Abstract
Classic histology still represents the gold standard in tumor tissue analytics. However, two-dimensional analysis of single tissue slides does not provide a representative overview of the inhomogeneous tumor physiology, and a detailed analysis of complex three-dimensional structures is not feasible with this technique. To overcome this problem, we applied multispectral fluorescence ultramicroscopy (UM) to the field of tumor analysis. Optical sectioning of cleared tumor specimen provides the possibility to three-dimensionally acquire relevant tumor parameters on a cellular resolution. To analyze the virtual UM tumor data sets, we created a novel set of algorithms enabling the fully automatic segmentation and quantification of multiple tumor parameters. This new postmortem imaging technique was applied to determine the therapeutic treatment effect of bevacizumab on the vessel architecture of orthotopic KPL-4 breast cancer xenografts at different time points. A significant reduction of the vessel volume, number of vessel segments, and branching points in the tumor periphery was already detectable 1 day after initiation of treatment. These parameters remained virtually unchanged in the center of the tumor. Furthermore, bevacizumab-induced vessel normalization and reduction in vascular permeability diminished the penetration behavior of trastuzumab-Alexa 750 into tumor tissue. Our results demonstrated that this newimaging method enables the three-dimensional visualization and fully automatic quantification of multiple tumor parameters and drug penetration on a cellular level. Therefore,UM is a valuable tool for cancer research and drug development. It bridges the gap between common macroscopic and microscopic imaging modalities and opens up new three-dimensional (3D) insights in tumor biology.
Introduction
The quantitative description of tumor growth, angiogenesis, and metastasis and also the treatment effects of new therapeutics on these steps are essential to evaluate the full potential of anticancer compounds. Therefore, different macroscopic and microscopic in vivo and ex vivo imaging modalities are used to monitor the complex interactions between a therapeutic drug and tumor biology [1,2].
Noninvasive in vivo imaging methods like computer tomography [3,4], positron-emission tomography [5,6], magnetic resonance imaging [7,8], fluorescence molecular tomography [9], and/or combinations thereof [10,11] offer the possibility to three-dimensionally monitor various tumor parameters quantitatively with a spatial resolution down to 50 µm [12–14]. However, to get more comprehensive information regarding the mode of action of antitumor substances, it is necessary to assess the different events of tumor progression at a cellular resolution [15].
Intravital microscopy in living animals can overcome these limitations and has provided cellular and/or molecular insights into tumorigenesis [16,17], angiogenesis [18–20], tumor microenvironment [21], and drug treatment response [15,22]. With a spatial resolution of nearly 500 nm, minimal invasive intravital confocal laser scanning microscopy and multiphoton laser scanning microscopy are eminently suitable for three-dimensional real-time observation of multiple biologic in vivo events down to cellular resolution. Nevertheless, the slight penetration depth into tissue of below 500 µm and a small field of view prevent entire tumor scanning [15,23–25]. Other intravital microscopy techniques, such as optical coherence tomography and optical frequency domain imaging, can measure a large field of view with penetration depths beyond 1 mm and a spatial resolution of about 10 to 50 µm. However, their basic principle (optical interferometry) of measuring the various optical scattering properties in tissue provides only morphologic and physiological tissue information [26–29]. Direct visualization of the labeled therapeutic agent and its penetration, accumulation, and interaction with the tumor target structure is not possible by using classic fluorochromes [9].
High spatial resolution over a large field of view coupled with the ability to provide morphologic and biologic information can currently only be provided by classic histology and immunohistochemistry (IHC). Therefore, conventional histology is still the gold standard in tumor analysis. Nevertheless, two-dimensional examination of a few tumor tissue slices may not provide a representative and comprehensive description of the complex and inhomogeneous tumor biology and is therefore prone to misinterpretation. The three-dimensional reconstruction of serial tissue slices is also not an appropriate alternative, because it is a very laborious and time-consuming process and the mechanical distortion of the tissue slices is difficult to overcome.
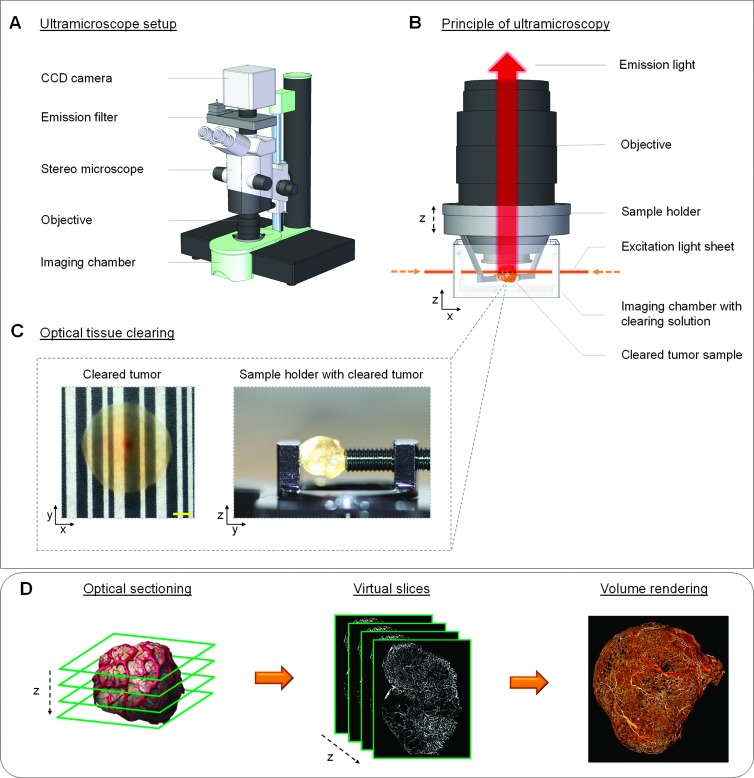
We applied multispectral fluorescence ultramicroscopy (UM) into the field of tumor analysis to overcome the limitations of conventional imaging techniques. For the last years, this ex vivo imaging method has already proven itself and became more and more relevant in different areas of application such as embryonic development [30–34], neuroscience [35–39], and immunology [40]. UM, as described by Dodt et al. [35], uses focused laser light for a double-side, in-focus plane illumination of the specimen (i.e., optical sectioning), and the fluorescent light is detected perpendicular to the illumination axis (Figure 1, A and B). Such principle of measurement prevents the formation of out-of-focus light and reduces photobleaching to a minimum. The problem of strong light absorption and scattering in tissue is solved by optical clearing. Thus, specimen obtains high transparency, and fluorescent light can traverse unrestricted for several millimeters into tissue [35,38,41]. This can be achieved by a chemical clearing procedure that removes the water from the biologic sample and replaces it by a liquid having approximately the same refractive index as solid tissue components, like lipids and proteins [42,43] (Figure 1C). UM in combination with optical tissue clearing enables the three-dimensional measurement of large tumor specimen up to 5 mm in diameter with a spatial resolution of about 5 µm [44]. By moving the transparent tissue sample vertical through the planar laser beam, a z stack of serial optical sections is produced. The virtual UM slices can subsequently be rendered as a maximum intensity projection (MIP) or volume [44,45] (Figure 1D).
Figure 1.
