Abstract
Retrospective studies have demonstrated that nearly 50% of patients with ovarian cancer with normal cancer antigen 125 (CA125) levels have persistent disease; however, prospectively distinguishing between patients is currently impossible. Here, we demonstrate that for one patient, with the first reported fibroblast growth factor receptor 2 (FGFR2) fusion transcript in ovarian cancer, circulating tumor DNA (ctDNA) is a more sensitive and specific biomarker than CA125, and it can also inform on a candidate therapeutic. For a 4-year period, during which the patient underwent primary debulking surgery and chemotherapy, tumor recurrences, and multiple chemotherapeutic regimens, blood samples were longitudinally collected and stored. Whereas postsurgical CA125 levels were elevated only three times for 28 measurements, the FGFR2 fusion ctDNA biomarker was readily detectable by quantitative real-time reverse transcription-polymerase chain reaction (PCR) in all of these same blood samples and in the tumor recurrences. Given the persistence of the FGFR2 fusion, we treated tumor cells derived from this patient and others with the FGFR2 inhibitor BGJ398. Only tumor cells derived from this patient were sensitive to FGFR2 inhibitor treatment. Using the same methodologic approach, we demonstrate in a second patient with a different fusion that PCR and agarose gel electrophoresis can also be used to identify tumor-specific DNA in the circulation. Taken together, we demonstrate that a relatively inexpensive, PCR-based ctDNA surveillance assay can outperform CA125 in identifying occult disease.
Highlights
Although clinical utility and cost-effectiveness of the differing circulating tumor DNA (ctDNA)-detecting strategies, and even the overall approach of using ctDNA as a biomarker, have yet to be determined, the daily reality faced by oncologists and patients is that more sensitive and specific biomarkers are needed. In many instances, currently Food and Drug Administration (FDA)-approved biomarkers may be providing an incomplete, hence, misleading, assessment of disease status. To our knowledge, this use of a personalized biomarker represents the first example in ovarian cancer demonstrating the presence of continuous occult disease in the face of negative clinical, radiologic, and biochemical examination. Moreover, the recent identification of fibroblast growth factor receptor (FGFR) fusions in other solid cancers suggests a broader application of this strategy beyond ovarian cancer.
Introduction
The measurement of ctDNA, the so-called liquid biopsy, suggests great potential for radically transforming our current diagnostic and prognostic abilities in human cancers [1]. The origins of the “liquid biopsy” have evolved from the first description of circulating free DNA (cfDNA) in human serum [2] to the original demonstration of increased cfDNA in cancer and other diseases [3]. More recently, a number of studies have demonstrated the ability to assess tumor dynamics and disease burden in patients undergoing treatment by following levels of ctDNA [4], as a quantitative marker for metastatic disease monitoring and treatment response [5] and even to monitor the evolution of tumor mutations and acquired resistance to treatment [6,7]. In some instances, structural variants present only in a patient's tumor and detected by massively parallel sequencing have provided an opportunity to generate highly specific, personalized tumor markers enabling monitoring of patients throughout their disease course [8,9]. We report the case of a 59-year-old female who was originally diagnosed and treated for stage III ovarian cancer and highlight the use of genomic technologies to deliver a tool for more accurate disease surveillance and, concurrently, identification of a candidate therapy.
Materials and Methods
Patients and Patient-Derived Samples
Blood and tumor samples were collected from patients at the Icahn School of Medicine at Mount Sinai (New York, NY) after obtaining appropriate informed consent as a part of our Institutional Review Board (IRB)-approved personalized ovarian cancer genomics program. For serum and plasma separation, blood samples were collected in BD Vacutainer SST Plus Blood Collection Tubes (BD Biosciences, San Jose, CA) and processed between 4 to 6 hours after collection. Blood samples were centrifuged at 2600 and 1200 rpm for 10 minutes at 4°C, for separation of serum and plasma, respectively. All samples were aliquoted and stored at -130°C until use.
