Abstract
Background
A successful development of herbivorous insects into plant tissues depends on coordination of metabolic processes. Plants have evolved complex mechanisms to recognize such attacks, and to trigger a defense response. To understand the transcriptional basis of this response, we compare gene expression profiles of two coffee genotypes, susceptible and resistant to leaf miner (Leucoptera coffella). A total of 22000 EST sequences from the Coffee Genome Database were selected for a microarray analysis. Fluorescence probes were synthesized using mRNA from the infested and non-infested coffee plants. Array hybridization, scanning and data normalization were performed using Nimble Scan® e ArrayStar® platforms. Genes with foldchange values +/-2 were considered differentially expressed. A validation of 18 differentially expressed genes was performed in infected plants using qRT-PCR approach.
Results
The microarray analysis indicated that resistant plants differ in gene expression profile. We identified relevant transcriptional changes in defense strategies before insect attack. Expression changes (>2.00-fold) were found in resistant plants for 2137 genes (1266 up-regulated and 873 down-regulated). Up-regulated genes include those responsible for defense mechanisms, hypersensitive response and genes involved with cellular function and maintenance. Also, our analyses indicated that differential expression profiles between resistant and susceptible genotypes are observed in the absence of leaf-miner, indicating that defense is already build up in resistant plants, as a priming mechanism. Validation of selected genes pointed to four selected genes as suitable candidates for markers in assisted-selection of novel cultivars.
Conclusions
Our results show evidences that coffee defense responses against leaf-miner attack are balanced with other cellular functions. Also analyses suggest a major metabolic reconfiguration that highlights the complexity of this response.
Keywords: Coffea arabica, Leaf miner, Microarray, Plant defense
Background
Once a plant recognizes a pathogen attack, the metabolism must balance demands for resources to support defense versus requirements for cellular maintenance, growth and reproduction [1-4]. Defense mechanisms involve a shift on metabolism, activating specific pathways such as synthesis of secondary metabolites, programmed cell death, ions translocation. Concomitantly, can occur a shutdown of other metabolic pathways not directly involved with defense response, such as those associated with growth and reproduction. A resistance and/or tolerance trait is attributed whenever this defense response is successful in controlling pathogen or herbivore attack.
Genetic control of metabolic re-programming is normally triggered by few resistance genes which are seek out to be transferred to other plants. However, resistance response involves changes in other genes, not usually identified, with determinant roles in the overall response. Therefore for an effective transference of resistance traits, to know how these genes interact during re-programming of plant metabolism is essential.
Among available methods for high-throughput analysis the microarray is a powerful tool for large-scale gene expression studies in many plant species with whole genome sequenced: potato [5,6], tomato [7,8], soybean [9,10], wheat [11], barley [12,13] maize [14,15], grape [16], pine [17], Arabidopsis [18-20]. The main advantage of microarray analyses is to evaluate the expression of large number of genes in different genotypes, organs, tissues, treatments, using the same set of genes. These genes can be compared during different biological situations allowing both an association with metabolic pathways and establishement of their role on resistance response. Several studies have been carried out using microarray analysis to identify genes associated with plant defense [21-25].
The leaf-miner, Leucoptera coffeella (Guérin-Méneville, 1842) (Lepidoptera-Lyonetiidae) is a specialist parasite of Coffea species. Upon oviposition on leaves, ecloded larvae feed directly from parenchyma tissues, leading to a reduction of foliar surface and eventual leaf drop [26]. This damage results in reduction of photosynthetic area and plant survival. In Brazilian breeding programs resistance genes from C. racemosa have been transferred to the susceptible C. arabica by controlled crosses, and so far a large number of hybrid progenies are under selection for resistance to leaf-miner [27]. Although defense mechanism to leaf-miner is not understood yet, genetic analysis demonstrated that resistance to the insect is dominant and controlled by two complimentary genes [28]. At the molecular level, there is little information regarding gene expression on coffee plants during defense response. Using subtractive hybridization methodology (SSH), Mondego et al. [29] found differentially expressed genes in coffee plants upon leaf-miner infestation, among which a miraculin-like encoding gene was significantly overexpressed in resistant coffee plants. Differential expression of defense-related genes such as lipoxygenase, glutathione transferase, protein-kinase receptor and glucanase was observed in response to leaf-miner infestation [30]. However, the expression profiles indicate that differences results from gene expression timing along insect infection rather than with gene regulation.