Basic principle of UM. (A) All UM measurements were performed with a commercially available light sheet microscope (LaVision BioTec). The sketch illustrates the basic setup of the applied imaging system. (B and C) To overcome the problem of light absorption and scattering, tumor specimen was chemically treated to change the tissue properties and make the sample optically transparent. The cleared tumor was fixed into a sample holder and placed in the imaging chamber. Afterwards, a single layer was illuminated perpendicular to the observation pathway by a thin laser light sheet. The emitted fluorescence light fromthe excited tumor plane is detected by a highly sensitive charge-coupled device (CCD) camera. (D) By stepping the cleared tumor sample vertical (z-axis) through the laser sheet, multiple optical sections are created. The received virtual slices of a specific tumor parameter, such as vessel architecture, can be rendered as MIP or volume. (C) Scale bar, 500 µm. (The illustration of the UM imaging hardware from A and B was kindly provided from LaVision BioTec.)
The virtual slices obtained from UM were compared with classic two-dimensional (2D) fluorescence histology and IHC to confirm the validity of this new method in tumor visualization. The results demonstrate the anatomic and morphologic accordance between both imaging techniques. To analyze the three-dimensional tumor data sets obtained from UM, we developed a new software that possesses excellent computational performance and enables a fully automatic segmentation and quantification of different tumor characteristics, vasculature, and drug penetration parameter. After successful evaluation and validation of this new imaging method, a proof-of-concept study in an orthotopic breast cancer xenograft model was performed to three-dimensionally quantify the antiangiogenic treatment effect of the monoclonal antibody bevacizumab at different time points on a cellular level. Furthermore, the influence of bevacizumab-induced vessel normalization on the subsequent penetration of fluorescent-labeled trastuzumab was also investigated.
Materials and Methods
Compounds and Labeling
The two monoclonal antibodies bevacizumab [recognizes and blocks vascular endothelial growth factor A (anti-VEGF-A)] and trastuzumab [recognizes and blocks human epidermal growth factor receptor 2 (anti-HER2)] were provided by Roche Diagnostics GmbH (Penzberg, Germany). Lectin from Bandeiraea simplicifolia, which has a high affinity for terminal α-d-galactosyl and N-acetyl-α-d-galactosaminyl residues and specifically binds to endothelial cells of the vessel network, was acquired from Sigma-Aldrich (Taufkirchen, Germany). Lectin and trastuzumab were labeled in-house with the fluorophore Alexa 647 and Alexa 750 (Invitrogen, Darmstadt, Germany). They were attached through monoreactive N-hydroxysuccinimide (NHS) ester to lysine residues with a labeling ratio of three fluorophores per molecule. Trastuzumab-Alexa 750 and lectin-Alexa 647 were purified by dialysis and size-exclusion chromatography, and afterwards, surface plasmon resonance analysis (GE Healthcare, BiaCore Life Sciences, Little Chalfont, United Kingdom) was performed to measure the binding characteristics of the antibody. There were no significant changes in the kon and koff constants and the resulting affinity between labeled and nonlabeled trastuzumab.
Cell Line and Culture
The human breast cancer cell line KPL-4 was kindly provided by J. Kurebayashi (Kawasaki Medical School, Kurashiki, Japan). The HER2-overexpressing tumor cells were incubated in Dulbecco's modified Eagle's medium (10% fetal calf serum and 2 mM l-glutamine) at 37°C and 5% CO2 until they reached the final cell concentration for injection. The viability of the KPL-4 cells was assessed using a Vi-CELL viability analyzer (Beckman Coulter, Krefeld, Germany) and reached nearly 95%.
Tumor Xenograft
We generated tumor xenograft by orthotopic injections of KPL-4 tumor cells [3 x 106 cells/20 µl of phosphate-buffered saline (PBS)] into the right penultimate inguinal mammary fat pad of anesthetized female severe combined immunodeficiency (SCID) beige mice (age, 6–8 weeks; weight, 20–25 g; Charles River Laboratories, Sulzfeld, Germany). The tumors grew until they reached a tumor size of 60 mm3 (measured by caliper). General animal conditions were controlled daily, and tumor volume and body weight were monitored once weekly. All experimental study protocols were reviewed and approved by the local government. The animals were handled according to guidelines [Gesellschaft für Versuchstierkunde-Society of Laboratory Animals (GV-SOLAS), Federation for Laboratory Animal Science Associations (FELASA), and TierSchG], and the animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Treatment of Animals
When tumor size reached a volume of about 60 mm3, mice were randomized and separated into two groups each consisting of 15 mice. The first group (treatment) was treated i.v. with a single dose of 10 mg/kg bevacizumab, and the second group (control) was injected i.v. with PBS. One, 3, and 7 days after bevacizumab treatment, five mice from the control and treatment groups were injected i.v. with 2 mg/kg trastuzumab-Alexa 750. Six hours thereafter, all animals were treated i.v. with 100 µg of lectin-Alexa 647 (100 µl), incubated for about 5 minutes, and then all tumors were explanted.
Fixation and Clearing
The explanted KPL-4 tumors were fixed in 10% buffered formalin (VWR International, Ismaning, Germany) for about 12 hours at 4°C in the dark and thereafter were dehydrated in a graded ethanol series (3 x 70%, 2 x 95%, and 2 x 100% for 30 minutes each). These steps were carried out using the Tissue-Tek VIP Vacuum Infiltration Processor (Sakura Finetek, Heppenheim, Germany). The dehydrated tumors were placed in a clearing solution consisting of one part benzylalcohol and two parts benzylbenzoate (Sigma-Aldrich) and incubated for 2 days at 4°C in the dark until the specimen became optically transparent.
Acquisition of UM Data Sets
The cleared specimens were scanned with a commercial ultramicroscope (LaVision BioTec, Bielefeld, Germany). The standard magnification of x0.63 in combination with a x2 objective lens (Mv PLAPO 2VC; Olympus, Hamburg, Germany) led to a final xy resolution of 5.1 µm, and the step size was set to the same value. We used this setting to scan specimens with a diameter up to about 5 mm. The integrated supercontinuum white light laser (SuperK EXTREME 80 mHz VIS; NKT Photonics, Cologne, Germany) covers the entire wavelength range of 400 to 2400 nm and enables the excitation of every commonly used fluorophore. The different optical properties of vital and necrotic tissue and the strong autofluorescence emission of the erythrocytes were used to create a detailed illustration of the tumor morphology (excitation range, 543/22 nm; emission range, 593/40 nm). Lectin-Alexa 647 (excitation range, 655/40 nm; emission range, 716/40 nm) and trastuzumab-Alexa 750 (excitation range, 710/40 nm; emission range, 775/46 nm) showed a strong binding affinity to their target structure and provided high signal intensities. Thus, the measurements were performed with short exposure times (100–400 ms per slice) resulting in a total acquisition time of about 30 to 40 minutes per tumor depending on its size. Thereafter, the generated tagged image file format (TIFF) slices were converted into Digital Imaging and Communications in Medicine (DICOM) files and visualized using the OsiriX software (Pixmeo, Bernex, Switzerland; open-source software). Furthermore, ImageJ (open-source software) and GNU Image Manipulation Program (GIMP; open-source software) were also used for the preparation of the individual images and videos included in this manuscript.
Quantification of UM Data Sets
Quantification of the raw data was performed with a set of custom developed image analysis algorithms implemented as a “Definiens Cognition Network Technology” rule set (Definiens AG, Munich, Germany). To increase performance, the skeletonization and branch point detection were written in C++ using the “Insight Segmentation and Registration Toolkit” and loaded as a custom plug-in. With Definiens requiring a six-pixel three-dimensional (3D) neighborhood, we modified an existing Insight Segmentation and Registration Toolkit implementation [46] so that the skeleton had no more diagonal pixels (26 neighborhood). On one central processing unit (CPU) core (Intel Xeon X5670 @ 2.93 GHz; Intel Corporation, Santa Clara, CA), it took between 10 minutes [peak random-access memory (RAM) usage, 8 GB] and 2 hours (peak RAM usage, 20 GB) to quantify a single tumor, depending on the tumor size and desired software settings. Multiple engines can run simultaneously if enough hardware resources and Definiens licenses are available, thus, enabling the timely and fully automatic quantification of large groups with our software. For this publication, the algorithm was configured to use the full resolution of the UM data sets. Furthermore, vasculature with a distance to the surface of the tumor smaller than 20% of the radius of a fictional sphere with same volume as the tumor was defined as periphery.