Tumor RNA Extraction and RNA Sequencing (RNA-Seq)
RNA was extracted from frozen tumor tissue using QIAzol according to manufacturer's instructions (Qiagen, Valencia, CA). Briefly, tissue was homogenized in QIAzol on ice. Chloroform was then added, mixed, and centrifuged to allow for separation and removal of the aqueous layer. RNA was precipitated in isopropanol overnight at -20°C. The suspension was centrifuged to pellet RNA, washed with 75% ethanol, and resuspended in RNAase-free water. RNA integrity numbers were analyzed (Agilent Bioanalyzer; Agilent Technologies, Santa Clara, CA), and only RNA samples with an RNA integrity number of >8.0 were submitted for next-generation sequencing.
Ovarian cancer transcriptomes were prepared for paired-end sequencing (100 bp) on the Illumina GAII platform and using the Roche NimbleGen capture kit (version 2.0; Madison, WI), always using the manufacturers' protocols but with a second size selection step to reduce ligation artifacts. Reads were aligned using Eland32 [provided with the Illumina sequencing platform (San Diego, CA)]. Expression levels were quantified by running ERANGE version 3.0.2 (http://woldlab.caltech.edu/rnaseq). For each gene, ERANGE reported the number of mapped reads per kilobase of exon per million mapped reads.
Identification of Gene Fusion Transcripts from RNA-Seq
We identified chimeric transcripts through examination of the spliced alignment of ∼30 million raw cDNA high-quality paired-end reads to a reference human genome (GRCh37/hg19), using two independent algorithm approaches. FusionHunter and TopHat-Fusion-Post with deFuse were used to parse through potential fusion candidates with a battery of stringent selection filters [10–12]. Fusion transcripts were confirmed by direct sequencing of tumor DNA and RNA and their tumor-specific expression by sequencing of paired peripheral blood mononuclear cell (PBMC) genomic DNA.
Isolation of Tissue Genomic and Circulating DNA
Genomic DNA was extracted from PBMCs and tumor samples using the DNeasy Blood and Tissue Kit according to the manufacturer's protocol (Qiagen). Circulating DNA was extracted from 200-µl aliquots of patient serum or plasma using the QIAamp MinElute Virus Spin Kit according to the manufacturer's recommended protocol (Qiagen). ctDNA was eluted with 100 µl of DNase- and RNase-free water.
Detection of FGFR2-FAM76A Fusion DNA in ctDNA
To detect the tumor-specific FGFR2-family with sequence similarity 76, member A (FAM76A) fusion in serum and/or plasma, we performed quantitative polymerase chain reaction (qtPCR) using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The primers used were as follows: fusion, forward—2 5′-GGATAAAGGAAGAGATTGCAC-3′ and fusion, reverse—2 5′ TGTGGGAGTTAAGTAAGAACT-3′. For each reaction, 5 µl of extracted ctDNA was used. Quantification was performed using a standard curve designed to assign quantities to each sample. The standard curve was prepared from a series of 10-fold dilutions covering a range of 101 to 106 copies of a purified FGFR2-FAM76A fusion gene fragment. DNA was used for the standard curve preparation, and PCR was performed on tumor genomic DNA using the following specific primers: fusion, forward—3 5′-TCTTATGCGTCTGACTGTGG-3′ and fusion, reverse—2 5′-TACTGGCATCACTTCCCTA-3′. The 186-bp PCR product was gel purified and quantified. PCR conditions used for amplification of this fragment were 94°C (5 minutes) for 1 cycle, 94°C (15 seconds), 55°C (15 seconds), and 72°C (20 seconds) for 35 cycles each, and a final extension of 72°C (10 minutes). Fusion copy number is expressed as the number of copies present in 5 µl of ctDNA.
