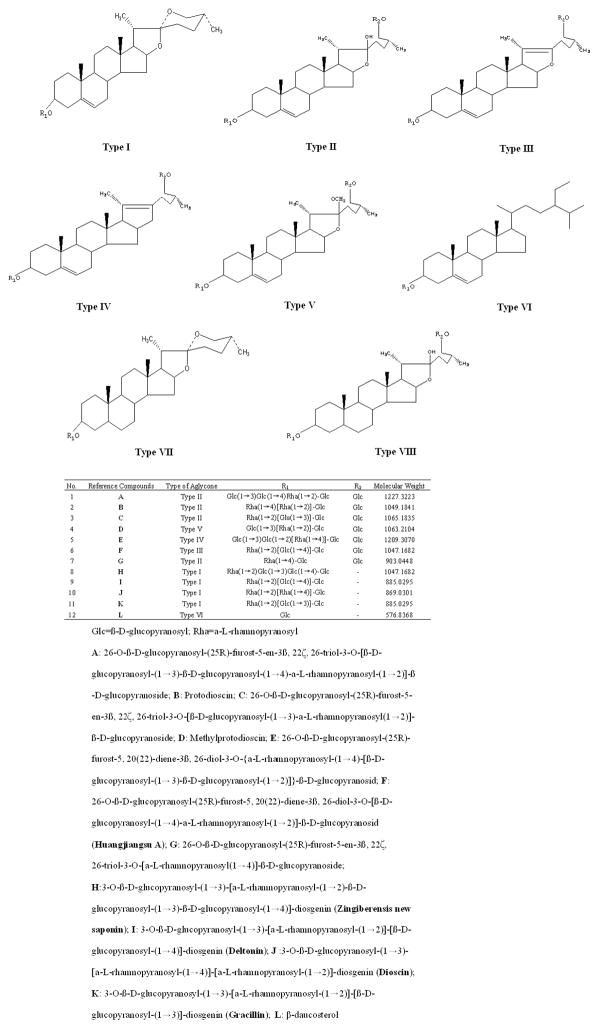
This paper describes two combined methods, a fingerprint using HPLC coupled with evaporative light scattering detector (HPLC-ELSD) and HPLC coupled with Q-TOF (HPLC-Q-TOF) for qualitation and quantification of the common characteristic peaks present in the fingerprint. Among the HPLC peaks, 12 known compounds were unambiguously confirmed by comparing their mass spectra and retention times with that of available reference compounds. Sugar chains of these reference compounds were summarized and divided into six different classes based on their aglycones in Fig. 1 and in some cases from the data reported in the literature. Other 56 unknown compounds were tentatively assigned either by matching empirical molecular formula with those of the published compounds or by elucidating quasi-molecular ions and fragment ions referring to the available literature information. The generated data thus provided valuable insight into the application of fingerprint and HPLC-MS in the analysis and quality control of DZW.
Figure 1.
Chemical structures of twelve known reference standard steroid saponins from Dioscorea zingiberensis C. H. Wright.
2. Experimental
2.1. Plant materials
In the present study 20 batches of DZW were collected in several different natural growth sites in Ankang County of Shaanxi Province in China. These herbs were identified by Professor Yazhou Wang (Northwest University, Xi’an 710069, China), and were spontaneously dried in the sun. Voucher specimens were deposited in the Biology and Medicine Key Laboratory of Shaanxi province, China.
2.2. Chemicals and reagents
HPLC grade acetonitrile (ACN) was obtained from Merck (Darmstadt, Germany). Deionized water (18 MΩ cm−1) was supplied with a Millipore Milli-Q water system (Milford, MA, USA). Other reagents were of analytical purity.
2.3. Standard compounds and preparation solutions
The following ten steroid saponins, were isolated and purified from DZW in the authors’ laboratory and their structures were elucidated by their spectral data (MS, and 13C NMR), and their purities were above 98% as determined by HPLC: they are (1) 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22ζ, 26-triol-3-O-[β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)-α-L-rhamnopyranosyl-(1→2)]-β-D-glucopyranoside (A), (2) 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22 ζ, 26-triol-3-O-[β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)]-β-D-glucopyranoside (C), (3) 26-O-β-D-glucopyranosyl-(25R)-furost-5, 20(22)-diene-3β, 26-diol-3-O-{α-L-rhamnopyranosyl-(1→4)-[β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→2)]}-β-D-glucopyranosid (E), (4) 26-O-β-D-glucopyranosyl-(25R)-furost-5, 20(22)-diene-3β, 26-diol-3-O-[β-D-glucopyranosyl-(1→4)-α-L-rhamnopyranosyl-(1→2)]-β-D-glucopyranosid (F, Huangjiangsu A), (5) 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22ζ, 26-triol-3-O-[α-L-rhamnopyranosyl-(1→4)]-β-D-glucopyranoside (G), (6) 3-O-β-D-glucopyranosyl-(1→3)-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)]-diosgenin (H, zingiberensis new saponin), (7) 3-O-β-D-glucopyranosyl-(1→3)-[α-L-rhamnopyranosyl-(1→2)]-[β-D-glucopyranosyl-(1→4)]-diosgenin (I, deltonin), (8) 3-O-β-D-glucopyranosyl-(1→3)-[α-L-rhamnopyranosyl-(1→4)]-[α-L-rhamnopyranosyl-(1→2)]-diosgenin (J, dioscin), (9) 3-O-β-D-glucopyranosyl-(1→3)-[α-L-rhamnopyranosyl-(1→2)]-[β-D-glucopyranosyl-(1→3)]-diosgenin (K, gracillin), (10) β-daucosterol (L). Standards of (11) protodioscin (B) and (12) methylprotodioscin (D) were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (NICPBP) (Beijing, China). These twelve stock samples: each accurately weighed and dissolved with HPLC grade methanol to prepare twelve reference compound of stock solutions (about 5 mg mL−1). Reference compounds mixture solution was prepared as follows: a certain amount of stock solutions of above twelve reference compounds were mixed and diluted with methanol to get a series of reference compound mixture solutions (520 μg mL−1 for A; 340 μg mL−1 for B; 560 μg mL−1 for C; 360 μg mL−1 for D; 420 μg mL−1 for E; 320 μg mL−1 for F; 300 μg mL−1 for G; 400 μg mL−1 for H; 380μg mL−1 for I; 200 μg mL−1 for J; 220 μg mL−1 for K; 240 μg mL−1 for L), while the corresponding working calibration solutions were prepared by successive serial dilution of the stock solution with methanol. All the solutions were stored in refrigerator at 4°C for subsequence use.
2.4. Sample preparation
Dried raw material of DZW (5 g) was powdered and refluxed thrice with 50 mL of 70% ethanol at 80°C, and each reflux time was 1 h. The ethanol solution was combined and evaporated to dryness under reduced pressure by a rotary evaporator, and the residue was redissolved in 30 mL of water, and subjected to centrifugation. In order to enrich the target components and remove impurities, the supernatant was separated on a D-101 macroporous resin column (2 cm × 15 cm), by eluting with water at first and next 20% ethanol until the effluent was colourless in every step. Then, the column was eluted with 70% ethanol, and 80 mL of effluent was collected. Finally, the collecting effluent was concentrated and decanted into a 25 ml volumetric flask, and then the methanol was added to make up to the scale. The final solution was passed through a 0.22 μm membrane prior to subsequent use. An aliquot of 10 μL sample solution was injected into the HPLC system for analysis.
2.5. HPLC apparatus
2.5.1. HPLC-ELSD analysis instrumentation
The HPLC fingerprinting analysis was carried out on a Waters Alliance 2695 equipment (Waters, Milford, MA, USA) with a vacuum degasser, a high pressure quaternary pump, an autosampler, and an Alltech 2000 evaporative light scattering detector. The separation of sample solution was performed on a Welchrom C18 column (250 × 4.6 mm, 5 μm), and evaluation and quantification were made on an Empower Workstation. The solvent flow rate was 1 mL min−1, and 10 μL of sample solution was injected in each run. A binary gradient elution system composed of acetonitrile as solvent A and water as solvent B was applied for the fingerprint analysis as follows: initial 25% A; 0–5 min linear gradient from 25% to 30% A; 5–20 min from 30% to 30% A, 20–35 min from 30% to 35% A, 35–45 min from 35% to 47% A, 45–47 min from 47% A to 70% A, 47–60 min from 70% to 70%. The effluent was introduced into the Alltech 2000 ELSD which the drift tube temperature was 90°C, and the gas flow rate was 2.8 L min−1.
2.5.2. HPLC-MS analysis instrumentation
The HPLC system described above was replaced with a Varian 212-LC equipped with a Q-TOF Premier, a quadrupole and orthogonal acceleration time-of-flight tandem mass spectrometer (Waters Co., USA), which was attached to an electrospray ionization interface. The MS data was recorded by the varian MS workstation software. High-purity nitrogen was used as the nebulizer and auxiliary gas. Argon was used as the collision gas. The same HPLC conditions including the column, elution program and flow-rate were applied for HPLC-ESI-MS analysis. A portion of the column effluent (0.2 mL min−1) was delivered into the ion source of mass spectrometry after a microsplit. The conditions of the ESI source were as follows: drying gas (N2) flow rate, 9.0 L min−1; drying gas temperature, 350°C; nebulizer, 35 psig; capillary voltage, 5000 V; scan spectra from m/z 300 to 1500.
2.6. Data fingerprint analysis
The data analysis was performed by the software named the Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (Version 2004A) according to the recommendation made from State Food and Drug Administration (SFDA) of China and mainly applied in the similarity study of chromatographic and spectral patterns. In this study, this software was used to synchronize the chromatographic peaks and to calculate the correlation coefficients between entire chromatographic profiles, as well as to compute and generate the mean chromatogram as a representative standard fingerprint chromatogram from a group of chromatograms. The similarities of different chromatographic patterns were analyzed among tested samples.
2.7. HPLC method validation
After the optimum conditions had been established, the method validation was performed to ensure the validity of this newly developed fingerprinting method. The mixture standard solution was analyzed under the optimal conditions six times both in 1 day for intra-day variation and in a day on 3 successive days for inter-day variation to evaluate the precision and stability. In order to check the repeatability, five solutions made by the same sample (S3) were measured. All the validations were determined by calculating their relative standard deviations (RSD) of the peak areas and retention times of reference standards. The linearity calibration curves were evaluated with at least six different concentrations of standard solutions by plotting logarithm of the peak area versus logarithm of the concentration, and so was the liner regression analysis by the external standard method. Each concentration was analyzed in triplicate. The limits of detection (LOD) and limits of quantification (LOQ) were measured with the signal-to-noise ratio of 3 and 10 as criteria to evaluate the sensitivity, respectively. The recovery of this method was using the standard addition method. This involved the addition of three different concentrations of known quantities of reference standard compounds to half the sample weight of DZW plant material (S4) in triplicate. The fortified samples were then extracted and analyzed as described before. The results were expressed as percentage recovery values. The data of the proposed method validation were listed in Tables 1–2. All results indicated that the method of HPLC fingerprint analysis was valid and satisfactory.
TABLE 1.
