Abstract
Background
Herpes simplex virus type 2 (HSV-2) is sexually transmitted, leading to blisters and ulcers in the genito-anal region. After primary infection the virus is present in a latent state in neurons in sensory ganglia. Reactivation and production of new viral particles can cause asymptomatic viral shedding or new lesions. Establishment of latency, maintenance and reactivation involve silencing of genes, continuous suppression of gene activities and finally gene activation and synthesis of viral DNA. The purpose of the present work was to study the genetic stability of the virus during these events.
Methods
HSV-2 was collected from 5 patients with true primary and recurrent infections, and the genes encoding glycoproteins B,G,E and I were sequenced.
Results
No nucleotide substitution was observed in any patient, indicating genetic stability. However, since the total number of nucleotides in these genes is only a small part of the total genome, we cannot rule out variation in other regions.
Conclusions
Although infections of cell cultures and animal models are useful for studies of herpes simplex virus, it is important to know how the virus behaves in the natural host. We observed that several glycoprotein gene sequences are stable from primary to recurrent infection. However, the virus isolates from the different patients were genetically different.
Keywords: Primary and recurrent infections of humans, Genetic stability of HSV-2
Background
Herpes simplex viruses (HSV) are widely distributed pathogens transmitted by close contact. Infection by type 1 (HSV-1) starts in childhood and increases during the following years so that more than 70% of the population is seropositive at an age of 40 years [1]. HSV-1 affects the orofacial region. Additionally it has become a common cause of genital herpes infections, presently responsible for at least 50% of the cases in some regions [2,3]. Encephalitis due to HSV-1 infection is a much more rare, but devastating disease [4-7].
Herpes simplex type 2 (HSV-2) is sexually transmitted, leading to blisters and ulcers in the anogenital region (for review, see ref. [8,9]). Prevalence of HSV-2 infections shows geographical variation and marked differences from one demographic group to another (reviewed in 1). It is high among prostitutes [10,11], related to number of sexual partners and to exposure to other sexually transmitted diseases [12,13]. The average prevalence is 17% in USA [14] and varies between 7% and 31% among European adults [1]. There is a well-documented relationship between infections with HSV-2 and HIV-1 [12,15-17]. HSV-2 seropositivity increases the risk for HIV acquisition by a factor of 3 [15].
Together with varicella zoster virus (VZV) and pseudorabies virus (PRV), HSV-1 and HSV-2 belong to the alphaherpesvirinae subfamily of herpesviridae. They are all neurotropic, and like many other members of the herpesviridae family cause latent infections. After primary infection in the skin/mucosal membranes the virus enters axons of innervating neurons, migrates retrograde and establishes latent infections in neuronal cells in sensory ganglia. Reactivation from latency is a commonly occurring event during which the virus migrates anterograde in the axon, leading to new infection in the periphery or to asymptomatic shedding of the virus. About 90% of the persons with primary HSV-2 infection have recurrence during the first year afterwards, and one fifth of them more than 10 times during this period [18-20]. Reactivation of HSV-2 is most frequent during the first years after primary infection. As much as 75% of these events may be short asymptomatic shedding lasting for approximately 12 hours [19].
The molecular processes involved during the various steps of primary and recurrent infection have been studied extensively and reviewed in recent reports [21,22]. Neither virus particles nor viral antigens are produced in the latent state, but latency-associated transcripts (LATs) and several micro RNAs are formed [23-27].
Major regulatory steps in these events are silencing of viral genes, continuous suppression of gene activities and finally stimulation of such activities followed by DNA synthesis. Several aspects of these processes have been studied in cell culture and in animal models, but with little focus on the genetic stability of the virus. The purpose of the present work was to study this stability, in the natural host, during these different series of events. We isolated HSV-2 from true primary and recurrent infections and sequenced a set of selected genes. The isolates were genetically different, but no sequence differences were observed between the initial infection and the reactivation.