For these studies, we also used two positive internal controls. First, and in addition to assaying for the fusion, we also tested for the presence of a DNA sequence [glyceraldehyde 3-phosphate dehydrogenase (GAPDH; actin)] present in both germ line and tumor DNA. Theoretically, if the control sequence was not detected, then the assay was repeated. This never occurred. Second, we also “spiked” the patient blood samples with a set concentration of plasmid DNA (pBluescript; Agilent Technologies) before ctDNA isolation. This allowed us to define reproducibility of the assay between sample runs and the efficiency of DNA extraction. Using quantitative real-time reverse transcription-PCR (qtRT-PCR), extractions resulted in a calculated recovery efficiency of between 30% to 75% of the spiked plasmid sample. All experiments were performed in triplicate and each done at least twice.
Generation of FGFR2-FAM76A Expression Constructs
To generate the fusion FGFR2-FAM76A-expressing construct, we amplified the full-length cDNA from patient tumor RNA using RT-PCR. Amplification was performed using the following set of primers: forward primer—5′-ATGGTCAGCTGGGGTCGTTTC-3′ (located at the translational start site of FGFR2 exon 1) and reverse primer—5′-TCATGGAGAGGTTATAGCTCCTG-3′ (present in the last exon of FAM76A). The native FAM76A stop codon was excluded to allow the generated product to express the vector's V5 epitope. The PCR cycling conditions were 94°C (5 minutes) for 1 cycle, 94°C (30 seconds), 55°C (1 minute), and 72°C (2 minutes) for 35 cycles each, and a final extension of 72°C (10 minutes). The amplified products were electrophoretically separated on an agarose gel, excised, and cloned into the pcDNA3.1/V5-His TOPO TA expression vector (Invitrogen, Carlsbad, CA). All constructs were confirmed by Sanger sequencing in both orientations before their use.
RNA Isolation and qtRT-PCR
Cell line RNA was extracted using RNeasy Mini Kits, including treatment with DNase according to the manufacturer's protocol (Qiagen). For RT-PCR, we used a total of 1 µg of RNA per reaction using iSCRIPT cDNA Synthesis following the manufacturer's protocol (Bio-Rad Laboratories). FGFR2-FAM76A RNA levels were measured using qtRT-PCR as described below using the following genomic specific primers: fusion, forward—one 5′-CAGAGACCAACGTTCAAGCAGT-3′ and fusion, reverse—5′-GGTTTTACTCTCCTGCTGGTACT-3′. All values were normalized either with GAPDH or hypoxanthine phosphoribosyltransferase 1 (HPRT) levels, and then the normalized value was used to calculate fold change compared with control. All experiments were done in triplicate and independently validated at least three times.
Western Blot Assay
Cell extracts were harvested in radioimmunoprecipitation assay buffer [Santa Cruz Biotechnology (Dallas, TX) standard protocol]. Equal amounts of protein (50 µg) as determined by the Bio-Rad DC Protein quantification assay were loaded and separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. We performed immunoblot analysis by using goat polyclonal antibody to β-actin (SC-1615) and monoclonal antibody to the V5 tag (Santa Cruz Biotechnology) present in the FGFR2-FAM76A constructs.
Cell Culture and Transfections
CP70 cell line [obtained from ATCC (Manassas, VA)] and patient-derived tumor cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin and grown at 37°C in 5% CO2. For the generation of CP70 stable cell lines expressing FGFR2-FAM76A and Lac-Z, cells were transfected 24 hours after plating with 10 µg of each construct using Lipofectamine 2000 according to the manufacturer's suggestions (Invitrogen). Twenty-four hours following transfection, cells were selected for in the presence of 2 mg/ml G418 (Life Technologies, Grand Island, NY). For these experiments, only early-passage (<passage 5) patient-derived tumor cell lines were used.
