Abstract
Hepatocellular carcinoma (HCC) is a highly virulent malignancy with diverse etiology. Identification of a common mediator of aggressive progression of HCC would be extremely beneficial not only for diagnostic/prognostic purposes but also for developing targeted therapies. AEG-1/MTDH/LYRIC gene is amplified in human HCC patients, and overexpression of AEG-1/MTDH/LYRIC has been identified in a high percentage of both hepatitis B virus and hepatitis C virus positive HCC cases, suggesting its key role in regulating hepatocarcinogenesis. Important insights into the molecular mechanisms mediating oncogenic properties of AEG-1/MTDH/LYRIC, especially regulating chemoresistance, angiogenesis, and metastasis, have been obtained from studies using HCC model. Additionally, analysis of HCC model has facilitated the identification of AEG-1/MTDH/LYRIC downstream genes and interacting proteins, thereby unraveling novel players regulating HCC development and progression leading to the development of novel interventional strategies. Characterization of a hepatocyte-specific AEG-1/MTDH/LYRIC transgenic mouse (Alb/AEG-1) has revealed novel aspects of AEG-1/MTDH/LYRIC function in in vivo contexts. Combination of AEG-1/MTDH/LYRIC inhibition and chemotherapy has documented significant efficacy in abrogating human HCC xenografts in nude mice indicating the need for developing effective AEG-1/MTDH/LYRIC inhibition strategies to obtain objective response and survival benefits in terminal HCC patients.
1. HEPATOCELLULAR CARCINOMA: ETIOLOGY, EPIDEMIOLOGY, AND PATHOGENESIS
Hepatocellular carcinoma (HCC) is an epithelial tumor arising from the primary resident cells of the liver, the hepatocytes (El-Serag & Rudolph, 2007). HCC is the most common type of liver cancer accounting for >80% of all cancers in the liver (El-Serag & Rudolph, 2007). It is one of the five most common cancers and the third most common cause of cancer-related deaths worldwide (El-Serag, 2011). The epidemiology of HCC shows two major geographical patterns. In central and south-east Asia, sub-Saharan Africa, and the Amazon basin, it is the most common cancer (El-Serag, 2011). Globally HCC causes 662,000 deaths per year, about half of which is in China. The etiology is predominantly viral hepatitis, either Hepatitis B virus (HBV) or Hepatitis C virus (HCV). In China, ~90% HCC cases are HBV positive, while in Japan 70% of HCC cases are associated with HCV (Umemura, Ichijo, Yoshizawa, Tanaka, & Kiyosawa, 2009). Contamination of food with fungus, such as Aspergillus flavus generating aflatoxin, is another major cause of HCC, especially in Africa (Bressac, Kew, Wands, & Ozturk, 1991). The incidence of HCC is two to four times higher in males than in females, and the age of onset is between 30 and 50 years. The early onset of the disease indicates hepatitis infection earlier in life, such as at birth.
In North America and Western Europe, HCC is a rare cancer. However, the incidence of HCC is rising in Western countries. The incidence of HCC in the United States has increased from 1.6 to 4.9 (more than 200% increase) cases per 100,000 of population from 1975 to 2005 (Yang & Roberts, 2010). In the United States, the number of new cases of HCC in 2012 was estimated to be 28,720, out of which 20,550 were expected to die (Siegel, Naishadham, & Jemal, 2012). HCV and HBV constitute 48% and 16% of cases of HCC in the United States, respectively. Chronic alcoholism leading to cirrhosis and metabolic syndromes, such as nonalcoholic fatty liver disease usually associated with obesity and type 2 diabetes, are the most common causes in the remaining cases (Calle, Rodriguez, Walker-Thurmond, & Thun, 2003; Donato et al., 2002; El-Serag, Tran, & Everhart, 2004; Starley, Calcagno, & Harrison, 2010). Other rare causes of HCC include hemochromatosis, α1-antitrypsin deficiency, autoimmune hepatitis, porphyrias, and Wilson disease (Dragani, 2010; El-Serag & Rudolph, 2007).
The pathogenesis of HCC usually follows two patterns, cirrhotic and noncirrhotic (Farazi & DePinho, 2006). Viral hepatitis triggers an immune response recruiting T-lymphocytes (Rehermann & Nascimbeni, 2005). Consequently, there is continuous hepatocyte necrosis, inflammation, regeneration, and fibrotic repair leading to cirrhosis. The endless cycle of necrosis/regeneration to restore the tissue architecture of the liver might initiate dysfunctional telomeres and genomic instability which, in turn, give rise to dysplastic lesions that progress to HCC. This scenario is more common for HCV infection and chronic alcoholism. The scenario might be different for HBV, which, unlike HCV, might be integrated into the genome resulting in microdeletion of genes (such as tumor suppressor genes), or the viral enhancer elements might upregulate genes, such as telomerase reverse transcriptase, platelet-derived growth factor receptor-β, and mitogen-activated protein kinase 1 (MAPK1), which favor limitless proliferation potential (Murakami et al., 2005). In HBV cases, HCC thus might develop in the absence of cirrhosis. However, chronic inflammation-induced cirrhotic changes are the predominant modality in HBV cases as well.
HCC might be either nodular or infiltrative macroscopically (Llovet, Bru, & Bruix, 1999). The nodular type might contain a large solitary mass or there might be multiple nodules. The nodules are not encapsulated but well circumscribed. The diffuse type is not well circumscribed and might infiltrate into the portal or hepatic vein. Microscopically, the tumor might be either well differentiated, in which the tumor cells resemble hepatocytes and form trabeculae, cords, and nests, or poorly differentiated, in which the malignant cells are pleomorphic, anaplastic, and giant. The tumor is usually compact with little stroma and contains a central necrotic core because of inadequate vascularization and hypoxia.
The incidence and mortality of HCC run parallel. HCC is a tumor with rapid growth and early vascular invasion (Llovet, Burroughs, & Bruix, 2003). It is also highly resistant to standard chemotherapy (Llovet et al., 2003; Pang et al., 2008; Poon et al., 2001). The treatment options for HCC depend upon the stages and grades of the disease (Llovet et al., 1999). With localized disease, surgical resection, radiofrequency ablation, and liver transplantations are the treatments of choice (Georgiades, Hong, & Geschwind, 2008; O'Neil & Venook, 2007). However, most HCC patients present with advanced symptomatic tumors with underlying cirrhotic changes that are not amenable to surgical resection or transplantation. Transarterial chemoembolization and systemic therapy with doxorubicin alone or a combination of cisplatin, interferon, doxorubicin, and 5-fluorouracil (PIAF) are being used for advanced disease with moderate improvement in overall survival (OS) duration varying between 6.8 and 8.6 months (Leung et al., 1999; Llovet & Bruix, 2003; Llovet et al., 2002; Yeo et al., 2005). Sorafenib, an inhibitor of c-Raf and B-Raf kinases as well as of vascular endothelial growth factor receptor family, has been approved by FDA for unresectable advanced HCC (Llovet et al., 2008). While the median survival for placebo-treated patients was approximately 7.9 months, sorafenib-treated patients survived 10.7 months. VEGF pathway inhibitor bevacizumab, either alone or in combination with chemotherapy, also demonstrates very limited response (O'Neil & Venook, 2007; Zhu et al., 2006). In view of this dismal scenario, understanding the molecular pathogenesis of HCC and developing targeted and effective treatments are mandatory to significantly increase the survival interval and ameliorate the sufferings of the patients.
2. HCC: THE MOLECULAR ABNORMALITIES
Multiple etiologies have been linked to HCC, and therefore, no consistent genomic abnormalities have been attributed to this disease. Chronic HBV infection results in HCC by integration of HBV DNA into the genome leading to chromosomal instability and by HBV × protein (HBx) that activates a plethora of proto-oncogenes and signaling pathways associated with HCC (Brechot, Pourcel, Louise, Rain, & Tiollais, 1980; Feitelson & Duan, 1997). Chronic HCV infection causes HCC via core HCV proteins, NS3 and NS5A that inhibit the cyclin-dependent kinase inhibitor p21 and interact with p53 (Kwun, Jung, Ahn, Lee, & Jang, 2001; Majumder, Ghosh, Steele, Ray, & Ray, 2001). Mutations in numerous proto-oncogenes and tumor suppressor genes, such as p53, p73, Rb, APC, DLC-1, DLC-2, PTEN, SOCS1, GSTP1, HCCS1, Smad2/4, AXIN1, IGF-2, β-catenin, c-myc, and cyclin D1, have been detected in HCC (Boyault et al., 2007; Mann et al., 2007; Teufel et al., 2007). Oncogenomic approaches have identified cIAP1, an inhibitor of apoptosis, and Yap, a transcription factor, as candidate oncogenes for HCC (Zender et al., 2006).
