Abstract
Background
Recent transcriptomic analysis of the bovine Y chromosome revealed at least six multi-copy protein coding gene families, including TSPY, HSFY and ZNF280BY, on the male-specific region (MSY). Previous studies indicated that the copy number variations (CNVs) of the human and bovine TSPY were associated with male fertility in men and cattle. However, the relationship between CNVs of the bovine Y-linked HSFY and ZNF280BY gene families and bull fertility has not been investigated.
Results
We investigated the copy number (CN) of the bovine HSFY and ZNF280BY in a total of 460 bulls from 15 breeds using a quantitative PCR approach. We observed CNVs for both gene families within and between cattle breeds. The median copy number (MCN) of HSFY among all bulls was 197, ranging from 21 to 308. The MCN of ZNF280BY was 236, varying from 28 to 380. Furthermore, bulls in the Bos taurus (BTA) lineage had a significantly higher MCN (202) of HSFY than bulls in the Bos indicus (BIN) lineage (178), while taurine bulls had a significantly lower MCN (231) of ZNF280BY than indicine bulls (284). In addition, the CN of ZNF280BY was positively correlated to that of HSFY on the BTAY. Association analysis revealed that the CNVs of both HSFY and ZNF280BY were correlated negatively with testis size, while positively with sire conception rate.
Conclusion
The bovine HSFY and ZNF280BY gene families have extensively expanded on the Y chromosome during evolution. The CN of both gene families varies significantly among individuals and cattle breeds. These variations were associated with testis size and bull fertility in Holstein, suggesting that the CNVs of HSFY and ZNF280BY may serve as valuable makers for male fertility selection in cattle.
Keywords: CNVs, HSFY, ZNF280BY, Male fertility, Testis size, Sire conception rate, Cattle
Background
Recent progress on sequencing of the mammalian Y chromosomes has revealed that the amplification of the male-specific region on the Y (MSY) is a unique phenomenon during the mammalian sex chromosome evolution [1]– [3]. Because this amplification process was lineage-dependent, it resulted in different structures and contents of DNA sequences and genes in the MSY regions in different lineages, leading to different sizes of MSYs (and Y chromosomes) in mammals. The MSY was found to be enriched in multi-copy gene families; and these multicopy gene families, irrespective of the lineage in which they originated, are all expressed predominantly or solely in testis and are involved in spermatogenesis and male fertility [1]– [8]. Although the copy number variations (CNVs) of the Y-linked gene families have not been well-studied in mammals because of the unavailability of the Y chromosome sequences, a limited number of studies have demonstrated that Y-linked CNVs are associated with male fertility in humans [9,10] and cattle [11,12].
The gene content of MSY in the bovine (Bos taurus) Y chromosome (BTAY) has recently been studied [2], which is significantly different from that of the human MSY [1]. There are at least six multi-copy protein-coding gene families on BTAY, including TSPY (testis-specific protein, Y-encoded), HSFY (heat-shock transcription factor, Y-linked), PRAMEY (preferentially expressed antigen in melanoma, Y-linked), ZNF280AY (zinc finger protein 280A, Y-linked), ZNF280BY (zinc finger protein 280B, Y-linked) and EGLY (envelope glycoprotein like, Y-linked) [2]. These gene families can be classified into two groups based on their evolutionary origins: proto-sex chromosome related genes, including TSPY, HSFY, and EGLY, of which TSPY and HSFY are conserved in some other mammalian species [13], and autosome-to-Y transposed genes, including ZNF280AY, ZNF280BY, and PRAMEY, which are bovid-specific [3,4]. Previous studies revealed that the bovine TSPY, HSFY, ZNF280AY, and ZNF280BY gene families were extensively amplified (up to 250 copies) on the Y during evolution [2,11,14]– [17], while the PRAMEY and EGLY gene families were less amplified (3-30 copies) [2,12]. Hamilton et al.[11] reported that the TSPY copy number (CN) was positively correlated with bull fertility and negatively correlated with the TSPY (mRNA) expression level in the testis. Yue et al.[12] has recently identified that the CNV of the bovine PRAMEY was negatively associated with testis size, non-return rate and percentage of normal sperm.
HSFY is a member of the heat shock transcriptional factor (HSF) family that is found in multiple copies on the Y chromosome and conserved in a number of species [17]. The HSFs regulate the expression of heat shock proteins (HSPs) through binding to the sequences located on the HSP genes, which are thought to have important roles in spermatogenesis [18]. The human HSFY is expressed predominantly in the testis [19]. Low or absent expression of HSFY in spermatogenic cells could lead to maturation arrest in men [18], and the deletion of HSFY on the human Y chromosome was associated with the unexplained cases of idiopathic male infertility [20]. In cattle, HSFY likely plays a similar role in spermatogenesis as it does in humans because the bovine HSFY is expressed specifically in the testis [2,17]. The CN of HSFY varies among species, for example, the human Y has 2 copies [1], while the feline Y possesses 8 copies [21]. A recent report found 70 copies of HSFY on BTAY with no CNV among 24 Holstein bulls [17]. However, another recent report found as many as 192 copies in a Hereford bull (L1 Domino 99375) that was used for the BTAY sequence project (GenBank acc. no. CM001061, Project ID: 20275) [2].
ZNF280BY is a newly identified BTAY-specific multi-copy gene family, which belongs to the family of zinc finger proteins [2,3]. Although zinc finger proteins are among the most abundant and functionally diverse proteins in mammalian genomes [22], little is known of the functions of ZNF280BY in mammals. In Drosophila, the ortholog of ZNF280BY, i.e. suppressor of Hairy-wing [Su(Hw)], encodes a protein that binds to an insulator element within a gypsy retrotransposon [23,24], implying that ZNF280BY may be involved in transcription regulation. The bovine ZNF280BY gene family was found to be expressed in different developmental stages of the testis with the sense RNA present in all cell types of seminiferous tubules while the antisense RNA present only in spermatids, signifying a role in spermatogenesis [3]. It was estimated that approximately 234 ZNF280BY genes are present on the Y chromosome of the Hereford bull, L1 Domino 99375 [2].
To date, there are few studies on the CNVs of either HSFY or ZNF280BY and their association with semen quality and male fertility traits in cattle. The objective of this study is to determine whether the bovine HSFY and ZNF280BY have any CNVs among individuals and breeds, and if their CNVs are associated with bull reproductive traits. Here, we provide evidence that the gene CN varies from 21 to 308 with a median of 197 for HSFY, while 28 to 380 with a median of 236 for ZNF280BY among cattle breeds. We found that the CNVs of both HSFY and ZNF280BY are associated with male reproductive performance.
Methods
Animal ethics statement
Animal care and seminal collection were carefully followed the Certified Semen Services (CSS) - Artificial Insemination Center (AIC) Animal Management Guidelines (Revised November 2011) (http://www.naab-css.org/about_css/practices2011.htm).