Precision, stability And Repeatability statistical results of the proposed method validation for the analytical method.
| Reference standards code | Intra-day (n=6) | Inter-day (n=6) | Repeatability (n=5, S4) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision |
Stability |
Precision |
Stability |
|
||||||
| (RSD of RT, %) | (RSD of PA, %) | (RSD of RT, %) | (RSD of PA, %) | (RSD of RT, %) | (RSD of PA, %) | (RSD of RT, %) | (RSD of PA, %) | (RSD of RT, %) | (RSD of PA, %) | |
| A | 0.21 | 3.21 | 0.17 | 2.53 | 0.19 | 2.51 | 0.11 | 2.66 | 0.13 | 2.56 |
| B | 0.38 | 3.03 | 0.24 | 3.64 | 0.27 | 2.72 | 0.19 | 3.37 | 0.22 | 2.67 |
| C | 0.24 | 3.48 | 0.16 | 2.67 | 0.18 | 3.22 | 0.23 | 2.55 | 0.20 | 2.79 |
| D | 0.25 | 3.62 | 0.23 | 3.55 | 0.11 | 2.73 | 0.27 | 2.83 | 0.16 | 2.77 |
| E | 0.27 | 2.84 | 0.19 | 3.77 | 0.24 | 2.69 | 0.14 | 3.09 | 0.18 | 3.03 |
| F | 0.24 | 2.76 | 0.11 | 2.61 | 0.26 | 2.97 | 0.17 | 2.59 | 0.12 | 2.98 |
| G | 0.13 | 2.59 | 0.20 | 2.73 | 0.25 | 3.56 | 0.18 | 3.45 | 0.11 | 3.66 |
| H | 0.21 | 1.98 | 0.26 | 3.69 | 0.23 | 3.66 | 0.26 | 3.67 | 0.24 | 3.01 |
| I | 0.19 | 1.79 | 0.22 | 2.54 | 0.15 | 2.76 | 0.15 | 2.87 | 0.26 | 3.28 |
| L | 0.17 | 2.01 | 0.10 | 3.06 | 0.22 | 3.77 | 0.09 | 3.39 | 0.21 | 3.56 |
| K | 0.18 | 2.32 | 0.18 | 2.98 | 0.09 | 2.83 | 0.22 | 3.62 | 0.19 | 2.99 |
| L | 0.14 | 2.46 | 0.09 | 3.66 | 0.21 | 3.52 | 0.11 | 2.99 | 0.23 | 3.77 |
Table 2.
Linearity, sensitivity, and recovery data for the analysis of the 12 reference compounds (Compounds A, B, and C by HPLC-MS/MS, while other reference compounds by HPLC-ELSD).
| Reference standard code | Amount in Sample (mg) | Add amount (mg) | Calculated amount (mg) | Mean recovery (mean ± SD, %) | Linear regression | sensitivity | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Linear regression | Correlation Coefficient(R2) | Linear range (μg) | LOD (μg, S/N=3) | LOQ (μg, S/N=10) | |||||
| A | 0.85 | 0.68 | 1.50 | ||||||
| 0.85 | 0.85 | 1.67 | 98.47 ± 2.14 | Y=1.8039X+6.2376 | 0.9989 | 0.325~5.2 | 4.22 × 10−2 | 7.67 × 10−2 | |
| 0.85 | 1.02 | 1.90 | |||||||
| B | 0.55 | 0.44 | 0.99 | ||||||
| 0.55 | 0.55 | 1.16 | 102.43 ± 1.78 | Y=1.4560X+4.7978 | 0.9984 | 0.213~3.4 | 3.89 × 10−2 | 8.04 × 10−2 | |
| 0.55 | 0.66 | 1.20 | |||||||
| C | 0.55 | 0.44 | 0.98 | ||||||
| 0.55 | 0.55 | 1.10 | 96.44 ± 1.97 | Y=0.8514X+6.1060 | 0.9980 | 0.35~5.6 | 3.94 × 10−2 | 8.13 × 10−2 | |
| 0.55 | 0.66 | 1.17 | |||||||
| D | 0.75 | 0.60 | 1.36 | ||||||
| 0.75 | 0.75 | 1.56 | 101.49 ± 1.28 | Y=1.7129X+4.2403 | 0.9993 | 0.225~3.6 | 4.12 × 10−2 | 7.97 × 10−2 | |
| 0.75 | 0.90 | 1.57 | |||||||
| E | 1.25 | 1.00 | 2.20 | ||||||
| 1.25 | 1.25 | 2.45 | 96.79 ± 1.24 | Y=1.3625X+5.3045 | 0.9995 | 0.263~4.2 | 4.32 × 10−2 | 7.84 × 10−2 | |
| 1.25 | 1.50 | 2.74 | |||||||
| F | 0.75 | 0.6 | 1.36 | ||||||
| 0.75 | 0.75 | 1.53 | 100.43 ± 1.79 | Y=1.7300X+6.1000 | 1.0000 | 0.2~3.2 | 3.69 × 10−2 | 8.55 × 10−2 | |
| 0.75 | 0.9 | 1.62 | |||||||
| G | 0.90 | 0.72 | 1.63 | ||||||
| 0.90 | 0.90 | 1.84 | 100.37 ± 1.98 | Y=1.9290X+5.8760 | 0.9990 | 0.188~3 | 4.01 × 10−2 | 8.62 × 10−2 | |
| 0.90 | 1.08 | 1.94 | |||||||
| H | 0.90 | 0.72 | 1.60 | ||||||
| 0.90 | 0.90 | 1.78 | 98.29 ± 1.57 | Y=1.8976X+5.3233 | 0.9992 | 0.25~4 | 3.90 × 10−2 | 8.13 × 10−2 | |
| 0.90 | 1.08 | 1.94 | |||||||
| I | 0.25 | 0.20 | 0.45 | ||||||
| 0.25 | 0.25 | 0.51 | 100.48 ± 1.33 | Y=1.6590X+5.7170 | 1.0000 | 0.238~3.8 | 4.52 × 10−2 | 7.90 × 10−2 | |
| 0.25 | 0.30 | 0.54 | |||||||
| J | 0.25 | 0.20 | 0.45 | ||||||
| 0.25 | 0.25 | 0.50 | 100.36 ± 1.84 | Y=1.2932X+5.4747 | 0.9990 | 0.333~2 | 3.85 × 10−2 | 8.01 × 10−2 | |
| 0.25 | 0.30 | 0.55 | |||||||
| K | 0.25 | 0.20 | 0.44 | ||||||
| 0.25 | 0.25 | 0.50 | 98.74 ± 1.26 | Y=1.5140X+5.6413 | 0.9991 | 0.138~2.2 | 4.19 × 10−2 | 7.74 × 10−2 | |
| 0.25 | 0.30 | 0.54 | |||||||
| L | 0.25 | 0.20 | 0.46 | ||||||
| 0.25 | 0.25 | 0.51 | 101.19 ± 1.46 | Y=0.5114X+6.5883 | 0.9990 | 0.15~2.4 | 3.17 × 10−2 | 8.32 × 10−2 | |
| 0.25 | 0.30 | 0.54 | |||||||
3. Results and discussion
3.1. HPLC fingerprint analysis of DZW
The above described HPLC-ELSD method was subsequently applied to analysis and quality evaluation of 20 batches of DZW which were collected from a variety of sources and conditions, including different naturally distributed areas, various growth environments, different harvesting times, etc. The results indicated that their chromatographic patterns were generally consistent although the absorbance intensity of peaks was different. The similarity of each chromatogram to their simulative mean chromatogram was 0.919 ± 0.022 (mean ± SD, n=20, Table 3). Our observation revealed that the chromatograms of different RS samples were associated with similar chemical components regardless of the morphological differences and variations in growth environments. In addition, structure determination of 12 reference compounds (Figure 2) was simultaneously accomplished. However, the reference compounds A, B and C displayed in Figure 3 failed to show clear separation with other sample peaks where their peaks were overlapped with the neighboring peaks. Therefore, quantification with the extracted ion chromatography (EIC) by HPLC-ESI-MS was applied to determine their structures not identified by HPLC-ELSD. The quantitative analytical results in Table 4 indicated that the variations of their contents existed, even in the samples from the same areas. Chinese medicine preparation with different pretreatment processes, manufacturing procedure, and dosage forms would be of different quality. Meanwhile, the content of bioactive markers was also affected by plant origins, sources, cultivated year, harvest time, geographical climate and environment. All of these could result in significant differences in quality of DZW.
Table 3.
The similarities of 20 sample chromatograms from Ankang country of Shannxi province of China.
| NO. | Year of Collection | Similaritiesa |
|---|---|---|
| S 1 | November, 2008 | 0.905 |
| S 2 | November, 2008 | 0.894 |
| S 3 | September, 2009 | 0.913 |
| S 4 | September, 2009 | 0.957 |
| S 5 | October, 2009 | 0.962 |
| S 6 | October, 2009 | 0.943 |
| S 7 | November, 2009 | 0.898 |
| S 8 | November, 2009 | 0.901 |
| S 9 | September, 2010 | 0.931 |
| S 10 | September, 2010 | 0.920 |
| S 11 | October, 2010 | 0.932 |
| S 12 | October, 2010 | 0.921 |
| S 13 | November, 2010 | 0.899 |
| S 14 | November, 2010 | 0.900 |
| S 15 | September, 2011 | 0.934 |
| S 16 | September, 2011 | 0.909 |
| S 17 | October, 2011 | 0.928 |
| S 18 | October, 2011 | 0.890 |
| S 19 | November, 2011 | 0.916 |
| S 20 | November, 2011 | 0.929 |
The reference fingerprint was developed with the median of all chromatograms
Figure 2.
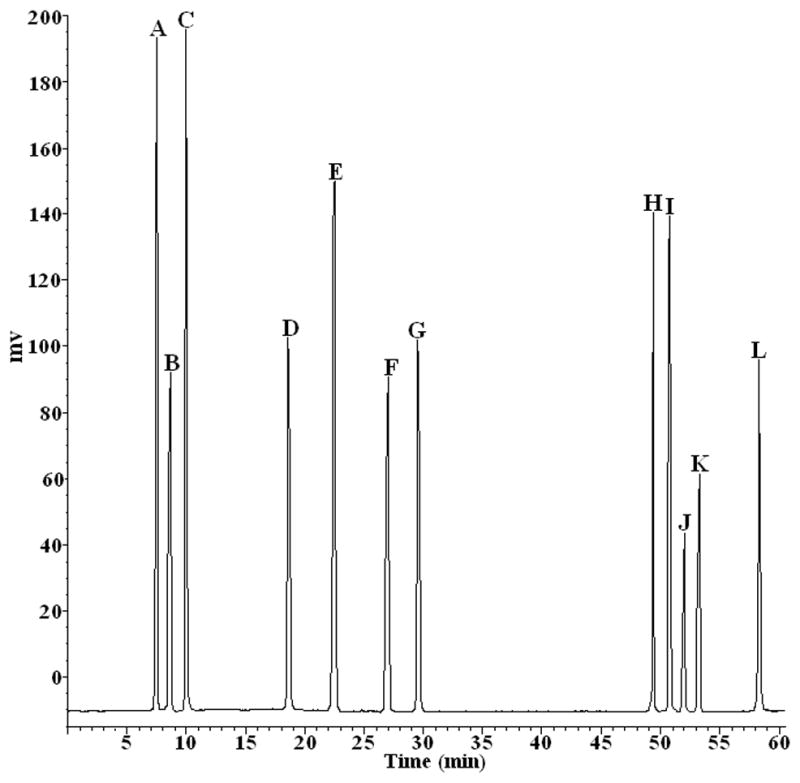
HPLC chromatograms of 12 reference steroid saponins. Conditions: column: Welchrom C18 column (250×4.6 mm, 5 μm); mobile phase: acetonitrile (Solvent A) water (Solvent B) as follows: initial 25% A; 0–5 min, 25% to 30% A; 5–20 min, 30% to 30% A; 20–35 min, 30% to 35% A; 35–45 min, 35% to 47% A; 45–47 min, 47% A to 70% A; 47–60 min, 70% to 70%. flow rate:1 mL min−1; Alltech 2000 ELSD condition: drift tube temperature: 90°C; gas flow rate 2.8 L min−1; column temperature: 25°C; injection volume: 10 μL. The names of these reference compounds corresponding to those given in Fig. 1.
Figure 3.