Cell Proliferation
Cell proliferation was determined at 24 and 48 hours by [3H] thymidine incorporation (1 mCi/ml; PerkinElmer, Waltham, MA). CP70 stable cell lines were plated at a density of 5000 cells per cm2 in 12-well dishes. At 24- and 48-hour time points, 1 µCi [3H] thymidine was added and incubated for 3 hours, then washed three times with cold phosphate-buffered saline, and fixed in methanol for 30 minutes at 4°C. Methanol removal and cell drying were performed, and cells were solubilized in 0.25% sodium hydroxide/0.25% sodium dodecyl sulfate and neutralized with hydrochloric acid (1 N). Disintegrations per minute were estimated by liquid scintillation counting. The proliferation index was calculated by dividing the counts obtained at 48 hours by the counts obtained at 24 hours. Each experiment was performed in triplicate and repeated three times.
Colony Formation Assay in Soft Agar
The ability of different transfectants to induce changes in proliferation in an anchorage-independent manner was quantified by standard soft agar assay. Approximately 5 x 105 cells were resuspended in 2.5 ml of Dulbecco's modified Eagle's medium supplemented with 20% FBS; then, 2.5 ml of 0.8% (wt/vol) SeaPlaque Agarose (Lonza, Rockland, ME) was added and carefully resuspended, and 1.5 ml of this mix was overlaid on top of three six-well plates containing 1% (wt/vol) agar. After 2 weeks of incubation at 37°C, colonies were visualized and counted after staining with 1 mg/ml p-iodonitotetrazonium violet. All experiments were performed in triplicate and repeated three times.
BGJ398 Treatment and 3-[4,5-Dimethylthiazol-2-yl]-2, 5-Diphenyltetrazolium Bromide (MTT) Assay
CP70 stables and patient-derived tumor cell lines were plated onto 96-well plates and, 24 hours later, incubated for 48 hours in the presence and absence of 4, 8, and 16 mM BGJ398 (No. S2183; Selleck, Houston, TX). CP70 stable cell lines were plated at a density of 10 x 103 cells per cm2 and patient-derived tumor cells at a density of 6 x 103 cell per cm2. Cell viability was measured using a standard MTT (thiazolyl blue) assay. Absorbance at 570 nm was measured using a microplate reader (Turner BioSystems Inc, Sunnyvale, CA). All experiments were performed in triplicate and repeated three times.
Nested PCR Detection of RANGAP1-PVALB Fusion
Nested PCR was performed for detection of the Ran GTPase Activating Protein 1 (RANGAP1)-Parvalbumin (PVALB) fusion in ctDNA extracted from patient serum and plasma. Two sequential rounds of PCR were performed. Primers for the first PCR round were given as follows: RANGAP1, forward 1—5′ GCTGGGATGATTTTTCATCCTTG and PVALB, reverse 1—5′ AGTCAGGAAATGGGCGTGAATTA. In the first round, 1 µl of extracted ctDNA was used for each reaction. Primers for the second PCR round were given as follows: RANGAP1, forward 2—5′ TCATTCACTCATTCATTCCCTCTC and PVALB, reverse 2—5′ TTACCTCTTCCATTGCCGGG. For each reaction, 1 µl of PCR product from the first PCR round was used. PCR conditions used for both PCR rounds were 94°C (5 minutes) for 1 cycle, 94°C (15 seconds), 55°C (15 seconds), and 72°C (20 seconds) for 30 cycles each, and a final extension of 72°C (10 minutes).
Results
At the time of the patient's original surgery 4 years ago, the serum level of the tumor marker cancer antigen 125 (CA125) was mildly elevated at 40 U/ml. At the conclusion of surgery, cytoreduction was considered optimal with no evidence of residual disease. The pathologic diagnosis was stage IIIC moderately differentiated papillary serous carcinoma. As part of our IRB-approved personalized cancer genomics program, fresh frozen tumor and blood samples were collected for deposit in a specialized tumor bank.