Collection of samples and relevant information
A total of 460 bulls from 15 breeds were analyzed in this study (Table 1). Of these, 257 are Holstein bulls used in artificial insemination (AI), which have phenotypic records. Out of 257 Holstein bulls, 140 have records in scrotal circumference (SC), age adjusted relative scrotal circumference (RLSC), post thaw motility (PTM), incubated motility (IM), percentage of normal sperm (PNS) and percentage of intact acrosome (PIA). In addition, 82 (of the 140) bulls have data on sire conception rate (SCR), which is a bull fertility evaluation system recently developed by USDA (http://aipl.arsusda.gov/reference/arr-scr1.htm), and 102 (of the 140) bulls have data on relative breeding efficiency (RBE), which is an in-house bull fertility evaluation parameter estimated by Select Sires Inc. (Plain City, OH, USA). RBE uses a similar methodology to SCR but contains only a one-year rolling database [12]. The remaining 117 Holstein bulls have data on non-return rate (NRR). NRR is a traditional methodology to evaluate bull fertility that a cow was inseminated, and was not called to re-service within a given amount of time (usually 60 days) for the first service [12]. The remaining animals were either from the bovine HapMap Project (13 breeds) [25] or from a Chinese local cattle breed (Qinchuan), which do not have any phenotypic records and pedigree information. The Hereford bull L1 Domino 99375 (American Hereford Association registration number 41170496) was sequenced in the BTAY sequencing project (https://www.hgsc.bcm.edu/content/y-chromosome-genome-project) and was used as a calibrator for Y-linked gene copy number estimation [12]. In addition, paternal pedigree data from 192 Holstein bulls were available from the public databases. These animals were descendants of 4 patrilineal founders (HOUSA1427381, HOUSA1428104, HOUSA1441440 and HOUSA1491007) that were born in the 1960s [12].
Table 1.
The median CN of HSFY and ZNF280BY in 15 cattle breeds
| Breed (full name) | Short name | Sample size (n)s | Median CN of HSFY (range) | Median CN of ZNF280BY (range) | Lineage |
|---|---|---|---|---|---|
| Angus |
ANG |
14 |
186 (126–216) |
257 (149–295) |
Bos taurus |
| Beefmaster |
BMA |
14 |
168 (135–218) |
259 (184–352) |
Composite |
| Brahman |
BRM |
10 |
153 (108–265) |
278 (152–337) |
Bos indicus |
| Brown Swiss |
BSW |
14 |
204 (48–291) |
247 (105–331) |
Bos taurus |
| Charolais |
CHL |
10 |
239 (160–308) |
300 (250–336) |
Bos taurus |
| Gir |
GIR |
14 |
185 (122–207) |
280 (244–341) |
Bos indicus |
| Hereford |
HFD |
15* |
202 (124–246) |
229 (165–311) |
Bos taurus |
| Holstein |
HOS |
257 |
204 (21–271) |
226 (28–295) |
Bos taurus |
| Jersey |
JER |
28 |
157 (122–219) |
259 (171–351) |
Bos taurus |
| Limousin |
LMS |
14 |
201 (57–238) |
280 (47–370) |
Bos taurus |
| Nelore |
NEL |
10 |
180 (85–209) |
289 (171–326) |
Bos indicus |
| Norwegian Red |
NRC |
9 |
209 (193–232) |
211 (168–376) |
Bos taurus |
| Qinchuan |
QC |
26 |
143 (102–197) |
161 (111–269) |
Composite |
| Red Angus |
RGU |
11 |
176 (110–247) |
201 (88–356) |
Bos taurus |
| Santa Gertrudis |
SGT |
14 |
175 (162–232) |
296 (234–380) |
Composite |
| Total | 460 | 197 (21–308) | 236 (28–380) |
*The bull used as the calibrator was not used for the median CN estimation for the Hereford breed.
Primer design and CNV estimation
The sequences of HSFY (GenBank acc. no. NM_001077006) and ZNF280BY (GenBank acc. no. NM_001078120) were aligned to the bovine Y chromosome draft sequence assembly (GenBank acc. no. CM001061, Project ID: 20275) using Splign [26]. A total of 192 HSFY loci and 234 ZNF280BY loci were predicted/annotated from the draft assembly of the BTAY sequence (GenBank acc. no. CM001061) [2]. The paralogous sequences for HSFY and ZNF280BY were retrieved and aligned by MEGA 5.0 [27]. Based on these alignments PCR primers were designed from conserved regions using the Primer Premier 5.0 program (http://www.premierbiosoft.com/). The binding sites of each primer on the draft BTAY sequence and the predicted sizes of each amplicon based on an electronic PCR (http://www.ncbi.nlm.nih.gov/tools/epcr/) were listed in Additional file 1: Table S1 and Additional file 2: Table S2. A single-copy gene DDX3Y (DEAD box polypeptide 3, Y-linked, GenBank acc. no. NT182066) was used as a reference. The primer information and primer efficiencies were listed in the Table 2. Subsequently, the confirmation of the Y chromosome specificity of the designed primers, quantitative real-time PCR (qPCR) and equations used for determining CN of HSFY or ZNF280BY were followed exactly as described by Yue et al.[12], except that PCR annealing temperatures were 59°C and 64°C for HSFY and ZNF280BY, respectively. Primer efficiencies were measured according to the equation E = 10[-1/slope] within every plate we run; and the slope was generated by a standard curve described earlier [12].
Table 2.
The information of primers used in qPCR
| Primer name | Sequence | Annealing Tm | Primer efficiency |
|---|---|---|---|
| HSFYF |
5′-CTCCTAAGGATGAATCAACTG-3′ |
59°C |
1.88 |
| HSFYR |
5′-CACAAGATCCTCAGACAAAGC-3′ |
||
| ZNF280BYF |
5′-GAAATACCACACCACCTGCC-3′ |
64°C |
1.88 |
| ZNF280BYR |
5′-GATCTGTAACTGCAAACCTGG-3′ |
||
| DDX3YF |
5′ -ATCGTGGGCGGAATGAGTGT-3′ |
59°C or 64°C | 1.95 at 59°C or 1.90 at 64°C |
| DDX3YR | 5′ -CTTGGTGGAAGCGGTTTTGA-3′ |
Association and statistical analysis
In order to minimize technical error and to have an accurate CN estimation, raw qPCR data that showed a coefficient of variation (CV) > 1% between the duplicates were excluded from further analysis. The normality of the CN data was assessed with the Kolmogorov-Smirnov and Shapiro-Wilk normality tests [28,29]. Box plot analyses on the CN data were conducted to detect the outliers in all the breeds (populations) as a whole and in the Holstein population. Multiple pair-wise comparisons of median copy number (MCN) between breeds were analyzed using a nonparametric Mann-Whitney U test [30] with a Bonferroni correction [31]. In addition, the Mann-Whitney U test was also used to compare the MCN between groups that were classified based on the origin and formation of the cattle breeds, i.e. BTA, BIN, and composite (COM).
Initial analysis of the association between CN and reproductive traits were performed for Holstein bulls by a Pearson’s correlation test in SPSS 17.0 (SPSS Inc., IL, USA). A three-way Analysis of Variance (ANOVA) was applied to investigate the impact of the CN and the founder (of a paternal pedigree) on the SC, RLSC, PTM, IM, PNS and PIA using a general linear model in SPSS 17.0. Furthermore, a mix model of SAS 9.2 (SAS Institute Inc., Cary, NC, USA) was applied to investigate the impact of the CNV of Y-linked genes on RLSC, SC, PTM, IM, PNS and PIA, in which the sire was included as a random effect. A P-value ≤ 0.05 was considered statistically significant for each test.
Results
CNVs of HSFY and ZNF280BY across cattle breeds
In order to validate the male specificity of primers used in this study, a routine PCR was run. The results demonstrated that every primer pair for the target genes, HSFY and ZNF280BY, or the single copy reference gene, DDX3Y, amplified a male-specific band with the expected fragment size, confirming that the designed primers are male-specific and can be used for the qPCR analysis in this study (Figure 1).
Figure 1.