The major signaling pathways activated in HCC are (i) MAPK that includes cascades of phosphorylation of ras, raf, mitogen-activated protein extracellular kinase (MEK), and extracellular signal-regulated kinase (ERK). Activation of this pathway has been well documented in HCC cell lines, in vivo HCC models, and human HCC specimens (Min, He, & Hui, 2011). (ii) Phosphotidyl-inositol-3-kinase (PI3K)/Akt/mTOR pathway (Llovet & Bruix, 2008). Activation of PI3K by growth factor receptors activates Akt that phosphorylates and inactivates proapoptotic proteins, such as Bad and caspase-9. Downstream of Akt is mTOR, a key regulator of cell translation machinery through two effector proteins, the eukaryotic initiation factor 4E-binding protein (4E-BP1) and the 40s ribosomal protein S6 kinase (p70s6k). These proteins regulate the translation of mRNAs of important genes regulating cell proliferation and angiogenesis, such as c-myc, cyclin D1, and HIF-1α. (iii) NF-κB pathway that might be activated by viral infection (Pikarsky et al., 2004). Persistent activation of NF-κB in the premalignant stage confers a survival advantage to hepatocytes that have acquired oncogenic mutations, thus favoring malignant transformation. (iv) Wnt/β-catenin signaling pathway (Ishizaki et al., 2004). The activation of multiple signaling pathways in different HCC makes it difficult to develop effective alternative therapies using small molecules. Identification of a key molecule that contributes to the activation of majority of these pathways would provide an important target for therapeutic intervention for HCC.
3. AEG-1/MTDH/LYRIC IS OVEREXPRESSED IN HCC
AEG-1/MTDH/LYRIC was initially identified as a HIV-1-inducible gene in primary human fetal astrocytes (Kang et al., 2005; Su et al., 2002). However, subsequent expression analysis documented that AEG-1 is overexpressed in a diverse array of cancers and AEG-1/MTDH/LYRIC is a downstream gene of oncogenic Ha-ras pathway, being transcriptionally regulated by c-Myc upon Ha-ras and PI3K activation, and AEG-1/MTDH/LYRIC cooperates with Ha-ras in promoting its transformation activity (Kang et al., 2005; Lee, Su, Emdad, Sarkar, & Fisher, 2006). Forced overexpression of AEG-1/MTDH/LYRIC increased proliferation and invasion, and activated NF-kB pathway by directly interacting with p65 subunit of NF-kB (Emdad et al., 2006; Sarkar et al., 2008). Additionally, AEG-1/MTDH/LYRIC was identified as a potent regulator of lung metastasis by breast cancer cells (Brown & Ruoslahti, 2004).
These initial studies prompted further analysis of AEG-1/MTDH/LYRIC in the context of HCC. Several studies analyzed AEG-1/MTDH/LYRIC expression and clinical correlation in human HCC patients and explored the potential molecular mechanism of AEG-1/MTDH/LYRIC overexpression. In the first study by Yoo et al., the expression of AEG-1/MTDH/LYRIC was analyzed by Western blotting in primary rat hepatocytes and human HCC cell lines HepG3, QGY-7703, SNU-423, Hep3B, Huh7, Sk-Hep-1, and Focus (Yoo, Emdad, et al., 2009). Among these cells, HepG3 do not form tumors in nude mice, while QGY-7703 HCC cells form aggressive tumors. Very low level of AEG-1/MTDH/LYRIC expression was detected in primary rat hepatocytes compared to all the human HCC cells. Interestingly, AEG-1/MTDH/LYRIC expression was higher in QGY-7703 cells compared to HepG3 cells. These findings were confirmed by immunohistochemistry in tissue microarrays (TMAs) containing 86 primary HCC, 23 metastatic HCC, and 9 normal adjacent liver samples. Very little to no AEG-1/MTDH/LYRIC immuno-staining was detected in the nine normal liver samples while significant AEG-1/MTDH/LYRIC staining was observed in HCC samples. AEG-1/MTDH/LYRIC expression was detected predominantly in the perinuclear region. Among the 109 HCC samples, only 7 scored negative for AEG-1/MTDH/LYRIC and the remaining 102 (93.58%) showed variable levels of AEG-1/MTDH/LYRIC that could be correlated with the stages of the disease based on the BCLC staging system. Expression of AEG-1/MTDH/LYRIC gradually increased with the stages from I to IV as well as with the grades of differentiation from well-differentiated to poorly differentiated, and a statistically significant correlation was obtained between AEG-1/MTDH/LYRIC expression level and the stage of HCC.
To interrogate the molecular mechanism of AEG-1/MTDH/LYRIC overexpression, AEG-1/MTDH/LYRIC mRNA expression was analyzed using a gene expression microarray (Affymetrix U133 plus 2.0) across 132 human samples in various stages of human hepatocarcinogenesis: normal liver (n=10), cirrhotic tissue (n=13), low-grade dysplastic nodules (n=10), high-grade dysplastic nodules (n=8), and HCC (n=91). Expression of AEG-1/MTDH/LYRIC in HCV-related HCC was significantly increased in comparison to normal liver and cirrhotic tissue. Mean upregulation in comparison to normal liver and cirrhosis were 1.7 (t-test, P=0.04)- and 1.65 (t-test, P<0.001)-fold increase, respectively.
Next, it was examined how many tumor samples had DNA copy gains at the AEG-1/MTDH/LYRIC locus located in chromosome 8q. To do so, the average copy number for the 52 SNP array probes within 250 kb on either side of AEG-1/MTDH/LYRIC was calculated. For a copy number cutoff >3, 27 of 103 tumors showed gains of chromosome 8q (26%). Nine of these same tumors had a copy number cutoff >4 (8.7%). Analysis of the pairwise correlation between copy number at the AEG-1/MTDH/LYRIC locus and the log-base-2 expression of every transcript on the U133 Plus 2.0 array, regardless of its position in the genome, revealed a significant statistical correlation between AEG-1/MTDH/LYRIC copy number and expression level (r=0.723; permutation P<0.004). AEG-1/MTDH/LYRIC was the 19th most significant gene on this candidate gene list in chromosome 8. Finally, the list of significantly overexpressed or underexpressed genes associated with AEG-1/MTDH/LYRIC copy number was analyzed using the Significance Analysis of Microarrays package. Among the 91 tumors with expression data, 24 had copy gains >3 and 8 had copy gains >4. AEG-1/MTDH/LYRIC was among the list of significantly overexpressed genes using copy number cutoffs >3 or >4 (FDR q-value<0.002). These findings were strengthened by fluorescence in situ hybridization (FISH) for AEG-1/MTDH/LYRIC in human HCC TMA (CF and DS, unpublished result). Copy number gains involving AEG-1/MTDH/LYRIC were demonstrated in 32% of the core HCC tissue samples, with amplification of AEG-1/MTDH/LYRIC detected in 4 cores (7%) and low-level gain of AEG-1/MTDH/LYRIC present in an additional 15 cores (25%). Of the remaining cores, 67% exhibited multiple copies (polysomy) of chromosome 8; cells with trisomy 8 were universally present in these samples, together with variable numbers of cells with four or more copies of chromosome 8. In summary, AEG-1/MTDH/LYRIC is significantly overexpressed in HCC when compared to normal liver. This overexpression is associated with elevated copy numbers of AEG-1/MTDH/LYRIC, predominantly due to gains of large regions of chromosome 8q.
Following the initial observations, two additional studies have analyzed AEG-1/MTDH/LYRIC expression in HCC samples and correlated with patients' prognosis (Gong et al., 2012; Zhu et al., 2011). In the study by Zhu et al., AEG-1/MTDH/LYRIC expression was assessed in TMA of 323 HCC patients. The immunohistochemistry results showed that AEG-1/MTDH/LYRIC was primarily located in the membrane and the cytoplasm. Most of the tumor tissues expressed significantly higher levels of AEG-1/MTDH/LYRIC than adjacent nontumorous tissues, with AEG-1/MTDH/LYRICHigh accounting for 54.2% (175 of 323) of all the patients. The Pearson χ2 test indicated that AEG-1/MTDH/LYRIC expression was closely associated with microvascular invasion (P<0.001), pathologic satellites (P=0.007), tumor differentiation (P=0.002), and TNM stage (P=0.001). These results suggest that tumors with more microvascular invasion or pathologic satellites, poorer differentiation, and TNM stages II and III are prone to exhibit higher AEG-1/MTDH/LYRIC expression. Expression of AEG-1/MTDH/LYRIC did not correlate with other clinicopathologic characteristics such as age, gender, liver cirrhosis, serum alpha-fetoprotein, tumor diameter, tumor encapsulation, or BCLC stage.
As of the last follow-up in March 2009, 54.2% (175 of 323) of the patients had suffered from recurrence and 51.1% (165 of 323) had died with local or distant recurrence. The 1-, 3-, and 5-year OS and cumulative recurrence rates in the whole cohort were 85.4% and 25.4%, 62.2% and 50.2%, 50.7% and 59.7%, respectively. Furthermore, the 1-, 3-, 5-year OS rates in the AEG-1/MTDH/LYRICHigh group were significantly lower than those in the AEG-1/MTDH/LYRICLow group (83.0% vs. 89.7%, 52.0% vs. 75.3%, 37.4% vs. 66.9%, respectively); the 1-, 3-, 5-year cumulative recurrence rates were markedly higher in the AEG-1/MTDH/LYRICHigh group than those in the AEG-1/MTDH/LYRICLow group (32.4% vs. 16.8%, 61.2% vs. 38.2%, 70.7% vs. 47.8%, respectively). Univariate and multivariate analyses revealed that along with tumor diameter, encapsulation, microvascular invasion, and TNM stage, AEG-1/MTDH/LYRIC was an independent prognostic factor for both OS (HR=1.870, P<0.001) and recurrence (HR=1.695, P<0.001).