Methods
Clinical HSV-2 isolates
HSV-2 isolates were collected from patients with genital lesions attending a clinic for sexually transmitted diseases in Bergen, Norway. Blood samples were analysed for the presence of HSV-2 antibodies as described previously [28]. Briefly, an oligopeptide corresponding to an antigen region in glycoprotein G of HSV-2 (gG-2) was used in an enzyme-linked immunoabsorbent assay (ELISA). Patients with primary infection were selected on the basis that there was no information of previous genital infection, and HSV-2-specific antibodies were not detected when presenting with blisters and/or ulcers for the first time. Five persons were included in the study. Description of them is given in Table 1. The patients were examined again at the first reactivation of genital herpes. Sterile Dacron swabs were used to collect lesional specimens. The swabs were stored in liquid virus transport medium. Virus from primary infection was confirmed as HSV-2 by using nested PCR targeting the type-specific promoter region of the gD-2 gene as described by Cinque et al. [29] and slightly modified [30]. The clinical isolates were analysed further at a low passage number (less than 5).
Table 1.
Patients included in the study
| Patient no * | Time between primary and recurrent infection |
Treatment (valaciclovir) |
|
|---|---|---|---|
| Primary infection | Recurrent infection | ||
| 1 |
4 months |
Yes |
No |
| 2 |
2 weeks |
Yes |
Yes |
| 3 |
3 weeks |
Yes |
No |
| 4 |
3 months |
Yes |
Yes |
| 5 | 6 weeks | Yes | Yes |
*All patients were women between 19 and 35 years old.
Ethics and consent statement
The study was approved by the Regional Committee for Medical and Health Research Ethics, Western-Norway, University of Bergen. All patients were informed about the various aspects of the project, including isolation of virus from the samples, analysis of the viruses and publication of the results. The samples and results were coded to by anonymous. Consent was obtained from all patients.
DNA isolation, PCR amplification and sequencing
Virus DNA was isolated using the Qiagen DNeasy Tissue Kit, as described by the manufacturer. The concentration varied from 25 to ≥ 1200 ng per μl. Three regions of the HSV-2 genome were amplified prior to sequencing. The UL27 gene (encoding glycoprotein B, gB-2) was amplified as a 2942-bp fragment spanning the region from 78 bp upstream of the start codon to 149 bp downstream of the termination codon (the positions refer to strain HG52). Several additional primers were used for sequencing (Table 2). Amplification of the US4 gene (encoding glycoprotein G, gG-2) and a fragment containing both the US7 and US8 genes (encoding glycoproteins gI and gE, gI-2 and gE-2, respectively) as well as noncoding sequences between the two genes were performed as described previously [31]. The fragment including the US4 gene contained 57 bp upstream and 39 bp downstream of the coding sequences. Similarly, the latter fragment contained 57 bp upstream of the start codon of the US7 gene and 47 bp downstream of the US8 gene. The primers used for amplification and sequencing of the two latter fragments have been published [31,32].
Table 2.