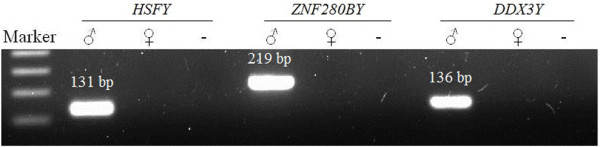
Gel electrophesis of PCR products of the bovine HSFY, ZNF280BY and DDX3Y. The primers of these three Y-linked genes amplified male specific bands with expected fragment size labeled above the band. Marker: 1 kb DNA ladder; ♂: the Hereford bull L1 Domino 99375 genomic DNA; ♀: female cattle genomic DNA; -: negative control (distilled water).
The CN of HSFY in the calibrator was estimated to be 204, which is close to the 192 copies estimated from the draft sequence assembly of the very same Y chromosome [2].The MCN of HSFY was 197, ranging from 21 to 308 among 459 bulls tested (Figure 2). We found that the CN data did not fit the normal distribution (P < 0.05) in either the Holstein population that is the largest population (257 AI bulls) in the study or all populations of 15 breeds analyzed based on Kolmogorov-Smirnov and Shapiro-Wilk normality tests. Pair-wise comparisons of the MCN of HSFY between breeds were listed in Table 3, which indicated a significant difference among breeds (Table 3). Brahman bulls had the lowest MCN of HSFY (153, ranging 108-265), whereas Charolais bulls possessed the highest MCN (239, 106-308) (Table 1). When we analyzed the CN data based on the taurine (BTA) and indicine (BIN) lineages irrespective of the breed origin, we found that bulls in the BTA lineage had significantly higher MCN (202) than bulls in the BIN lineage (178) and the composite breeds (Beefmaster, Santa Gertrudis and Qinchuan, 170) (Figure 3).
Figure 2.
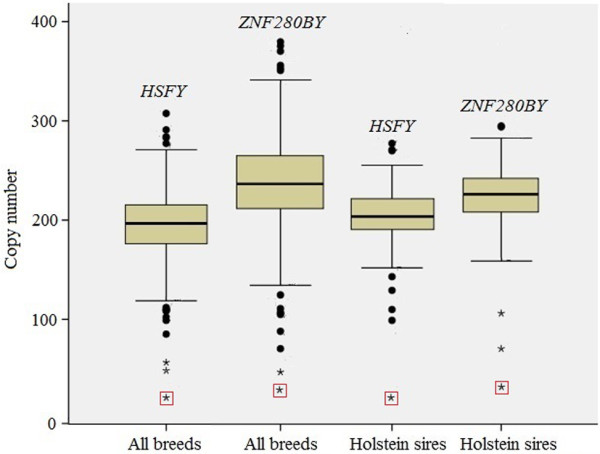
Box plot analysis of the HSFY and ZNF280BY CN in cattle. The outliers were indicated by a solid circle or an asterisk (extremely low CN), which include 12 bulls in all breeds and 9 bulls in Holstein for HSFY, while 14 and 6 for ZNF280BY, respectively. The animal that has the lowest CN of HSFY and ZNF280BY was marked with a red square.
Table 3.
Comparisons of the median CN of HSFY among 15 cattle breeds
| ANG | BMA | BRM | BSW | CHL | GIR | HFD | JER | LMS | NEL | NRC | QCT | RGU | SGT | HOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| BMA |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
| BRM |
* |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
|
|
| BSW |
nsd |
nsd |
* |
- |
|
|
|
|
|
|
|
|
|
|
|
| CHL |
** |
** |
*** |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
| GIR |
nsd |
nsd |
** |
nsd |
** |
- |
|
|
|
|
|
|
|
|
|
| HFD |
nsd |
* |
** |
nsd |
nsd |
* |
- |
|
|
|
|
|
|
|
|
| JER |
nsd |
nsd |
nsd |
** |
*** |
* |
*** |
- |
|
|
|
|
|
|
|
| LMS |
nsd |
* |
** |
nsd |
nsd |
nsd |
nsd |
** |
- |
|
|
|
|
|
|
| NEL |
nsd |
nsd |
nsd |
nsd |
** |
nsd |
* |
nsd |
nsd |
- |
|
|
|
|
|
| NRC |
* |
** |
** |
nsd |
nsd |
*** |
nsd |
*** |
nsd |
** |
- |
|
|
|
|
| QCT |
nsd |
nsd |
* |
nsd |
** |
nsd |
** |
nsd |
*** |
nsd |
*** |
- |
|
|
|
| RGU |
nsd |
nsd |
nsd |
nsd |
* |
nsd |
nsd |
nsd |
nsd |
nsd |
* |
nsd |
- |
|
|
| SGT |
nsd |
nsd |
* |
nsd |
** |
nsd |
* |
* |
* |
nsd |
** |
nsd |
nsd |
- |
|
| HOS | ** | *** | *** | nsd | * | *** | nsd | *** | nsd | ** | nsd | *** | ** | *** | - |
Note: nsd: no significant difference; * P < 0.05; ** P < 0.01; *** P < 0.001. The full name of each breed is listed in Table 1.
Figure 3.
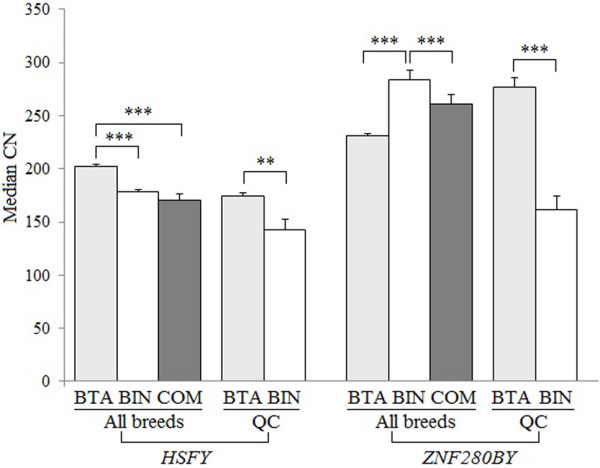
Comparison of the CN of HSFY and ZNF280BY between the BTA and BIN lineages. The CN of HSFY and ZNF280BY was significantly different between the lineages (P < 0.01).The MCN of HSFY in taurine cattle (BTA) was significant higher than that of indicine cattle (BIN) and composite cattle (COM). In contrast, the MCN of ZNF280BY in BIN was significant higher than that in BTA and composite cattle (COM). In the Qinchuan cattle, BTA-derived bulls had a significantly higher MCN of HSFY and ZNF280BY than BIN-derived bulls (P < 0.01). Y-axis represents the MCN of HSFY and ZNF280BY. The error bars represent standard errors. ** P < 0.01; *** P < 0.001.
The CN of ZNF280BY in the calibrator was estimated to be 239, almost identical to the CN (234) annotated from the Y chromosome sequence data [2]. The MCN was 236 with a range from 28 to 380 among the bulls tested. Like the HSFY gene family, the CN of ZNF280BY also did not fit the normal distribution in Holstein and other breeds with several bulls having CN in outliers (Figure 2); the MCN between breeds was significantly varied (Table 4); and Red Angus had the lowest MCN of 201, whereas Charolais bulls had the highest MCN of 300 (Table 1). However, unlike HSFY, ZNF280BY showed significantly higher MCN in the BIN lineage (284) than that in the BTA lineage (231), and the three composite cattle breeds had an intermediate MCN of 262 (Figure 3).
Table 4.