Despite the efforts of breeding programs to develop novel coffee cultivars bearing leaf-miner resistance, selection of progenies homozygotes for this trait is difficult [27], as advanced generations are still producing susceptible plants. Therefore, information regarding molecular control of resistance response as well as identification of candidate-genes associated with these processes will contribute with assisted-selection.
In this context, the aim of our study was to explore transcriptomic differences throughout insect infestation, in susceptible and resistant C. arabica plants challenged by L. coffeella, using microarray technology. The arrays were developed using coffee-specific oligoprobes designed based on gene sequences available at the Brazilian Coffee Genome Project [31]. The database contains a collection of around 32,000 gene sequences, covering most of the C. arabica genome [32]. Besides this, we selected a group of candidate-genes to be used as molecular markers for assisted-selection. As far as we know, this is the first report of a large-scale transcriptional profile analysis used to study gene expression changes in coffee plants in the presence of an herbivore insect.
Results
Microarray analyses were performed to characterize large-scale gene expression profiles during leaf-miner development on coffee leaves. The analyses included a hybridization of a 135 K array with 6 different samples, corresponding to time-course infestation stages in both resistant and susceptible plants. The arrays contain sequences of around 33 K genes identified in EST libraries prepared from different physiological and metabolic situations [31]. A minimum of 6 - 24mer match probes for each selected gene were used for the array set up. The arrays were hybridized with probes corresponding to 3 treatments of both susceptible (S) and resistant (R) leaves: non-infestated (T0), after oviposition and egg-eclosion (T1) and damaged by insect feeding (T2).
Initially, differential expression patterns were identified using statistical analysis, and specific transcriptional profiles were established for each evaluated interaction. In a second approach, genes exhibiting differential expression among genotypes and treatments were submitted to in silico evaluations to classify and categorize those genes regarding their possible molecular functions and metabolic pathways. Finally, a group of 19 genes involved with defense-related mechanisms, exhibiting regulated expression, were further characterized using qRT-PCR.
Microarray and statistical analysis
A total of 2141, 2359 and 2257 differentially expressed genes were identified from in silico analyses of raw hybridization data considering 3 interactions: T0R X T0S, T1R X T1S and T2R X T2S (Table 1). Comparing T0R X T0S we observed higher differential expression levels where foldchange values range from 400 and 1000 times in up-regulated genes, and from 200 to 400 times in down-regulated genes in resistant leaves. The other interactions exhibited foldchange values ranging from 150 and 350 times in down-regulated, and from 10 to 15 times in up-regulated genes (Table 2).
Table 1.
Distribution of regulated genes in each interaction analyzed
| Genes |
T0R_T0S |
T1R_T1S |
T2R_T2S |
|---|---|---|---|
| (non infested plants) | (Egg hatching) | (Egg eclosion) | |
| Up regulated |
1268 |
1231 |
889 |
| Down regulated |
873 |
1128 |
1368 |
| No differences | 19057 | 18837 | 18939 |
Table 2.
Survey of differentially expressed genes in all considered interactions including number and fold-change values range
| T0R_T0S | T1R_T1S | T2R_T2S | Range of “fold change” |
|---|---|---|---|
| 14 |
9 |
10 |
1000-100 |
| 32 |
17 |
23 |
99-40 |
| 57 |
51 |
39 |
39-20 |
| 103 |
96 |
125 |
19-10 |
| 1062 |
1058 |
1171 |
9-2 |
| 858 |
1117 |
880 |
(-)2- (-)9 |
| 15 |
11 |
9 |
(-)10- (-)499 |
| Total 2141 | Total 2359 | Total 2257 |
Different regulation profiles for defense response were observed among differentially expressed genes. The first group includes chitinase and polygalacturonase genes, regulated differently in resistant and susceptible leaves from T0 to T2. The second group includes genes that were up or down regulated in control resistant or susceptible leaves, but throughout insect development inverted their expression pattern. Example of this group is the gene encoding the enzyme polyphenol oxidase, up regulated at T0 in resistant plants and down regulated at T1 and T2. And a third group includes genes that were up and/or down regulated in response to the leaf-miner infestation, either in susceptible or resistant leaves.