The Gong et al. study showed that AEG-1/MTDH/LYRIC expression levels were elevated in HBV-related HCC tissues compared to normal liver tissues. There was a trend for gradually increased AEG-1/MTDH/LYRIC expression from normal liver tissue to hepatitis B and HBV-related HCC tissues. Furthermore, a statistical analysis revealed that AEG-1/MTDH/LYRIC expression significantly correlated with the American Joint Committee on Cancer (AJCC, seventh edition) stage (P=0.020), T classification (P=0.007), N classification (P=0.044), vascular invasion (P=0.006), and histological differentiation (P=0.020) in the HBV-related HCC patients. In addition, patients with high AEG-1/MTDH/LYRIC levels had shorter survival times compared to those with low AEG-1/MTDH/LYRIC expression (P=0.001).
miR-375 that targets AEG-1/MTDH/LYRIC has been shown to be downregulated in human HCC. Analysis of 60 pairs of HCC and matched adjacent nontumor tissues revealed that miR-375 was significantly suppressed in HCC tissues, and the abundance of miR-375 was inversely correlated with that of AEG-1/MTDH/LYRIC. Transcription of AEG-1/MTDH/LYRIC is induced by direct binding of c-myc to AEG-1/MTDH/LYRIC promoter upon activation of Ha-ras/PI3K/Akt signaling. Since PI3K/Akt is activated and c-myc is overexpressed in human HCC, they might also regulate AEG-1/MTDH/LYRIC transcription in HCC. Thus, multiple mechanisms, such as genomic amplification as well as transcriptional and posttranscriptional regulation, might be responsible for AEG-1/MTDH/LYRIC overexpression in human HCC.
4. BIOLOGICAL CONSEQUENCE OF AEG-1/MTDH/LYRIC OVEREXPRESSION IN HCC
The availability of HepG3 cells, poorly aggressive HCC cells with low level of AEG-1/MTDH/LYRIC, and QGY-7703 cells, strongly aggressive HCC cells with high level of AEG-1/MTDH/LYRIC, facilitates the analysis of AEG-1/MTDH/LYRIC function by “gain-of-function” and “loss-of-function” approaches. Stable clones of HepG3 cells (Hep-AEG-1-8 and Hep-AEG-1-14) overexpressing AEG-1/MTDH/LYRIC demonstrated increased proliferation, colony formation, anchorage-independent growth, and Matrigel invasion in vitro and formed highly aggressive, metastatic, and angiogenic tumors in nude mice compared to the control clones (Hep-pc-4) (Yoo, Emdad, et al., 2009). Hep-AEG-1 clones expressed higher levels of proangiogenic factors, such as FGFα, VEGF, and pIGF compared to Hep-pc-4 clone. As a corollary, an adenovirus expressing AEG-1/MTDH/LYRIC siRNA significantly abrogated tumor formation by QGY-7703 cells in nude mice (Yoo, Emdad, et al., 2009). Stable clones of LM3 and 97H human HCC cells expressing AEG-1/MTDH/LYRIC shRNA demonstrated significant abrogation of migratory ability, as revealed by scratch assay and transwell migration assay, and upon orthotopic implantation in the liver of nude mice, these clones developed significantly less number of pulmonary and abdominal metastases (Zhu et al., 2011). These knockdown clones exhibited a switch in the EMT phenotype characterized by upregulation of E-cadherin, downregulation of N-cadherin and Snail, and increased cytoplasmic accumulation of β-catenin (Zhu et al., 2011).
Analysis of signal transduction pathway revealed activation of MEK/ERK, p38 MAPK, Akt, and NF-κB pathways in Hep-AEG-1 clones compared to control Hep-pc-4 clone (Yoo, Emdad, et al., 2009). Inhibition of MEK/ERK and p38 MAPK pathways by their specific inhibitors PD98059 and SB203580, respectively, did not significantly affect increased proliferation conferred by AEG-1/MTDH/LYRIC; however, it markedly inhibited AEG-1/MTDH/LYRIC-induced Matrigel inhibition and anchorage-independent growth, indicating that MEK/ERK and p38 MAPK pathways might mediate more aggressive phenotype conferred by AEG-1/MTDH/LYRIC.
5. AEG-1/MTDH/LYRIC DOWNSTREAM GENES
To identify the downstream genes mediating the effects of AEG-1/MTDH/LYRIC in HCC cells, an Affymetrix oligonucleotide microarray (Human U133 plus 2.0) was performed between Hep-pc-4 and Hep-AEG-1-14 clones (Yoo, Emdad, et al., 2009). With a 1.5-fold cutoff, expressions of 5180 different oligonucleotides, that include ESTs and multiple oligonucleotides belonging to the same gene, were modulated in Hep-AEG-1-14 clone compared to Hep-pc-4 clone. One cluster of genes that were significantly modulated belongs to the Wnt signaling pathway. LEF-1, the transcription factor activated by Wnt signaling, was induced by 12.35-fold, while two negative regulators of Wnt signaling CTBP2 and APC were downregulated by 33.76- and 2.32-fold, respectively, in Hep-AEG-1-14 clone compared to Hep-pc-4 clone.
The second cluster of genes that were upregulated in Hep-AEG-1-14 clone is associated with chemoresistance. These genes include drug-metabolizing enzymes, such as dihydropyrimidine dehydrogenase (DPYD), principal enzyme inactivating 5-fluorouracil (5-FU), cytochrome P4502B6 (CYP2B6), involved in metabolism of multiple drugs, and dihydrodiol dehydrogenase (AKR1C2), conferring resistance to doxorubicin and cisplatin. The ATP-binding cassette transporter, ABCC11/MRP8, that causes efflux of multiple chemotherapeutics including 5-FU, was significantly induced in Hep-AEG-1-14 clone. Expression of LSF/TFCP2/LBP1-1c which activates the transcription of thymidylate synthase (TS), target of 5-FU, was significantly upregulated in Hep-AEG-1-14 clone.
Genes associated with invasion, such as claudin 4 and tetraspanin 8 (TSPAN8), were upregulated, and transgelin (TAGLN, a suppressor of MMP-9) was downregulated significantly in Hep-AEG-1-14 clone. IGFBP7, a secreted protein involved in senescence induction, was markedly downregulated in Hep-AEG-1-14 clone. Pyruvate kinase, a key enzyme of the glycolytic pathway, was also upregulated in Hep-AEG-1-14 clone. The up- or downregulation of majority of these genes by AEG-1/MTDH/LYRIC were confirmed by Taqman quantitative PCR and correlated well with the findings of microarray. Immunohistochemical analysis of tissue sections of matched normal liver and HCC samples revealed concomitant upregulation of AEG-1/MTDH/LYRIC, LEF-1, LSF, and DPYD and downregulation of IGFBP7 in HCC in 13 out of 18 patients further strengthening the potential regulation of these genes by AEG-1/MTDH/LYRIC.
5.1. LEF-1 and Wnt/β-catenin signaling
Activation of Wnt/β-catenin signaling plays an important role in hepatocarcinogenesis (Ishizaki et al., 2004; Thompson & Monga, 2007). The marked induction in LEF-1 and concomitant downregulation of CTBP2 prompted further analysis of AEG-1/MTDH/LYRIC-induced activation of Wnt/β-catenin pathway (Yoo, Emdad, et al., 2009). LEF-1 and its downstream target c-Myc were significantly upregulated, and LEF-1-regulated luciferase reporter activity was significantly higher in Hep-AEG-1 clones compared to Hep-pc-4 clone (Yoo, Emdad, et al., 2009). Knocking down LEF-1 by siRNA significantly abrogated AEG-1/MTDH/LYRIC-induced Matrigel invasion. Nuclear accumulation of β-catenin was significantly augmented in Hep-AEG-1 clones compared to Hep-pc-4 clone. β-Catenin is phosphorylated by GSK3β and undergoes proteasomal degradation while phosphorylation of GSK3β inactivates it and allows nuclear translocation of β-catenin. Since AEG-1/MTDH/LYRIC activates ERK42/44, it was hypothesized that ERK42/44 might phosphor-ylate GSK3β. The level of phosphorylated GSK3β was significantly higher in Hep-AEG-1-14 clone compared to Hep-pc-4 clone. As a consequence, the level of phosphorylated β-catenin was downregulated resulting in an increase in total β-catenin. Treatment with PD98059 decreased phosphorylated GSK3β level thus activating it, resulting in an increase in phosphorylated β-catenin level and a decrease in total β-catenin. These findings indicate that AEG-1/MTDH/LYRIC activates the Wnt signaling pathway by directly inducing LEF-1 level and indirectly by activating ERK42/44 thus facilitating nuclear translocation of β-catenin (Fig 7.1).
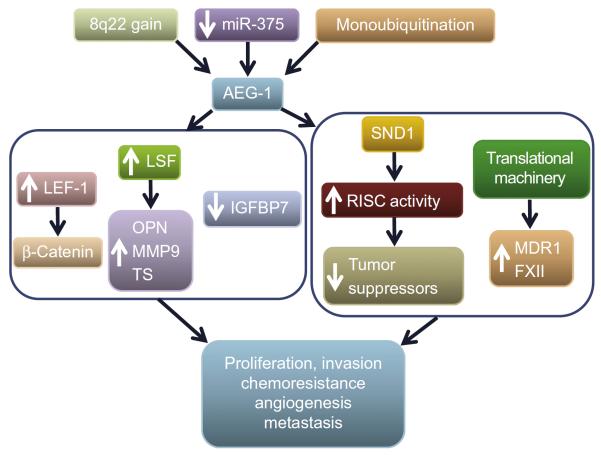
Figure 7.1.