Comparisons of the median CN of ZNF280BY among 15 cattle breeds
| ANG | BMA | BRM | BSW | CHL | GIR | HFD | JER | LMS | NEL | NRC | QCI | RGU | SGT | HOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| BMA |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
| BRM |
nsd |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
|
|
| BSW |
nsd |
nsd |
nsd |
- |
|
|
|
|
|
|
|
|
|
|
|
| CHL |
** |
** |
nsd |
* |
- |
|
|
|
|
|
|
|
|
|
|
| GIR |
* |
** |
nsd |
nsd |
nsd |
- |
|
|
|
|
|
|
|
|
|
| HFD |
nsd |
nsd |
nsd |
nsd |
* |
* |
- |
|
|
|
|
|
|
|
|
| JER |
nsd |
nsd |
nsd |
nsd |
** |
* |
nsd |
- |
|
|
|
|
|
|
|
| LMS |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
- |
|
|
|
|
|
|
| NEL |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
nsd |
- |
|
|
|
|
|
| NRC |
nsd |
nsd |
nsd |
nsd |
** |
** |
nsd |
nsd |
* |
nsd |
- |
|
|
|
|
| QC |
nsd |
* |
nsd |
nsd |
nsd |
nsd |
* |
nsd |
nsd |
nsd |
** |
- |
|
|
|
| RGU |
nsd |
nsd |
nsd |
nsd |
** |
** |
nsd |
* |
* |
* |
nsd |
** |
- |
|
|
| SGT |
** |
*** |
nsd |
* |
nsd |
nsd |
** |
** |
nsd |
nsd |
** |
nsd |
*** |
- |
|
| HOS | ** | ** | nsd | nsd | *** | *** | nsd | *** | *** | *** | nsd | *** | nsd | *** | - |
Note: nsd: no significant difference; * P < 0.05; ** P < 0.01; *** P < 0.001. The full name of each breed is listed in Table 1.
We analyzed a total of 26 bulls from a native Chinese cattle population of Qinchuan in the present study; 16 of which had a BTA Y chromosome, while the remaining 10 had a BIN Y [32]. We found that the BTA derived Y chromosome had a significant higher MCN of HSFY (174) and ZNF280BY (277) than the BIN derived Y chromosome (143 and 161) (Figure 3).
Box plot analyses of the CN data revealed that a small number of animals (~ 3%) were outliers who had significantly higher or lower CN (Figure 2). Of those, 7 and 4 outliers were identified for HSFY and ZNF280BY in the Holstein population, respectively (Figure 2). None of the outliers was part of the Holstein bulls with the SCR record. One of the outliers in Holstein had 21 copies of HSFY and 28 copies of ZNF280BY on its Y chromosome, the lowest copy number in both gene families among all individuals tested (highlighted in Figure 2).
The correlation between the CN of HSFY and ZNF280BY on the bovine Y chromosome
To investigate the relationship of CN between HSFY and ZNF280BY, we included a third Y-linked gene family, PRAMEY, as a reference in our Pearson’s correlation analysis because PRAMEY is less amplified (compared to HSFY and ZNF280BY) on BTAY [2], and the CNV of PRAMEY was assessed with the same groups of animals in our previous study [12].We found that the CN of ZNF280BY was positively correlated to that of HSFY and PRAMEY (P < 0.001) in all bulls, irrespective of their origin. However, the CN of HSFY was not significantly correlated to the CN of PRAMEY (P = 0.637) (Table 5). When the cattle breeds were considered separately, the CN of these three genes was positively correlated to each other in only Holstein and Limousin (P < 0.0001), but not to other breeds (Table 5). The correlation coefficients were 0.488, 0.297, and 0.301 for ZNF280BY-HSFY, ZNF280BY-PRAMEY, and HSFY-PRAMEY, respectively, in the Holstein population, while 0.877, 0.862, and 0.886 in the Limousin population (Table 5). Furthermore, the CN of ZNF280BY was positively correlated to that of HSFY in Nelore and Qinchuan bulls, and that of PRAMEY in Brown Swiss and Gir bulls. In contrast, the CN of HSFY was negatively correlated to CN of PRAMEY in Jersey bulls. When the data were analyzed based on the BTA and BIN lineages, we observed that the CN of ZNF280BY was positively correlated with that of HSFY in both BTA (P < 0.0001), and BIN lineages (P = 0.006), while only positively correlated to that of PRAMEY in the BTA lineage (P < 0.0001) (Table 5).
Table 5.
The correlation between the CNV of PRAMEY , HSFY and ZNF280BY
| Groups/breeds |
ZNF280BY
-
HSFY
|
ZNF280BY
-
PRAMEY
|
HSFY
-
PRAMEY
|
|||
|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |
| Angus |
0.385 |
0.273 |
0.094 |
0.825 |
0.484 |
0.111 |
| Beefmaster |
0.184 |
0.548 |
0.324 |
0.280 |
0.148 |
0.630 |
| Brahman |
0.284 |
0.458 |
0.078 |
0.855 |
-0.033 |
0.934 |
| Brown Swiss |
-0.215 |
0.526 |
0.739 |
0.015 |
-0.134 |
0.694 |
| Charolais |
0.223 |
0.565 |
-0.294 |
0.442 |
0.350 |
0.396 |
| Gir |
-0.031 |
0.921 |
0.997 |
0.003 |
0.283 |
0.717 |
| Hereford |
0.394 |
0.230 |
-0.222 |
0.511 |
0.185 |
0.610 |
| Holstein |
0.488 |
<0.0001 |
0.297 |
<0.0001 |
0.301 |
<0.0001 |
| Jersey |
0.278 |
0.249 |
-0.264 |
0.432 |
-0.728 |
0.017 |
| Limousin |
0.877 |
<0.0001 |
0.862 |
<0.0001 |
0.886 |
<0.0001 |
| Nelore |
0.858 |
<0.0001 |
-0.252 |
0.748 |
0.012 |
0.998 |
| Norwegian Red |
-0.320 |
0.485 |
-0.586 |
0.127 |
0.410 |
0.313 |
| Qinchuan |
0.755 |
<0.0001 |
-0.082 |
0.780 |
0.261 |
0.328 |
| Red Angus |
-0.469 |
0.172 |
0.302 |
0.468 |
-0.467 |
0.205 |
| Santa Gertrudis |
-0.239 |
0.432 |
0.327 |
0.527 |
0.101 |
0.829 |
|
Bos taurus |
0.252 |
<0.0001 |
0.455 |
<0.0001 |
0.122 |
0.055 |
|
Bos indicus |
0.478 |
0.006 |
0.081 |
0.766 |
0.008 |
0.977 |
| All breeds | 0.181 | <0.001 | 0.443 | <0.001 | -0.026 | 0.637 |
Association of the HSFY and ZNF280BY CNV with male reproductive traits in Holstein bulls
We performed the association analysis of CNVs of HSFY and ZNF280BY and their logarithm (base 10) transformation data with the three types of reproductive traits from 257 Holstein bulls, including testis size (RLSC and SC), semen quality (PTM, IM, PNS and PIA) and male fertility (SCR, NRR and RBE). The association analysis revealed that the RLSC was negatively associated with CNV (r = -0.249, P = 0.008) and Log10CNV of HSFY (r = -0.267, P = 0.004) (Table 6), and SC was negatively associated with Log10CNV of HSFY (r = -0.202, P = 0.025), while tended to be negatively associated with CNV of HSFY (r = -0.169, P = 0.073). For the ZNF280BY, RLSC is significantly associated with log10CNV of ZNF280BY (r = -0.239, P = 0.025), and tend to be associated with its CNV (r = -0.174, P = 0.055). SC was neither associated with CNV nor log10CNV of ZNF280BY (Table 6). These results suggested that a lower CN of HSFY and ZNF280BY is associated with a larger testis size (Figure 4). The results also demonstrated that the CNVs or Log10CNVs of HSFY and ZNF280BY were positively correlated to SCR, suggesting that a higher CN of HSFY and ZNF280BY is associated with a higher SCR (Figure 4). Furthermore, we investigated the relationship between reproductive traits and the sum of HSFY and ZNF280BY CNV (sCNV) and Log10sCNV. We found that sCNV and Log10sCNV of HSFY and ZNF280BY were negatively associated with RLSC, whereas positively associated with SCR. The remaining traits were not significantly associated with either sCNV or Log10sCNV (Table 6). Interestingly, the Holstein bull (mentioned above) who had the lowest copy number in both HSFY and ZNF280BY gene families had the largest RLSC among all animals tested, strongly supporting our association results that a lower CN of the two Y-linked genes is associated with a larger testis.