Immunohistochemistry
Following 3D scanning, the cleared specimens were shortly incubated in 100% xylene to remove residues from the clearing solution. Then, tumor samples were incubated in paraffin (3 x 1 hour) and embedded into paraffin blocks. All incubation steps were carried out using the Tissue-Tek VIP Vacuum Infiltration Processor (Sakura Finetek). For verification of the 3D imaging method, we compared the optical sectioning results from UM with classic 2D histology. Therefore, different layers (between the top and center) of a paraffin embedded KPL-4 tumor were randomly selected. Four serial tissue slices with a thickness of 5 µm were created at each tumor layer. Afterwards, the obtained tissue slices were analyzed by conventional fluorescence histology [4′,6-diamidino-2-phenylindole (DAPI), lectin-Alexa 647, and trastuzumab-Alexa 750] and IHC [hematoxylin and eosin (H&E), anti-cluster of differentiation 34 (CD34), and anti-HER2 staining]. In the preclinical efficacy study (day 7), three different layers of the tumor (between the top and center) were randomly selected. Four serial tissue slices with a thickness of 2.5 µm were prepared at each tumor region and analyzed by IHC (H&E, anti-CD34, anti-HER2, and Ki67 staining). All tumor tissue slices were deparaffinized in a descending xylene and ethanol series. Visualization of tumor tissue was accomplished by DAPI mounting medium (Vector Laboratories, Burlingame, CA) and classic H&E staining (Carl Roth, Karlsruhe, Germany). Tumor vessels were stained with a primary rat anti-mouse CD34 antibody (Hycult Biotech, Uden, Netherlands), followed by a secondary goat anti-rat ImmPRESS HRP antibody (Vector Laboratories) and colored with DAB (Dako, Hamburg, Germany). The staining of the extracellular HER2 expression was performed using a primary rabbit anti-human HER2 antibody (Thermo Fisher Scientific, Rockford, IL), followed by a secondary goat anti-rabbit ZytoChem Plus AP polymer antibody (Zytomed Systems, Berlin, Germany). Slides were colored with permanent red (Zytomed Systems). Finally, proliferating cells were detected using Ki67. The staining was performed using a primary rabbit anti-human Ki67 antibody (Ventana Medical Systems, Tucson, AZ), followed by the secondary OmniMap antirabbit HRP antibody (Ventana), and colored with DAB (Ventana). All tissue slides were mounted with a cover slide and subsequently digitalized using a slide scanner with a magnification of x20 (Pannoramic 250; 3DHISTECH, Budapest, Hungary).
Statistical Analysis
The analysis was performed using the JMP software (version 8.0.2.2; SAS Institute, Cary, NC). We used a classic two-tailed, unpaired Student's t test to determine significant differences for each time point. We considered P values of ≤.05 as significant. The significance levels are indicated by asterisks (*P < .05, **P < .01, and ***P < .001). All data are presented as means ± SD.
Results
Visualization of Tumor Morphology
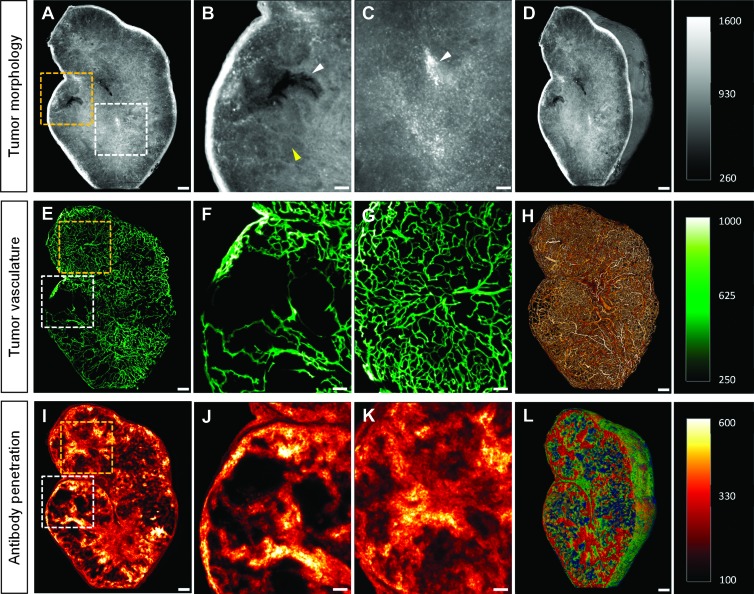
For the visualization of tumor morphology, the cleared tumor specimen were excited with green laser light (λ = 543/22 nm), which induced a strong tissue autofluorescence signal. The variation of light absorption caused by differences in the structural compositions of the tissue resulted in autofluorescence-rich contrast, thereby allowing tissue components to be roughly differentiated (Figure 2A). We observed that the necrotic areas had a weaker autofluorescence signal because of lower tissue density and were therefore seen as darker regions. This allowed us to differentiate vital from necrotic regions in the tumor tissue (Figure 2B). Conventional H&E staining verified these necrotic areas of tissue (Figure W1). Erythrocytes had very high autofluorescence emission levels at this wavelength range, distinguishing them clearly from the surrounding tissue. This information was used to visualize their distribution pattern and also for identifying the presence of blood pools in the tumor tissue (Figure 2C). Compared to conventional 2D histology where only a few tissue slices are commonly analyzed, UM enables the three-dimensional visualization of the tumor morphology (Figure 2D and Video W1A).
Figure 2.
Visualization of tumor morphology, vessel architecture, and drug penetration. (A) The detection of tissue autofluorescence provides detailed information about tumor morphology. (B) The strong tissue contrast allowed the differentiation between vital (yellow arrowhead) and necrotic (white arrowhead) tumor areas. (C) Erythrocytes have a strong autofluorescence signal in the autofluorescence range (white arrowhead). As a result of this, their distribution and accumulation in the tumor tissue were clearly visible. (D) Volume rendering of the tumor morphology. (E) The complex and chaotic structures of the lectin-Alexa 647-stained tumor vessels were clearly visible in the MIP of 30 virtual single UM slices. (F and G) The blow-up images of the MIP from (E) showed the heterogeneity of the tumor vessel architecture. Some tumor areas possess a very dense vascularization (G), whereas others provide only a sparse or absent vasculature (F). (H) Volume rendering of the vessel architecture. (I) The penetration and binding behavior of trastuzumab-Alexa 750 after an incubation time of 6 hours were visualized in the MIP of 30 virtual single UM slices. (J and K) The inhomogeneous penetration and distribution of the labeled antibody in the tumor tissue were clearly visible in the blow-up images from I. Most parts of the tumor areas were extensively penetrated (K), although there were tumor areas that had only a weak or completely absent antibody signal (J). (L) Volume rendering of the antibody penetration. (A–C) Single slice, 5-µm diameter. (E–G and I–K) MIP, 30 slices with 5-µm diameter per slice. Scale bar, 250 µm and 100 µm (blowup).