AEG-1/MTDH/LYRIC promotes hepatocarcinogenesis. In HCC genomic amplification (8q22 gain), downregulation of miRNA-375 and stabilization of AEG-1/MTDH/LYRIC protein by monoubiquitination have been identified as potential mechanisms of AEG-1/MTDH/LYRIC overexpression. Microarray analysis identified AEG-1/MTDH/LYRIC downstream genes (left box). AEG-1/MTDH/LYRIC increases LEF-1 with resultant activation of β-catenin pathway. AEG-1/MTDH/LYRIC induces the transcription factor late SV40 factor (LSF) which transcriptionally regulates osteopontin (OPN), matrix metalloproteinase-9 (MMP9), and thymidylate synthase (TS). AEG-1/MTDH/LYRIC downregulates insulin-like growth factor-binding protein-7 (IGFBP7), an inducer of senescence. Right box shows AEG-1/MTDH/LYRIC-interacting proteins identified in HCC cells. AEG-1/MTDH/LYRIC interacts with staphylococcal nuclease domain containing-1 (SND1) and together these two proteins increase RNA-induced silencing complex (RISC) activity that facilitates oncogenic miRNA-mediated degradation of tumor suppressor mRNAs. AEG-1/MTDH/LYRIC interacts with the translational machinery that potentially facilitates association of specific mRNAs, such as multidrug resistance 1 (MDR1) and coagulation factor XII (FXII), to polysome increasing their translation. The net effects of these events are augmentation of hallmarks of cancer.
5.2. LSF, DPYD, and resistance to 5-FU
5-FU is a common chemotherapeutic for HCC. 5-FU is converted intracellularly into 5′-fluoro-2′-deoxyuridine by thymidine phosphorylase with subsequent phosphorylation by thymidine kinase into the active metabolite 5-fluoro-2′-deoxyuridine 5′-monophosphate (FdUMP) (Longley, Harkin, & Johnston, 2003). FdUMP inhibits TS which reduces the thymidine pool and increases the uracil pool leading to the inhibition of DNA synthesis. 5-FU is converted into its inactive metabolite fluoro-5,6-dihydrouracil (FUH2) by DPYD. TS and DPYD gene expression and/or activity are major determinants of the efficacy of 5-FU (Oguri et al., 2005; Yoshinare et al., 2003).
The transcription factor late SV40 factor (LSF), also known as TFCP2, functions as a transcriptional activator or repressor. It activates transcription of serum amyloid A3, IL-4, α-globin, α-A crystallin, TS, and PAX6 in different vertebrate species (Hansen, Owens, & Saxena, 2009; Santhekadur, Rajasekaran, et al., 2012; Veljkovic & Hansen, 2004). In cell-free extracts, it activates RNA polymerase II transcription by binding to basal promoter factor TFIIB. LSF also inhibits transcription of HIV LTR by binding to YY1 and histone deacetylase 1. A major cellular target of LSF is the TS gene, which encodes the rate-limiting enzyme in production of dTTP, required for DNA synthesis. LSF binds to the TS promoter and upregulates TS mRNA at the G1/S transition (Powell, Rudge, Zhu, Johnson, & Hansen, 2000). Inhibition of LSF by a dominant-negative construct (LSFdn) inhibits TS induction and induces apoptosis while addition of thymidine in the medium protects the cells from inhibition of DNA synthesis and induction of apoptosis. As a consequence, LSF plays an important role in DNA synthesis and cell survival. However, even though LSF has been studied for more than 25 years, no study was performed to dissect its potential role in carcinogenesis.
Expression of DPYD and LSF and its target gene TS was significantly higher in Hep-AEG-1 clones compared to Hep-pc-4 clone (Yoo, Gredler, et al., 2009). Nuclear accumulation of LSF and LSF-dependent luciferase reporter activity were also significantly higher in Hep-AEG-1 clone compared to Hep-pc-4 clone. Knocking down AEG-1/MTDH/LYRIC by siRNA resulted in significant downregulation of DPYD, LSF, and TS level while an LSFdn also induced downregulation of TS (Yoo, Gredler, et al., 2009). AEG-1/MTDH/LYRIC-overexpressing cells showed significant resistance to 5-FU which could be overcome by inhibiting AEG-1/MTDH/LYRIC, LSF, or DPYD. A lentivirus delivering AEG-1/MTDH/LYRIC siRNA in combination with 5-FU markedly inhibited growth of QGY-7703 HCC cells xenotransplanted in athymic nude mice when compared to either agent alone (Yoo, Gredler, et al., 2009).
Several follow-up studies highlighted the importance of LSF in hepatocarcinogenesis. Analysis of 109 human HCC samples by immunohistochemistry documented the overexpression of LSF in 100 samples compared to the normal liver which was associated with polyploidy of chromosome 12, where LSF gene is located, in 68% cases (Yoo, Emdad, et al., 2010). LSF expression is almost undetectable in normal hepatocytes, is very low in HepG3 cells, and is markedly high in most of the human HCC cell lines including QGY-7703. Forced overexpression of LSF in HepG3 cells increased proliferation, colony formation, Matrigel invasion, and anchorage-independent growth in vitro and resulted in highly aggressive, angiogenic, multiorgan metastatic tumors in nude mice upon subcutaneous xenograft assays, and tail vein injection (Yoo, Emdad, et al., 2010). Conversely, inhibition of LSF by a dominant-negative construct in QGY-7703 cells significantly abrogated in vitro proliferation, colony formation, Matrigel invasion, anchorage-independent growth, in vivo tumor growth, and metastasis. Affymetrix microarray identified many genes associated with invasion, angiogenesis, chemoresistance, and metastasis to be modulated by LSF, and the most robust induction was observed for osteopontin (OPN), a known mediator for tumor progression and metastasis (Bellahcene, Castronovo, Ogbureke, Fisher, & Fedarko, 2008). It was documented that LSF transcriptionally regulates OPN, and inhibition of OPN significantly abrogated the oncogenic functions of LSF (Yoo, Emdad, et al., 2010). OPN, induced by LSF, activates c-Met via interaction with CD44 receptor, and inhibition of c-Met significantly abrogated LSF-induced tumorigenesis and metastasis (Yoo, Gredler, et al., 2011). LSF transcriptionally upregulates MMP-9 that plays an important role in LSF-induced angiogenesis (Santhekadur, Gredler, et al., 2012). LSF also activates two important protumorigenic signaling, ERK and NF-κB (Yoo, Emdad, et al., 2010). Since the majority of HCC arises in the background of HBV or HCV infection, the activation of NF-κB by LSF indicates that LSF might also be involved in the inflammatory component of hepatocarcinogenesis. Indeed, a number of inflammatory cytokines induce LSF expression. Thus LSF promotes cell survival, chemoresistance, angiogenesis, and metastasis, all important hallmarks of cancer.
A high-throughput screening identified small-molecule inhibitors of LSF DNA binding and the prototype of these molecules, Factor quinolinone inhibitor 1 (FQI1), profoundly inhibited cell viability and induced apoptosis in human HCC cells without exerting harmful effects to normal immortal human hepatocytes and primary mouse hepatocytes (Grant et al., 2012). In nude mice xenograft studies, FQI1 markedly inhibited growth of human HCC xenografts as well as angiogenesis without exerting any toxicity. Thus, the original studies analyzing AEG-1/MTDH/LYRIC downstream genes identified a novel oncogene, its downstream pathways, and a class of novel anti-HCC therapeutics with promising efficiency.
5.3. IGFBP7: A tumor suppressor for HCC
IGFBP7, also known as mac25 or IGFBP-related protein-1 (IGFBP-rP1), is a secreted protein belonging to the IGFBP family. IGFBPs regulate the bio-availability of IGFs and insulin by physical binding and thereby limit access of IGFs or insulin to their corresponding receptors inhibiting the activity (Hwa, Oh, & Rosenfeld, 1999). IGFBP7 differs from the other six members of this family by lacking the C-terminus and having 100 times lower affinity for IGF-I but significantly higher affinity for insulin. IGFBP7 also exerts insulin/IGF-independent action where it induces senescence and apoptosis by inhibiting BRAF/MEK/ERK signaling cascade. IGFBP7 has been implicated to be a tumor suppressor for breast, prostate, colorectal cancers, and melanoma (Burger et al., 1998; Ruan et al., 2007; Sprenger, Damon, Hwa, Rosenfeld, & Plymate, 1999; Wajapeyee, Serra, Zhu, Mahalingam, & Green, 2008). IGFBP7 was identified as the most robustly downregulated gene by AEG-1/MTDH/LYRIC (Yoo, Emdad, et al., 2009). Using an immunohistochemical approach in TMA, high IGFBP7 expression was documented in normal human liver, while in 104 HCC patients, there was a statistically significant gradual decrease in IGFBP7 expression with the stages of HCC (Chen et al., 2011). Moreover, in each stage, IGFBP7 expression was much lower in poorly differentiated grades compared to moderately differentiated grades. FISH in TMA revealed loss of heterozygosity for IGFBP7 locus in 26% of HCC patients (Chen et al., 2011). IGFBP7 mRNA and protein expression was significantly lower in human HCC cells compared to THLE-3 cells, which are normal human hepatocytes immortalized by SV40 T/t Ag. Inverse correlation between AEG-1/MTDH/LYRIC and IGFBP7 expression was observed in normal liver and matched HCC samples in 13 out of 18 patients. A separate study analyzing 104 patients documented negative IGFBP7 staining in 35.6% HCC patients which statistically correlated with large tumor size, increased vascular invasion, poor OS, and disease-free survival rates (Tomimaru et al., 2012). Thus IGFBP7 downregulation might be a clinically relevant prognostic marker for aggressive HCC. Thus IGFBP7 downregulation might be a clinically relevant prognostic marker for aggressive HCC.