Table 6.
The Pearson’s correlation between the CNV of HSFY and ZNF280BY and the reproductive traits
| Traits* |
HSFY
CNV |
Log
10
(
HSFY
CNV) |
ZNF280BY
CNV |
Log
10
(
ZNF280BY
CNV) |
Sum CNV |
Log
10
sCNV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | r | P | r | P | r | P | |
| SC |
-0.169 |
0.073 |
-0.190 |
0.044 |
-0.079 |
0.394 |
-0.113 |
0.218 |
-0.144 |
0.258 |
-0.158 |
0.117 |
| RLSC |
-0.249 |
0.008 |
-0.267 |
0.004 |
-0.174 |
0.055 |
-0.202 |
0.025 |
-0.218 |
0.030 |
-0.250 |
0.012 |
| SCR |
0.294 |
0.021 |
0.294 |
0.021 |
0.339 |
0.008 |
0.341 |
0.008 |
0.366 |
0.007 |
0.361 |
0.008 |
| NRR |
-0.005 |
0.961 |
-0.019 |
0.852 |
-0.162 |
0.106 |
-0.154 |
0.126 |
-0.118 |
0.267 |
-0.119 |
0.264 |
| RBE |
0.112 |
0.309 |
0.077 |
0.485 |
0141 |
0.210 |
0.075 |
0.508 |
0.164 |
0.162 |
0.096 |
0.414 |
| PTM |
-0.013 |
0.892 |
-0.031 |
0.748 |
0.045 |
0.632 |
0.002 |
0.981 |
-0.006 |
0.958 |
-0.024 |
0.818 |
| IM |
-0.061 |
0.533 |
-0.045 |
0.647 |
-0.087 |
0.359 |
-0.082 |
0.388 |
-0.115 |
0.272 |
-0.079 |
0.450 |
| PNS |
0.144 |
0.243 |
0.058 |
0.550 |
0.016 |
0.863 |
-0.004 |
0.969 |
0.048 |
0.647 |
0.017 |
0.874 |
| PIA | -0.019 | 0.846 | -0.002 | 0.986 | -0.014 | 0.866 | -0.007 | 0.940 | -0.046 | 0.667 | -0.014 | 0.895 |
*See full name in the Methods section.
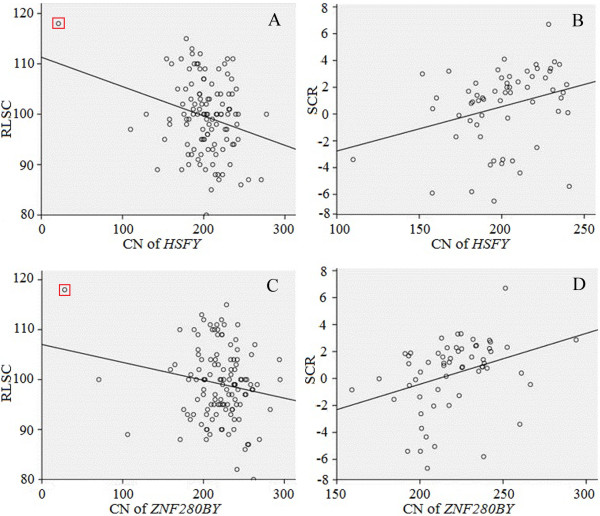
Figure 4.
Correlations between the CNV of HSFY and ZNF280BY and male reproductive traits in Holsteins. The CNV of HSFY was (A) negatively associated with RLSC (r = -0.249, P = 0.008) and (B) positively associated with SCR (r = 294, P = 0.021). The ZNF280BY CNV was (C) negatively associated with RLSC (r = -0.174, P = 0.055) and (D) positively associated with SCR (r = 0.339, P = 0.008). Each circle represents one individual. The animal that has the lowest CN of HSFY and ZNF280BY was marked with a red square.
In order to check whether the outlier bulls identified in the box plot analysis have any significant effect on the association analyses, we re-calculated the Pearson’s correlation coefficient by excluding the outliers from the data set. The results indicated that these outliers had considerable effect on the association results. Taken away the outliers from the analysis led to a non-significant association between testis size and CNVs of ZNF280BY and HSFY (P > 0.05, Table 7).
Table 7.
The Pearson’s correlation between the CNV of HSFY and ZNF280BY and the reproductive traits after excluding the outliers from the data set
| Traits* |
HSFY
CNV |
ZNF280BY
CNV |
||
|---|---|---|---|---|
| r | P | r | P | |
| SC |
-0.085 |
0.187 |
-0.079 |
0.197 |
| RLSC |
-0.150 |
0.057 |
-0.131 |
0.078 |
| NRR |
-0.037 |
0.714 |
-0.153 |
0.131 |
| RBE |
0.126 |
0.261 |
0.182 |
0.111 |
| PTM |
-0.064 |
0.527 |
0.122 |
0.206 |
| IM |
-0.011 |
0.910 |
-0.005 |
0.958 |
| PNS |
0.119 |
0.239 |
0.097 |
0.314 |
| PIA | -0.018 | 0.858 | 0.044 | 0.654 |
*See full name in the Methods section.
To further investigate the effect of low and high CN of HSFY and ZNF280BY on the bull performance, we simply grouped the140 Holstein bulls (whose phenotypic data were available for SC, RLSC, PTM, IM, PNS and PIA) into two groups based on a cut-off threshold of MCN for ZNF280BY (223) and HSFY (203) in these 140 bulls (Table 8). Statistical analysis indicated that the difference between the low and high groups of the HSFY CN was significant in RLSC (P = 0.008) and closed to be significant in SC (P = 0.063), but not significant in the remaining four traits (PTM, IM, PNS and PIA) (P > 0.05) (Table 7). For the ZNF280BY, there is no significant difference between the low and high CN groups (Table 8). These results, from a different angle, confirmed that bulls with a low CN of HSFY tend to have a large testis size.
Table 8.
Reproductive performance of Holstein bulls with a high and low MCN of HSFY and ZNF280BY
| Reproductive traits* |
HSFY
|
ZNF280BY
|
||||
|---|---|---|---|---|---|---|
|
Low MCN |
High MCN |
P value |
Low MCN |
High MCN |
P value | |
| (CN < 203, n = 56) | (CN ≥ 203, n = 58) | (CN < 223, n = 60) | (CN ≥ 223, n = 62) | |||
| SC (cm) |
40.72 ± 0.44 |
39.62 ± 0.38 |
0.063 |
40.11 ± 0.43 |
39.81 ± 0.39 |
0.616 |
| RLSC (%) |
101.41 ± 0.99 |
97.74 ± 0.90 |
0.008 |
100.11 ± 0.97 |
98.24 ± 0.89 |
0.157 |
| PTM (%) |
77.59 ± 0.28 |
77.62 ± 0.41 |
0.962 |
77.51 ± 0.29 |
77.88 ± 0.38 |
0.444 |
| IM (%) |
34.37 ± 0.27 |
34.05 ± 0.46 |
0.540 |
34.22 ± 0.30 |
34.15 ± 0.42 |
0.900 |
| PNS (%) |
77.92 ± 1.09 |
78.38 ± 1.04 |
0.762 |
77.18 ± 1.00 |
79.52 ± 1.02 |
0.106 |
| PIA (%) | 80.34 ± 0.39 | 80.16 ± 0.44 | 0.440 | 80.24 ± 0.36 | 80.31 ± 0.42 | 0.891 |
*See full name in the Methods section.
