Abstract
By combining pseudo-random bead-based aptamer libraries with conjugation chemistry, we have created next-generation aptamers, X-aptamers (XAs). Several X ligands can be added in a directed or random fashion to the aptamers to further enhance their binding affinities to the target proteins. Here we describe the addition of a drug (N-acetyl-2,3-dehydro-2-deoxyneuraminic acid) demonstrated to bind to CD44-HABD, to a complete monothioate backbone substituted aptamer to increase its binding affinity to the target protein by up to 23-fold, while increasing the drugs’ binding 1-million fold.
Keywords: X-Aptamer, thioaptamer, aptamer selection, bead-based library, CD44-HABD
Aptamers are single stranded RNA and DNA oligonucleotides that can bind a specific molecular target and exhibit high binding affinity. Aptamers are thus emerging as viable alternatives to small molecules and antibodies for many applications in research, diagnostics, imaging, and therapeutics.1–2
A diverse range of modifications have been reported1,3–5 for either enhancing aptamers’ nuclease resistance or expanding their chemical functionalities. Here we present the first example of X-aptamers which was endowed with both properties by adding drug-like molecules to 5-positions of certain uridines on a complete monothiophosphate-backbone substituted oligonucleotide aptamer. By combining our one-bead, one-sequence thioaptamer selection method6,7 with the incorporation of pseudo-randomly placed bases containing chemical linkers, additional X-ligands can be appended onto aptamers or thioaptamers to create a next-generation, X-aptamer library, and the best binding X-aptamers can be selected from this large pool of sequences. In a previous report8, we described thioaptamers substituted with monothiophosphates on the 5’ side of dA that bind to the hyaluronic acid binding domain of CD44 (CD44-HABD) (KD = 187–295 nM). Based on the primary sequence of several of these thioaptamers and observed variations, we synthesized a pseudo-random, one-bead-one-sequence bead library of aptamers using an automated four-column, split-pool synthesizer9. This synthesis produced a library with >1 million (410) unique X-aptamer sequences (Figure 1) in which X = 5-(aminoethyl-3-acrylimido)-deoxyuridine (amino-dU) or later, a conjugated drug-like appendage. The bead-based library consists of a 5’-primer region, a 30 nucleotide pseudo-random sequence (9 split/pool steps), and a 3’-primer region that is covalently linked by a non-cleavable hexaethyleneglycol linker to a 65-m polystyrene bead (ChemGenes). The unique sequence on a given bead may contain zero to 12 X-positions, but three or four Xs is most likely. In order to select highly nuclease-resistant X-aptamers, the library was prepared with a fully monothiophosphate (permonothioated) backbone. This original library served as the base library from which a variety of additional X-aptamer libraries were derived by conjugation with NHS-ester forms of drug-like molecules. To select small molecule ligands as binding affinity enhancers that could be attached to the X-aptamer base library, we carried out in silico screening using AMBER910 and DOCK6.411 programs (see the Supporting Information for details). ADDA (N-acetyl-2,3-dehydro-2-deoxyneuraminic acid) was selected as the lead compound (Figure S3B of the Supporting Information), and several other compounds are under investigation. ADDA binds to CD44-HABD with an equilibrium dissociation constant of 2.22 ± 0.82 mM (Supporting Information). The docking experiments suggested that ADDA binds into part of the hyaluronic acid binding pocket which is believed also to be the binding site of the originally selected thioaptamers (Figure 2A and Figure S3 of the Supporting Information).
Figure 1.

Four sequences used in the split-pool, complete monothioate XA library synthesis and one example of resulting bead sequence. X = 5-(aminoethyl-3-acrylimido)-deoxyuridine created by incorporation of “Amino Modifier C2 dT” (Glen Research). Dashes represent the presence of a split and pool synthesis step. The example sequence in Figure 1 above would result from a bead that followed the column path (from 3’ to 5’) 3-4-2-4-1-3-2-4-2-1, depicted as shaded parts, during the split-pool method.