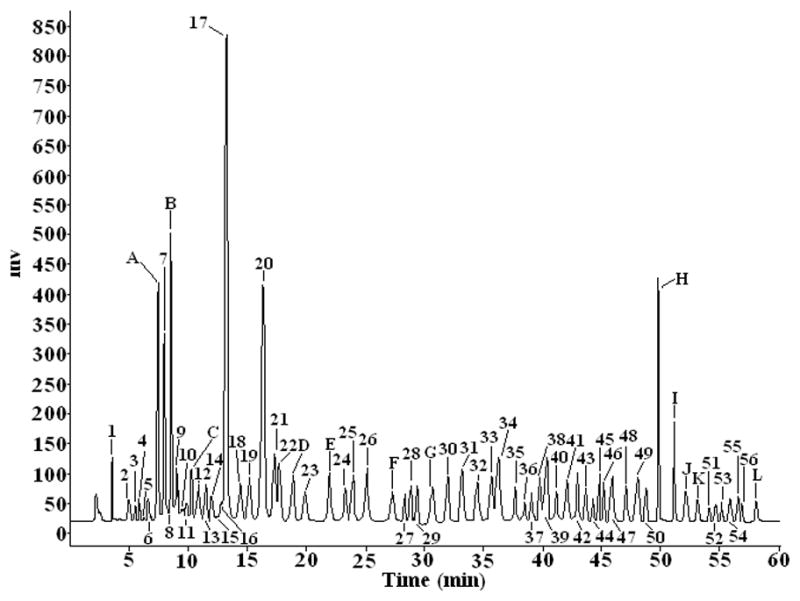
The common pattern HPLC fingerprint of steroid saponins from Dioscorea zingiberensis C. H. Wright based on 20 samples. The HPLC conditions as in Fig. 2. The peak numbers of compounds corresponding to those in Table 3.
Table 4.
Content values of 12 reference standards present in the crude extraction in 20 batches samples.
| Sample NO. | A | B | C | D | E | F | G | H | I | J | K | L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | mean ± SD (mg) | |
| S1 | 2.58 ± 0.25 | 2.72 ± 0.39 | 1.66 ± 0.38 | 2.82 ± 0.86 | 2.94 ± 1.03 | 2.23 ± 1.01 | 2.53 ± 0.69 | 2.75 ± 0.78 | 1.48 ± 0.65 | 1.55 ± 0.62 | 1.43 ± 0.54 | 1.31 ± 0.51 |
| S2 | 2.56 ± 0.31 | 2.64 ± 0.48 | 1.91 ± 0.43 | 2.69 ± 0.63 | 3.01 ± 1.26 | 2.22 ± 0.66 | 2.47 ± 0.67 | 2.91 ± 0.81 | 1.46 ± 0.64 | 1.71 ± 0.65 | 1.65 ± 0.58 | 1.56 ± 0.61 |
| S3 | 1.43 ± 0.36 | 1.78 ± 0.31 | 1.14 ± 0.28 | 1.77 ± 0.85 | 2.27 ± 0.99 | 1.03 ± 0.54 | 1.72 ± 0.52 | 1.70 ± 0.57 | 0.70 ± 0.49 | 0.67 ± 0.44 | 0.68 ± 0.39 | 0.50 ± 0.41 |
| S4 | 1.71 ± 0.43 | 1.09 ± 0.17 | 1.11 ± 0.27 | 1.56 ± 0.94 | 2.50 ± 0.95 | 1.40 ± 0.50 | 1.81 ± 0.54 | 1.79 ± 0.59 | 0.54 ± 0.46 | 0.56 ± 0.42 | 0.44 ± 0.34 | 0.50 ± 0.42 |
| S5 | 1.95 ± 0.35 | 2.15 ± 0.38 | 0.80 ± 0.21 | 1.94 ± 0.99 | 2.35 ± 0.91 | 1.83 ± 0.59 | 2.19 ± 0.62 | 2.00 ± 0.64 | 0.72 ± 0.49 | 0.70 ± 0.45 | 0.69 ± 0.39 | 0.48 ± 0.45 |
| S6 | 1.51 ± 0.25 | 2.49 ± 0.45 | 1.06 ± 0.27 | 2.32 ± 0.79 | 2.28 ± 0.90 | 1.55 ± 0.53 | 1.45 ± 0.47 | 1.93 ± 0.62 | 0.63 ± 0.48 | 0.87 ± 0.48 | 0.88 ± 0.43 | 0.77 ± 0.38 |
| S7 | 2.56 ± 0.47 | 2.23 ± 0.40 | 1.32 ± 0.32 | 2.76 ± 0.88 | 2.68 ± 0.99 | 2.21 ± 0.66 | 2.61 ± 0.70 | 2.27 ± 0.68 | 1.62 ± 0.67 | 1.61 ± 0.63 | 1.43 ± 0.54 | 1.38 ± 0.59 |
| S8 | 2.87 ± 0.53 | 2.40 ± 0.43 | 1.27 ± 0.31 | 2.26 ± 0.78 | 2.79 ± 1.01 | 2.19 ± 0.66 | 2.48 ± 0.67 | 2.15 ± 0.65 | 1.88 ± 0.73 | 1.56 ± 0.62 | 1.52 ± 0.57 | 1.41 ± 0.57 |
| S9 | 1.40 ± 0.31 | 1.39 ± 0.29 | 0.81 ± 0.22 | 1.01 ± 0.53 | 2.18 ± 0.88 | 0.82 ± 0.45 | 1.12 ± 0.40 | 1.35 ± 0.50 | 0.29 ± 0.41 | 0.43 ± 0.39 | 0.56 ± 0.36 | 0.30 ± 0.45 |
| S10 | 1.29 ± 0.23 | 1.58 ± 0.38 | 0.54 ± 0.16 | 2.03 ± 0.74 | 1.76 ± 0.79 | 0.87 ± 0.46 | 1.23 ± 0.43 | 1.24 ± 0.48 | 0.41 ± 0.43 | 0.47 ± 0.45 | 0.60 ± 0.36 | 0.57 ± 0.50 |
| S11 | 2.31 ± 0.43 | 2.36 ± 0.53 | 0.75 ± 0.55 | 2.05 ± 0.74 | 2.18 ± 0.88 | 1.51 ± 0.60 | 1.95 ± 0.57 | 2.36 ± 0.70 | 0.63 ± 0.48 | 1.23 ± 0.64 | 0.96 ± 0.42 | 1.24 ± 0.64 |
| S12 | 2.05 ± 0.46 | 1.96 ± 0.45 | 0.99 ± 0.78 | 2.26 ± 0.78 | 2.03 ± 0.85 | 1.63 ± 0.62 | 2.14 ± 0.61 | 2.09 ± 0.65 | 1.12 ± 0.57 | 1.22 ± 0.59 | 1.05 ± 0.46 | 1.18 ± 0.63 |
| S13 | 2.59 ± 0.57 | 2.86 ± 0.63 | 1.71 ± 0.64 | 2.57 ± 0.84 | 2.02 ± 0.84 | 2.35 ± 0.60 | 2.91 ± 0.76 | 2.73 ± 0.78 | 1.59 ± 0.67 | 1.87 ± 0.68 | 1.74 ± 0.60 | 1.65 ± 0.72 |
| S14 | 2.54 ± 0.47 | 2.77 ± 0.61 | 1.78 ± 0.66 | 2.54 ± 0.84 | 1.87 ± 0.81 | 2.14 ± 0.58 | 2.47 ± 0.67 | 1.82 ± 0.58 | 1.52 ± 0.66 | 1.98 ± 0.70 | 1.80 ± 0.61 | 1.67 ± 0.71 |
| S15 | 1.74 ± 0.31 | 1.67 ± 0.39 | 0.54 ± 0.71 | 1.32 ± 0.59 | 2.05 ± 0.85 | 1.20 ± 0.39 | 1.49 ± 0.47 | 2.08 ± 0.65 | 0.37 ± 0.42 | 0.32 ± 0.38 | 0.67 ± 0.38 | 0.44 ± 0.47 |
| S16 | 1.55 ± 0.24 | 1.52 ± 0.36 | 0.33 ± 0.77 | 1.75 ± 0.68 | 1.95 ± 0.83 | 1.19 ± 0.39 | 1.61 ± 0.50 | 1.67 ± 0.65 | 0.31 ± 0.41 | 0.62 ± 0.43 | 0.81 ± 0.41 | 0.55 ± 0.50 |
| S17 | 2.33 ± 0.40 | 1.93 ± 0.43 | 1.09 ± 0.62 | 2.64 ± 0.86 | 2.83 ± 1.01 | 1.62 ± 0.47 | 2.51 ± 0.69 | 2.32 ± 0.69 | 1.04 ± 0.56 | 1.51 ± 0.62 | 1.35 ± 0.52 | 1.14 ± 0.62 |
| S18 | 2.39 ± 0.41 | 2.84 ± 0.63 | 1.63 ± 0.75 | 2.58 ± 0.85 | 2.73 ± 0.99 | 1.58 ± 0.47 | 1.95 ± 0.57 | 2.08 ± 0.37 | 1.32 ± 0.61 | 1.43 ± 0.60 | 1.98 ± 0.64 | 1.03 ± 0.60 |
| S19 | 3.10 ± 0.55 | 2.91 ± 0.64 | 1.58 ± 0.57 | 1.97 ± 0.72 | 2.65 ± 0.97 | 1.60 ± 0.43 | 2.36 ± 0.65 | 2.61 ± 0.69 | 1.57 ± 0.64 | 1.22 ± 0.55 | 1.67 ± 0.58 | 1.37 ± 0.66 |
| S 20 | 2.90 ± 0.51 | 3.02 ± 0.66 | 1.42 ± 0.43 | 1.89 ± 0.71 | 2.49 ± 0.93 | 1.59 ± 0.44 | 2.45 ± 0.67 | 2.27 ± 0.67 | 1.39 ± 0.59 | 1.59 ± 0.63 | 1.43 ± 0.54 | 1.29 ± 0.65 |
3.2. Investigation of the fragmentation patterns of standard steroidal saponins
In order to qualitatively express the chemical constituents in the fingerprints of DZW, the on-line ESI-MS technique was used to identify their structures. Preliminarily, both positive and negative ion modes were tested to ionize the standards and the fragmentation patterns of the standards were summarized. In the positive ion mode the MS/MS spectra of the [M+H]+ ions also displayed ions arising from the eliminated sugar moieties substituted at C (3) or C (26) from the aglycone. This was followed by the characteristic fission of aglycone to form the diagnostic ions such as [Aglycone+H]+, [Aglycone+H-H2O]+, [Aglycone+H-144]+ and so on. This positive ion method was found to be more suitable for the identification of steroidal saponins than negative mode. Therefore, ESI in the positive mode was selected for the follow-up analysis.
In this experiment, we investigated the fragmentation patterns for these standard steroid saponins with different aglycone core rings displayed in Figure 4. They showed various rich characteristic fragment ions which were produced by the loss of branched glycoside chains or side chains, or by dehydration. They were classified into six types as summarized in Figure 1.