Relationship between bull pedigrees and their HSFY and ZNF280BY CNVs and reproductive traits in Holstein
Among the 257 Holstein bulls investigated, 192 had paternal pedigree information [12]. These bulls were descendants of only 4 patrilineal founders (HOUSA1427381, HOUSA1428104, HOUSA1441440 and HOUSA1491007) that were born in the 1960s. The MCN of HSFY and ZNF280BY for each founder lineage was re-calculated (Additional file 3: Figure S1). The MCN of HSFY among the descendants was 203,198 and 205, for the founder HOUSA1427381, HOUSA1441440 and HOUSA1491007, respectively (the fourth founder had only one descendant in the tested populations), while the corresponding MCN of ZNF280BY for these founders was 221, 220 and 223. The MCN variations were not significant between these founder lineages (P > 0.05).
To investigate whether the founder effect has any impact on our gene CNV and male reproductive trait association analysis, we performed three-way ANOVA within the two relative large descendant population HOUSA1427381 (40 individuals) and HOUSA1491007(81 individuals). Our results (Table 9) revealed that the founders had no significant effects on the reproductive traits, and the interactions between the founders and the CNVs of the Y-linked genes had no significant effects either.
Table 9.
The P values of the three-way ANOVA
| Reproductive traits* | Founder | HSFY | ZNF280BY | HSFY × Founder | ZNF280BY × Founders | ZNF280BY × HSFY | Founder × HSFY × ZNF280BY |
|---|---|---|---|---|---|---|---|
| SC |
0.428 |
0.373 |
0.094 |
0.493 |
0.363 |
0.401 |
0.485 |
| RLSC |
0.270 |
0.691 |
0.490 |
0.580 |
0.075 |
0.152 |
0.926 |
| PTM |
0.500 |
0.245 |
0.578 |
0.279 |
0.619 |
0.346 |
0.326 |
| IM |
0.438 |
0.822 |
0.673 |
0.971 |
0.963 |
0.190 |
0.721 |
| PNS |
0.673 |
0.836 |
0.922 |
0.057 |
0.829 |
0.130 |
0.955 |
| PIA | 0.502 | 0.858 | 0.779 | 0.367 | 0.255 | 0.165 | 0.294 |
*See full name in the Methods section.
Out of the 192 bulls, 94 were half-sib brothers from 27 sires. In order to exclude the sire effect on the above CNV and testis size association analyses, we included sire in the mix model as a random effect (see Methods). We found that the CNV of HSFY had a significant effect on SC (P = 0.022) and RLSC (P = 0.002), whereas the CNV of ZNF280BY had no significant on testis size (P = 0.09 on RLSC and P = 0.511 on SC).
Discussion
The CN of the HSFY and ZNF280BY gene families are variable across cattle breeds
In this study, we investigated the CN of two Y-linked gene families, HSFY and ZNF280BY, among 15 cattle breeds by a modified qPCR method [12] that has some advantages over the previously described method [16]. We estimated that 204 and 239 copies of HSFY and ZNF280BY are present on the Y chromosome of a Hereford bull (L1 Domino 99375), the calibrator in this study, whose DNA was used for the bovine Y sequence project (NCBI Project ID: 20275). These results were in close agreement with the 192 and 234 copies (of HSFY and ZNF280BY) annotated from the draft assembly of the BTAY sequence (GenBank acc. no. CM001061), indicating the reliability of our qPCR method. For the first time, we were able to show that the Y-linked HSFY and ZNF280BY gene families, like the TSPY family, were extensively amplified on the bovine Y chromosome. The CN varies significantly from 21 to 308 for HSFY, and 28 to 380 for ZNF280BY among all bulls, with a variable MCN among 15 breeds tested. We noticed that the estimation of 197 copies (MCN) for the bovine HSFY in this work was significantly higher than the ~70 copies reported by Hamilton et al. [17]. Despite the possible influence from the cattle breeds as the two studies used different number of bulls/breeds, we believe that the most important reason for the HSFY CN discrepancy between the two studies is the HSFY gene PCR primer design as the draft BTAY sequence was not available when Hamilton et al. carried out their study. After carefully aligning the PCR primer sequences against the draft assembly of the BTAY sequence (GenBank acc. no. CM001061), we understood that the HSFY primer pairs designed by Hamilton et al. matched approximately 120 loci (with a 100% identity), suggesting that their primer pairs targeted ~ 60% of all HSFY loci on the sequenced Y chromosome. Further, our data did not support the previous report that found no CNV of HSFY among 24 randomly selected Holstein bulls [17]. In contrast, we found that the CNV of HSFY was not only present, but also with the largest variation range (21-271, Table 1) in the Holstein population. In addition to genetic factors that may determine the variation, the largest CNV range of HSFY observed in this group of Holsteins was most likely due to a much larger number of Holstein bulls tested in the present study.
We further analyzed the CNV among three patrilineal lineages in the Holstein population and found considerable variations among the Y chromosomes within any given Y-lineage even though the median CN was not significantly different among these three Y-lineages. These results were expected as both HSFY and ZNF280BY gene families are located in the palindrome-like repeats (see discussion below) [2,3], which provide a genetic mechanism (i.e. intrachromosomal recombination between repeated homologous sequences) for deletion (or amplification) of the Y-linked genes (or DNA segments). We believe that our current observation in the bovine Y CNV is similar to the earlier reports on the human Y chromosome P1 (Palindrome 1) region where the AZFc (Azoospermia Factor c) is located [6,33,34]. AZFc is the most common known genetic cause of severe spermatogenic failure in men [34]. Frequent deletions in this region have been identified in patients with azoo and/or oligospermia and account for ~ 15% of all infertile cases [33]. Within a human Y-lineage, de novo deletion in the AZFc region was estimated at a rate of 1.1 × 10-5 to 1.4 × 10-4 per father-to-son transmission of the Y chromosome [34]. As a consequence of lack of recombination during meiosis in the MSY region, these de novo AZFc-related deletions will be kept in the son’s Y-lineage. Although there is no detailed research on the rate of the bovine Y mutation (deletion, insertion, etc.), results from the human Y chromosome provide a reference for the future research in cattle. It is worth noting that the Holstein bull that had the lowest CN of both HSFY and ZNF280BY on its Y chromosome had the largest relative testis size (RLSC) among all bulls tested (Figure 4A and C). It is likely that this particular Y chromosome has a large deletion in the HSFY- and ZNF280BY- related region, which, from a different angle, supports our finding that less CN is associated with larger testis in Holsteins.
The amplification of the HSFY and ZNF280BY gene families are different between the taurine and indicine Y chromosomes
The previous studies indicated that bovine Y ampliconic region consists of ∼ 80 palindrome-like repeat units (∼ 420 kb per unit) and that each unit contains one to three copies of the corresponding gene families (TSPY, HSFY, ZNF280BY and ZNF280AY) [2,3]. The TSPY gene family has been evidenced to have undergone extensive amplification (50-200 copies) across the entire ampliconic region (~ 30 Mb) during evolution [2,16]. The CNV of HSFY and ZNF280BY observed in current study further confirmed the extensive amplification of the HSFY and ZNF280BY on BTAY. In addition, we found that CN between HSFY and ZNF280BY was positively correlated (Table 5), which was accordant with previous finding that HSFY and ZNF280BY were amplified together on BTAY [2].