Visualization of Tumor Vessel Architecture
Lectin-Alexa 647, which binds specifically to the sugar residues of vascular endothelial cells, was injected i.v. into a KPL-4 tumor-bearing animal. After tumor explantation and tissue clearance, we measured the specimen three-dimensionally with UM. High binding specificity of the vessel marker in combination with the fluorescent properties of the applied near-infrared fluorochrome resulted in a high signal-to-noise ratio, and thus, the tumor vasculature was clearly visible in the virtual UM slices. A MIP of 30 optical vascular slices clearly visualized the chaotic, irregular, and dramatically branched vascular structures typical of a tumor [15] (Figure 2E). The heterogeneity of the vascular network was also clearly demonstrated. Some tumor areas displayed only sparse or absent vascular density (Figure 2F), whereas vascularization in other regions was high (Figure 2G). The complexity of the whole tumor vessel architecture at cellular level was perfectly shown in the 3D volume rendering (Figure 2H and Video W1B and Video W2). Furthermore, if it is not necessary to capture the entire vasculature of the tumor, local xy resolutions can be increased up to 0.5 µm. This allowed us to differentiate between regular blood vessels and tumor vessels (Figure W2A), and even individual vascular endothelial cells could be clearly displayed (Figure W2B).
Visualization of Antibody Penetration in Tumor Tissue
Trastuzumab-Alexa 750, which has a high affinity for the extracellular domain of HER2, was injected i.v. into the HER2-overexpressing KPL-4 xenograft model, to monitor the antibody penetration and binding behavior in tumor tissue. An MIP of 30 optical slices showed the heterogeneous penetration and binding of the labeled antibody after 6 hours of injection (Figure 2I). Some areas of the tumor had only very low or no antibody signals (Figure 2J), and in other regions, very strong and homogeneous trastuzumab distribution was detected (Figure 2K). In addition, UM opens up the possibility to three-dimensionally visualize drug penetration inside a whole tumor at a cellular level (Figure 2L and Video W1C).
Combination of Different Tumor Parameters
UM allows the simultaneous visualization of antibody penetration and tumor vessel architecture. The relationship between the density and morphology of the tumor vessels and the antibody penetration was clearly demonstrated (Figure 3A and Video W3A). Highly vascularized areas of the tumor displayed very strong and homogeneous antibody distribution, whereas areas with low vascular density were only slightly or not at all penetrated (Figure 3, B and C). Morphologic abnormalities or changes in the tumor tissue (such as necrosis) could also be correlated to the vascular network (Figure 3D and Video W3B). Tissue areas with a vascular density below a certain level were apparently no longer supplied with oxygen and/or nutrients, inevitably leading to tissue necrosis (Figure 3E). Accumulation of RBCs in the tumor tissue allows an indication of vessel permeability (Figure 3F). The combination of tissue information with antibody penetration indicated how the drug was penetrating various tissue areas (Figure 3G and Video W3C). We further observed that trastuzumab-Alexa 750, as a tumor-targeting drug, generally penetrated only marginally into necrotic areas of the tissue (Figure 3H), whereas solid tumor tissue with sufficient vascular density had a very strong antibody signal (Figure 3I).
Figure 3.
Combination of different tumor parameters. (A) The overlay of the vascular (green) and antibody (red) channels showed that the differing density of the vessels leads to an uneven distribution of the antibody. (B) Tumor areas with low vascular density showed only very slight or absent antibody signals, whereas (C) in areas with a high vascular density, there was strong and homogeneous antibody penetration. (D) The combination of the tissue (gray) and vascular information clearly showed the correlation between vascular density and the development of necrosis. (E) Areas with low or absent vascular connections developed necrosis, whereas (F) those with a high vascular density had a solid tissue structure. In the latter, erythrocytes (white spots) were also more prevalent. (G) The overlapping of the tissue and antibody channel showed the penetration of the trastuzumab-Alexa 750 in the different areas of tissue. (H) Necrotic areas of the tumor were hardly penetrated by the labeled antibody at all. (I) Solid tumor tissue, however, demonstrated very strong and homogeneous antibody penetration and distribution. (A–I) MIP, 30 slices with 5-µm diameter per slice. Scale bar, 250 µm and 100 µm (blowup).
Validation of UM Data Sets by Conventional Histology
Because UM was being used for the first time for multispectral analysis of postmortem tumor tissue, we validated the virtual slices using classic 2D histology (Figure 4A). Therefore, subsequent to UM measurement, a cleared KPL-4 xenograft tumor was paraffin embedded and conventionally cut at different tumor regions. Four serial tissue slices were created at each plane. The first tumor tissue slice was DAPI stained, and afterwards, the including fluorescence signals (DAPI, lectin-Alexa 647, and trastuzumab-Alexa 750) were visualized (Figure 4, C, F, and I). Furthermore, H&E and different IHC stainings, such as anti-CD34 and anti-HER2, were performed with the other serial slices (Figure 4, D, G, and J). The direct comparison between the virtual and conventionally prepared slices of the same tumor tissue layer showed an excellent correlation. Both the tissue morphology and vascular architecture of the tumor and the penetration and binding of the therapeutic antibody were shown to be identically rendered with UM and classic histology (Figure 4, B–J).
Figure 4.
Validation of UM data sets by conventional histology. (A) Optical slicing of the cleared KPL-4 tumor sample was performed by UM to receive a stack of single virtual tumor tissue slices. Thereafter, the tumor specimen was paraffin embedded and conventionally cut into serial tissue slices. Different targets of the tumor tissue slices were stained by classic IHC. The virtual slices of the different fluorescence channel were compared to conventional histology to show the validity of UM. (B–D) The direct comparison between tumor tissue information from the virtual autofluorescence UM slice (B) and conventional fluorescence histology (C; DAPI) and classic IHC staining (D; H&E) showed nearly identical tissue structures. (E–G) The virtual UM slice of the lectin-Alexa 647-stained tumor vasculature from E provided also a great morphologic accordance to the conventional fluorescence histology (F; lectin-Alexa 647) and classic IHC vessel staining (G; anti-CD34). (H–J) The penetration and binding behavior of trastuzumab-Alexa 750 in the tumor tissue after 6 hours of incubation showed nearly identical distribution pattern in virtual UM (H) and conventional fluorescence histology (I). (J) The IHC staining of the extracellular HER2 receptor domain (anti-HER2, SP3) provided the expression level of all available HER2 tumor cell receptors. (B–J) Single slice, 5-µm diameter. Scale bar, (A) 500 µm and (B–J) 250 µm.
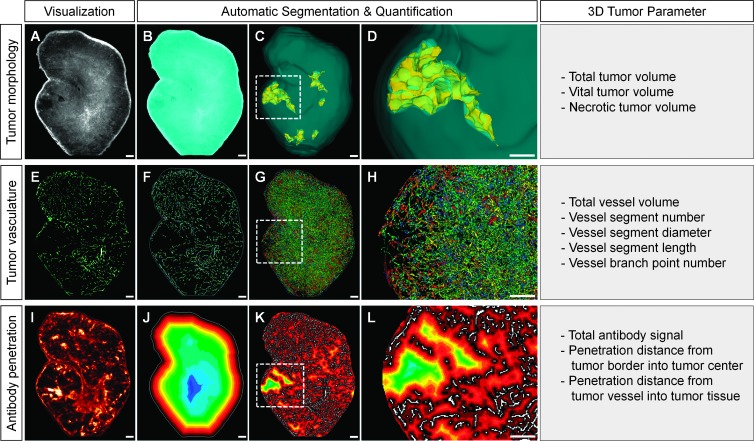
Quantification of UM Data Sets
To analyze the different virtual UM tumor data sets, we created a novel set of algorithms enabling the fully automatic segmentation and quantification of multiple tumor composition, vessel architecture, and drug penetration parameters without any manual user interference. The first step was a segmentation of the tumor sample from the background using automatic thresholding techniques on the autofluorescence channel (Figure 5, A and B). The software then generated a binary image of the vessels inside the tissue region. Vessel-free areas inside the tumor with a minimum distance to the surrounding vasculature were classified as necrotic tissue (Figure W3, A–C). This parameter was determined on the basis of the results of conventional histology and enabled the 3D quantification of tumor necrosis (Figure 5, C and D). Next, the skeletonization of the binary vascular tree was executed using an iterative 3D-thinning algorithm [47]. Thereafter, a 3D kernel found the branch points and segments (Figure 5, E and F). The individual skeleton segments were then extended back to the full vessel diameter using a pixel-based object-growing algorithm that ensured that the enlarged segments were connected to each other only by their branch points. Multiple single-segment parameters and aggregate vessel statistics were then calculated for the whole tumor. This allowed the color-coded three-dimensional visualization of different vessel features, for example segment diameter (Figure 5, G and H).