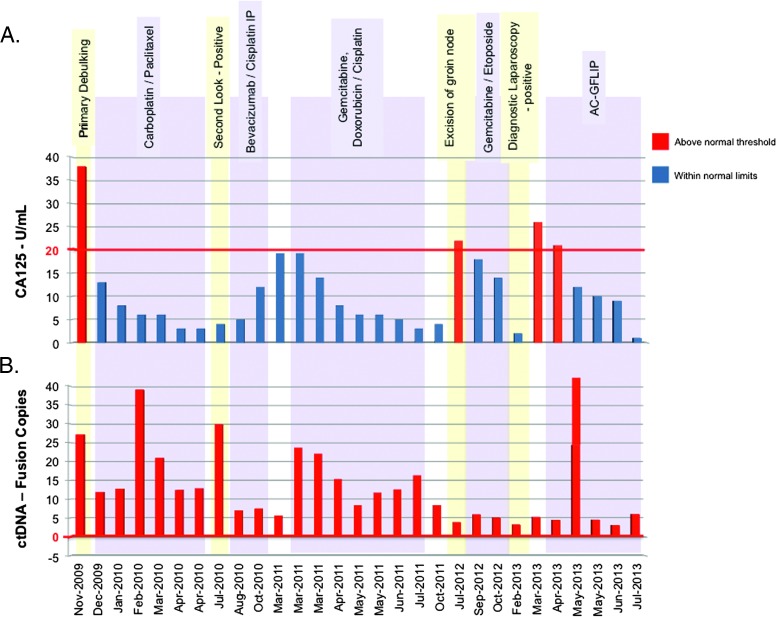
CA125 is the only biomarker recommended for both monitoring of therapy and detection of recurrence in ovarian cancer. For this past 4 years, and following the original surgery, CA125 levels only exceeded upper limits of normal in 3 of 28 tests. Despite this, the patient has had a series of biopsy-proven tumor recurrences even in the face of normal CA125 levels. These recurrences were managed with interval debulking and the empirical use of second- and third-line chemotherapy treatments (Figure 1, top panel). Given this history, we wondered whether a genomics-based approach could provide a more sensitive surveillance strategy than reliance on CA125 levels. We therefore used high-throughput RNA sequencing of the patient's primary and initial recurrent tumor, coupled with bioinformatic analysis, to search for tumor-specific gene fusions. The chimeric junction sequences were used for developing tumor-specific, qtRT-PCR primers.
Figure 1.
Detection of fusion sequences in ctDNA. (A) The surgical (highlighted in yellow) and chemotherapy (purple) events associated with the patient's history. Serum CA125 levels (red), determined at the time blood samples were originally drawn, and FGFR2-FAM76A copy number present in ctDNA in plasma are plotted. ctDNA copy number was divided by 33.3 to allow CA125 levels and ctDNA values to be plotted on the same graph. Fusion levels were determined using qtRT-PCR. The red line represents the upper limit of normal for CA125 (20 U/ml; Quest Diagnostics, Madison, NJ).
In this patient, both fusion detection algorithms that we used in our sequence analysis pipeline, FusionHunter and TopHat-Fusion-Post with deFuse {[10–12], respectively}, supported identification of an in-frame, interchromosomal FGFR2(chr10)-FAM76A(chr1) translocation with excellent statistical confidence in both the primary and recurrent tumor samples (Figure W1). These RNA-Seq findings were verified by qtPCR and Sanger sequencing (Figure W2). We mapped the genomic breakpoint using long-range PCR and identified the intervening introns between these two genes (Figure W3). Of particular interest, and suggesting that this may represent a relatively frequent event in human cancers, the ovarian FGFR2 genomic breakpoint is identical to that present in several solid human tumor types that were recently reported [13]. The breakpoints, using the University of California Santa Cruz (UCSC) Genome Browser hg19 coordinates, are FGFR2(chr10), position 123243211 and FAM76A(chr1), 28053982. As would be expected for a tumor-specific event, the FGFR2-FAM76A ovarian fusion was present only in tumor-derived DNA but not germ line DNA. The fusion preserves almost the entire length of the FGFR2 protein, including the terminal tyrosine kinase domains, and maintains the FAM76A coding sequence in-frame beginning in exon 2.
qtRT-PCR primers were designed to amplify the sequence of the genomic breakpoint. ctDNA was extracted from 200 µl of either plasma or serum from each of a total of 28 samples collected and stored frozen since the patient's original surgery 4 years ago. Significantly, the tumor-specific fusion is consistently detectable in the circulation throughout the course of the patient's postsurgical and multiple chemotherapy treatments, indicating the continued presence of occult tumor cells (Figure 1, bottom panel). In direct contrast to the presence of pathologic ctDNA, CA125 levels were elevated only three times following the initial surgery. Taken together, these findings suggest the persistence of residual disease during this 4-year period.