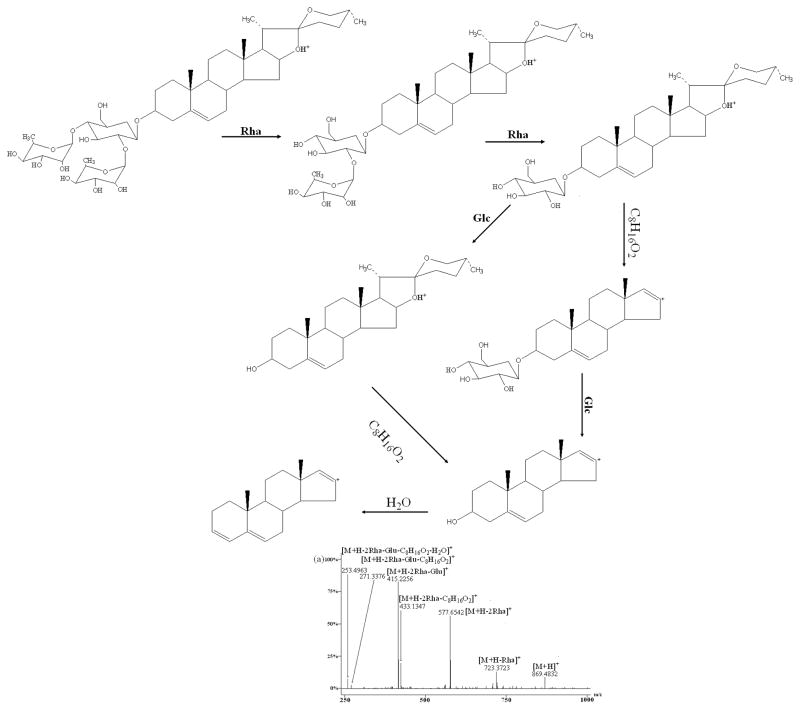
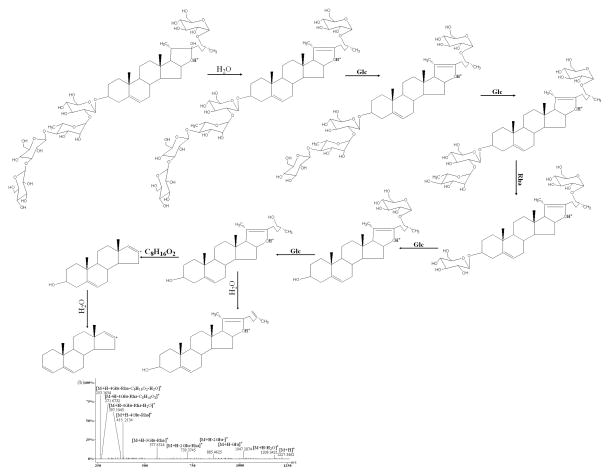
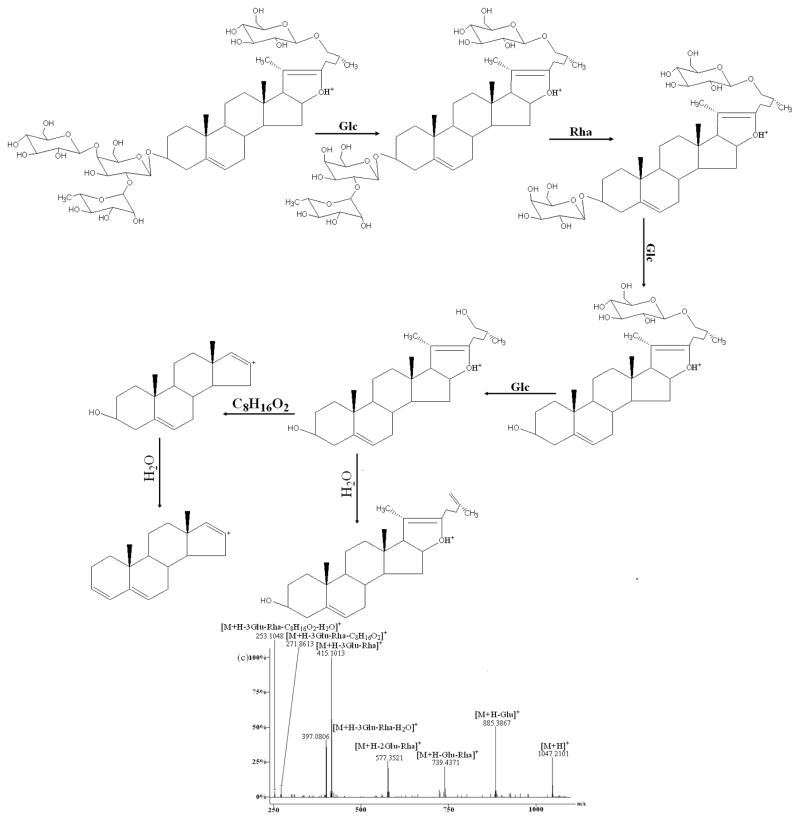
Figure 4.
Figure 4a–4f. The proposed MS and MS/MS spectra of reference standards with six type aglycone skeletons. (4a): Reference J with type I; (4b): Reference A with type II; (4c): Reference F with type III; (4d): Reference E with type IV; (4e): Reference D with type V; (4f): Reference L with type VI. The MS detection parameters as follows: drying gas (N2) flow rate, 9.0 L min−1; drying gas temperature, 350 °C; nebulizer, 35 psig; capillary voltage, 5000 V; scan spectra from m/z 100 to 1500.
3.2.1 Type I
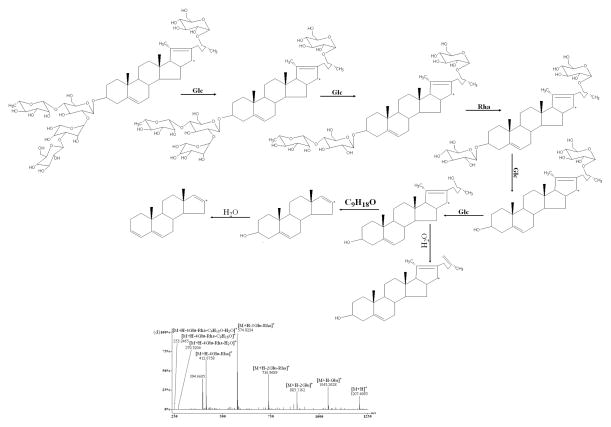
As the four reference standards (H, I, J, and K) shared the same aglycone skeleton and similar fragmentation pattern, dioscin was used as an example to discuss the fragmentation patterns for these reference standards in detail. In (+)ESI-MS/MS, observed were the fragment ions of the dioscin at m/z 723.3723, 577.6542, 433.1347, 415.2256, 271.3376 and 253.4963 (Fig. 4a) which were attributed to the loss of two rhamnosyls, one glucosyl, and one molecule of water from the protonated molecular ion [M+H]+ (m/z 869.4832). Loss of 144 Da from the fragment ion at m/z 577.6542 produced the fragment ion at m/z 433.1347. The fragment ions at m/z 271.3376 and 253.4963 may result from the consecutive loss of 144 and 18 Da from the fragment ion at m/z 415.2256. The elimination of 144 Da (fragment C8H16O2) might be produced by the cleavage of E-ring of the aglycone. The 18 Da units was derived from the loss of a molecule of water [20–24]. At the same time, the ions at m/z 433.13, 415.22, 271.33 and 253.49 from other three standards were also found as those from dioscin. Therefore, the fragment ions at m/z 433.13, 415.22, 271.33 and 253.49 can be considered as diagnostic ions for this type of spirostanol steroidal saponins.
3.2.2 Type II
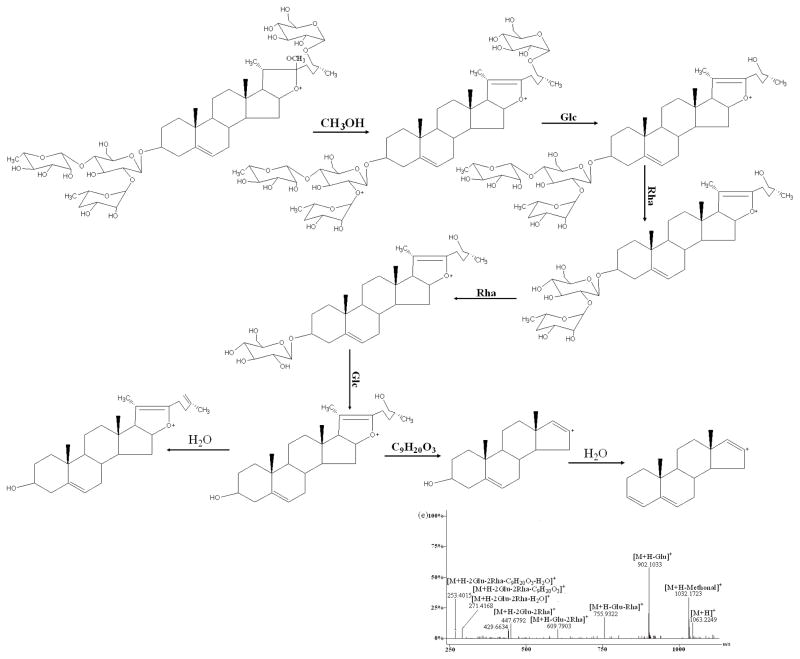
In (+)ESI-MS/MS, reference standard A produced ions at m/z 1209.3421, 1047.2874, 885.4625, 739.3745, 577.6314, 415.2134, 397.1045, 271.6732, 253.1654 (Fig. 4b) which were attributed to the sequential loss of one molecule of water, two glucosyls, one rhamnosyl, two glucosyls, one molecule of water, 144 Da (formula C8H16O2) and one molecule of water from the protonated molecular ion [M+H]+ (1227.3462). This fragmentation pattern that the [M+H]+ yielded the ion at the m/z [M+H-H2O]+ via loss of one molecule of water, was a common feature for this kind of furostanol steroidal saponins, suggesting the presence of a hydroxyl group at the C-22 position of the aglycone as reported [23, 24, 25–27]. However, the fragment ion at m/z 433 was not observed which was attributed to neutral loss of C8H16O2 (144 Da) directly from the fragment ion at m/z 577.6314. Zhu et al. [20, 24] concluded that the neutral loss of C8H16O2 directly from the molecular ion would occur in spirostanol saponins, while the sugar moiety present in C-26 position was preferentially eliminated in furostanol saponins. What’s more, B, C, and G produced almost the same ions except for the sugar moiety, so the fragment ions at m/z 577.63, 415.21, 397.10, 271.67, 253.16 together with the ion at m/z [M+H-H2O]+ can also be considered as the diagnostic ions for this type of furostanol steroidal saponins in which the sugar moiety was present in C-26 position. At the same time, the similar MS/MS behaviors of this type of saponins were also reported in the literature.