Figure 5.
Quantification of UM data sets. (A) The raw data from the tissue channel were used to determine the total tumor volume. (B) Segmentation between tissue and background. (C and D) Three-dimensional visualization of the volume (blue) and necrotic areas (yellow) of the tumor. (E) The virtual tissue slices formed the basis of the vascular analysis. (F) Three-dimensional segmentation of the binary vascular tree. (G and H) Growth of the skeletal segments and quantification of the different segments and branching points. The different diameters of the vascular segments are represented by different colors. (I) The information from the antibody channel is used to determine the total signal and penetration of trastuzumab-Alexa 750. (J) We used the color-coded segmented 3D distance map shown here to calculate the penetration of the antibody from the tumor margin to its center. (K and L) We used another color-coded 3D distance map to calculate the penetration of the antibody from the tumor vessels into the surrounding tissue. (A, B, E, F, and I–L) Single slice, 5-µm diameter. Scale bar, 250 µm.
To determine the penetration behavior of a fluorescence-labeled substance (e.g., trastuzumab-Alexa 750) into the tumor tissue, the software initially subtracted the computed background signal from the raw data. Thereby, the accumulated antibody signal and the average signal intensity of trastuzumab-Alexa 750 in the entire tumor area were calculated (Figure 5I). Furthermore, the distribution pattern of the fluorescence signals within the sample was analyzed. For this, the software calculated three-dimensional distance maps to the tumor border and the nearest vessel and then segmented these into different color-coded bins (Figure 5, J–L). Thereafter, the antibody signal intensities were quantified in every bin, resulting in penetration gradients of the antibody (Figure W3, D–H). This process allowed us to analyze the penetration behavior of the antibody from the tumor margins to its center (Figure 5J) and the penetration behavior of the therapeutic drug from the vessels into the surrounding tumor tissue (Figure 5, K and L).
Monitoring of Antiangiogenic Treatment Response
Having demonstrated the utility of this new imaging method for the visualization and automatic quantification of different tumor parameters and drug penetration, we performed a preclinical efficacy study to examine the effect of an antiangiogenic compound in the KPL-4 xenograft model. The treatment group received a single dose of bevacizumab, and control mice were treated with PBS. After 1, 3, and 7 days, respectively, we selected five animals from both groups and injected trastuzumab-Alexa 750. Six hours later, lectin-Alexa 647 was applied to tag the tumor vessels. Subsequently, the tumors were explanted, cleared, and scanned by UM. Afterwards, the tumors were embedded in paraffin, and conventional histology and IHC were performed.
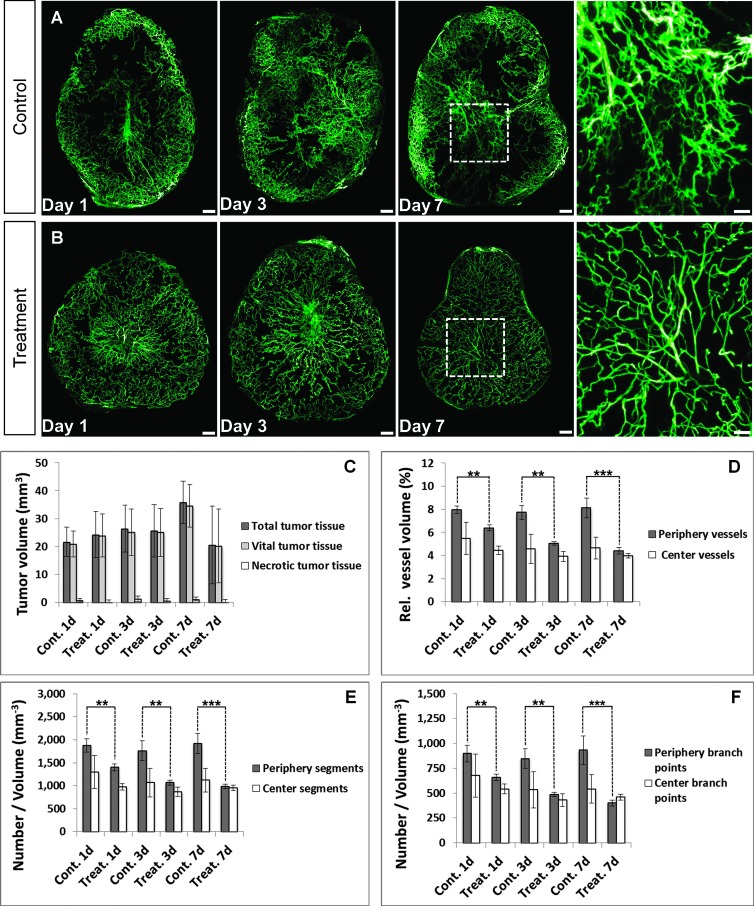
The UM results of the tumor vessels in the control group showed chaotic and inhomogeneous vascularity. Large sections of the tumor periphery appeared to be highly vascularized, whereas some areas of the tumor had only isolated vessels, and others were completely devoid of vascularization (Figure 6A). An effect of bevacizumab on the tumor vessels was already detectable 1 day after initiation of treatment. We found a significant reduction of the vessel volume (P < .01), number of vessel segments (P < .01), and branching points (P < .01) in the tumor periphery, whereas these parameters remained virtually unchanged in the center of the tumor (Figure 6, D–F). The treatment also resulted in vessel normalization, which confirms extensive studies performed by Rakesh Jain's group [48], and the vessel architecture appeared to be more homogeneous. Both effects became more pronounced as the length of treatment was extended (Figure 6B). Bevacizumab exerted its antiangiogenic effect significantly on vessels with a diameter between 10 and 30 µm, whereas vessels greater than 30 µm were not significantly affected by the therapy (Figure W4). The normalized vessel network obviously allowed an improved blood supply to the tumor, thereby reducing the interstitial fluid pressure [49], and thus provides a more constant supply of oxygen and nutrients [15]. In concordance to this, we found proliferating tumor cells evenly distributed in the tissues analyzed 7 days after start of treatment with bevacizumab (Figure W5; H&E and Ki67). Compared to the PBS-treated mice, necrotic areas were reduced by an average of 69% in the bevacizumab-treated tumors. In contrast to these changes, we did not observe a significant reduction of tumor volume even after 7 days of treatment (Figure 6C).
Figure 6.