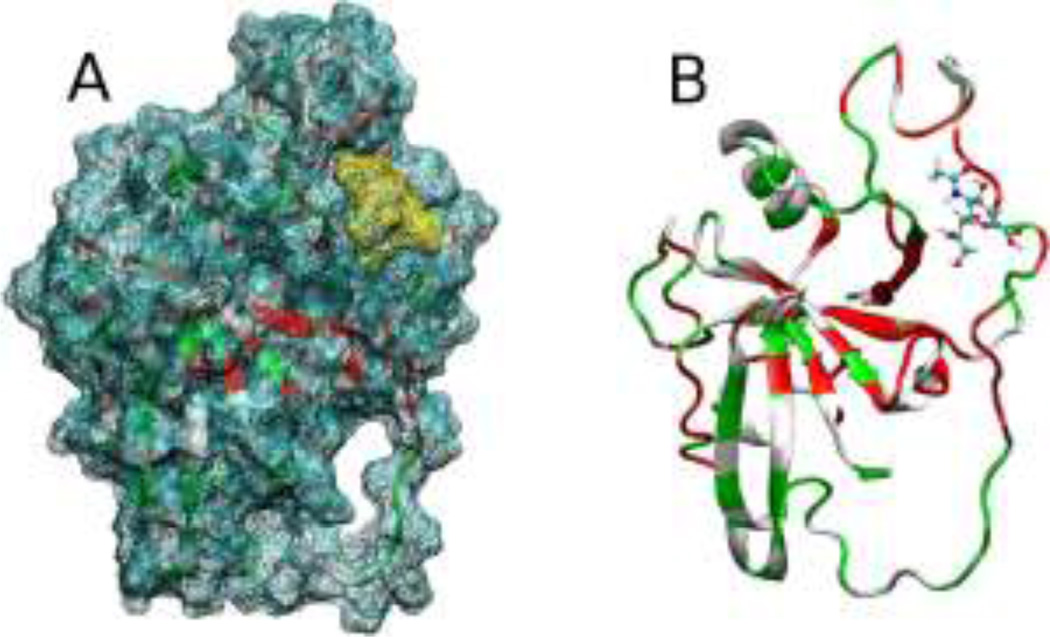
Figure 2.
(A) Predicted docking of ADDA to CD44-HABD. (B) Chemical shift changes observed in an 1H,15N-HSQC experiment mapped onto the structure of CD44-HABD.12 Large changes are indicated in red, small changes in white, and unassigned residues in green.
The 15N-HSQC NMR experiments (Figure 2B and Figure S4A of the Supporting Information) also support these calculations, as shown by large changes in amide proton chemical shifts13 surrounding the purported binding pocket.
ADDA was conjugated to the amino-dU of the original XA library by using amine-carboxy coupling. A bead-based selection method was conducted to find the XAs specific for CD44-HABD (see the Supporting Information for details). The selected positive X-aptamer beads were used for ‘one-bead one-PCR’ amplification process6,7. The X positions in the sequence were determined based on the original library design, using the adjacent bases as “bar-coding”. Table S1 (Supporting Information) lists the XA sequences obtained.
Nitrocellulose-nylon filter binding assays were used to determine equilibrium dissociation constants (KD) of the selected XAs to CD44-HABD, and are listed in Table S2 (Supporting Information). Saturation binding curves were generated by curve fits assuming a single binding site.
Although our previously reported8 thioaptamer had a KD value of 191 ± 25 nM, the sequence was monothioated adjacent to only the dAs, with nuclease-susceptible phosphate backbones at all remaining positions. When the same sequence was permonothioated to enhance nuclease resistance, it bound to CD44-HABD with similar affinity (KD = 230 ± 47 nM, Table 1). However, the selected permonothioated amino X-aptamers and ADDA-modified X-aptamers exhibited KD values around 60–80 nM (Table S2 of the Supporting Information), approximately three- to four-fold improvement from the unmodified permonothioate thioaptamers.
Table 1.