3.2.3 Type III IV V and VI
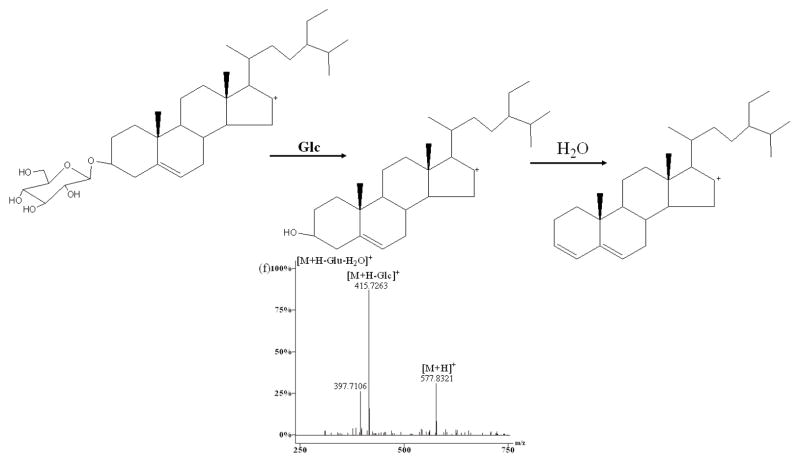
As shown in Figure 4, D, E, F, and L reference standards all showed that the [M+H]+ ion would lose sugar rings consecutively to form the ions of the type [Aglycone+H]+ in the (+)ESI-MS/MS mode. F produced main fragment ions at m/z 885.3867, 739.4371, 577.3521, 415.1013, 397.0860, 271.8613, and 253.1048 (Fig. 4c) which resulted from the consecutive loss of one glucosyl, one rhamnosyl, two glucosyls, one molecule of water, 144 Da and one molecule of water from the protonated molecular ion [M+H]+ (m/z 1047.2101). Comparing the difference between the aglycones of type II and type III, only one change exists in the E-ring. Therefore, it was reasonable to deduce that the fragment ion at m/z 270.7382 from m/z 415.1013 could be generated via elimination of the neutral formula C8H16O2 (144 Da). E produced ions at m/z 1045.2628, 883.1162, 736.9689, 574.8224, 412.6758, 394.6605, 270.5204, and 253.2465 (Fig. 4d) by the sequential loss of two glucosyls, one rhamnosyl, two glucosyls, one molecule of water (142 Da), and one molecule of water from the protonated molecular ion [M+H]+ (m/z 1207.4093). Considering the difference between the aglycones of type III and type IV, it might be concluded that the fragment ion at m/z 270.5204 from m/z 411.8157 could be produced by a loss of the neutral formula C9H18O (142 Da) which was 2 Da smaller than formula C8H16O2 (144 Da). D generated ions at m/z 1032.1723, 902.1033, 755.9322, 609.7903, 447.6792, 429.6634, 271.4168, and 253.4015 (Fig. 4e) resulted from the consecutive loss of one molecule of methanol, one glucosyl, two rhamnosyls, one glucosyl, one molecule of water, (176 Da) and one molecule of water from the protonated molecular ion [M+H]+ (m/z 1063.2249). Regarding the different structures between the aglycones of type II and type V, it might be deduced that the fragment ion at m/z 289.4397 originated from m/z 447.6792 could be produced by loss of the neutral formula C9H20O3 (176 Da) which was 14 Da larger than formula C8H16O2 (144 Da). However, two major fragmental ions at 415.7263 and 397.7106 resulted from the protonated molecular ion [M+H]+ (m/z 577.8321) (Fig. 4f) which attributed to loss of one glucosyl and gain of one molecule of water in compound L. Therefore, these ions originated from [Aglycone+H]+, in which all sugar moieties were lost, could be regarded as the diagnostic ions of these types. In order to elucidate structures of the saponins in this report, these ions may offer some information serving as reference ions.
3.3. Identification of the structures of unknown common characteristic peaks in the fingerprints
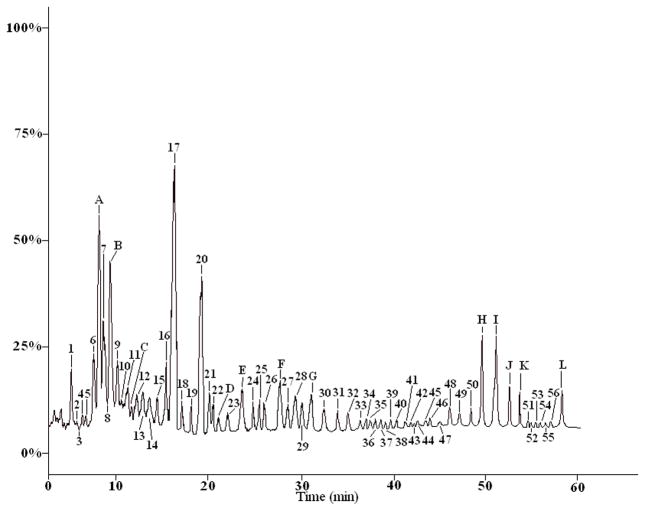
As shown in Figures 3 and 5, there existed a total of 68 common characteristic peaks in the HPLC-ELSD fingerprint and HPLC-MS chromatogram. Among those, twelve characteristic peaks were assigned by comparing their retention times and the MS and MS/MS data in the literature with those of the reference compounds in peaks A, B, C, D, E, F, G, H, I, J, K and L. Other unknown peaks corresponding to Table 5 were tentatively assigned based on their on-line MS fragmentation behaviors obtained by LC-MS/MS.
Figure 5.
The representative total ion chromatograms of steroid saponins from Dioscorea zingiberensis C. H. Wright by HPLC-ESI-Q/TOF. The chromatographic conditions and MS detection parameters were described in section 2.5.2. Peak numbers of compounds corresponding to those in Table 3.
Table 5.
The identification results of unknown steroidal saponins in the crude extracts from DZW by HPLC/Q-TOF-MS/MS.
| Peak NO. | RT (min) | Proposed molecular Formula | m/z Experiment | m/z Calculate | Error (mDa) | (+)ESI-MS/MS m/z [M+H]+ | Proposed fragmentation Pathway | Possible type of aglycone |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.544 | C57H92O27 | 1209.3274 | 1209.3242 | 3.2 | 1210.3353,1064.1941,902.0535, 739.9124, 577.7723, 433.5609, 415.5456,271.3342,253.3189 | [M+H-Rha-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Rha-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 2 | 5.517 | C51H82O22 | 1047.1809 | 1047.1836 | −2.7 | 1048.1888,886.0482,723.9076, 577.7664,433.5550,415.6258, 271.4144,253.3991 | [M+H-Glu-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 3 | 5.444 | C45H71O18 | 900.0338 | 900.0344 | −0.6 | 901.0417,738.9011,576.7605 414.6200,396.6047,271.4164, 253.4012 |
[M+H-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 4 | 5.873 | C51H82O23 | 1063.1841 | 1063.1830 | 1.1 | 1064.1920,902.0514,739.9108, 577.7702,433.5588,415.5435, 271.3320,253.3168 | [M+H-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 5 | 6.331 | C57H92O27 | 1209.3230 | 1209.3242 | −0.8 | 1210.3337,1048.1931,902.0519, 739.9113,577.7707,433.5593, 415.5440,271.3326,253.3173 | [M+H-Glu-Rha-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 6 | 6.720 | C57H92O27 | 1209.3230 | 1209.3242 | −0.8 | 1210.3309,1048.1903,886.0497, 739.9085,577.7679,433.5565, 415.5412,271.3298,253.3145 | [M+H-Glu-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 7 | 6.905 | C51H82O23 | 1063.1823 | 1063.1830 | −0.7 | 1064.1902,902.0496,739.9090, 577.7684,433.5570,415.5417, 271.3303,253.3150 | [M+H-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 8 | 7.420 | C51H84O22 | 1049.1979 | 1049.1995 | −1.6 | 1050.0079,887.8673,741.7261, 579.5855,435.3741,417.4449, 273.2335,255.2182 | [M+H-Glu-Rha-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-Glu-C8H16O2-Glu-H2O]+ | Type VII (Spirostanol) [Ref 24] |
| 9 | 7.967 | C51H81O22 | 1046.1766 | 1046.1756 | 1.0 | 1047.1845,901.0440,738.9033,
576.7621,414.6215,396.6062 271.4180,253.4028 |
[M+H-Rha-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 10 | 9.429 | C57H94O28 | 1227.3380 | 1227.3395 | −1.5 | 1228.3459,1210.3307,1048.1901, 902.0489,739.9083,577.7677, 415.6271,397.6118,271.4156, 253.4003 | [M+H-H2O-Glu-Rha-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 11 | 9.617 | C57H92O27 | 1209.3216 | 1209.3242 | −2.6 | 1210.3295,1048.1889,886.0483, 739.9071,577.7665,433.5551, 415.5398,271.3284,253.3131 | [M+H-Glu-Glu-Rha-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Rha-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 12 | 10.259 | C57H94O27 | 1211.3425 | 1211.3401 | 2.4 | 1212.3504,1050.2098,888.0692, 741.9280,579.7874,435.5760, 417.5607,273.3493,255.3340 | [M+H-Glu-Glu-Rha-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Rha-Glu-C8H16O2-Glu-H2O]+ | Type VII (Spirostanol) [Ref 24] |
| 13 | 10.811 | C39H62O13 | 738.9037 | 738.9018 | 1.9 | 739.2037,576.7010,414.6304, 396.6151,271.4269,253.4117 | [M+H-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 14 | 11.623 | C51H82O23 | 1063.1802 | 1063.1830 | −2.8 | 1064.1881,902.0475,739.9069, 577.7663,433.5549,415.5396, 271.3282,253.3129 | [M+H-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 15 | 11.938 | C39H62O13 | 738.9048 | 738.9018 | 3.0 | 739.9127,577.7721,433.5607, 415.5454,271.3340,253.3187 | [M+H-Glu-Glu-C8H16O2-H2O]+Or [M+H-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 16 | 12.397 | C51H84O23 | 1065.1994 | 1065.1989 | 0.5 | 1066.2073,1048.1921,886.0515,
739.9103,577.7697,415.6291 397.6118,271.4176,253.4023 |
[M+H-H2O-Glu-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 17 | 12.771 | C45H72O17 | 885.0426 | 885.0430 | −0.4 | 886.0505,723.9099,577.7687, 433.5573,415.5420,271.3306, 253.3153 | [M+H-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 18 | 13.216 | C51H82O22 | 1047.1841 | 1047.1836 | 0.5 | 1048.1893,886.0487,723.9081, 577.7669,433.5555,415.5402, 271.3288,253.3135 | [M+H-Glu-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 19 | 14.181 | C39H62O13 | 738.9059 | 738.9018 | 4.1 | 739.9138,577.7732,433.5618, 415.5465,271.3351,253.3198 | [M+H-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 20 | 14.882 | C45H74O19 | 919.0565 | 919.0577 | −1.2 | 920.0644,902.0492,739.9086, 577.7680,415.6274,397.6121, 271.4160,253.4006 | [M+H-H2O-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 21 | 16.338 | C51H86O23 | 1067.2123 | 1067.2147 | −2.4 | 1068.2202,1050.2050,904.0644, 742.9238,579.7826,417.6420, 399.6267,273.4305,255.4152 | [M+H-H2O-Rha-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type VIII (Furostanol) [Ref 24] |
| 22 | 17.295 | C57H92O28 | 1225.3221 | 1225.3236 | −1.5 | 1226.3300,1064.1894,902.0488, 739.9082,577.7676,433.5562, 415.5409,271.3295,253.3142 | [M+H-Glu-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 23 | 19.846 | C51H82O23 | 1063.1845 | 1063.1830 | 1.5 | 1064.1924,902.0518,739.9112, 577.7064,433.5592,415.5439, 271.3325,253.3172 | [M+H-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 24 | 21.317 | C45H72O17 | 885.0467 | 885.0430 | 3.7 | 886.0546,739.9134,577.7728, 433.5614,415.5461,271.3347, 253.3194 | [M+H-Rha-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Rha-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 25 | 22.920 | C51H82O22 | 1047.1839 | 1047.1836 | 0.3 | 1048.1918,902.0506,739.9100, 577.7694,433.5580,415.5427, 271.3313,253.3160 | [M+H-Rha-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Rha-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 26 | 23.873 | C51H82O23 | 1063.1868 | 1063.1830 | 3.8 | 1064.1947,902.0541,739.9135, 577.7729,433.5615,415.5462, 271.3348,253.3195 | [M+H-Glu-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 27 | 27.349 | C51H82O21 | 1031.1860 | 1031.1842 | 1.8 | 1032.1939,870.0533,723.9121, 577.7709,433.5595,415.5442, 271.3328,253.3175 | [M+H-Glu-Rha-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 28 | 30.690 | C51H82O21 | 1031.1818 | 1031.1842 | −2.4 | 1032.1897,886.0485,723.9079 577.7667,433.5553,415.5400, 271.3286,253.3133 |
[M+H-Rha-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Rha-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 29 | 31.197 | C51H81O22 | 1046.1761 | 1046.1756 | 0.5 | 1047.1840,901.0428,738.9022,
576.7616,414.6210,396.6057 271.4175,253.4015 |