Primers used for amplification and sequencing of the gB gene
| Nucleotide position | Type | Sequence |
|---|---|---|
|
53254–53273 |
AS |
5′-CCGTTAGCACATGTCTGCAT |
| 53437–53456 |
S |
5′-AGGTACTCTCCGCTCCACAA |
| 53522–53541 |
AS |
5′-TCTTTCTGGCCTTGTGTTCC |
| 53710–53739 |
S |
5′-CTACGTCCTGCAACTGCAAC |
| 53801–53820 |
AS |
5′-CGAAGGGGTTGGACATAAAG |
| 54013–54032 |
S |
5′-CTGCTGGACTACACGGAGGT |
| 54108–54128 |
AS |
5′-AGGTCGATGAAGGTGCTGAC |
| 54299–54319 |
S |
5′-GCTTTCGGTACGAAGACCAG |
| 54371–54390 |
AS |
5′-AGTTCTGCACGATCACGTTG |
| 54510–54529 |
S |
5′-ACCACGAGCTGACTCTCTGG |
| 54730–54749 |
AS |
5′-TACTCCCGCACGTACAGCTC |
| 54943–54962 |
S |
5′-ATCTCGACCACCTTCACCAC |
| 55027–55046 |
AS |
5′-CACTTGGTCATGGTGCAGAC |
| 55232–55252 |
S |
5′-TTGTGTACATGTCCCCGTTTT |
| 55372–55391 |
AS |
5′-TACTTGAGGTCGGTGGTGTG |
| 55481–55500 |
S |
5′-CAAGTACGTGCGGAACAACA |
| 55643–55662 |
S |
5′-CAAATTCAAGGCCACCATGT |
| 55648–55667 |
AS |
5′-GTGGCCTTGAATTTGTACGG |
| 55914–55923 |
AS |
5′-CTTTTTGGTTTTCCGCTTCC |
| 55914–55923 |
S |
5′-GGAAGCGGAAAACCAAAAAG |
| 56176–56195 | S | 5′-CCATCCTCTACTCGGTCCTG |
Nucleotide positions are given for the reference strain HG52, GenBank accession number Z 86099.2. Boldface indicates primers used for amplification, the rest were used for sequencing. S: sense. AS: antisense.
PCR and gel electrophoresis of the amplified fragments were performed as described previously [30]. Briefly, Tfl DNA polymerase (Epicentre, Madison, USA) and buffer solution GN (Epicentre, Madison, USA) were used in a total volume of 50 μl containing 5 μl of diluted, purified DNA extract. The incubation steps were 5 min initial denaturation at 96°C, 30 cycles of denaturation at 95°C for 1 min, annealing of primers for 1 min at 57°C for the US7-US8 fragment or at 60°C for the UL27 and US4 fragments, elongation for 3 min at 68°C and a final extension cycle at 68°C for 15 min.
Sets of overlapping primers as shown in Table 2 and elsewhere [31,32] were used for sequencing. PCR products were treated with Exo-SAP (Affymetrix, Ca. USA) and sequenced using the Big-Dye kit ver.3.1 (Life Tech., Ca. USA) Unincorporated dyes were removed using the Big-Dye X-terminator purification kit (Life Tech., Ca. USA) and the samples were analysed on an ABI 3730 sequencer (Life Tech., Ca. USA).
Sequence analysis
The sequences were analysed using the SeqScape software (Life Tech., Ca. USA), using JN561323.1 in the NCBI GenBank as a reference sequence. Sequences for the reference strain and for the clinical isolates were converted to FASTA (http://www.ebi.ac.uk/cgi-bin/readseq.cgi) and alignment performed using Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Both programs are connected to the EMBL-EBI database.
The genome of HSV-2 HG52 has two accession numbers in the GenBank, Z 86099.2 and JN 561323.2, respectively. The latter was submitted more recently than the former. Since previous published primers were also used in the present study, and their nucleotides were numbered with reference to Z 86099.2, the same reference was used for the new primers (Table 2). However, when comparing the database sequences of various clinical isolates with the reference strain, accession number JN 561323.2 was used.
Results and discussion
To analyse for potential variation of the virus from primary to recurrent infection, we focused on genes reported to show polymorphism. They encode glycoprotein B (gB), glycoprotein G (gG), glycoprotein I (gI) and glycoprotein E (gE), respectively. The nucleotide diversity of these genes can be studied by comparing the sequences of clinical isolates of HSV-2 with the reference strain HSV-2 HG52. In the NCBI GenBank each of these genes have been sequenced in a number of isolates, varying from 48 to 69. The results of this comparison are shown in Table 3. Substitutions affecting 2 isolates or more are specified.
Table 3.