To analyze the role of IGFBP7 downregulation in mediating AEG-1/MTDH/LYRIC function, stable IGFBP7-overexpressing clones were established in Hep-AEG-1-14 background (Chen et al., 2011). In in vitro assays, these clones showed small but significant inhibition of proliferation. However, in subcutaneous xenograft assays, the IGFBP7-overexpressing clones demonstrate profound inhibition of tumor growth. This marked in vivo inhibitory effect of IGFBP7 could be attributed to its ability to induce senescence as revealed by senescence-associated β-galactosidase (SA β-gal) and senescence-associated heterochromatin foci assays, and inhibit angiogenesis as revealed by human vascular endothelial cell (HUVEC) differentiation and chicken chorioallantoic membrane (CAM) assays. Reduced activation of IGF-IR, Akt, and ERK was observed in IGFBP7-overexpressing clones. Thus, IGFBP7 downregulation might play an important role in promoting AEG-1/MTDH/LYRIC function. The molecular mechanism by which AEG-1/MTDH/LYRIC downregulates IGFBP7 remains to be determined.
The profound growth inhibitory properties of IGFBP7 suggest it to be a potential therapeutic for HCC. A replication incompetent adenovirus expressing IGFBP7 (Ad.IGFBP7) profoundly inhibited viability and induced apoptosis in multiple human HCC cell lines by inducing reactive oxygen species (ROS) and activating a DNA damage response (DDR) and p38 MAPK (Chen et al., 2013). In orthotopic xenograft models of human HCC in athymic nude mice, intravenous administration of Ad. IGFBP7 profoundly inhibited primary tumor growth and intrahepatic metastasis. In a nude mice subcutaneous model, xenografts from human HCC cells were established in both flanks and only left-sided tumors received intratumoral injection of Ad.IGFBP7. Growth of both left-sided injected tumors and right-sided uninjected tumors was markedly inhibited by Ad.IGFBP7 with profound suppression of angiogenesis. These findings indicate that Ad.IGFBP7 might be a potent therapeutic eradicating both primary HCC and distant metastasis and might be an effective treatment option for terminal HCC patients either alone or in combination with other agents.
6. AEG-1/MTDH/LYRIC-INTERACTING PROTEINS
Apart from having a transmembrane domain and three nuclear/nucleolar localization signals, AEG-1/MTDH/LYRIC protein does not have any known domains and motifs. However, AEG-1/MTDH/LYRIC is localized in the nucleus and nucleolus, in the ER/perinuclear region, abundantly in the cytoplasm, and also in the membrane. In the nucleus, multiple interacting partners of AEG-1/MTDH/LYRIC have been identified, such as p65 subunit of NF-κB, CBP, and PLZF, exerting diverse levels of transcriptional regulation (Emdad et al., 2006; Sarkar et al., 2008; Thirkettle, Mills, Whitaker, & Neal, 2009). These findings strongly suggest that depending upon its subcellular localization AEG-1/MTDH/LYRIC might interact with different protein complexes, thereby mediating the oncogenic functions of AEG-1/MTDH/LYRIC. With that end in view, the identification of AEG-1/MTDH/LYRIC-interacting protein was endeavored using two approaches. The first approach involved yeast two-hybrid (Y2H) screening. The N-terminal (a.a. 1–57) and C-terminal (a.a. 68–582) regions of AEG-1/MTDH/LYRIC that precedes and follows the transmembrane domain, respectively, were used as baits to separately screen a human liver cDNA library using the technology of Hybrigenics (http://www.hybrigenics-services.com) (Yoo, Santhekadur, et al., 2011). The C-terminal region showed autoactivator function, thereby complicating the assay. However, using selective medium containing 20 mM of 3-aminotriazole, the inhibitor of the reporter gene product, the assay could be optimized. Despite these efforts, only five known proteins with moderate confidence in the interaction were identified, namely, staphylococcal nuclease domain-containing 1 (SND1), chaperonin-containing TCP1, subunit 3 (gamma) (CCT3), extra-cellular matrix protein 2 (ECM2), inter-alpha (globulin) inhibitor H4 (ITIH4), and solute carrier family 22, member 1 (SLC22A1) (Yoo, Santhekadur, et al., 2011). A number of unknown proteins were also identified by this assay.
For the N-terminal interaction, 11 known proteins were identified. Arginase (ARG1) and 24-dehydrocholesterol reductase showed high and prostaglandin reductase 1 showed good confidence of interaction, respectively. The eight proteins showing moderate confidence of interaction were ATPase, class VI, type 11C (ATP11C), cytochrome P450, family 2, subfamily C (CYP2C8), ITIH3, MAP/microtubule affinity-regulating kinase 2, quinoid dihydropteridine reductase, retinoid X receptor, beta, tenascin N, and tripartite motif-containing 8. Three previously uncharacterized proteins were also identified as potential AEG-1/MTDH/LYRIC-interacting protein.
The relatively modest result of the Y2H screening prompted the employment of alternative strategy of coimmunoprecipitation coupled with mass spectrometry. Cell lysates from Hep-AEG-1-14 and Hep-pc-4 cells were subjected to immunoprecipitation using protein A agarose conjugated with anti-HA antibody (anti-HA agarose) (Yoo, Santhekadur, et al., 2011). The immunoprecipitates were eluted using HA peptide and were run in a SDS-PAGE gel. The gel was stained with Coomassie blue and the stained bands, that were present only in Hep-AEG-1-14 immunoprecipitates but not in Hep-pc-4 immunoprecipitates, were cut and were subjected to LC-MS/MS analysis after in-gel trypsin digestion. A total of 182 potential AEG-1/MTDH/LYRIC-interacting proteins were thus identified. However, the most represented proteins were AEG-1/MTDH/LYRIC and SND1.
6.1. Staphylococcal nuclease domain-containing 1
The identification of SND1 as an AEG-1/MTDH/LYRIC-interacting protein by both genomic and proteomic approaches strongly suggested further exploration. SND1, also known as p100 coactivator or Tudor-SN, is a multifunctional protein modulating transcription, mRNA splicing, RNAi function, and mRNA stability (Caudy et al., 2003; Paukku, Kalkkinen, Silvennoinen, Kontula, & Lehtonen, 2008; Paukku, Yang, & Silvennoinen, 2003; Yang et al., 2002, 2007). In the cytoplasm, SND1 functions as a nuclease in the RNA-induced silencing complex (RISC) in which small RNAs (such as siRNAs or miRNAs) are complexed with ribonucleo-proteins to ensue RNAi-mediated gene silencing (Caudy et al., 2003). Immunofluorescence analysis using human HCC cell lines and HCC tissue samples demonstrated AEG-1/MTDH/LYRIC and SND1 interaction in the cytoplasm which was confirmed by coimmunoprecipitation analysis (Yoo, Santhekadur, et al., 2011). It was documented that AEG-1/MTDH/LYRIC also interacts with Argonaute 2, the major nuclease in the RISC, and thus AEG-1/MTDH/LYRIC is a bona fide component of RISC. Using a luciferase-based reporter assay and AEG-1/MTDH/LYRIC or SND1 overexpressing and knockdown clones, it was documented that both AEG-1/MTDH/LYRIC and SND1 are required for optimum RISC activity. Immunohistochemical analysis of TMA containing 109 HCC samples documented higher SND1 expression in 81 cases (~74%) compared to the normal liver, and SND1 expression gradually increased with the stages and grades of the disease. As a corollary, higher RISC activity was observed in human HCC cells compared to normal immortal hepatocytes THLE-3 cells. Increased RISC activity, conferred by AEG-1/MTDH/LYRIC or SND1, resulted in increased degradation of tumor suppressor mRNAs that are the target of oncomiRs. These mRNAs include PTEN, target of miR-221 and miR-21; CDKN1C (p57), target of miR-221; CDKN1A (p21), target of miR-106b; SPRY2, target of miR-21; and TGFBR2, target of miR-93. Overexpression of AEG-1/MTDH/LYRIC or SND1 downregulates, while knockdown of AEG-1/MTDH/LYRIC or SND1 upregulates all these mRNA levels in HCC cells thus supporting the hypothesis that increased RISC activity might contribute to hepatocarcinogenesis by augmenting oncomiR-mediated degradation of tumor suppressor mRNAs.