It was interesting to see that the CN between PRAMEY and ZNF280BY was only positively correlated in the BTA lineage, but not in BIN lineage. Recent studies demonstrated that PRAMEY and ZNF280BY, unlike the TSPY and HSFY that are present in other mammalian Y chromosomes, were bovid-specific and were derived from the transposition of the ZNF280B/ZNF280A/PRAME block on BTA17 and amplified separately thereafter on BTAY [3,4]. The positive correlation of CN between PRAMEY and ZNF280BY observed in this work further supports the previous findings [3]. Our data on HSFY revealed that bulls in the BTA lineage had a significantly higher MCN of HSFY than bulls in the BIN lineage (202 copies vs. 178 copies). In contrast, bulls in the BTA lineage had a significantly lower MCN of ZNF280BY than those bulls in the BIN lineage (231 vs. 284). Though the mechanism behind variations of gene CN is still largely unknown, it is believed that gene CN may originally depend on functional requirement and natural selection [35]. If so, HSFY could be one of the examples for functional selection. As its name indicated, HSFY may play an important role in heat reaction for zebu to adapt to the high temperatures in the tropical countries by reducing the CN of HSFY.
As a consequence of the bovid evolution, the only visible morphological difference identified between BTA and BIN genome at a cytogenetic level is the Y chromosome. The BTA Y chromosome is metacentric, while the BIN Y is acrocentric [36]. This morphological difference is believed to be caused by a Y chromosome rearrangement [37]. Obviously, the difference observed in the CN correlation of the three Y-linked gene families (PRAMEY, HSFY and ZNF280BY) between BTA and BIN lineages in this study (Table 5) is related to the origin of the BTA and BIN Y chromosomes. Since the current researches are focused mainly on the BTA Y chromosome, including the BTAY sequencing project, the data from this study clearly indicate that more research is needed on the BIN Y lineage in order to understand the mechanism underlying the Y-linked gene expansion and their function in male reproduction.
The CNVs of Y-linked gene families could be used as potential DNA markers for male fertility selection
The majority of the genes on the Y chromosome is found to be involved in male spermatogenesis and, hence, are closely associated with male fertility [1]. Previous studies have shown that CNV of TSPY was associated with male fertility in human and cattle [9]– [11]. The CNV of a bovid-specific Y-linked gene, PRAMEY, was found to be negatively associated with testis size, PNS and NRR in cattle [12]. Concerning the relationship of CNVs and reproductive traits in this study, the CNVs of HSFY and ZNF280BY are negatively correlated with testis size (SC and RLSC), but positively associated with SCR. SCR is a newly developed method to evaluate the fertility of bull in the US, and includes several components in the evaluation model, i.e. the expanded service sire term, inbreeding of the bull, inbreeding of the embryo from the mating, age of the bull, AI organization combined with year of the mating, and fertility of the cow a bull is mated with (http://aipl.arsusda.gov/reference/arr-scr1.htm). In contrast, SC and RLSC are simply and directly quantitative traits. In a natural mating system, SC is important for fertility (more sperm with larger testicles) [38]. This may not be true in AI because semen is diluted to allow many breeding from a single ejaculate. Thus, pregnancy rates (like SCR) achieved by different sires are minimized [39]. Therefore, it is not surprising to observe this conflict tendency (positive vs. negative) in association analysis.
The molecular mechanism by which a CNV can affect transcription, translation, or even a phenotype is largely unknown. Previous studies revealed that CNVs can be inversely correlated to their mRNA expression [11,40,41]. However, how the CNV of ZNF280BY or HSFY impacts the transcription and translation of the gene family has not been studied yet. The human Y chromosome comprises 2 copies of HSFY. A deletion on one or both copies can lead to severe impacts on fertility [18,20]. In contrast, cattle have about 200 copies of HSFY and ZNF280BY which may allow for many “backups” in case one or more of the copies undergo mutations, thereby minimizing/eliminating harmful effects on fertility as suggested by Hamilton et al.[17].
Testicular size is a very unique trait for a bull and has been used as a predictive indicator for output of sperm cells for yearling bulls [42]. However, very few genetic markers have been identified to date for a selection purpose due to the lack of molecular genetic study in bull fertility. The CNVs of HSFY and ZNF280BY found in this study, and that of PRAMEY found in our previous study [12], were all negatively associated with testis size. These results indicated that the CN of these Y-linked genes may provide a new insight for testis selection in an early age in cattle.
Conclusion
We confirmed here that the two Y-linked gene families, HSFY and ZNF280BY, were extensively and differentially expanded on the BTA and BIN Y chromosomes. The copy number of these gene families is highly variable among individuals and breeds. The CNVs of HSFY and ZNF280BY are negatively associated with testis size, which may serve as valuable markers for male fertility selection in an early age.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WSL and XPY conceived the experiments; XPY carried out the experiments; XPY, CD and TCC analyzed the data; XPY and WSL wrote the manuscript. JMD and CEM collected the semen samples and the phenotypic data. TCC, CZL, JMD and CEM revised the manuscript. All co-authors read and approved the final manuscript.
Supplementary Material
The binding sites of the HSFY primers against the bovine Y chromosome draft sequence assembly.
The binding sites of the ZNF280BY primers against the bovine Y chromosome draft sequence assembly.
The pedigree information of 140 Holstein bulls whose phenotypic data were available for this study.
Contributor Information
Xiang-Peng Yue, Email: yux13@psu.edu.
Chad Dechow, Email: cdd1@psu.edu.
Ti-Cheng Chang, Email: disonchang@hotmail.com.
James Melton DeJarnette, Email: DeJarnette@selectsires.com.
Clifton Eugene Marshall, Email: cmarshall@selectsires.com.
Chu-Zhao Lei, Email: leichuzhao1118@126.com.
Wan-Sheng Liu, Email: wul12@psu.edu.
Acknowledgements
We would like to thank Select Sires Inc. and Semex Alliance for providing bull semen samples and phenotypic records. We are grateful to Dr. Lee Alexander and Dr. Clare Gill for sharing DNA samples of the Hereford bull L1 Domino 99375 and bulls used in the Bovine HapMap Project. This project was supported by a grant (no. 2010-65205-20362) from USDA NIFA and a research grant from Select Sires to WSL. XPY was supported by a scholarship from the Chinese Scholarship Council for his joint-training PhD program at Penn State.