[M+H-Rha-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 30 | 31.926 | C45H72O17 | 885.0409 | 885.0430 | −2.2 | 886.0488,723.9082,577.7670, 433.5556,415.5403,271.3289, 253.3136 | [M+H-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 31 | 33.236 | C63H102O31 | 1355.4660 | 1355.4654 | 0.6 | 1356.4739,1194.3333,1048.1921, 886.0515,723.9109,577.7697, 433.5583,415.5430,271.3316, 253.3163 | [M+H-Glu-Rha-Glu-Glu-Rha-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-Glu-Glu-Rha-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 32 | 34.945 | C57H92O27 | 1209.3261 | 1209.3242 | 1.8 | 1210.3340,1048.1934,902.0522, 739.9116,577.7710,433.5596, 415.5443,271.3329,253.3176 | [M+H-Glu-Rha-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Rha-Glu-Glu-C8H16O2-H2O]+ | Type I (Spirostanol) |
| 33 | 35.507 | C45H76O19 | 921.0751 | 921.0735 | 1.6 | 922.0830,904.0678,741.9272, 579.7860,417.6454,399.6301, 273.4339,255.4186 | [M+H-H2O-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type VIII (Furostanol) [Ref 24] |
| 34 | 35.847 | C45H74O18 | 903.0598 | 903.0583 | 1.5 | 904.0677,886.0525,739.9118, 577.7706,415.6300, 397.6147, 271.4185, 253.4032 | [M+H-H2O-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 35 | 37.618 | C45H72O18 | 901.0460 | 901.0423 | 3.7 | 902.0539,739.9133,577.7727, 433.56138,415.5460,271.3346, 253.3193 | [M+H-Glu-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 36 | 37.928 | C45H76O19 | 921.0713 | 921.0735 | −2.2 | 922.0792,904.0640,741.9234, 579.7828,417.6422,399.6269, 273.4307,255.4154 | [M+H-H2O-Glu-Glu-Glu-H2O-C8H16O2-H2O]+ | Type VIII (Furostanol) [Ref 24] |
| 37 | 38.559 | C45H71O17 | 884.0347 | 884.0350 | −0.3 | 885.0426,738.9020,576.7608, 414.6202,396.6049,271.4167, 253.4015 | [M+H-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 38 | 39.509 | C45H71O17 | 884.0346 | 884.0350 | −0.4 | 885.0425,738.9019,576.7607, 414.6201,396.6048,271.4166, 253.4014 | [M+H-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 39 | 39.987 | C39H62O13 | 738.9020 | 738.9018 | 0.2 | 739.9099,577.7693,433.5579, 415.5426,271.3312,253.3159 | [M+H-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 40 | 40.588 | C39H62O13 | 738.9067 | 738.9018 | 4.9 | 739.9146,577.7740,433.5626, 415.5473,271.3359,253.3206 | [M+H-Glu-Glu-C8H16O2-H2O]+ Or [M+H-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 41 | 41.126 | C33H52O8 | 576.7602 | 576.7612 | −1.0 | 577.7681,433.5567,415.5412, 271.3300,253.3147 | [M+H-Glu-C8H16O2-H2O]+ Or [M+H-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 42 | 41.686 | C39H64O14 | 756.9175 | 756.9171 | 0.4 | 757.9254,739.9102,577.7690, 415.6284,397.6131,271.4169, 253.4016 | [M+H-H2O-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 43 | 41.931 | C45H72O17 | 885.0436 | 885.0430 | 0.6 | 886.0515,739.9109,577.7697, 433.5583,415.5430,271.3316, 253.3163 | [M+H-Rha-Glu-Glu-C8H16O2-H2O]+ Or] [M+H-Rha-Glu-C8H16O2-Glu-H2O]+ | Type I (Spirostanol) |
| 44 | 42.441 | C46H76O19 | 933.0850 | 933.0842 | 0.8 | 934.0850,902.0431,771.9444, 609.8038,447.6632,429.6479, 271.4099,253.3946 | [M+H-Methanol-Glu-Glu-Glu-H2O-C9H20O3-H2O]+ | Type V (Furostanol) |
| 45 | 43.007 | C45H74O18 | 903.0577 | 903.0583 | −0.6 | 904.0656,886.0504,739.9092, 577.7686,415.6280,397.6127, 271.4165,253.4012 | [M+H-H2O-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 46 | 43.635 | C40H66O14 | 770.9423 | 770.9436 | −1.3 | 771.1423,739.1004,609.5954,
447.6387,429.6234,271.4854 253.4701 |
[M+H-Methanol-Glu-Glu-H2O-C9H20O3-H2O]+ | Type V (Furostanol) |
| 47 | 44.238 | C45H71O17 | 884.0364 | 884.0350 | 1.4 | 885.0443,738.9031,576.7625, 414.6219,396.6066,271.4184, 253.4032 | [M+H-Rha-Glu-Glu-H2O-C8H16O2-H2O]+ | Type III (Furostanol) |
| 48 | 48.800 | C46H73O16 | 882.0639 | 882.0622 | 1.7 | 883.0718,736.9312,574.7900, 412.6494,394.6341,271.4188, 253.4035 | [M+H-Rha-Glu-Glu-H2O-C9H18O-H2O]+ | Type IV (Furostanol) |
| 49 | 49.391 | C46H73O16 | 882.0636 | 882.0622 | 1.4 | 883.0715,736.9303,574.7891, 412.6485,394.6332,271.4179, 253.4026 | [M+H-Rha-Glu-Glu-H2O-C9H18O-H2O]+ | Type IV (Furostanol) |
| 50 | 49.823 | C40H66O14 | 770.9457 | 770.9436 | 2.1 | 771.1051,739.1231,609.5632, 447.6803,429.6650,271.4270, 253.4117 | [M+H-Methanol-Glu-Glu-H2O-C9H20O3-H2O]+ | Type V (Furostanol) |
| 51 | 54.117 | C39H64O14 | 756.9164 | 756.9171 | −0.7 | 757.9243,739.9091,577.7685, 415.6279,397.6126,271.4164, 253.4011 | [M+H-H2O-Glu-Glu-H2O-C8H16O2-H2O]+ | Type II (Furostanol) |
| 52 | 54.861 | C39H66O13 | 758.9318 | 758.9329 | −1.1 | 759.9397,741.9245,579.7839, 417.6431,399.6280,273.4316, 255.4163 | [M+H-H2O-Glu-Glu-H2O-C8H16O2-H2O]+ | Type VIII (Furostanol) [Ref 24] |
| 53 | 55.183 | C41H70O11 | 738.9866 | 738.9879 | −1.3 | 739.1866,577.8763,415.2328, 397.3749 | [M+H-Glu-Glu-H2O]+ | Type VI (daucosterol) |
| 54 | 56.021 | C41H70O11 | 738.9860 | 738.9879 | −1.9 | 739.0860,577.9111,415.6065, 397.4489 | [M+H-Glu-Glu-H2O]+ | Type VI (daucosterol) |
| 55 | 57.103 | C40H63O12 | 735.9230 | 735.9210 | 2.0 | 736.9309,574.7903,412.6497, 394.6344,271.4191,253.4038 | [M+H-Glu-Glu-H2O-H2O-C9H18O-H2O]+ | Type IV (Furostanol) |
| 56 | 57.630 | C41H70O10 | 723.4411 | 723.4406 | 0.5 | 724.1411,577.2871,415.6325, 397.6016 | [M+H-Rha-Glu-H2O]+ | Type VI (daucosterol) |
3.3.1. Characterization of peaks 1–2, 4–7, 11, 14–15, 17–19, 22–28, 30–32, 35, 39–41, and 43
Based on the characteristic ions (at m/z 577.7, 433.5, 415.5, 271.3, and 253.3), compounds of these peaks showed the same fragmentation behaviors as reference compound H. Thus, they were considered sharing the same aglycone skeleton of Type I saponins.
Peaks 1, 5, 6, 11, and 32 shared the same protonated molecular ion [M+H]+ (m/z 1210.3) and molecular formula. Through the major fragments, there were four glucosyls and one rhamnosyl in these five compounds. Peaks 5 and 32 had the same five main fragments (at m/z 1048.19, 902.05, 739.91, 577.77, and 415.54) attributed to the consecutive loss of one glucosyl, one rhamnosyl and three glucosyls from the protonated molecular ion [M+H]+ (m/z 1210.33). Hence they were characterized to be a pair of structural isomers whose difference existed in the sequence of sugar units of the saponins and location variation of the glycosidic bond. Peaks 6 and 11 showed another five main fragments (at m/z 1048.19, 886.04, 739.90, 577.76, and 415.54) via sequence of two glucosyls, one rhamnosyl and two glucosyls from the protonated molecular ion [M+H]+ (m/z 1210.33). So, they were also considered to be a pair of structural isomers whose difference existed in the sequence of sugar units of the saponins and location variation of the glycosidic bond. While, seen from the fragments of peak 1 formed by the loss of one rhamnosyl and four glucosyls, they had different sugar units linkage.
Peaks 2, 18, and 25 possessed the identical molecular formula. Their fragmental patterns indicated that there were one rhamnosyl and three glucosyls in their structures. Peaks 2 and 18 displayed similar fragmental behavior with the reference compound zingiberensis saponin, and they were tentatively identified as a pair of structural isomers of zingiberensis saponin with the same sugar residues [24]. Although having the same sugar groups with peaks 2 and 8, peak 25 showed five different ion behaviors by generating the protonated molecular ion [M+H]+. Considering these fragments, it was tentatively identified as 3-O-β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)-[α-L-rhamn opyranosyl-(1→4)]-β-D-glucopyranoside which had been isolated from Dioscorea parviflora, the same genus with DZW [28].
Peaks 4, 7, 14, 23, and 26 generated four major fragmentation ions (at m/z 902.05, 739.91, 577.7, and 415.54) from the protonated molecular ion [M+H]+ (m/z 1064.19) suggesting that a saccharide chain including four glucosyls connecting to the hydroxyl group at C-3 position of the aglycone was present in their structures. Consequently, they were tentatively identified as structural isomers of each other with the same sugar residues. Their structural difference could be deduced to the different sugar units linkage and location variation of the glycosidic bond.
The major ions of peaks 27 (at m/z 870.0533, 723.9121, 577.7790, and 415.5442) and 28 (at m/z 886.0485, 723.9079, 577.7667, 415.5400) in the (+)ESI MS/MS mode could be deduced to represent two rhamnosyls and two glucosyls groups in their structures. The distinction between these two compounds existed in the sequence of sugar moieties linkage.
According to the main ions by the loss of certain fragments from the protonated molecular ion [M+H]+ (m/z 886.05), peaks 17, 24, 30, and 43 suggested the presence of one rhamnosyl and two glucosyls in their structures. Three major fragmental ion behaviors (at m/z 723.90, 577.76, and 415.54) were observed in peaks 17 and 30, while three different fragmental ions (at m/z 739.91, 577.77, and 415.54) were observed in peaks 24 and 43. Therefore, these four compounds can form two groups, each being tentatively identified as structural isomers of the two reference compounds of zingiberensis new saponin and deltonin, respectively, whose structural difference could be deduced to be the sequence of sugar units in the aglycone [24].
Two main fragments (at m/z 577.7 and 415.5) were obtained from protonated molecular ion [M+H]+ (m/z 739.9) of peaks 15, 19, 39, and 40 suggesting the presence of two glucosyls group in these four compounds. For this reason, they were tentatively identified as structural isomers of the diosgenin diglucoside that had the same molecular formula and ion behavior patterns [24].
Peak 35 yielded the main fragments (at m/z 739.9133, 577.7727, and 415.5460) resulted from the consecutive loss of three glucosyls from the protonated molecular ion [M+H]+ (m/z 902.0539). It was tentatively identified as a structural isomer of the diosgenin triglucoside [24]. Peak 41 whose ions chiefly included two fragments at m/z 577.7681 and 415.5412, had only one glucosyl residue in its structure. As it had only one sugar moiety, it was identified as the same structure with trillin. Peak 22 produced main fragments (m/z 1064.7894, 902.0488, 739.9082, 577.7676, and 415.5409) attributed to consecutive loss of five glucosyls. Peak 31 showed seven fragments resulted from the loss of two rhamnosyls and three glucosyls. To our knowledge, steroid saponin with six sugar moieties was reported for the first time in Dioscorea zingiberensis C. H. Wright. Since the accurate structure of this unknown compound couldn’t be elucidated by the MS data alone, further investigation must be carried out.