Inhibition of SND1 enzymatic activity using 3′, 5′-deoxythymidine bisphosphate (pdTp) significantly abrogated proliferation and colony formation by Hep-AEG-1 clones, indicating that AEG-1/SND1 interaction is required to mediate AEG-1/MTDH/LYRIC function (Yoo, Santhekadur, et al., 2011). Stable overexpression of SND1 in Hep3B cells, expressing low level of SND1, increased in vitro proliferation and colony formation and in vivo tumor formation in nude mice (Yoo, Santhekadur, et al., 2011). Stable knockdown of SND1 by shRNA in QGY-7703 cells, expressing high level of SND1, significantly abrogated the aforementioned in vitro and in vivo phenotypes. Conditioned medium from Hep3B–SND1 cells augmented while that from QGY-SND1si cells significantly inhibited angiogenesis as analyzed by CAM assay and HUVEC differentiation assay (Santhekadur, Das, et al., 2012). A linear pathway was unraveled in which SND1-induced activation of NF-κB resulted in induction of miR-221 and subsequent induction of angiogenic factors angiogenin and CXCL16 (Santhekadur, Das, et al., 2012). Inhibition of either of these components resulted in significant inhibition of SND1-induced angiogenesis thus highlighting the importance of this molecular cascade in regulating SND1 function. As SND1 regulates NF-κB and miR-221, two important determinants of HCC controlling the aggressive phenotype, SND1 inhibition might be an effective strategy to counteract this fatal malady.
Apart from its role in mRNA degradation as a component of RISC, SND1 has also been documented to bind to 3′-UTR of specific mRNAs, such as angiotensin II type 1 receptor (AT1R), and increase translation by reducing decay (Paukku et al., 2008). It was documented that by increasing the level of AT1R by posttranscriptional stabilization of its mRNA, SND1 activates AT1R downstream signaling, such as activation of ERK and SMAD2, resulting in activation of TGFβ signaling that regulates EMT, migration, and invasion of human HCC cells (PKS and DS, unpublished results). A significant positive correlation between SND1 and AT1R expression level was observed in human HCC samples validating the regulation of AT1R by SND1. Angiotensin-converting enzyme inhibitors and AT1R blockers inhibit hepatic fibrosis and subsequent hepatocarcinogenesis. The results of this study unravel a novel mechanism of AT1R activation in HCC, brought forth by SND1 overexpression, thereby strengthening the rationale of using AT1R blockers as adjuvant therapy for HCC.
6.2. Translational machinery and chemoresistance
One striking phenotype conferred by AEG-1/MTDH/LYRIC is chemoresistance. Upregulation of mRNA of DPYD, LSF, and TS by AEG-1/MTDH/LYRIC underlies the mechanism of 5-FU resistance (Yoo, Gredler, et al., 2009). However, AEG-1/MTDH/LYRIC employs a different mechanism to induce resistance to anthracycline compounds, such as doxorubicin. AEG-1/MTDH/LYRIC upregulated the expression of multidrug resistance gene 1 (MDR1/ABCB1) at the protein level but not at the mRNA level resulting in increased efflux and decreased accumulation of doxorubicin in HCC cells (Yoo, Chen, et al., 2010). Overexpression of AEG-1/MTDH/LYRIC, either forced or endogenous, provided significant resistance to doxorubicin which could be overcome either by knocking down AEG-1/MTDH/LYRIC with siRNA or by blocking MDR1 with siRNA or chemical inhibitors. A lentivirus expressing AEG-1/MTDH/LYRIC shRNA significantly augmented the efficacy of doxorubicin in inhibiting growth of human HCC cells in subcutaneous xenograft assays in athymic nude mice (Yoo, Chen, et al., 2010). It was demonstrated that AEG-1/MTDH/LYRIC facilitates association of MDR1 mRNA to polysomes thus facilitating MDR1 translation. The increased association of MDR1 mRNA to polysome could be blocked by LY294002, indicating that activation of PI3K/Akt pathway by AEG-1/MTDH/LYRIC is necessary to induce this effect. To provide support for this observation, a low dose of LY294002, which itself did not exert significant toxicity, significantly increased cytotoxicity of doxorubicin. Additionally, AEG-1/MTDH/LYRIC inhibited ubiquitination and proteasomal degradation of MDR1 protein resulting in further accumulation of MDR1 protein (Yoo, Chen, et al., 2010).
The observation that AEG-1/MTDH/LYRIC increases translation of MDR1 prompted further analysis of regulation of the translational machinery by AEG-1/MTDH/LYRIC. The phosphorylation and activation status of key cap-dependent translational initiation factors, namely, eukaryotic translation initiation factor 4E (eIF4E), eukaryotic translation initiation factor 4G (eIF4G), and 4E binding protein (4E-BP) that are regulated by the PI3K/Akt/mTOR pathway in Hep-pc-4 cells and Hep-AEG-1-14 cells untreated or treated with LY294002 were analyzed (Yoo, Chen, et al., 2010). Marked phosphorylation of eIF4G was observed in Hep-AEG-1-14 cells compared to Hep-pc-4 cells, which was not affected by LY294002 treatment. The phosphorylation of eIF4E and 4E-BP was not modulated by AEG-1/MTDH/LYRIC. Hypophosphorylated 4E-BP binds to the 5′ cap binding protein of eIF4E, thereby preventing its interaction with eIF4G and inhibiting translation. Phosphorylation of 4E-BP via the mTOR pathway releases 4E-BP from eIF4E resulting in the recruitment of eIF4G to the 5′ cap and thereby allowing translation initiation to proceed. As 4E-BP phosphorylation by AEG-1/MTDH/LYRIC was not detected, the phosphorylation of eIF4G and subsequent augmentation of translation by AEG-1/MTDH/LYRIC might be independent of the mTOR pathway. One possible mechanism by which AEG-1/MTDH/LYRIC facilitates translation initiation might be direct involvement of AEG-1/MTDH/LYRIC in the translational machinery. AEG-1/MTDH/LYRIC is a highly basic protein that might function as a scaffold facilitating binding of nucleic acids, including mRNA. Indeed, several eukaryotic translation initiation factors, translation elongation factors, and ribosomal proteins were identified as potential AEG-1/MTDH/LYRIC-interacting proteins, and AEG-1/MTDH/LYRIC was implicated to be a potential RNA-binding protein in other systems (Meng et al., 2012; Yoo, Santhekadur, et al., 2011). The mechanism by which AEG-1/MTDH/LYRIC increases translation of specific mRNAs remains to be elucidated.
7. THE ROLE OF AEG-1/MTDH/LYRIC IN HEPATOCARCINOGENESIS: NOVEL INSIGHTS FROM A MOUSE MODEL
The oncogenic properties of AEG-1/MTDH/LYRIC have been validated through numerous studies employing human patient samples, in vitro cell culture system, and nude mice xenograft models. When overexpressed in normal immortal human cells, AEG-1/MTDH/LYRIC significantly protects from serum starvation-induced apoptosis (Lee et al., 2008). Overexpression of AEG-1/MTDH/LYRIC in normal immortal cloned rat embryo fibroblasts results in aggressive tumor formation in nude mice (Emdad et al., 2009). These studies indicate that AEG-1/MTDH/LYRIC could confer transforming properties to normal immortal cells. However, whether AEG-1/MTDH/LYRIC alone might induce transformation and evoke an explicit carcinogenic phenotype was not clear. To better comprehend the role of AEG-1/MTDH/LYRIC in hepatocarcinogenesis and to decipher the underlying molecular mechanism(s) in an in vivo context, a transgenic mouse with hepatocyte-specific expression of AEG-1/MTDH/LYRIC (Alb/AEG-1) was generated (Srivastava et al., 2012). The hepatocyte-specific expression was engineered by using a mouse albumin promoter/enhancer element to drive human AEG-1/MTDH/LYRIC expression. Alb/AEG-1 mice did not develop spontaneous HCC when followed for a period of 1 year. However, when treated with a hepatocarcinogen, N-nitrosodiethylamine (DEN), at 28 weeks of age, only two out of 11 WT animals showed a few very small nodules in the liver, whereas all of the 17 Alb/AEG-1 mice livers harbored numerous nodules of different sizes. Histological analysis of the liver of WT mice showed a few dysplastic, hyperchromatic nuclei indicating that with time HCC would eventually develop. In Alb/AEG-1 mice, a marked increase in dysplastic, hyperchromatic nuclei was observed in both the nodules and the adjacent normal liver. The most striking feature was observed in the hepatic nodules of Alb/AEG-1 mice, showing profound steatotic phenotypes with large lipid droplets in the hepatocytes. A moderate level of steatosis was also observed in the adjacent normal liver in Alb/AEG-1 mice. There was a significant increase in hepatic enzymes, such as AST, ALT, and alkaline phosphatase, in the sera of Alb/AEG-1 mice versus the sera of WT mice. At 32 weeks of age, the WT mice developed hepatic nodules; however, the nodules developed in Alb/AEG-1 mice were markedly larger. These findings indicate that AEG-1/MTDH/LYRIC significantly accelerated the hepatocarcinogenic process in DEN-treated animals.