Sequence differences between HSV-2 HG52 and clinical isolates of HSV-2 in the database
|
Gene |
UL27/gB (2715nt) |
US4/gG (2100nt) |
US7/gI (1119nt) |
US8/gE (1638nt) |
||||||||
| Total no. of isolates |
69 |
64 |
49 |
48 |
||||||||
| Nucleotide diversity (%) affecting: | ||||||||||||
| 1 isolate |
1.14 |
2.24 |
1.70 |
1.40 |
||||||||
| ≥2 isolates |
0.81 |
1.52 |
0.80 |
0.55 |
||||||||
| |
Position
|
No. of isolates
|
Substitutions
,
comments
|
Position
|
No. of isolates
|
Substitutions
,
comments
|
Position
|
No. of isolates
|
Substitutions
,
comments
|
Position
|
No. of isolates
|
Substitutions
,
comments
|
| |
19 |
21 |
A ⇒ G |
104 |
58 |
G ⇒ A |
39 |
5 |
C ⇒ T |
131 |
46 |
ins. GGC CGG AGG |
| |
64-72 or 76-84* |
66 |
del. GCC CCG GCG or GCG GCC CCG |
274 |
26 |
T ⇒ C |
338 |
2 |
A ⇒ G |
341 |
2 |
G ⇒ T |
| |
104 |
2 |
G ⇒ A |
329 |
17 |
G ⇒ A |
530 |
2 |
C ⇒ T |
392 |
11 |
T ⇒ G |
| |
106 |
2 |
G ⇒ A |
405 |
8 |
C ⇒ T |
618 |
3 |
A ⇒ G |
605 |
6 |
C ⇒ A |
| |
117 |
6 |
C ⇒ G |
432 |
22 |
G ⇒ C |
642 |
2 |
A ⇒ C |
1146 |
8 |
A ⇒ G |
| |
146 |
48 |
G ⇒ A |
611 |
10 |
C ⇒ T |
643 |
13 |
C ⇒ T |
1211 |
14 |
A ⇒ C |
| |
179 |
13 |
A ⇒ G |
635 |
13 |
G ⇒ A |
644 |
6 |
C ⇒ T |
1245 |
2 |
C ⇒ T |
| |
210 |
8 |
C ⇒ T |
872 |
3 |
A ⇒ G |
716 |
45 |
T ⇒ G |
1293 |
2 |
C ⇒ T |
| |
211 |
13 |
A ⇒ G |
878-880 |
16 |
del. TCG |
717 |
11 |
A ⇒ C |
1621 |
44** |
C ⇒ T |
| |
850 |
2 |
C ⇒ T |
891 |
4 |
G ⇒ A |
|
|
|
|
|
|
| |
989 |
16 |
G ⇒ A |
930 |
50 |
C ⇒ T |
|
|
|
|
|
|
| |
1186 |
35 |
G ⇒ C |
982 |
2 |
G ⇒ A |
|
|
|
|
|
|
| |
2247 |
47 |
A ⇒ C |
993 |
2 |
G ⇒ T |
|
|
|
|
|
|
| Specification of substitutions affecting ≥2 isolates | 2533 |
29 |
G ⇒ C |
1045 |
19 |
G ⇒ A |
|
|
|
|
|
|
| |
|
|
1048 |
64 |
A ⇒ G |
|
|
|
|
|
|
|
| |
|
|
1116 |
64 |
A⇒ G |
|
|
|
|
|
|
|
| |
|
|
1125 |
2 |
C ⇒ A |
|
|
|
|
|
|
|
| |
|
|
1268 |
37 |
T ⇒ C |
|
|
|
|
|
|
|
| |
|
|
1282-1285 or 1284-1286* |
63 |
del.GCG or GGC |
|
|
|
|
|
|
|
| |
|
|
1324 |
7 |
T ⇒ C |
|
|
|
|
|
|
|
| |
|
|
1419 |
2 |
A ⇒ G |
|
|
|
|
|
|
|
| |
|
|
1470 |
7 |
G ⇒ A |
|
|
|
|
|
|
|
| |
|
|
1510 |
3 |
A ⇒ G |
|
|
|
|
|
|
|
| |
|
|
1722 |
2 |
G ⇒ T |
|
|
|
|
|
|
|
| |
|
|
1758 |
2 |
T ⇒ C |
|
|
|
|
|
|
|
| |
|
|
1761 |
7 |
G ⇒ C |
|
|
|
|
|
|
|
| |
|
|
1853 |
4 |
G ⇒ A |
|
|
|
|
|
|
|
| 1994 | 2 | T ⇒ G | ||||||||||
*Alternative alignments. **Lacking sequences in the 3′- end of a few isolates. Nt nucleotides, Del Deletion, Ins insertion. Positions are numbered from the first nucleotide in the start codon.