3.3.2. Characterization of peaks 10, 16, 20, 34, 42, 45, and 51
According to the characteristic ions (at m/z 577.7, 415.6, 397.61, 271.41, and 253) and the main fragmentation pattern [M+H-H2O]+ as the similar ion behaviors with reference compound A, these peaks were considered to have the same aglycone skeleton of Type II whose structures shared a hydroxy group at C-22, and were assigned as furostanol saponins.
Peak 10 was dominated by the main fragment ion at 1210.3307, 1048.1901, 902.0489, 739.9083, 577.7677, and 415.6271 which may be resulted from the consecutive loss of one molecule of water, one glucosyl, one rhamnosyl and three glucosyls from the protonated molecular ion [M+H]+ (m/z 1228.3459) in (+)ESI-MS. According to the fragmentation patterns, reference data and comparison with known compounds in the literature, peak 10 was tentatively identified as the isomer of parvifloside which had been isolated from D. zingiberensis [24] and Dioscorea parviflora [28]. The difference instructure between these two isomers could be in the sequence of sugar units in the aglycone.
As shown in Table 5, peak 16 readily gave five major fragmentation ions (m/z at 1048.1921, 886.0515, 739.9103, 577.7697, and 415.6291) from the protonated molecular ion [M+H]+ (m/z at 1066.2073) via consecutive loss of one molecule of water, one glucosyl, one rhamnosyl and two glucosyls. Based on the MS/MS data and the known structure reported in the literature, this compound was tentatively identified as the isomer of deltoside or protodeltonin which had been isolated from Dioscorea parviflora and D. zingiberensis [28], respectively.
In the (+)ESI-MS mode, peak 20 showed four fragments (at m/z 902.0492, 739.9086, 577.7608, and 415.6274) by the sequential loss of one molecule of water and three glucosyls. Considering the fragmental behavior and reported structure in the literature, it was tentatively identified as an isomer of 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22ξ, 26-triol-3-O-β-D-glucopyranosyl-(1→4)-β-D-glucopyranoside [24], whose structural difference was only at the location of glycosidic bond, probably C1–C4, C1–C3, or C1–C2, between these two glucosyls attaching to C-3 position of aglycone.
Peaks 34 and 45 shared the same formula and protonated molecular ion [M+H]+ (at m/z 904.06), and showed the same four main fragmentation patterns which were attributed to successive loss of one molecule of water, one rhamnosyl, and two glucosyls. Comparing with ions and structure reported in the literature, these two compounds were tentatively characterized as an isomer of 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22α, 26-triol-3-O-[α-L-rhamnopyranosyl-(1→4)-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22ξ, 26-triol-3-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranoside (Protobioside) [29] whose structural difference existed in the sequence of sugar linkage between rhamnosyl and glucosyl connecting to the hydroxyl group at C-3 position of the aglycone.
Peaks 42 and 51 showed three primary fragmentation ions (at m/z 739.9, 577.76, 415.62) originated from the protonated molecular ion [M+H]+ (at m/z 757.92) suggesting the presense of two glucosyls in the aglycone. As the aglycone was furostanol type, one glucosyl was connected to the hydroxyl groups at C-3, while the other connected to the hydroxyl groups at C-26. Therefore, they were tentatively characterized as 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-3β, 22ξ, 26-triol-3-O-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-(25S)-furost-5-en-3β, 22ξ, 26-triol-3-O-β-D-glucopyranoside. The variation between them could attribute to the different stereoscopic position of C-25 methyl.
3.3.3. Characterization of peaks 3, 9, 13, 29, 37–38, and 47
These compounds showed the same fragmental diagnostic ions (at m/z 576.7, 414.62, 396.6, 271.4, and 253.4) as reference F. Thus, they may be sharing the same aglycone skeleton as the type III saponins.
Peaks 37, 38, and 47 generated three major fragment ions at m/z 738.90, 576.76, and 414.62 from the protonated molecular ion [M+H]+ (at m/z 885.04) and could be deduced to represent the loss of one rhamnosyl and two glucosyls in their structures. Taking into account the characteristic of furostanol saponins and comparison with the data in the literature, these compounds were tentatively designated as 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[α-L-rhamnopyranosyl-(1→4)]-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[α-L-rhamnopyranosyl-(1→3)]-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[α-L-rhamnopyranosyl-(1→2)]-β-D-glucopyranoside whose structural difference was only present in the location of the glycosidic bond between the rhamnosyl and glucosyl connecting to the hydroxyl group at C-3 position of the aglycone [20, 24, 30].
Two peaks, namely 9 and 29, had the same formula and four similar fragments (at m/z 901.04, 738.90, 576.76, and 414.621) resulted from the consecutive loss of one rhamnosyl and three glucosyls from the protonated molecular ion [M+H]+ (m/z 1047.18). Therefore, they were tentatively characterized to be a pair of isomers whose structural difference existed in the sequence of sugar units of the saponins.
Peak 3 generated three fragmental ions (at m/z 738.9011, 576.7605, and 414.6200) via consecutive loss of three glucosyls from the protonated molecular ion [M+H]+ (m/z 901.0417). Considering the characteristic of this type of aglycone and linkage between sugar moieties, this compound was tentatively regarded as 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[β-D-glucopyranosyl-(1→4)]-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[β-D-glucopyranosyl-(1→3)]-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-[β-D-glucopyranosyl-(1→2)]-β-D-glucopyranoside whose structural difference was only present in the location of glycosidic bond between two glucosyls connecting to the hydroxyl group at C-3 position of the aglycone.
Peak 13 produced two fragment ions at 576.7010 and 414.6304 resulted from the sequential loss of two glucosyls from the protonated molecular ion [M+H]+ (m/z 739.2037). So, it was tentatively identified as 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-β-D-glucopyranoside.
3.3.4. Characterization of peaks 48, 49 and 55
Considering the similar fragment ions (at m/z 574.7, 412.64, 394.63, 271.41, and 253.40) resulted from the loss of certain moieties from the protonated molecular ion [M+H]+ as the reference E, it was reasonable to deduce that these peaks shared the same aglycone skeleton as the type IV saponins.
Peaks 48 and 49 had the same protonated molecular ion [M+H]+ (at m/z 883.07) and three major fragments (at m/z 736.93, 574.7, and 412.64) attributed to the sequential loss of one rhamnosyl and two glucosyls. They were characterized to be a pair of isomers whose structural difference existed in the sequence of sugar units of the saponins.
Peak 55 showed two fragment ions (at m/z 574.7903 and 412.6497) resulted from the protonated molecular ion suggesting that one glucosyl was connected to the hydroxyl groups at C-3 and the other to the hydroxyl groups at C-26 in its structure. It was tentatively identified as 26-O-β-D-glucopyranosyl-3β, 26-diol-25(R)-Δ5, 20(22)-dien-furost-3-O-β-D-glucopyranoside.
3.3.5. Characterization of peaks 44, 46 and 50
These three peaks generated several major and diagnostic fragment ions (at m/z 447.6, 429.6, 271.4, and 253.4) resulted from the consecutive loss of neutral formula C9H18O2 and one molecular of water with the same behavior as reference D. Therefore, it could be considered that they possessed the same aglycone skeleton as the type V saponins whose structures shared a methoxy group at C-22, hence they were designated as furostanol saponins.
Peaks 46 and 50 generated the identical formula and three main similar fragment ions (at m/z 739.1, 609.5, and 447.6) resulted from the same protonated molecular ion suggesting the presence of two glucosyls in their structures (at m/z 771.1). As the aglycone was of furostanol type, there might be one glucosyl connected to the hydroxyl groups at C-3, while the other connected to the hydroxyl groups at C-26. Because of this, these two compounds were characterized as a pair of isomers and tentatively designated as 26-O-β-D-glucopyranosyl-(25R)-furost-5-en-22-methyl-3β, 26-diol-3-O-β-D-glucopyranoside or 26-O-β-D-glucopyranosyl-(25S)-furost-5-en-22-methyl-3β, 26-diol-3-O-β-D-glucopyranoside. The difference between these compounds could be derived from the different stereoscopic positions of C-25 methyl.
Three primary fragment ions (at m/z 771.9444, 609.8038, and 447.6632) were generated from the protonated molecular ion (at m/z 934.0850) in peak 44. This ion behavior suggested the presence of three glucosyls in its structure. According to the report of furostanol aglycone type, a glucosyl was connected to the hydroxyl group at the C-26 position and a saccharide chain including two glucosyls was connected to the hydroxyl group at C-3 position of the aglycone.
Based on the data in the literature, the compounds with the aglycone of type VI were also found in Dioscorea zingiberensis C. H. Wright for the first time. As we all know that the MS data alone couldn’t supply enough information to determine the exact structure, further research, for instance NMR, will be requisite.
3.3.6. Characterization of peaks 53, 54 and 56
The characteristic ions and similar fragmentation patterns (at m/z 577, 415, and 397) as reference L indicated that these peaks had the same aglycone skeleton as the type VI saponins.
Peaks 53 and 54 had the same protonated molecular ion [M+H]+ (at m/z 739) and three major fragments (at m/z 577, 415, and 397) resulted from the consecutive loss of two glucosyls. They were considered to be a pair of isomers whose structural difference existed in the sequence of sugar linkage between two glucosyls connected to the hydroxyl groups at C-3 in aglycone of type VI. Comparing the formulas between peaks 53, 54 and 56, their difference only existed in the number of oxygen atoms which could be deduced to represent one rhamnosyl and one glucosyl in peak 56. To our knowledge, these three compounds with the aglycone of type VI were also found in Dioscorea zingiberensis C. H. Wright for the first time, and the precise structures of these new saponins can’t be determined by MS data alone. Therefore, further investigation is required.
3.3.7. Characterization of peaks 8 and 12
Comparing the data reported in the literature [23] and the readily diagnostic ions (at m/z 579, 435, 417, 273, and 255), these two compounds could be deduced to possess the same aglycone of type VII.
The major ions (at m/z 887.8673, 741.7261, 579.5855, 435.3741 for peak 8; at m/z 1050.2098, 888.0692, 741.9208, 579.7874, and 435.5760 for peak 12) originated from the protonated molecular ion [M+H]+ (at m/z 1050.0079 and 1212.3504, respectively) may have lost one glucosyl, one rhamnosyl, and two glucosyls in the structure forming peak 8, while the loss of two glucosyls, one rhamnosyl, and two glucosyls resulted to form peak 12. In order to elucidate the definite structures, further investigation must be needed.
3.3.8. Characterization of peaks 21, 33, 36, and 52
Comparing the data reported in the literature [23], the characteristic ions (at m/z 579.78, 417.64, 399.6, 273.43, and 255.41) and the primary fragmental pattern [M+H-H2O]+ whose structures share a hydroxy group at C-22 elucidated as furostanol saponins, they may share the skeleton aglycone as the type VIII.
Peaks 33 and 36 showed the same protonated molecular ion [M+H]+ (at m/z 922.0) and four main fragments (at m/z 904.06, 741.92, 579.78, and 417.64) by the consecutive loss of three glucosyls. They were considered to be the isomers of 26-O-β-D-glucopyranosyl-furost-3β, 22, 26-triol-3-O-β-D-glucopyranosyl(1→2)-β-D-glucopyranoside [31, 32] whose structural difference existed in the sequence of glycosidic bond of sugar moieties.