Monitoring of antiangiogenic treatment response. (A and B)Wevisualized the vascular development of KPL-4 tumors in the control and treatment groups for a period of 7 days using UM. Treatment with bevacizumab reduced, homogenized, and normalized the tumor vasculature, whereas the vessel network of the control group remained unchanged. The effect was proportional to the length of treatment (days 1, 3, and 7; n = 5 per day). MIP, 60 slices with 5-µm diameter per slice. Scale bar, 250 µm and 100 µm (blowup). (C) Quantification results of the total, vital, and necrotic tumor tissue volume for the control and treatment groups at different time points. Even after 7 days of treatment, there was no significant reduction in tumor volume. (D–F) The quantification of the UM data sets showed a significant reduction of vessel volume (D), vessel segments (E), and branching points (F) in the tumor periphery, whereas these parameters remained virtually unchanged in the core of the tumor [relative vessel volume (%) = (tumor vessel vol/total tumor tissue vol) * 100]. Vasculature with a distance to the surface of the tumor smaller than 20% of the radius of a fictional sphere with the same volume as the tumor was defined as periphery. Control group, days 1, 3, and 7; n = 5 per day. Treatment group, days 1, 3, and 7; n = 5 per day. All values are given as means ± SD. *P < .05, **P < .01, and ***P < .001; t test.
Drug Penetration after Antiangiogenic Pretreatment
Besides analyzing the direct therapeutic effect of bevacizumab on the vascular architecture and the tumor volume, we also investigated the influence of an antiangiogenic pretreatment on the subsequent penetration of a tumor-targeting antibody into tumor tissue. Therefore, we combined the UM information of tumor vasculature from the previous chapter with the penetration behavior of trastuzumab-Alexa 750.
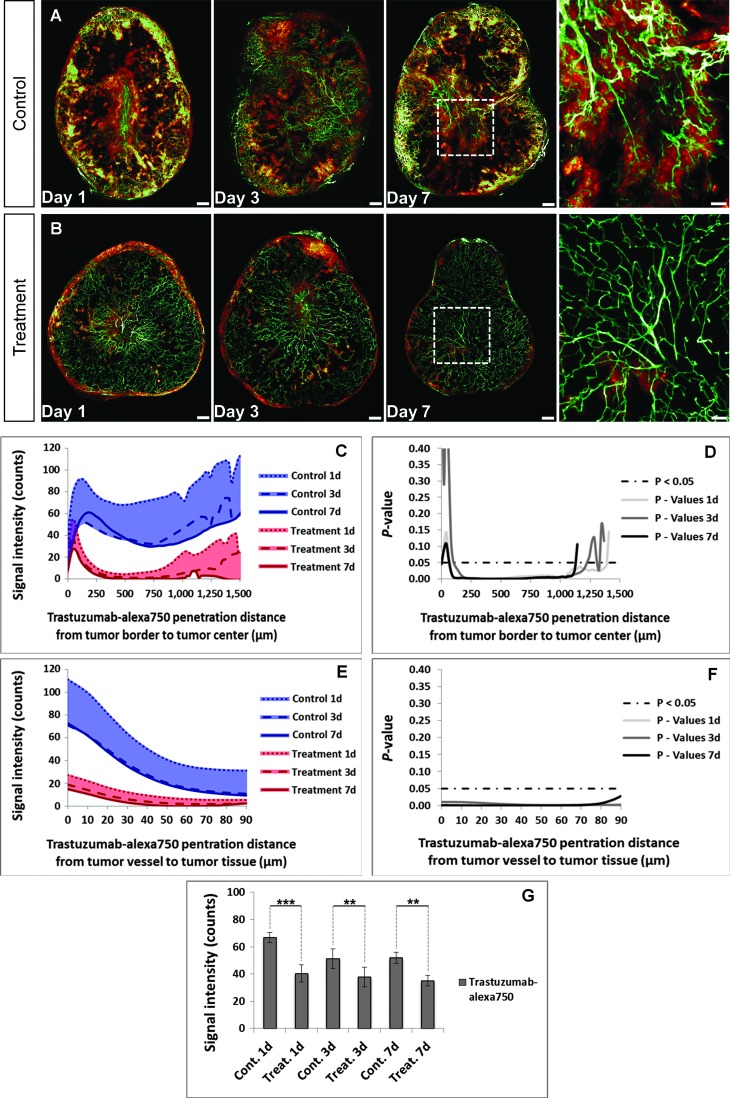
Control mice treated with PBS demonstrated a strong penetration of trastuzumab-Alexa 750 in highly vascularized tumor areas (Figure 7A). In contrast, explanted tumors from mice pretreated with bevacizumab showed a significant reduction in the trastuzumab-Alexa 750 penetration (P < .001) 1 day after initiation of treatment (Figure 7B). Furthermore, the antiangiogenic therapy decreased significantly the penetration of trastuzumab-Alexa 750 from the tumor border to its center (Figure 7, C and D) and also the penetration from the tumor vessels into the surrounding tumor tissue (Figure 7, E and F). These results demonstrated that bevacizumab diminishes deposition of trastuzumab in the tumor tissue for all tested time points (Figure 7G). To ensure that the antiangiogenic pretreatment did not affect the HER2 expression level, we performed IHC staining on tumor tissue sections. Both study groups (PBS and bevacizumab) demonstrated high extracellular HER2 expression (3+) on the surface of the KPL-4 tumor cells (Figure W5; HER2), and thus, we were able to rule out an internalization and/or shedding of the extra-cellular domains from the tumor cell surface.
Figure 7.
Monitoring of antibody penetration after antiangiogenic pretreatment. (A and B) The control group showed a strong penetration behavior of trastuzumab-Alexa 750 in the KPL-4 xenograft (A), whereas the labeled antibody penetration was significantly reduced by antiangiogenic pretreatment with bevacizumab (B). MIP, 60 slices with 5-µm diameter per slice. Scale bar, 250 µm and 100 µm (blowup). (C–G) Pretreatment with bevacizumab led to a significant reduction in trastuzumab-Alexa 750 penetration (G). This was seen in both the reduced penetration of trastuzumab-Alexa 750 from the tumor border to its center (C and D) and in the reduced labeled antibody penetration from the tumor vessels into the surrounding tumor tissue (E and F). Control group, blue; mean of days 1, 3, and 7; n = 5 per day. Treatment group, red; mean of days 1, 3, and 7; n = 5 per day. All values are given as means ± SD. *P < .05, **P < .01, and ***P < .001; t test.
Therefore, the diminished penetration of trastuzumab-Alexa 750 can only be attributed to the reduction in vascular permeability promoted by bevacizumab. The reduced pore size in the normalized tumor vessels made it more difficult for trastuzumab-Alexa 750 to penetrate due to its high molecular weight, and as a result, more of the antibody is confined to the vessel compartment [18,50] (Figure W6).
Discussion
In this work, we applied multispectral fluorescence UM to the field of tumor analysis and thus were able to create unprecedented three-dimensional and quantitative insights into whole tumors with cellular resolution.
This ex vivo imaging technique uses optical tissue clearing in combination with near-infrared fluorochromes to reduce light absorption and scattering in tissue to a minimum and therefore increase the penetration depth and image quality. The resultant high signal-to-noise ratio enabled a fast measurement of tumor samples up to 5 mm in diameter with an almost isotropic resolution of 5.1 µm. The applied optical clearing procedure did not give rise to any appreciable reduction of the fluorescence signals in the tumor samples (Figure W7) and had also no recognizable influence on the performance of conventional IHC staining. Various routine tissue stainings, such as H&E, Ki67, CD34, and HER2, could be conducted without any problems subsequently after UM measurements and illustrated perfect staining results. Furthermore, a direct comparison of the virtual data sets created by UM and conventional 2D histology demonstrated an excellent morphologic and anatomic correlation between both techniques.