A consistent feature was that whenever a substitution was observed at a given position, it was identical in all isolates affected, regardless of the number of isolates subjected to change. This was the situation for all 4 genes. Except for deletion of 9 nucleotides in UL27 (gB), and of three nucleotides at two different positions in US4 (gG), the remaining changes were single nucleotide substitutions. Reiteration of sequences may cause alternative alignment, as observed both for the gB and the gG gene. Thus, alternative positions for substitutions may be indicated by the computer. From positions 62 to 88 in the former gene the sequence CGG CGG CCC is repeated three times. Similarly, CGG is repeated three times from position 1277 to 1285 in the gG gene. Identical variations in the same position in two or more clinical isolates indicate recombination. This was observed at numerous sites in all genes and is consistent with the fact that the isolates are from studies demonstrating different clades of virus and recombination [31,33]. However, neither sequences nor positions for nucleotide substitutions were given in these reports. In addition to substitutions generated by recombination, a number of single nucleotide substitutions affected only one clinical isolate. Such changes could be generated by random mutations or by recombination, but one cannot distinguish between these possibilities.
Nucleotide diversity affecting one isolate only varied from 1.14% (UL27/gB) to 2.24% (US4/gG). Diversity affecting two isolates or more was lower, but still highest for the US4 gene. These results indicate that the 4 selected genes might be suitable candidates for detection of potential genetic variation among viruses included in the present study.
All viral isolates were from females between 19 and 35 years old (Table 1). The time between collection of the first virus sample (primary infection) and the second one (recurrent infection) varied between 2 weeks and 4 months. All patients received treatment with valaciclovir at the first episode and 3 of them also at the second one. One patient might have had a recurrent infection between the episodes without attending the hospital clinic.
The four glycoprotein genes were sequenced in all HSV-2 isolates. Sequencing spanned the entire open reading frames and a variable number of nucleotides in the 5′- and 3′- untranscribed regions of each gene. When analysing the results from each patient separately, comparing sequences from primary to recurrent infection, no difference was detected in any patient, as indicated in Figure 1. One might not expect a substantial variation due to mutation, since proof reading during synthesis of HSV-DNA has been reported to be efficient and the rate of nucleotide substitution estimated to be 3 × 10-8 per site per year [34]. Similar genetic stability of HSV-2 has been observed under other conditions. Terhune et al. [35] studied isolates of HSV-2 propagated in cell culture and did not observe sequence variation in any of the genes encoding gB, gC and gD, respectively. Figure 1 also shows the genetic relationship between the viruses isolated from the 5 patients. All 5 isolates are clearly different.
Figure 1.
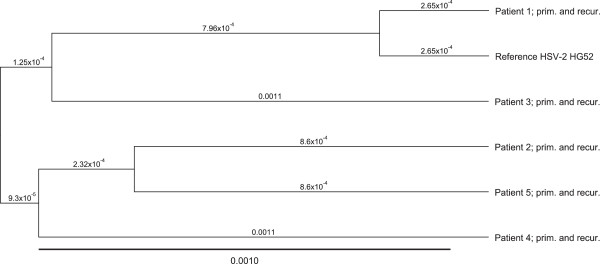
Genetic relationship between the viruses included in the study. The tree is based upon the open reading frame nucleotide sequences of the genes encoding gB, gG, gI and gE.