The main fragmental ions (at m/z 1050.2050, 904.0644, 742.9238, 579.7826, and 417.6420 for peak 21; at m/z 741.9245, 579.7839, and 417.6431 for peak 52) resulted from sequential loss of one molecule of water and four glucosyls formed peak 21, whereas the loss of one molecule of water and two glucosyls formed peak 52, both from the protonated molecular ion [M+H]+. Comparison with the structure reported in the literature, peak 52 was tentatively identified as 26-O-β-D-glucopyranosyl-furost-3β, 22, 26-triol-3-O-β-D-glucopyranoside in which one glucosyl was attached to the C-3 position of aglycone by a hydroxyl group, whereas the other was connected to the C-26 position of aglycone by a hydroxyl group.
4. Conclusions
An HPLC-ELSD fingerprinting and an HPLC-ESI-Q/TOF methods were simultaneously established for comprehensive analysis of multiple steroid saponins in Dioscorea zingiberensis C. H. Wright with the advantage of avoiding the tedious purification of compounds from the crude extracts. As a result, 68 common characteristic peaks including 22 new steroid saponins with eight aglycone skeletons in the fingerprint were detected. However, it should be noted that like other mass spectrometric methods, this analytical method still has some limitations in the linkage position of sugar moieties. To determine the definite structures of the unknown compounds, further investigation, for example NMR experiment, is requisite. Overall, this research sets a good example for quality evaluation and the consistency check of DZW collected from different sources, and structural characterization of steroid saponin constituents in the crude extracts. Fingerprinting combined with the on-line HPLC/MS technique, could become a powerful tool in the quality control of TCMs and chemical constituents analysis.
Acknowledgments
The authors thank Prof. Zhongfu Wang of the Key Laboratory of Resource Biology and Biotechnology in western China and Juan Gao for assistance in ESI-MS experiments.
References
- 1.Yan SK, Xin WF, Luo GA, Wang YM, Cheng YY. An approach to develop two-dimensional fingerprint for the quality control of Qingkailing injection by high-performance liquid chromatography with diode array detection. J Chromatogr A. 2005;1090:90–97. doi: 10.1016/j.chroma.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 2.Yang LW, Wu DH, Tang X, Peng W, Wang XR, Ma Y, Su WW. Fingerprint quality control of Tianjihuang by high-performance liquid chromatography-photodiode array detection. J Chromatogr A. 2005;1070:35–42. doi: 10.1016/j.chroma.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 3.Xu SJ, Yang L, Tian RT, Wang ZT, Liu ZJ, Xie PS, Feng QR. Species differentiation and quality assessment of Radix Paeoniae Rubra (Chi-shao) by means of high-performance liquid chromatographic fingerprint. J Chromatogr A. 2009;1216:2163–2168. doi: 10.1016/j.chroma.2008.04.064. [DOI] [PubMed] [Google Scholar]
- 4.Chang YX, Ding XP, Qi J, Cao J, Kang LY, Zhu DN, Zhang BL, Yu BY. The antioxidant-activity-integrated fingerprint: An advantageous tool for the evaluation of quality of herbal medicines. J Chromatogr A. 2008;1208:76–82. doi: 10.1016/j.chroma.2008.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Zhang H, Xiao W, Yong ZP, Bai N. High-performance liquid chromatographic fingerprint analysis for different origins of sea buckthorn berries. J Chromatogr A. 2007;1154:250–259. doi: 10.1016/j.chroma.2007.03.097. [DOI] [PubMed] [Google Scholar]
- 6.Li SL, Shen H, Zhu LY, Xu J, Jia XB, Zhang HM, Lin G, Cai H, Cai BC, Chen SL, Xu HX. Ultra-high-performance liquid chromatography-quadrupole/time of flight mass spectrometry based chemical profiling approach to rapidly reveal chemical transformation of sulfur-fumigated medicinal herbs, a case study on white ginseng. J Chromatogr A. 2012;1231:31–45. doi: 10.1016/j.chroma.2012.01.083. [DOI] [PubMed] [Google Scholar]
- 7.Qi LW, Wang HY, Zhang H, Wang CZ, Li P, Yuan CS. Diagnostic ion filtering to characterize ginseng saponins by rapid liquid chromatography with time-of-flight mass spectrometry. J Chromatogr A. 2012;1230:93–99. doi: 10.1016/j.chroma.2012.01.079. [DOI] [PubMed] [Google Scholar]
- 8.Liang X, Zhang L, Zhang X, Dai WX, Li HY, Hu LW, Liu H, Su J, Zhang WD. Qualitative and quantitative analysis of traditional Chinese medicine Niu Huang Jie Du Pill using ultra performance liquid chromatography coupled with tunable UV detector and rapid resolution liquid chromatography coupled with time-of-flight tandem mass spectrometry. J Pharm Biomed Anal. 2010;51:565–571. doi: 10.1016/j.jpba.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Li SP, Wang YT, Chen XJ, Tu PF. Differentiation of Herba Cistanches by fingerprint with high-performance liquid chromatography-diode array detection-mass spectrometry. J Chromatogr A. 2009;1216:2156–2162. doi: 10.1016/j.chroma.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 10.Wang YH, Qiu C, Wang DW, Hu ZF, Yu BY, Zhu DN. Identification of multiple constituents in the traditional Chinese medicine formula Sheng-Mai San and rat plasma after oral administration by HPLC-DAD-MS/MS. J Pharm Biomed Anal. 2011;54:1110–1127. doi: 10.1016/j.jpba.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Lu GH, Chan K, Liang YZ, Leung K, Chan CL, Jiang ZH, Zhao ZZ. Development of high-performance liquid chromatographic fingerprints for distinguishing Chinese Angelica from related umbelliferae herbs. J Chromatogr A. 2005;1073:383–392. doi: 10.1016/j.chroma.2004.11.080. [DOI] [PubMed] [Google Scholar]
- 12.Pellati F, Orlandini G, Benvenuti S. Simultaneous metabolite fingerprinting of hydrophilic and lipophilic compounds in Echinacea pallida by high-performance liquid chromatography with diode array and electrospray ionization-mass spectrometry detection. J Chromatogr A. 2012;1242:43–58. doi: 10.1016/j.chroma.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Lucio-Gutiérrez JR, Garza-Juárez A, Coello J, Maspoch S, Salazar-Cavazos ML, Salazar-Aranda R, Waksman de Torres N. Multi-wavelength high-performance liquid chromatographic fingerprints and chemometrics to predict the antioxidant activity of Turnera diffusa as part of its quality control. J Chromatogr A. 2012;1235:68–76. doi: 10.1016/j.chroma.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 14.Qiu LL, Niu H, Huang W. Ultrasonic and fermented pretreatment technology for diosgenin production from Diosorea zingiberensis C.H. Wright. Chem Eng Res Des. 2011;89:239–247. [Google Scholar]
- 15.Qin Y, Wu XH, Huang W, Gong GH, Li D, He Y, Zhao YL. Acute toxicity and sub-chronic toxicity of steroidal saponins from Dioscorea zingiberensis C.H.Wright in rodents. J Ethnopharmacol. 2009;126:543–550. doi: 10.1016/j.jep.2009.08.047. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Dong YS, Xiu ZL. Three-liquid-phase extraction of diosgenin and steroidal saponins from fermentation of Dioscorea zingibernsis C.H. Wright. Process Biochem. 2010;45:752–756. [Google Scholar]
- 17.Li H, Ni JR. Treatment of wastewater from Dioscorea zingiberensis tubers used for producing steroid hormones in a microbial fuel cell. Bioresour Technol. 2011;102:2731–2735. doi: 10.1016/j.biortech.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Dong YS, Teng H, Qi SS, Liu L, Wang H, Zhao YK, Xiu ZL. Pathways and kinetics analysis of biotransformation of Dioscorea zingiberensis by Aspergillus oryzae. Biochem Eng J. 2010;52:123–130. [Google Scholar]
- 19.Wang YH, Kang AL, Fan BJ, Sun WJ. HPLC-ELSD simultaneous determination of three components in root of Dioscorea zingiberensis C.H. Wright. Chin J Pharm Anal. 2009;29:739–742. [Google Scholar]
- 20.Li R, Zhou Y, Wu ZJ, Ding LS. ESI-Qq TOF-MS/MS and APCI-IT-MS/MS analysis of steroid saponins from the rhizomes of Dioscorea panthaica. Mass Spectrom. 2006;41:1–22. doi: 10.1002/jms.988. [DOI] [PubMed] [Google Scholar]
- 21.Kang LP, Yu K, Zhao Y, Liu YX, Yu HS, Pang X, Xiong CQ, Tan DW, Gao Y, Liu C, Ma BP. Characterization of steroidal glycosides from the extract of Paris Polyphylla var. Yunnanensis by UPLC/Q-TOF MSE. J Pharm Biomed Anal. 2012;62:235–249. doi: 10.1016/j.jpba.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 22.Lu YY, Luo JG, Xu DR, Huang XF, Kong LY. Characterization of spirostanol saponins in solanum torvum by high-performance liquid chro-matography/evaporative light scattering detector/electrospray ionization with multi-stage tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:2447–2452. doi: 10.1002/rcm.3630. [DOI] [PubMed] [Google Scholar]
- 23.Kang LP, Zhao Y, Pang X, Yu HS, Xiong CQ, Zhang J, Gao Y, Yu K, Liu C, Ma BP. Characterization and identification of steroidal saponins from the seeds of Trigonella foenum-graecum by ultra high-performance liquid chromatography and hybrid time-of-flight mass spectrometry. J Pharm Biomed Anal. 2013;74:257–267. doi: 10.1016/j.jpba.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Zhu JB, Guo XJ, Fu SP, Zhang XL, Liang XM. Characterization of steroidal saponins in crude extracts from Dioscorea zingiberensis C. H. Wright by ultra-performance liquid chromatography/electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J Pharm Biomed Anal. 2010;53:462–474. doi: 10.1016/j.jpba.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Lin SH, Wang DM, Yang DP, Yao JH, Tong Y, Chen JP. Characterization of steroidal saponins in crude extracts from Dioscorea nipponica Makino by liquid chromatography tandem multi-stage mass spectrometry. Anal Chim Acta. 2007;599:98–106. doi: 10.1016/j.aca.2007.07.070. [DOI] [PubMed] [Google Scholar]
- 26.Liang F, Li LJ, Abliz Z, Yang YC, Shi JG. Structural characterization of steroidal saponins by electrospray ionization and fast-atom bombardment tandem mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:1168–1173. doi: 10.1002/rcm.690. [DOI] [PubMed] [Google Scholar]
- 27.Liu YZ, Liang F, Cui LJ, Xia M, Zhao LY, Yang YC, Shi JG, Abliz Z. Multi-stage mass spectrometry of furostanol saponins combined with electrospray ionization in positive and negative ion modes. Rapid Commun Mass Spectrom. 2004;18:235–238. doi: 10.1002/rcm.1310. [DOI] [PubMed] [Google Scholar]
- 28.Yang SL, Ma YH, Liu XK. Steroidal constituents from Dioscorea parviflora. Acta Pharm Sin. 2005;40:145–149. [PubMed] [Google Scholar]
- 29.Yang RT, Tong HY. Research of steroidal saponins from the fresh rhizomes of Dioscorea zingiberensis. Chin Med Mat. 2010;33:62–64. [PubMed] [Google Scholar]
- 30.Dong M, Wu LJ, Chen Q, Wang BX. Isolation and identification of steroidal saponins from Dioscorea Panthaica prain et burkill. Acta Pharm Sin. 2001;36:42–45. [PubMed] [Google Scholar]
- 31.Yang YC, Huang SY, Shi JG. Two new furostanol glycosides from Asparagus cochinchinensis. Chin Chem Lett. 2002;13:1185–1188. [Google Scholar]
- 32.Shen Y, Chen HS, Wang Q. Studies on chemical constituents of Asparagus cochinchinensis. Acad J Sec Mil Med Univ. 2007;28:1241–1244. [Google Scholar]