X-aptamer sequences and equilibrium dissociation constants of binding to CD44 hyaluronic acid binding domain
| Dissociation constant (nM) | |||||
|---|---|---|---|---|---|
| Parent a | Full-length sequence, partially monothioated | 191 ± 25 b | |||
| Parent a | Full-length sequence permonothioated | 230 ± 47 | |||
| Phospho X-aptamers | Thiophospho X-aptamers | ||||
| Motif Sequence c | X = amino-dU | X = ADDA-dU d | X= amino-dU | X = ADDA-dU d | |
| Motif 1 e | 10.3 ± 1.3 | 15.0 ± 2.0 | |||
| Motif 2 | 43.1 ± 9.5 | 2.0 ± 0.6 | 48.0 ± 18.0 | 15.5 ± 3.2 | |
| Motif 3 | 27.6 ± 3.5 | 19.5 ± 3.2 | 81.2 ± 30.9 | 64.8 ± 13.7 | |
| Motif 4 | 6.8 ± 1.8 | 2.1 ± 0.2 | 35.4 ± 7.4 | 13.6 ± 3.0 | |
| Motif 5 | 13.8 ± 4.1 | 3.9 ± 1.0 | 18.0 ± 3.7 | 10.1 ± 2.6 | |
The sequence of the full-length parent aptamer is shown in the Col. 1 part of Figure 1 and corresponds to TA1 from reference 3;
from reference 3;
alternating shading indicates split-pool sequence sections in Figure 1 for these aligned sequences;
ADDA-dU is the ADDA adduct with 5-(aminoethyl-3-acrylimido)-deoxyuridine;
Motif 1, which contains no X, was included to compare the phosphoaptamer and the thioaptamer forms.
Secondary structure predictions performed using MFold14 suggested that all selected XA sequences can form hairpin loop structures in which the random regions form loops and the primers form stem regions (Figure S1 of the Supporting Information). Based on these predicted structures, we identified several binding motifs and smaller constructs of various stem-loop regions. The equilibrium binding constants of these small XA constructs (Table 1) were also determined by filter binding assays.
Remarkably, coupling ADDA with smaller stem-loop constructs from the best X-aptamer sequences, motifs 2 and 4(ADDA adduct) have ~2 nM affinity to CD44-HABD, which is an increase in binding affinity of ~115-fold between the full-length permonothioated parent sequence and the final ADDA-conjugated XA (phosphoform) and ~23-fold between the full-length sequence complete monothioates and motif 5 (thiophosphoform). Moreover, in every case in Table 1, the ADDA-conjugated XA showed increased affinity compared to the unconjugated XA. In the best case, ADDA conjugation increased affinity ~22-fold. ADDA is a weaker binder (~2 mM) to CD44-HABD. By conjugation to an aptamer, the binding affinity of ADDA modified XAs has improved the affinity 1 million fold (2 nM).
By introducing a protein binding small drug molecule, ADDA, into the 5-position of dU residues at random positions of the aptamers and/or replacing one of the non-bridging phosphate oxygen atoms with sulfur atoms, we are able to select an X-aptamer with <10 nM affinity to CD44-HABD through a non-iterative bead-based selection from large combinatorial libraries of X-aptamers. Our bead-based method is compatible with both monothiophosphate- and dithiophosphate-modified thioaptamers, and even complete monothiophosphate modification of the backbone, as reported here, which is not possible with traditional SELEX methods15,16. In addition, only one or two rounds of aptamer selection are required in contrast to the 10–15 rounds necessary in traditional SELEX.
As expected, the effect of ADDA as binding affinity enhancer is location dependent within the aptamer. The process to find optimal position of the ligand was part of our X-aptamer selection since ADDA was attached to the aptamers at various positions. By simultaneously selecting optimal sequence of the aptamer scaffold and orientation and position of the small drug presented by the optimal scaffold, we enhance affinities. The incorporation of ADDA not only expands the XA’s chemical diversity but also the surface area of binding, thus the XA can also offer enhanced specificity.
While the present work describes conjugation with one specific drug at a time, multiple drug hits can be randomly attached as well, to provide enhanced combinations of binding moieties. More than one ligand can be attached by pausing the DNA synthesis for the addition of a ligand and subsequent DNA synthesis and coupling reactions. By using two or more chemical linkers in one root library, multiple drugs can be selectively incorporated. Our methodology can be applied to most target proteins with a variety of small molecules to create highly chemically modified X-aptamers that have the combined characteristics of drug molecules, proteins and nucleic acids.