There was no indication of recombination, but we cannot exclude the possibility that it might have occurred, as discussed below. For recombination to occur, at least two different viruses have to infect the same host. Such infection could be in previously uninfected sensory neurons since only a small portion of neurons harbour virus DNA in the latent state. For HSV-1 this has been shown to be between 2% and 11% [36]. However, two different viruses could also infect a single neuron, as shown for HSV-1 and VZV [37]. Since reactivation of HSV-2 is a commonly occurring event, several of them being asymptomatic [18-20], the patients could repeatedly be exposed to HSV-2 during the study, through their partners. The partners were not included in the study. Absence of detectable recombination could suggest that, in a given couple, the same type of virus was present at all times. Alternatively, the dose of a second virus could have been too low to establish a new infection [38].
All genes in the present work encode glycoproteins present in the viral envelope. Among their important roles are involvement of gB in binding virus to the cell membrane, and gE and gI acting in axonal transport of capsids and/or virions [21]. Since gG of HSV-2 is larger than that of HSV-1, it has been used as a tool for serodiagnosis of HSV-2 infection [28]. The total number of nucleotides in the open reading frames of these genes is approximately 5% of the total HSV-2 genome. Thus, the possibility should be left open that sequence variation, including recombination, might have occurred in the period from primary to recurrent infection in parts of the viral genome outside the analysed portions.
Conclusions
Although infections of cell cultures and animal models are very useful for studies of herpes simplex virus (and other viruses), it is important to know how the virus behaves in the natural host. We have studied lesional virus isolates from humans with genital HSV-2 infection and observed that several glycoprotein gene sequences are stable from primary to recurrent infection. However, isolates from the different patients were genetically different.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AN collected blood samples and lesional material from the patients. LH was responsible for growing virus. Sequencing was a collaboration between PK and LH. NL was involved in design of the study and further discussions, and in the preparation of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Lars Haarr, Email: lars.haarr@k2.uib.no.
Arvid Nilsen, Email: arvid.nilsen@helse-bergen.no.
Per M Knappskog, Email: per.knappskog@helse-bergen.no.
Nina Langeland, Email: Nina.Langeland@mofa.uib.no.
Acknowledgements
We are grateful for the excellent technical assistance of Guri Matre, and want to thank Ove Bruland and Øyvind Kommedal for useful discussion. A special thank goes to Reidun Hægland for her patience and commitment and for her conscientious and skilful work in the laboratory.
References
- Smith JS, Robinson J. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J Infect Dis. 2002;14(Suppl 1):S3–28. doi: 10.1086/343739. [DOI] [PubMed] [Google Scholar]
- Lafferty WE, Downey L, Celum C, Wald A. Herpes simplex virus type 1 as a cause of genital herpes: impact on surveillance and prevention. J Infect Dis. 2000;14:1454–1457. doi: 10.1086/315395. [DOI] [PubMed] [Google Scholar]
- Nilsen A, Myrmel H. Changing trends in genital herpes simplex virus infection in Bergen, Norway. Acta Obstet Gynecol Scand. 2000;14:693–696. [PubMed] [Google Scholar]
- Aurelius EB, Johansson B, Skoldenborg B, Forsgren M. Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or type 2 as determined by type-specific polymerase chain reaction and antibody assay of cerebrospinal fluid. J Med Virol. 1993;14:179–186. doi: 10.1002/jmv.1890390302. [DOI] [PubMed] [Google Scholar]