References
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Chang T, Yang Y, Retzel EF, Liu W-S. The male-specific region of the bovine Y chromosome is gene-rich with a high transcriptomic activity in testis development. Proc Natl Acad Sci U S A. 2013;110:12373–12378. doi: 10.1073/pnas.1221104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chang T-C, Yasue H, Bharti AK, Retzel EF, Liu W-S. ZNF280BY and ZNF280AY: autosome derived Y-chromosome gene families in Bovidae. BMC Genomics. 2011;12:13. doi: 10.1186/1471-2164-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-C, Yang Y, Yasue H, Bharti AK, Retzel EF, Liu W-S. The expansion of the PRAME gene family in Eutheria. PLoS One. 2011;6:e16867. doi: 10.1371/journal.pone.0016867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJI, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. The multicopy gene Sly represses the Sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- Reynard LN, Cocquet J, Burgoyne PS. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol Reprod. 2009;81:250–257. doi: 10.1095/biolreprod.108.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka R, Vrtel R, Dusek L, Singh AR, Krizova K, Svacinova V, Horinova V, Dostal J, Oborna I, Brezinova J, Sobek A, Santavy J. TSPY gene copy number as a potential new risk factor for male infertility. Reprod Biomed Online. 2007;14:579–587. doi: 10.1016/S1472-6483(10)61049-8. [DOI] [PubMed] [Google Scholar]
- Giachini C, Nuti F, Turner DJ, Laface I, Xue Y, Daguin F, Forti G, Tyler-Smith C, Krausz C. TSPY1 copy number variation influences spermatogenesis and shows differences among Y lineages. J Clin Endocrinol Metab. 2009;94:4016–4022. doi: 10.1210/jc.2009-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CK, Verduzco-Gómez AR, Favetta LA, Blondin P, King WA. Testis-specific protein Y-encoded copy number is correlated to its expression and the field fertility of Canadian Holstein bulls. Sex Dev. 2012;6:231–239. doi: 10.1159/000338938. [DOI] [PubMed] [Google Scholar]
- Yue X-P, Chang T-C, DeJarnette JM, Marshall CE, Lei C-Z, Liu W-S. PRAMEY copy number variation across breeds and its association with male fertility in Holstein sires. J Dairy Sci. 2013;96(12):8024–8034. doi: 10.3168/jds.2013-7037. [DOI] [PubMed] [Google Scholar]
- Bhowmick BK, Satta Y, Takahata N. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 2007;17:441–450. doi: 10.1101/gr.5734907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Dechend F, Manz E, Jung C, Jakubiczka S, Fehr S, Schmidtke J, Schnieders F. Organization and expression of bovine TSPY. Mamm Genome. 1997;8:491–496. doi: 10.1007/s003359900482. [DOI] [PubMed] [Google Scholar]
- Verkaar ELC, Zijlstra C, van ’t Veld EM, Boutaga K, van Boxtel DCJ, Lenstra JA. Organization and concerted evolution of the ampliconic Y-chromosomal TSPY genes from cattle. Genomics. 2004;84:468–474. doi: 10.1016/j.ygeno.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Hamilton CK, Favetta LA, di Meo GP, Floriot S, Perucatti A, Peippo J, Kantanen J, Eggen A, Iannuzzi L, King WA. Copy number variation of testis-specific protein, Y-encoded (TSPY) in 14 different breeds of cattle (Bos taurus) Sex Dev. 2009;3:205–213. doi: 10.1159/000228721. [DOI] [PubMed] [Google Scholar]
- Hamilton CK, Revay T, Domander R, Favetta LA, King WA. A large expansion of the HSFY gene family in cattle shows dispersion across Yq and testis-specific expression. PLoS One. 2011;6:e17790. doi: 10.1371/journal.pone.0017790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshida K, Shinka T, Nozawa S, Nakahori Y, Iwamoto T. Altered expression pattern of heat shock transcription factor, Y chromosome (HSFY) may be related to altered differentiation of spermatogenic cells in testes with deteriorated spermatogenesis. Fertil Steril. 2006;86:612–618. doi: 10.1016/j.fertnstert.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Shinka T, Sato Y, Chen G, Naroda T, Kinoshita K, Unemi Y, Tsuji K, Toida K, Iwamoto T, Nakahori Y. Molecular characterization of heat shock-like factor encoded on the human Y chromosome, and implications for male infertility. Biol Reprod. 2004;71:297–306. doi: 10.1095/biolreprod.103.023580. [DOI] [PubMed] [Google Scholar]
- Vinci G, Raicu F, Popa L, Popa O, Cocos R, McElreavey K. A deletion of a novel heat shock gene on the Y chromosome associated with azoospermia. Mol Hum Reprod. 2005;11:295–298. doi: 10.1093/molehr/gah153. [DOI] [PubMed] [Google Scholar]
- Pearks Wilkerson AJ, Raudsepp T, Graves T, Albracht D, Warren W, Chowdhary BP, Skow LC, Murphy WJ. Gene discovery and comparative analysis of X-degenerate genes from the domestic cat Y chromosome. Genomics. 2008;92:329–338. doi: 10.1016/j.ygeno.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Modolell J, Bender W, Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983;80:1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 2007;8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Taylor JF, van Tassell CP, Barendse W, Eversole KA, Gill CA, Green RD, Hamernik DL, Kappes SM, Lien S, Matukumalli LK, McEwan JC, Nazareth LV, Schnabel RD, Weinstock GM, Wheeler DA, Ajmone-Marsan P, Boettcher PJ, Caetano AR, Garcia JF, Hanotte O, Mariani P, Skow LC, Sonstegard TS, Williams JL, Diallo B, Hailemariam L, Martinez ML, Morris CA, Silva LOC. et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–532. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- Justel A, Peña D, Zamar R. A multivariate Kolmogorov-Smirnov test of goodness of fit. Stat Probab Lett. 1997;35:251–259. doi: 10.1016/S0167-7152(97)00020-5. [DOI] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of Two random variables is stochastically larger than the other. Ann Math Stat. 1947;18:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- Dunn OJ. Multiple comparisons among means. J Am Stat Assoc. 1961;56:52–64. doi: 10.1080/01621459.1961.10482090. [DOI] [Google Scholar]
- Chang Z, Wei L, Zhang R, Guo B, He C, Lan X, Chen H, Lei C. Genetic diversity and origin based on Y-SNPs in Chinese cattle. Acta Vet Zootech Sin. 2011;42:1537–1542. [Google Scholar]
- Krausz C, Degl’Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci J Virtual Libr. 2006;11:3049–3061. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, Oates RD, Silber SJ, Ardlie K, Page DC. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet. 2012;91:890–896. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci U S A. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldammer T, Brunner RM, Schwerin M. Comparative analysis of Y chromosome structure in Bos taurus and B. indicus by FISH using region-specific, microdissected, and locus-specific DNA probes. Cytogenet Cell Genet. 1997;77:238–241. doi: 10.1159/000134584. [DOI] [PubMed] [Google Scholar]
- Meo GPD, Perucatti A, Floriot S, Incarnato D, Rullo R, Jambrenghi AC, Ferretti L, Vonghia G, Cribiu E, Eggen A, Iannuzzi L. Chromosome evolution and improved cytogenetic maps of the Y chromosome in cattle, zebu, river buffalo, sheep and goat. Chromosome Res. 2005;13:349–355. doi: 10.1007/s10577-005-2688-4. [DOI] [PubMed] [Google Scholar]
- Brito LFC, Silva AEDF, Barbosa RT, Kastelic JP. Testicular thermoregulation in Bos indicus, crossbred and Bos taurus bulls: relationship with scrotal, testicular vascular cone and testicular morphology, and effects on semen quality and sperm production. Theriogenology. 2004;61:511–528. doi: 10.1016/S0093-691X(03)00231-0. [DOI] [PubMed] [Google Scholar]
- Amann RP, DeJarnette JM. Impact of genomic selection of AI dairy sires on their likely utilization and methods to estimate fertility: a paradigm shift. Theriogenology. 2012;77:795–817. doi: 10.1016/j.theriogenology.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Schuster-Böckler B, Conrad D, Bateman A. Dosage sensitivity shapes the evolution of copy-number varied regions. PLoS One. 2010;5:e9474. doi: 10.1371/journal.pone.0009474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavaré S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson TJ. Evaluation of fertility and infertility in natural service bulls. Vet J Lond Engl. 1997;168(2004):215–229. doi: 10.1016/j.tvjl.2003.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The binding sites of the HSFY primers against the bovine Y chromosome draft sequence assembly.
The binding sites of the ZNF280BY primers against the bovine Y chromosome draft sequence assembly.
The pedigree information of 140 Holstein bulls whose phenotypic data were available for this study.