Supplementary Material
ACKNOWLEDGMENT
We thank the W. M. Keck Foundation and the John S. Dunn, Sr. Foundation for supporting the John S. Dunn, Sr. Gulf Coast Consortium for Magnetic Resonance, which purchased the 800 MHz NMR spectrometer used in these studies. The authors also thank Sean Moran (Rice University NMR).
Funding Sources
Supported by the Welch Foundation (AU-1296), NCI (CA151668), NIAID (HHSN272200800048C and AI054827), NHLBI (HHSN268201000037C), NICHD (NO1-HD-80020), NIGMS (RC2GM092599ARRA and GM076695), and DoD (W81XWH-09-1-0212 and W81XWH-09-2-0139).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Additional experimental details, MD protocols, and Supplemental figures and tables. This material is available free of charge via the Internet at http://pubs.acs.org
Author Contributions
DV, AS, VT and DG designed the bead library; JE synthesized the bead library; ME and CC designed and conducted docking and MD experiments; WH conjugated drugs to the library; WH and RD designed and conducted bead selection experiments; WH, XL and GL performed binding assays. All authors contributed to writing the manuscript.
Notes
Dr. Gorenstein and the University of Texas Health Science Center at Houston have research related financial interests in AptaMed Inc. and AM Biotechnologies LLC, Houston, TX, USA.
REFERENCES
- 1.Yang X, Li N, Gorenstein DG. Expert Opin Drug Discov. 2011;6:75–87. doi: 10.1517/17460441.2011.537321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song KM, Lee S, Ban C. Sensors. 2012;12:612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody EN, Gold L, Lawn RM, Walker JJ, Zichi D. Expert Rev Mol Diagn. 2010;10:1013–1022. doi: 10.1586/erm.10.89. [DOI] [PubMed] [Google Scholar]
- 4.Battersby TR, Ang DN, Burgstaller P, Jurczyk SC, Bowser MT, Buchanan DD, Kennedy RT, Benner SA. J. Am. Chem. Soc. 1999;121:9781–9789. doi: 10.1021/ja9816436. [DOI] [PubMed] [Google Scholar]
- 5.Keefe AD, Cload ST. Current Opinion in Chemical Biology. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, Ellington AD, Gorenstein DG. Nucleic Acids Research. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, Herzog NK, Aronson J, Shope RE, Leary JF, Gorenstein DG. Nucleic Acids Research. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somasunderam A, Thiviyanathan V, Tanaka T, Li X, Neerathilingam M, Lokesh GLR, Mann A, Peng Y, Ferrari M, Klostergaard J, Gorenstein DG. Biochemistry. 2010;49:9106–9112. doi: 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelhardt JP, Gorenstein DG, Luxon B, Herzog N. 7,576,037 U.S. Patent. 2009
- 10.Case DA, Darden T, III, TEC, Simmerling C, Wang J, Merz KM, Wang B, Pearlman DA, Duke RE, Crowley M, Brozell S, Luo R, Tsui V, Gohlke H, Mongan J, Hornak V, Caldwell JW, Ross WS, Kollman PA. AMBER9. University of California; San Francisco: 2006. [Google Scholar]
- 11.Lang PT, Brozell SR, Mukherjee S, Pettersen ET, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. RNA. 2009;15:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, Lowe E, Mahoney DJ, Tammi MI, Kahmann JD, Campbell ID, Day AJ, Jackson DG. Mol. Cell. 2004;13:483–496. doi: 10.1016/s1097-2765(04)00080-2. [DOI] [PubMed] [Google Scholar]
- 13.Takeda M, Terasawa H, Sakakura M, Yamaguchi Y, Kajiwara M, Kawashima H, Miyasaka M, Shimada I. J. Biomol. NMR. 2004;29:97–98. doi: 10.1023/B:JNMR.0000019465.12250.e0. [DOI] [PubMed] [Google Scholar]
- 14.Zuker M. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.