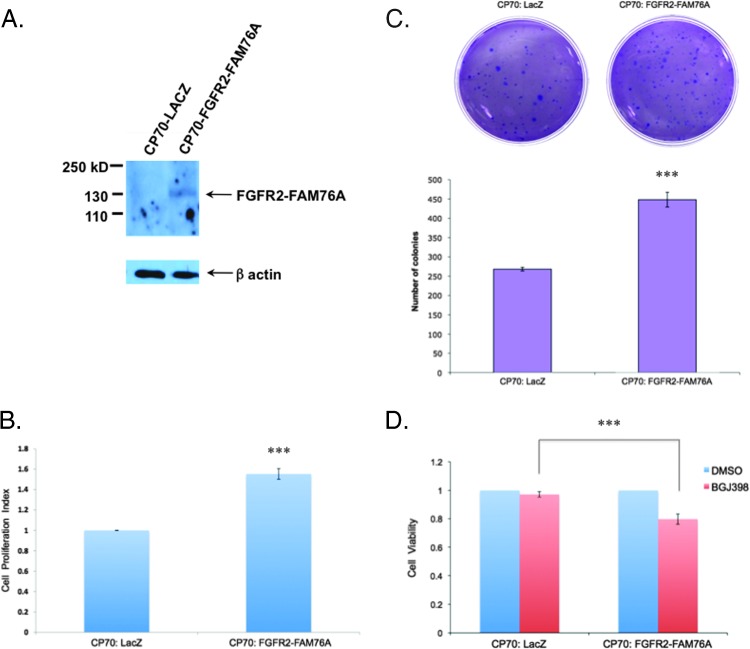
The constancy of the FGFR2-FAM76A fusion in each of the subsequently isolated biopsy samples (surgical dates are highlighted in yellow in Figure 1) further suggested a possible biologic pressure to maintain this transcript throughout tumor evolution and spread. Therefore, to begin assessing its potential functional role, we first constructed CP70 cisplatin-resistant ovarian cancer cell lines stably expressing the fusion protein to examine both the effect of FGFR2 fusion expression and inhibition on tumor cell behavior. Expression of the FGFR2 fusion (Figure 2A) resulted in increasing proliferation of the ovarian cancer cells by more than 50% (Figure 2B). Similarly, expression of the fusion increased the anchorage-independent growth of these cells in soft agar by nearly 50% (Figure 2C).
Figure 2.
FGFR2-FAM76A fusion increases cell proliferation. (A) Western blot analysis demonstrates stable ectopic expression of FGFR2-FAM76A fusion in the generated CP70 cell line. β-Actin, loading control. (B and C) Expression of FGFR2-FAM76A increases cellular proliferation and anchorage-independent growth, respectively, when the fusion is stably expressed in the cisplatin-resistant cell line CP70 and compared to the forced expression of the control protein Lac-Z. ***P <.001. (D) CP70 cells expressing the FGFR2-FAM76A fusion are more sensitive than control cells to cell death when treated with the pan-FGFR inhibitor BGJ398.