- Sköldenberg B, Aurelius E, Hjalmarsson A, Sabri F, Forsgren M, Andersson B, Linde A, Strannegård Ö, Studahl M, Hagberg L, Rosengren L. Incidence and pathogenesis of clinical relapse after herpes simplex encephalitis. J Neurol. 2006;14:163–70. doi: 10.1007/s00415-005-0941-6. [DOI] [PubMed] [Google Scholar]
- Tyler KL. Herpes simplex virus infections of the central nervous system: encephalitis and meningitis, including Mollaret’s. Herpes. 2004;14(Suppl. 2):57A–64A. [PubMed] [Google Scholar]
- Rozenberg F, Deback C, Agut H. Herpes simplex encephalitis: from virus to therapy. Infect Disord Drug Targets. 2011;14:235–250. doi: 10.2174/187152611795768088. [DOI] [PubMed] [Google Scholar]
- Schiffer JT, Corey L. New concepts in understanding genital herpes. Curr Infect Dis Rep. 2009;14:457–464. doi: 10.1007/s11908-009-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle DK, Corey L. Herpes simplex: insights on pathogenesis and possible vaccines. Ann Rev Med. 2008;14:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- Hashido M, Lee FK, Nahmias AJ, Tsugami H, Isomura S, Nagata Y, Sonoda S, Kanawa T. An epidemiologic study of herpes simplex virus type 1 and 2 infection in Japan based on type-specific serological assays. Epidemiol Infect. 1998;14:179–186. doi: 10.1017/S095026889700856X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Salas F, Hernandéz-Avila M, Juárez-Figueroa L, Conde-Glez CJ, Uribe-Zúniga P. Risk factors for herpes simplex type 2 infection among female commercial sex workers in Mexico City. Int J STD AIDS. 1999;14:105–111. doi: 10.1258/0956462991913727. [DOI] [PubMed] [Google Scholar]
- Nilsen A, Mwakagile D, Chalamila G, Langeland N, Matre R, Haarr L. Demographic and behavioural factors in Tanzanian and Norwegian patients with sexually transmitted infections. Acta Derm Venereol. 2006;14:320–328. doi: 10.2340/00015555-0100. [DOI] [PubMed] [Google Scholar]
- Cherpes TL, Meyn LA, Krohn MA, Hillier SL. Risk factors for infection with herpes simplex virus type 2: role of smoking, douching, uncircumcised males, and vaginal flora. Sex Transm Dis. 2003;14:405–410. doi: 10.1097/00007435-200305000-00006. [DOI] [PubMed] [Google Scholar]
- Xu F, Sternberg MR, Kottirie B, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz L. Trends in herpes simplex virus type 1 and 2 seroprevalence in the United States. JAMA. 2006;14:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- Freeman EE, Weiss HA, Glynn JR, Cross P, Whitworth JA, Hayes RJ. Herpes simplex virus type 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;14:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- Brown EL, Wald A, Hughes JP, Morrow RA, Krantz E, Mayer K, Buchbinder S, Koblin B, Celum C. High risk of human immunodeficiency virus in men who have sex with men with herpes simplex virus type 2 in the EXPLORE study. Am J Epidemiol. 2006;14:733–741. doi: 10.1093/aje/kwj270. [DOI] [PubMed] [Google Scholar]
- Abu-Raddad LJ, Magaret AS, Celum C, Wald A, Longini IM, Self SG, Corey L. Genital herpes has played a more important role than any other sexually transmitted infection in driving HIV prevalence in Africa. PLos One. 2008;14:e2230. doi: 10.1371/journal.pone.0002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti J, Corey L, Ashley R. Recurrence rates in genital herpes after symptomatic first-episode infection. Ann Intern Med. 1994;14:847–854. doi: 10.7326/0003-4819-121-11-199412010-00004. [DOI] [PubMed] [Google Scholar]
- Mark K, Wald A, Magaret AS, Selke S, Olin L, Huang M-L, Corey L. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J Infect Dis. 2008;14:1141–1149. doi: 10.1086/591913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald A, Zeh J, Selke S, Warren T, Runcarz AJ, Ashley R, Krieger JN, Corey L. Reactivation of genital herpes simplex type 2 virus infection in asymptomatic seropositive persons. N Engl J Med. 2000;14:844–850. doi: 10.1056/NEJM200003233421203. [DOI] [PubMed] [Google Scholar]