The fully automated 3D analysis of the various UM data sets, such as tumor morphology, vessel architecture, and drug penetration, was carried out with a novel set of custom-developed image analysis algorithms that were implemented into a quantification software. This new quantification tool enables the automatic segmentation of different tumor parameters from each fluorescence channel and analyzed their specific value without any manual user interference. The complete quantification process for all parameters took less than 2 hours per tumor on a single processor, whereas the number of concurrent tumor quantifications is only limited by available hardware resources. Multiple engines can run simultaneously if enough hardware resources and Definiens licenses are available, thus, enabling the timely and fully automatic quantification of large groups. This aspect allows a very rapid, detailed, and cost-efficient implementation and analysis of preclinical studies on a large scale.
After successful validation, we applied this new imaging method in preclinical cancer research for the three-dimensional and quantitative analysis of the therapeutic treatment effect of bevacizumab on the tumor vasculature. We further investigated the influence of induced vessel normalization on the subsequent penetration behavior of fluorescent-labeled trastuzumab. Besides the determination of the total, vital, and necrotic tumor tissue volume, a quantitative analysis of the tumor vessel architecture was for the first time realized in a three-dimensional way at cellular resolution. A significant reduction of the tumor vessel volume, number of vessel segments, and branching points could be detected by UM even 1 day after initiation of treatment (P < .01). The antiangiogenic treatment effect of bevacizumab on vessel diameter between 10 and 30 µm was mainly executed in the tumor periphery (P < .01), whereas the established vasculature in the tumor center remained virtually unchanged. Furthermore, strong normalization and homogenization of the vessel network became more pronounced as the length of treatment extended.
It has been proposed by others that vessel normalization improves the supply of blood, nutrients, and oxygen into the tumor [15], reduces the interstitial fluid pressure [49], and thereby improves the delivery and penetration of therapeutics into tumor tissue [49,51,52]. In contrast to this, our results showed that bevacizumab-induced vessel normalization and reduction in vascular permeability diminished the penetration behavior of trastuzumab-Alexa 750 into tumor tissue. These results are in accordance to published data [50,53]. In consequence of this, a clear rational for scheduling of large therapeutic drugs, such as proteins and antibodies, in combination treatment with antiangiogenic compounds can be optimized. To receive optimal tumor penetration of both therapeutic agents, the tumor cell-targeting antibody must be injected before starting the antiangiogenic therapy.
The work presented here illustrates the huge potential of this new ex vivo imaging method for the three-dimensional and fully quantitative endpoint analysis of tumors at cellular resolution. By assessing multiple tumor parameters simultaneously, UM enables a unique, accurate, and comprehensive 3D overview of the heterogeneous tumor biology and drug penetration. In preclinical cancer research and drug development, we consider UM as a new link between current in vivo imaging methods and classic two-dimensional histology. The outstanding combination possibility and complementary behavior of fluorescent-based in vivo and ex vivo imaging techniques and classic histology enable a detailed pharmacokinetic and pharmacodynamic analysis of tumor biology and drug behavior from a macroscopic down to a microscopic scale. Comprehensive information about these complex mechanisms will help to efficiently design new drug formats and select suitable candidates more quickly. Moreover, because fluorescent-labeled proteins are becoming more and more relevant for clinical applications regarding tumor diagnosis, either for intraoperative or catheter-based procedures, we assume that UM can be used for three-dimensional analysis of human tissue samples in the near future [54,55].
In summary, multispectral fluorescence UM in combination with our novel automatic quantification software is a new postmortem imaging technique that provides the possibility for three-dimensional and fully quantitative endpoint analysis of multiple tumor parameters and drug penetration at cellular resolution. These unique and holistic three-dimensional insights will help to understand the complex tumor biology in more detail and should accelerate the various stages of preclinical drug development.
Supplementary Material
Acknowledgments
We thank J. Kurebayashi (Kawasaki Medical School) for kindly providing the human breast cancer cell line KPL-4. We also thank A. Wessner and R. Vogel for fluorescence labeling, U. Jucknischke for performing the Biacore studies, U. Haupt for animal handling and histologic support, and F. Osl for cell culturing and statistical support. We are indebted to T. Poschinger and O. Tonn for helpful discussion and S. Bader for supporting statistical analysis. The constant support of M. Weidner and especially of C. Meisel and K. Bosslet is very much appreciated.
Footnotes
This work is dedicated to our dear colleague S.S. who died from the consequences of an accident after the finalization of the manuscript. The authors M.D., W.S., and S.S. are employees of Roche Diagnostics GmbH. No conflict of interest was disclosed by V.N.
This article refers to supplementary materials, which are designated by Figures W1 to W7 and Videos W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 2.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savai R, Langheinrich AC, Schermuly RT, Pullamsetti SS, Dumitrascu R, Traupe H, Rau WS, Seeger W, Grimminger F, Banat A. Evaluation of angiogenesis using micro-computed tomography in a xenograft mouse model of lung cancer. Neoplasia. 2009;11:48–56. doi: 10.1593/neo.81036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalender WA. CT: the unexpected evolution of an imaging modality. Eur Radiol. 2005;15:D21–D24. doi: 10.1007/s10406-005-0128-3. [DOI] [PubMed] [Google Scholar]
- 5.Blankenberg FG, Levashova Z, Sarkar SK, Pizzonia J, Backer MV, Backer JM. Noninvasive assessment of tumor VEGF receptors in response to treatment with pazopanib: a molecular imaging study. Transl Oncol. 2010;3:56–64. doi: 10.1593/tlo.09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng JR, Stuart D, Aardalen K, Kaplan A, Aziz N, Hughes NP, Gambhir SS. Use of DNA microarray and small animal positron emission tomography in preclinical drug evaluation of RAF265, a novel B-Raf/VEGFR-2 inhibitor. Neoplasia. 2011;13:266–275. doi: 10.1593/neo.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyszka JM, Fraser SE, Jacobs RE. Magnetic resonance microscopy: recent advances and applications. Curr Opin Biotechnol. 2005;16:93–99. doi: 10.1016/j.copbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Loveless ME, Lawson D, Collins M, Nadella MVP, Reimer C, Huszar D, Halliday J, Waterton JC, Gore JC, Yankeelov TE. Comparisons of the efficacy of a Jak1/2 inhibitor (AZD1480) with a VEGF signaling inhibitor (cediranib) and sham treatments in mouse tumors using DCE-MRI, DW-MRI, and histology. Neoplasia. 2012;14:54–64. doi: 10.1593/neo.111478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razansky D, Deliolanis NC, Vinegoni C, Ntziachristos V. Deep tissue optical and optoacoustic molecular imaging technologies for pre-clinical research and drug discovery. Curr Pharm Biotechnol. 2012;13:504–522. doi: 10.2174/138920112799436258. [DOI] [PubMed] [Google Scholar]
- 10.Jarzyna PA, Deddens LH, Kann BH, Ramachandran S, Calcagno C, Chen W, Gianella A, Dijkhuizen RM, Griffioen AW, Fayad ZA, et al. Tumor angiogenesis phenotyping by nanoparticle-facilitated magnetic resonance and near-infrared fluorescence molecular imaging. Neoplasia. 2012;14:964–973. doi: 10.1593/neo.121148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampath D, Oeh J, Wyatt SK, Cao TC, Koeppen H, Eastham-Anderson J, Robillard L, Ho CC, Ross J, Zhuang G, et al. Multimodal microvascular imaging reveals that selective inhibition of class I PI3K is sufficient to induce an antivascular response. Neoplasia. 2013;15:694–711. doi: 10.1593/neo.13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissleder R. Scaling down imaging: molecular mapping of cancer in mice. Nat Rev Cancer. 2002;2:11–18. doi: 10.1038/nrc701. [DOI] [PubMed] [Google Scholar]