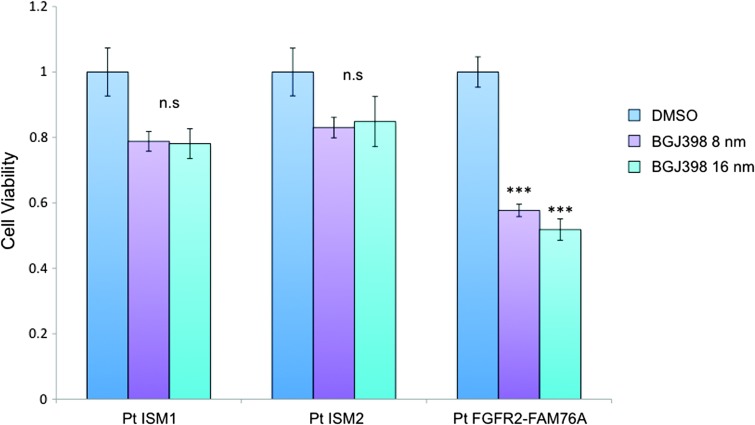
Given these findings demonstrating a functional effect of the FGFR2-FAM76A fusion, we wanted to next determine the effect of targeted inhibition of this fusion protein. We therefore assessed the antiproliferative effect of the pan-FGFR inhibitor BGJ398, currently in phase I clinical trials for patients with advanced solid malignancies and FGFR2 amplification. As shown (Figure 2D), this inhibitor had a magnified effect on decreasing cell proliferation in the tumor cells ectopically expressing the FGFR2 fusion. To gain more direct evidence of the effect of the FGFR2-FAM76A fusion on tumor cell growth and its possible therapeutic candidacy, we next analyzed the effect of the inhibitor on patient-derived tumor cell lines. Specifically, the effect of FGFR inhibition was compared between patient-derived ascites cell lines from this patient and two other patients (ISM1 and ISM2) who had also presented with stage IIIC primary ovarian cancer (Figure 3). The FGFR2-FAM76A fusion was present, in both DNA and RNA, exclusively in the tumor cells from our original patient but not in the other two patient-derived cell lines ISM1 and ISM2. Treatment with BGJ398, depending on dose, significantly reduced cell viability by more than 40% in the FGFR fusion-expressing cell line. In the other two cell lines, treatment with the FGFR inhibitor had no significant effect on cell viability.
Figure 3.
Targeted treatment of patient-derived cell lines. Early-passage ascites cells from three different patients, all with chemonaive, primary stage III ovarian cancer, were grown and treated with the pan-FGFR inhibitor BGJ398. Only tumor cells expressing the FGFR2-FAM76A fusion are sensitive to treatment. ns, not significant. ***P <.001.
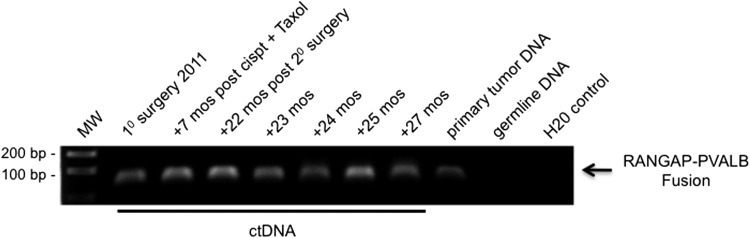
We tested this overall approach in a second patient in whom we had detected a different fusion. This patient's pathologic diagnosis following primary surgery in 2011 was also stage IIIC moderately differentiated papillary serous carcinoma with optimal surgical debulking. RNA-Seq analysis identified an intrachromosomal deletion (chr22) leading to the generation of the fusion transcript RANGAP1-PVALB (Figure W4). ctDNA containing this specific fusion was retrospectively identified in blood samples stored and frozen for a 2-year period using PCR and agarose gel electrophoresis (Figure 4). Direct Sanger sequencing of the resultant amplicons confirmed the fusion. All seven of the serum samples collected, and which spanned the time frame from original surgery to now, demonstrated the presence of the fusion product in ctDNA. This finding was consistent with the recent recurrence of clinically demonstrable disease and the fact that the fusion was present in the recurrent tumor's genomic DNA.
Figure 4.