Interaction between resistant and susceptible genotypes without insect infestation (T0R x T0S)
Since higher values for differential gene expression were observed when comparing T0R X T0S samples, we chose this interaction for further analyses and selection of candidate-genes for validation. In this interaction, 2141 genes exhibited differential expression, 1268 were up regulated and 873 were down regulated.
Regulated genes from the T0 samples were functionally characterized into three gene ontology categories – molecular function, component cellular and biological function - and grouped according their metabolic categories (Figures 1 and 2). A description of the first one hundred most variable genes, both up and down regulated, is shown on Tables 3 and 4 and illustrated in Figures 3 and 4. Several contigs had no correspondence with defined categories. A larger number of genes are up-regulated rather than down-regulated, and associated with primary and cellular metabolism, which included functions such as ion, protein and nucleic acid binding, hydrolase and transferase activities, among others. As these differences were observed in non-infested leaves, possibly a different transcriptional programming takes place in resistant leaves. This may result in a pre-defense status, enabling resistant plants to a faster defense response upon leaf-miner attack.
Figure 1.
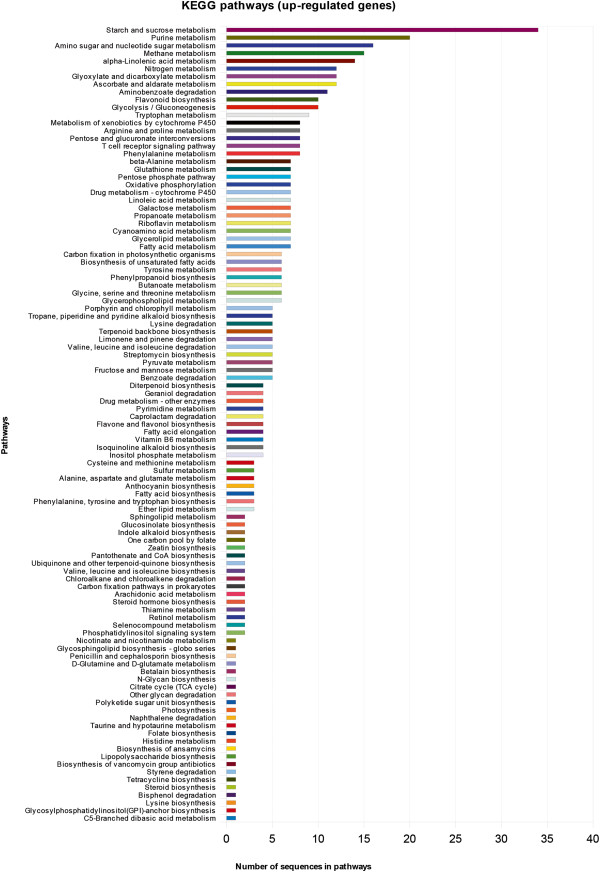
Pathways from the top 100 up-regulated genes in T0 interaction. Pathways were identified considering T0 interaction. Amount of genes belonging to pathways is specified in each line.
Figure 2.
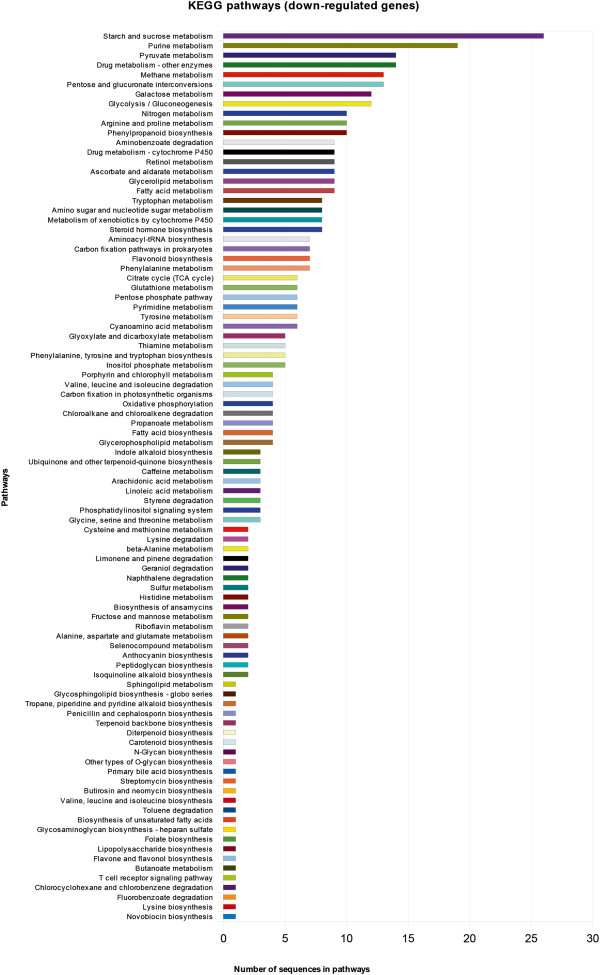
Pathways from the top 100 down-regulated genes in T0 interaction. Pathways were identified considering T0 interaction. Amount of genes belonging to pathways is specified in each line.