- Smith G. Herpesvirus transport to the nervous system and back again. Ann Rev Microbiol. 2012;14:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll MP, Proenca JT, Efstathiou S. The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev. 2012;14:6894–705. doi: 10.1111/j.1574-6976.2011.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;14:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Zabolotny JM, Krummenacher C, Fraser NW. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;14:4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rødahl E, Haarr L. Analysis of the 2-kilobase latency–associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structures. J Virol. 1997;14:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;14:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak K, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;14:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen A, Ulvestad E, Marsden H, Langeland N, Myrmel H, Matre R, Haarr L. Performance characteristics of a glycoprotein G-based oligopeptide (peptide 55) and two different methods using the complete glycoprotein as assays for detectionm of anti-HSV-2 antibodies in human sera. J Virol Methods. 2003;14:63–70. doi: 10.1016/s0166-0934(02)00185-4. [DOI] [PubMed] [Google Scholar]
- Cinque P, Vago L, Dahl H, Brytting M, Terreni MR, Fornara C, Racca S, Castagna A, Monforte AD, Wahren B, Lazzarin A, Linde A. Polymerase chain reaction on cerebrospinal fluid for diagnosis of virus-associated opportunistic diseases of the central nervous system in HSV-infected patients. AIDS. 1996;14:951–58. doi: 10.1097/00002030-199610090-00004. [DOI] [PubMed] [Google Scholar]
- Nilsen A, Kasubi MJ, Mohn SC, Mwakagile D, Langeland N, Haarr L. Herpes simplex virus infection and genital ulcer disease among patients with sexually transmitted infections in Dar es Salaam, Tanzania. Acta Derm Venerol. 2007;14:355–359. doi: 10.2340/00015555-0241. [DOI] [PubMed] [Google Scholar]
- Norberg P, Kasubi MJ, Haarr L, Bergström T, Liljeqvist J-Å. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J Virol. 2007;14:13158–13167. doi: 10.1128/JVI.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeqvist J-Å, Svennerholm B, Bergström T. Herpes simplex type 2 glycoprotein G-negative clinical isolates are generated by single frameshift mutations. J Virol. 1999;14:9796–9802. doi: 10.1128/jvi.73.12.9796-9802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Chanasit J, Bialonski A, Heinemann P, Ulrich RG, Günther S, Rabenau HF, Doerr HW. A 12-year molecular survey of clinical herpes simplex virus type 2 isolates demonstrates the circulation of clade A and B strains in Germany. J Clin Virol. 2010;14:208–2011. doi: 10.1016/j.jcv.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Sakaoka H, Kurita H, Iida Y, Takada S, Umene K, Kim YT, Ren CS, Nahmias AJ. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J Gen Virol. 1994;14:513–527. doi: 10.1099/0022-1317-75-3-513. [DOI] [PubMed] [Google Scholar]
- Terhune SS, Coleman KT, Sekulovich R, Burke RL, Spear PG. Limited variability of glycoprotein gene sequences and neutralizing targets in herpes simplex virus type 2 isolates and stability on passages in cell culture. J Infect Dis. 1998;14:8–15. doi: 10.1086/515590. [DOI] [PubMed] [Google Scholar]
- Wang K, Lau TY, Morales M, Mont EK, Straus S. Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella zoster DNA in human trigeminal ganglia at the single-cell level. J Virol. 2005;14:14079–14087. doi: 10.1128/JVI.79.22.14079-14087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil D, Patipovic I, Derfuss T, Herberger S, Strupp M, Arbusow V, Brandt T. Dually infected (HSV-1/VZV) single neurons in human trigeminal ganglia. Ann Neurol. 2003;14:678–681. doi: 10.1002/ana.10746. [DOI] [PubMed] [Google Scholar]
- Hoshio Y, Qin J, Follmann D, Cohen JI, Straus SE. The number of herpes simplex virus-infected neurons and the number of viral genome copies per neuron correlate with the latent load in ganglia. Virology. 2008;14:56–63. doi: 10.1016/j.virol.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]