Detection of ctDNA fusion by PCR and gel electrophoresis. Fusion sequences can also be detected in serum/plasma using standard PCR and gel electrophoresis. The RANGAP-PVALB fusion was detected using nested PCR (a total of 60 amplification cycles) in all serum/plasma samples from patient No. 2. As expected, the fusion was not detected in the patient's germ line DNA isolated from PBMC. The patient's time line from which blood samples were originally drawn and stored away frozen is shown above the gel.
Discussion
In general, levels of cfDNA—not distinguishing between tumor or normal cell origin—have previously been shown to be elevated in high-grade, advanced stage serous ovarian carcinomas, suggesting the possibility of noninvasive screening and surveillance [14]. Given the large number of deletions, insertions, and chromosomal translocations inherent in ovarian cancer [15], this cancer type may be ideally suited for generating tumor-specific sequences on the basis of the detection of structural variants that can be serially detected with theoretically exquisitely high specificity from blood samples. In general, levels of cfDNA has previously been shown to be elevated in high-grade, advanced stage serous ovarian carcinomas. Of particular interest, with respect to the robustness of our approach, is that minimal amounts of serum/plasma (less than 0.5 ml) were used for DNA extraction for the ctDNA assays. Moreover, many of these samples were stored frozen for several years. The approach we have outlined is also highly flexible with regard to methodology used for the surveillance assays. For example, and as demonstrated in the second patient, standard, nonqualitative PCR could also be used to provide an even more inexpensive and rapid assay when compared to qtRT-PCR. Taken together, we believe that the specificity and flexibility of this overall approach provides yet another step toward the goal of providing precision cancer diagnostics and eventually improving quality of life and survival.
In our patient with FGFR2-FAM76A, RNA-Seq analysis at the time of her primary surgery identified a highly specific ctDNA biomarker, and in particular, the analysis identified a candidate growth driver and drug target. What was initially unexpected, but biologically consistent given the patient's tumor recurrences, was that every ctDNA assay was positive, consistent with the existence of residual disease. This was in direct contrast to an overwhelming number of normal intervening CA125 levels and interspersed normal radiologic studies. As has been previously suggested [8], if these ctDNA assays could be performed in real time following a patient's original surgery, the detection of residual disease would trigger more aggressive/different treatment options at an earlier time point, cessation of ineffective, systemically toxic treatments, and/or directed participation in a clinical trial. For this patient, the complete range of precision medicine goals are in evidence starting with the development of PCR-based, rapid (<24-hour turnaround) and inexpensive disease surveillance assays coupled with the identification of a candidate-tailored therapeutic strategy. In a setting where up to 50% of patients with ovarian cancer with normal CA125 levels following chemotherapy are known to have persistent disease [16], the value and limitations of ctDNA-based biomarkers in this disease need to be further explored if we are to avoid losing opportunities for improving management and treatment.
Supplementary Material
Acknowledgments
The authors thank the Gordon family and the Derald H. Ruttenberg Foundation for their generous financial support which funded, in part, these studies.
Footnotes
The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med. 2013;5:207ps14. doi: 10.1126/scitranslmed.3006305. [DOI] [PubMed] [Google Scholar]
- 2.Mandel P, Métais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Acad Sci Paris. 1948;142:241–243. (Fre). [PubMed] [Google Scholar]
- 3.Koffler D, Agnello V, Winchester R, Kunkel HG. The occurrence of single-stranded DNA in the serum of patients with systemic lupus erythematosus and other diseases. J Clin Invest. 1973;52:198–204. doi: 10.1172/JCI107165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 6.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 7.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 8.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, Hämäläinen E, Stebbings LA, Andersson LC, Flanagan AM, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062–1069. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Chien J, Smith DI, Ma J. FusionHunter: identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011;27:1708–1710. doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamat AA, Sood AK, Dang D, Gershenson DM, Simpson JL, Bischoff FZ. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann NY Acad Sci. 2006;1075:230–234. doi: 10.1196/annals.1368.031. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network, author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bast RC., Jr Commentary: CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer. 2010;116:2850–2853. doi: 10.1002/cncr.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.