Affymetrix microarray analysis of DEN-treated WT and Alb/AEG-1 livers revealed upregulation of several genes that include HCC marker α-feto protein (Afp); invasion- and metastasis-associated genes Tspan8 and Lcn2; several genes associated with fat metabolism, such as, Scd2, Lpl, Apoa4, and Apoc2; and genes regulating angiogenesis, such as TFF3. Tspan8 was also found to be upregulated by AEG-1/MTDH/LYRIC in human HCC cells indicating a potential key role in mediating AEG-1/MTDH/LYRIC function.
Analysis of hepatocytes from WT and Alb/AEG-1 hepatocytes provided several important insights into the function of AEG-1/MTDH/LYRIC. Alb/AEG-1 hepatocytes demonstrated marked resistance to doxorubicin and 5-FU treatment when compared to their WT littermates. Primary mouse hepatocytes, cultured in the presence of growth factors, do not divide and show decreasing viability after ~4 days as they enter senescence. The viability of Alb/AEG-1 hepatocytes in complete growth media was significantly higher than that of WT hepatocytes, as monitored by standard MTT assay over a 7-day period. Upon removal of growth factors, the WT hepatocytes started losing viability within 1 day and by 3 days more than 50% of the cells were dead. In contrast, Alb/AEG-1 hepatocytes were significantly resistant to removal of growth factors, and even after 7 days in basal media, the cell viability was only reduced by 20%. These observations indicate that AEG-1/MTDH/LYRIC might autonomously activate growth factor-induced signaling and might inhibit pathways mediating senescence. Indeed, Alb/AEG-1 hepatocytes exhibited higher levels of activated (phosphorylated) ERK, Akt, and p38 MAPK and antiapoptotic proteins, Bcl-2 and Mcl-1 but not Bcl-xL, when compared to the WT hepatocytes.
Very interestingly, Alb/AEG-1 hepatocytes displayed significant resistance to senescence. At day 7 following isolation, the WT hepatocytes became large and vacuolated and ~55% cells were positive for SA β-gal while only 3% Alb/AEG-1 hepatocytes stained positive for SA β-gal. WT hepatocytes had higher levels of ROS and displayed activation of a DDR, characterized by activation of ATM, ATR, CHK1, CHK2, and p53 and upregulation of p21. In contrast, the ROS level in Alb/AEG-1 hepatocytes was significantly lower and the DDR response was also markedly dampened. Thus, activation of growth factor signaling, upregulation of antiapoptotic proteins, and abrogation of ROS-induced DDR response and senescence might underlie the mechanism by which AEG-1/MTDH/LYRIC promotes a prosurvival response.
Conditioned media (CM) from Alb/AEG-1 hepatocytes induced a significant angiogenic response, as revealed by HUVEC differentiation and CAM assays, when compared to WT hepatocytes. Proteomic analysis of the CM identified upregulation of several components of the coagulation pathway, including Fibrinogen α and β chains, Factor XII (FXII), plasminogen, and prothrombin, that are known to play significant roles in cancer angiogenesis, metastasis, and invasion in Alb/AEG-1 hepatocytes compared to the WT hepatocytes. The role of FXII, showing 56-fold upregulation in Alb/AEG-1 hepatocytes over WT hepatocytes, and TFF3, identified by Affymetrix microarray, was analyzed by knocking them down by siRNA in Alb/AEG-1 hepatocytes and subjecting the CM to angiogenesis assays. Knocking down either FXII or TFF3 markedly abrogated the angiogenic response by AEG-1/MTDH/LYRIC although the effect of FXII siRNA was much more pronounced indicating that FXII might play a pivotal role in mediating AEG-1/MTDH/LYRIC-induced angiogenesis. FXII cross talks with EGFR-activating MAPK and Akt signaling to promote proliferation and differentiation of endothelial cells. HUVECs were treated with CM from Alb/AEG-1 hepatocytes transfected with either control siRNA or FXII siRNA. While CM from control siRNA-treated HUVEC maintained activation of EGFR, Akt, ERK, and p38 MAPK, absence of FXII in the CM from FXII knockdown cells significantly abrogated the activation of EGFR, Akt, ERK, and p38 MAPK in HUVEC further highlighting the importance of FXII in AEG-1/MTDH/LYRIC action. Compared to the changes in the protein level, FXII mRNA level showed little increase in Alb/AEG-1 hepatocytes over WT hepatocytes. It was demonstrated that similar to MDR1, FXII mRNA also showed increased association with polysomes upon AEG-1/MTDH/LYRIC overexpression. The level of miRNA-181a, that targets FXII, was similar between WT and Alb/AEG-1 hepatocytes, indicating that the translational, and not miRNA-mediated, regulation is the molecular mechanism by which AEG-1/MTDH/LYRIC increases FXII expression.
8. CONCLUSION
HCC is a highly radio- and chemoresistant cancer, and there is virtually no therapy for advanced HCC. The only FDA-approved targeted drug, the Raf kinase inhibitor sorafenib, provides a survival benefit of only 2.8 months in nonresectable HCC patients (Llovet et al., 2008). As such there is a dire need of developing new modalities of treatment providing lasting disease-free survival benefit to the patients. Our extensive studies in human cell lines and Alb/AEG-1 mice firmly establish that even though AEG-1/MTDH/LYRIC may not induce HCC it is required for subsequent progression of the disease. Thus, AEG-1/MTDH/LYRIC inhibition, in combination with other modalities of therapy, might be an effective way to counteract HCC. As AEG-1/MTDH/LYRIC itself does not catalyze any reaction and functions most likely as a scaffold protein facilitating formation of protein complexes, developing small molecule inhibitors for AEG-1/MTDH/LYRIC by a standard high-throughput library screening approach may not be tenable. Resolution of crystal structure of AEG-1/MTDH/LYRIC is thus mandatory to identify key regions facilitating protein–protein interaction and develop peptidomimetic inhibitors using fragment-based drug discovery approach (Bembenek, Tounge, & Reynolds, 2009; Chen et al., 2007). While AEG-1/MTDH/LYRIC is detected primarily in the nucleus in normal hepatocytes, in HCC cells it is detected mainly in the cytoplasm, suggesting that in hepatocytes AEG-1/MTDH/LYRIC might serve unique functions that are different from its role in HCC cells. Liver is a specialized organ intimately involved in metabolic and hormonal regulation. Whether AEG-1/MTDH/LYRIC plays any role in regulating the specialized functions of liver remains to be determined. An AEG-1/MTDH/LYRIC knockout mouse model is thus required to be developed to understand the role of this unique molecule in normal and diseased states.
REFERENCES
- Bellahcene A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Nature Reviews. Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek SD, Tounge BA, Reynolds CH. Ligand efficiency and fragment-based drug discovery. Drug Discovery Today. 2009;14:278–283. doi: 10.1016/j.drudis.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- Burger AM, Zhang X, Li H, Ostrowski JL, Beatty B, Venanzoni M, et al. Down-regulation of T1A12/mac25, a novel insulin-like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene. 1998;16:2459–2467. doi: 10.1038/sj.onc.1201772. [DOI] [PubMed] [Google Scholar]
- Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. The New England Journal of Medicine. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Chen D, Siddiq A, Emdad L, Rajasekaran D, Gredler R, Shen X-N, et al. Insulin-like growth factor binding protein-7 (IGFBP7): A promising gene therapeutic for hepatocellular carcinoma (HCC) Molecular Therapy. 2013;21(4):758–766. doi: 10.1038/mt.2012.282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen D, Yoo BK, Santhekadur PK, Gredler R, Bhutia SK, Das SK, et al. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clinical Cancer Research. 2011;17:6693–6701. doi: 10.1158/1078-0432.CCR-10-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Stebbins JL, Zhang X, Hoffman R, Moore A, et al. A fragment-based approach for the discovery of isoform-specific p38alpha inhibitors. ACS Chemical Biology. 2007;2:329–336. doi: 10.1021/cb700025j. [DOI] [PubMed] [Google Scholar]
- Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, Albertini A, et al. Alcohol and hepatocellular carcinoma: The effect of lifetime intake and hepatitis virus infections in men and women. American Journal of Epidemiology. 2002;155:323–331. doi: 10.1093/aje/155.4.323. [DOI] [PubMed] [Google Scholar]
- Dragani TA. Risk of HCC: Genetic heterogeneity and complex genetics. Journal of Hepatology. 2010;52:252–257. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma. The New England Journal of Medicine. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Tran T, Everhart JE. Diabetes increases the risk of chronic liver disease and hepatocellular carcinoma. Gastroenterology. 2004;126:460–468. doi: 10.1053/j.gastro.2003.10.065. [DOI] [PubMed] [Google Scholar]
- Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche H, Sarkar D, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21300–21305. doi: 10.1073/pnas.0910936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emdad L, Sarkar D, Su ZZ, Randolph A, Boukerche H, Valerie K, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Research. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nature Reviews. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. The American Journal of Pathology. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- Georgiades CS, Hong K, Geschwind JF. Radiofrequency ablation and chemoembolization for hepatocellular carcinoma. Cancer Journal. 2008;14:117–122. doi: 10.1097/PPO.0b013e31816a0fac. [DOI] [PubMed] [Google Scholar]
- Gong Z, Liu W, You N, Wang T, Wang X, Lu P, et al. Prognostic significance of metadherin overexpression in hepatitis B virus-related hepatocellular carcinoma. Oncology Reports. 2012;27:2073–2079. doi: 10.3892/or.2012.1749. [DOI] [PubMed] [Google Scholar]