- 13.Rudin M, Weissleder R. Molecular imaging in drug discovery and development. Nat Rev Drug Discov. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- 14.Weissleder R. Molecular imaging in cancer. Science. 2006;312:1168–1171. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 15.Fukumura D, Duda DG, Munn LL, Jain RK. Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation. 2010;17:206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman RM. Visualization of GFP-expressing tumors and metastasis in vivo. Biotechniques. 2001;30:1016–1022. doi: 10.2144/01305bi01. [DOI] [PubMed] [Google Scholar]
- 17.Alencar H, Mahmood U, Kawano Y, Hirata T, Weissleder R. Novel multiwavelength microscopic scanner for mouse imaging. Neoplasia. 2005;7:977–983. doi: 10.1593/neo.05376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan F, Salehi HA, Boucher Y, Vasthare US, Tuma RF, Jain RK. Vascular permeability and microcirculation of gliomas and mammary carcinomas transplanted in rat and mouse cranial windows. Cancer Res. 1994;54:4564–4568. [PubMed] [Google Scholar]
- 19.Padera TP, Stoll BR, So PT, Jain RK. Conventional and high-speed intravital multiphoton laser scanning microscopy of microvasculature, lymphatics, and leukocyte-endothelial interactions. Mol Imaging. 2002;1:9–15. doi: 10.1162/15353500200200004. [DOI] [PubMed] [Google Scholar]
- 20.Vajkoczy P, Ullrich A, Menger MD. Intravital fluorescence video-microscopy to study tumor angiogenesis and microcirculation. Neoplasia. 2000;2:53–61. doi: 10.1038/sj.neo.7900062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin GR, Jain RK. Noninvasive measurement of interstitial pH profiles in normal and neoplastic tissue using fluorescence ratio imaging microscopy. Cancer Res. 1994;54:5670–5674. [PubMed] [Google Scholar]
- 22.Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, Munn LL, Tearney GJ, Fukumura D, Jain RK, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15:1219–1223. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown EB, Campbell RB, Tsuzuki Y, Xu L, Carmeliet P, Fukumura D, Jain RK. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nat Med. 2001;7:864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- 24.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 25.Masters B, So P. Handbook of Biomedical Nonlinear Optical Microscopy. Oxford, UK: Oxford University Press; 2008. [Google Scholar]
- 26.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, et al. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto JG, Pitris C, Boppart SA, Brezinski ME. Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia. 2000;2:9–25. doi: 10.1038/sj.neo.7900071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun SH, Tearney GJ, de Boer JF, Iftimia N, Bouma BE. High-speed optical frequency-domain imaging. Opt Express. 2003;11:2953–2963. doi: 10.1364/oe.11.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vakoc BJ, Fukumura D, Jain RK, Bouma BE. Cancer imaging by optical coherence tomography: preclinical progress and clinical potential. Nat Rev Cancer. 2012;12:363–368. doi: 10.1038/nrc3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunohistochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- 31.Keller PJ, Stelzer EHK. Quantitative in vivo imaging of entire embryos with Digital Scanned Laser Light Sheet Fluorescence Microscopy. Curr Opin Neurobiol. 2008;18:624–632. doi: 10.1016/j.conb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Huisken J, Stainier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DYR. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 34.Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, Wittbrodt J, Stelzer EHK. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgansberger W, Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 36.Holekamp TF, Turaga D, Holy TE. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron. 2008;57:661–672. doi: 10.1016/j.neuron.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Jährling N, Becker K, Schönbauer C, Schnorrer F, Dodt H-U. Three-dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Front Syst Neurosci. 2010;4:1. doi: 10.3389/neuro.06.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erturk A, Mauch CP, Hellal F, Förstner F, Keck T, Becker K, Jährling N, Steffens H, Richter M, Hübener M, et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat Med. 2012;18:166–171. doi: 10.1038/nm.2600. [DOI] [PubMed] [Google Scholar]
- 39.Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brede C, Friedrich M, Jordán-Garrote AL, Riedel SS, Bäuerlein CA, Heinze KG, Bopp T, Schulz S, Mottok A, Kiesel C, et al. Mapping immune processes in intact tissues at cellular resolution. J Clin Invest. 2012;122:4439–4446. doi: 10.1172/JCI65100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 42.Spalteholz W. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten. Leipzig: S. Hirzel; 1914. [Google Scholar]
- 43.Genina EA, Bashkatov AN, Tuchin VV. Tissue optical immersion clearing. Expert Rev Med Devices. 2010;7:825–842. doi: 10.1586/erd.10.50. [DOI] [PubMed] [Google Scholar]
- 44.Jährling N, Becker K, Dodt H-U. 3D-reconstruction of blood vessels by ultramicroscopy. Organogenesis. 2009;5:145–148. doi: 10.4161/org.5.4.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomer R, Khairy K, Keller PJ. Shedding light on the system: studying embryonic development with light sheet microscopy. Curr Opin Genet Dev. 2011;21:558–565. doi: 10.1016/j.gde.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Homann H. Implementation of a 3D thinning algorithm. The Insight Journal. 2007 Jul-Dec;:1–4. [Google Scholar]
- 47.Lee TC, Kashyap RL, Chu CN. Building skeleton models via 3-D medial surface/axis thinning algorithms. Cvgip: Graphical Models and Image Processing. 1994;56:462–478. [Google Scholar]
- 48.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 49.Tong RT, Boucher Y, Kozin SV, Winkler F, Hicklin DJ, Jain RK. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer Res. 2004;64:3731–3736. doi: 10.1158/0008-5472.CAN-04-0074. [DOI] [PubMed] [Google Scholar]
- 50.Pastuskovas CV, Mundo EE, Williams SP, Nayak TK, Ho J, Ulufatu S, Clark S, Ross S, Cheng E, Parsons-Reponte K, et al. Effects of anti-VEGF on pharmacokinetics, biodistribution, and tumor penetration of trastuzumab in a preclinical breast cancer model. Mol Cancer Ther. 2012;11:752–762. doi: 10.1158/1535-7163.MCT-11-0742-T. [DOI] [PubMed] [Google Scholar]
- 51.Wildiers H, Guetens G, De Boeck G, Verbeken E, Landuyt B, Landuyt W, de Bruijn EA, van Oosterom AT. Effect of antivascular endothelial growth factor treatment on the intratumoral uptake of CPT-II. Br J Cancer. 2003;88:1979–1986. doi: 10.1038/sj.bjc.6601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang AD, Bauer TW, Camp ER, Somcio R, Liu WB, Fan F, Ellis LM. Improving delivery of antineoplastic agents with anti-vascular endothelial growth factor therapy. Cancer. 2005;103:1561–1570. doi: 10.1002/cncr.20942. [DOI] [PubMed] [Google Scholar]
- 53.Van der Veldt AA, Lubberink M, Bahce I, Walraven M, de Boer MP, Greuter HN, Hendrikse NH, Eriksson J, Windhorst AD, Postmus PE, et al. Rapid decrease in delivery of chemotherapy to tumors after anti-VEGF therapy: implications for scheduling of anti-angiogenic drugs. Cancer Cell. 2012;21:82–91. doi: 10.1016/j.ccr.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 54.van Dam GM, Themelis G, Crane LM, Harlaar NJ, Pleijhuis RG, Kelder W, Sarantopoulos A, de Jong JS, Arts HJ, van der Zee AG, et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-α targeting: first in-human results. Nat Med. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 55.Scheuer W, van Dam GM, Dobosz M, Schwaiger M, Ntziachristos V. Drug-based optical agents: infiltrating clinics at lower risk. Sci Transl Med. 2012;4:1–5. doi: 10.1126/scitranslmed.3003572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.