- Grant TJ, Bishop JA, Christadore LM, Barot G, Chin HG, Woodson S, et al. Antiproliferative small molecule inhibitors of transcription factor LSF reveal oncogene addiction to LSF in hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4503–4508. doi: 10.1073/pnas.1121601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U, Owens L, Saxena UH. Transcription factors LSF and E2Fs: Tandem cyclists driving G0 to S? Cell Cycle. 2009;8:2146–2151. doi: 10.4161/cc.8.14.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocrine Reviews. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Ikeda S, Fujimori M, Shimizu Y, Kurihara T, Itamoto T, et al. Immunohistochemical analysis and mutational analyses of beta-catenin, Axin family and APC genes in hepatocellular carcinomas. International Journal of Oncology. 2004;24:1077–1083. [PubMed] [Google Scholar]
- Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ, Fisher PB. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. The Journal of General Virology. 2001;82:2235–2241. doi: 10.1099/0022-1317-82-9-2235. [DOI] [PubMed] [Google Scholar]
- Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF, Fisher PB. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene. 2008;27:1114–1121. doi: 10.1038/sj.onc.1210713. [DOI] [PubMed] [Google Scholar]
- Leung TW, Patt YZ, Lau WY, Ho SK, Yu SC, Chan AT, et al. Complete pathological remission is possible with systemic combination chemotherapy for inoperable hepatocellular carcinoma. Clinical Cancer Research. 1999;5:1676–1681. [PubMed] [Google Scholar]
- Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Seminars in Liver Disease. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Real MI, Montana X, Planas R, Coll S, Aponte J, et al. Arterial embolisation or chemoembolisation versus symptomatic treatment in patients with unresectable hepatocellular carcinoma: A randomised controlled trial. Lancet. 2002;359:1734–1739. doi: 10.1016/S0140-6736(02)08649-X. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. The New England Journal of Medicine. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: Mechanisms of action and clinical strategies. Nature Reviews. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. Journal of Virology. 2001;75:1401–1407. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann CD, Neal CP, Garcea G, Manson MM, Dennison AR, Berry DP. Prognostic molecular markers in hepatocellular carcinoma: A systematic review. European Journal of Cancer. 2007;43:979–992. doi: 10.1016/j.ejca.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhu D, Yang S, Wang X, Xiong Z, Zhang Y, et al. Cytoplasmic Metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. The Journal of Biological Chemistry. 2012;287:4485–4491. doi: 10.1074/jbc.C111.291518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, He B, Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Seminars in Cancer Biology. 2011;21:10–20. doi: 10.1016/j.semcancer.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Brechot C, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–1168. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguri T, Achiwa H, Bessho Y, Muramatsu H, Maeda H, Niimi T, et al. The role of thymidylate synthase and dihydropyrimidine dehydrogenase in resistance to 5-fluorouracil in human lung cancer cells. Lung Cancer. 2005;49:345–351. doi: 10.1016/j.lungcan.2005.05.003. [DOI] [PubMed] [Google Scholar]
- O'Neil BH, Venook AP. Hepatocellular carcinoma: The role of the North American GI Steering Committee Hepatobiliary Task Force and the advent of effective drug therapy. The Oncologist. 2007;12:1425–1432. doi: 10.1634/theoncologist.12-12-1425. [DOI] [PubMed] [Google Scholar]
- Pang RW, Joh JW, Johnson PJ, Monden M, Pawlik TM, Poon RT. Biology of hepatocellular carcinoma. Annals of Surgical Oncology. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- Paukku K, Kalkkinen N, Silvennoinen O, Kontula KK, Lehtonen JY. p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Research. 2008;36:4474–4487. doi: 10.1093/nar/gkn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Molecular Endocrinology. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Poon RT, Fan ST, Lo CM, Ng IO, Liu CL, Lam CM, et al. Improving survival results after resection of hepatocellular carcinoma: A prospective study of 377 patients over 10 years. Annals of Surgery. 2001;234:63–70. doi: 10.1097/00000658-200107000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Rudge TL, Zhu Q, Johnson LF, Hansen U. Inhibition of the mammalian transcription factor LSF induces S-phase-dependent apoptosis by down-regulating thymidylate synthase expression. The EMBO Journal. 2000;19:4665–4675. doi: 10.1093/emboj/19.17.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nature Reviews. Immunology. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- Ruan W, Xu E, Xu F, Ma Y, Deng H, Huang Q, et al. IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. Cancer Biology & Therapy. 2007;6:354–359. doi: 10.4161/cbt.6.3.3702. [DOI] [PubMed] [Google Scholar]
- Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. The Journal of Biological Chemistry. 2012;287:13952–13958. doi: 10.1074/jbc.M111.321646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur PK, Gredler R, Chen D, Siddiq A, Shen XN, Das SK, et al. Late SV40 factor (LSF) enhances angiogenesis by transcriptionally up-regulating matrix metalloproteinase-9 (MMP-9) The Journal of Biological Chemistry. 2012;287:3425–3432. doi: 10.1074/jbc.M111.298976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur PK, Rajasekaran D, Siddiq A, Gredler R, Chen D, Schaus SE, et al. The transcription factor LSF: A novel oncogene for hepatocellular carcinoma. American Journal of Cancer Research. 2012;2:269–285. [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Research. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Sprenger CC, Damon SE, Hwa V, Rosenfeld RG, Plymate SR. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) is a potential tumor suppressor protein for prostate cancer. Cancer Research. 1999;59:2370–2375. [PubMed] [Google Scholar]
- Srivastava J, Siddiq A, Emdad L, Santhekadur P, Chen D, Gredler R, et al. Astrocyte elevated gene-1 (AEG-1) promotes hepatocarcinogenesis: Novel insights from a mouse model. Hepatology. 2012;56(5):1782–1791. doi: 10.1002/hep.25868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: A weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao W, Volsky DJ, et al. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World Journal of Gastroenterology. 2007;13:2271–2282. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkettle HJ, Mills IG, Whitaker HC, Neal DE. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene. 2009;28:3663–3670. doi: 10.1038/onc.2009.223. [DOI] [PubMed] [Google Scholar]
- Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- Tomimaru Y, Eguchi H, Wada H, Kobayashi S, Marubashi S, Tanemura M, et al. IGFBP7 downregulation is associated with tumor progression and clinical outcome in hepatocellular carcinoma. International Journal of Cancer. 2012;130:319–327. doi: 10.1002/ijc.25994. [DOI] [PubMed] [Google Scholar]
- Umemura T, Ichijo T, Yoshizawa K, Tanaka E, Kiyosawa K. Epidemiology of hepatocellular carcinoma in Japan. Journal of Gastroenterology. 2009;44(Suppl. 19):102–107. doi: 10.1007/s00535-008-2251-0. [DOI] [PubMed] [Google Scholar]
- Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Onco-genic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. The EMBO Journal. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nature Reviews. Gastroenterology & Hepatology. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, et al. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Research. 2007;35:4485–4494. doi: 10.1093/nar/gkm470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W, Mok TS, Zee B, Leung TW, Lai PB, Lau WY, et al. A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. Journal of the National Cancer Institute. 2005;97:1532–1538. doi: 10.1093/jnci/dji315. [DOI] [PubMed] [Google Scholar]
- Yoo BK, Chen D, Su Z-Z, Gredler R, Yoo J, Shah K, et al. Molecular mechanism of chemoresistance by Astrocyte Elevated Gene-1 (AEG-1) Cancer Research. 2010;70:3249–3258. doi: 10.1158/0008-5472.CAN-09-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Emdad L, Gredler R, Fuller C, Dumur CI, Jones KH, et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8357–8362. doi: 10.1073/pnas.1000374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. The Journal of Clinical Investigation. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Gredler R, Chen D, Santhekadur PK, Fisher PB, Sarkar D. c-Met activation through a novel pathway involving osteopontin mediates oncogenesis by the transcription factor LSF. Journal of Hepatology. 2011;55:1317–1324. doi: 10.1016/j.jhep.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Gredler R, Vozhilla N, Su ZZ, Chen D, Forcier T, et al. Identification of genes conferring resistance to 5-fluorouracil. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, Bhutia SK, et al. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocelllular carcinoma. Hepatology. 2011;53:1538–1548. doi: 10.1002/hep.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinare K, Kubota T, Watanabe M, Wada N, Nishibori H, Hasegawa H, et al. Gene expression in colorectal cancer and in vitro chemosensitivity to 5-fluorouracil: A study of 88 surgical specimens. Cancer Science. 2003;94:633–638. doi: 10.1111/j.1349-7006.2003.tb01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AX, Blaszkowsky LS, Ryan DP, Clark JW, Muzikansky A, Horgan K, et al. Phase II study of gemcitabine and oxaliplatin in combination with bevacizumab in patients with advanced hepatocellular carcinoma. Journal of Clinical Oncology. 2006;24:1898–1903. doi: 10.1200/JCO.2005.04.9130. [DOI] [PubMed] [Google Scholar]
- Zhu K, Dai Z, Pan Q, Wang Z, Yang GH, Yu L, et al. Metadherin promotes hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clinical Cancer Research. 2011;17:7294–7302. doi: 10.1158/1078-0432.CCR-11-1327. [DOI] [PubMed] [Google Scholar]