Table 3.
List of up-regulated genes observed for the T0R_T0S interaction with respective annotation and fold-change values
| Name | Blast Sol network genomics | e-value | Annotation* | Fold change (T0R X T0S) |
|---|---|---|---|---|
| 0U1 |
SGN-E628893 |
0 |
---NA--- |
1000,27 |
| 0U2 |
SGN-E1352064 |
5e-89 |
Caffeine synthase |
642,35 |
| 0U3 |
SGN-E1326397 |
0 |
Acidic endochitinase se2 |
447,45 |
| 0U4 |
SGN-E1310344 |
0 |
Metallothionein-like protein |
280,91 |
| 0U5 |
SGN-E1334735 |
0 |
Kunitz trypsin inhibitor |
238,17 |
| 0U6 |
SGN-E1316291 |
0 |
Acid phosphatase |
236,53 |
| 0U7 |
SGN-E661231 |
1e-175 |
Polygalacturonase-1 non-catalytic subunit beta |
236,37 |
| 0U8 |
SGN-E1327615 |
0 |
Kunitz trypsin inhibitor |
168,12 |
| 0U9 |
SGN-E1337775 |
0 |
Organ-specific protein |
155,92 |
| 0U10 |
SGN-E659257 |
0 |
Polygalacturonase-1 non-catalytic subunit beta |
152,17 |
| 0U11 |
SGN-E1352070 |
0 |
Caffeine synthase |
148,75 |
| 0U12 |
SGN-E642649 |
0 |
Cytokinin oxidase |
143,25 |
| 0U13 |
SGN-E833713 |
1e-146 |
Asr1 protein |
119,38 |
| 0U14 |
SGN-E667484 |
0 |
Protein |
115,77 |
| 0U15 |
SGN-E1319644 |
0 |
pr-10 type pathogenesis-related protein |
98,14 |
| 0U16 |
NM** |
|
Protein |
97,12 |
| 0U17 |
NM |
|
Protein |
94,57 |
| 0U18 |
SGN-E1321440 |
0 |
Acid phosphatase |
93,94 |
| 0U19 |
NM |
|
Invertase pectin methylesterase inhibitor family protein |
82,40 |
| 0U20 |
SGN-E1312621 |
0 |
---NA--- |
76,83 |
| 0U21 |
SGN-E1309331 |
0 |
mpbq msbq methyltransferase 2 |
75,15 |
| 0U22 |
SGN-E837532 |
2e-31 |
Class iii chitinase |
75,05 |
| 0U23 |
SGN-E1316252 |
0 |
Heat shock |
73,55 |
| 0U24 |
SGN-E1325880 |
0 |
Swib complex baf60b domain-containing protein |
72,42 |
| 0U25 |
SGN-E832873 |
1e-119 |
Kunitz trypsin inhibitor |
63,26 |
| 0U26 |
SGN-E1322100 |
1e-73 |
Polyphenol oxidase |
60,83 |
| 0U27 |
SGN-E660241 |
0 |
Polyphenol oxidase |
57,50 |
| 0U28 |
SGN-E671322 |
0 |
60s acidic ribosomal protein p0 |
56,95 |
| 0U29 |
SGN-E1128614 |
8e-11 |
Protein |
53,42 |
| 0U30 |
SGN-E682004 |
0 |
Lipid transfer protein |
50,27 |
| 0U31 |
SGN-E1334549 |
0 |
Type ii proteinase inhibitor family protein |
49,57 |
| 0U32 |
SGN-E1337715 |
0 |
Protein |
49,57 |
| 0U33 |
SGN-E640935 |
0 |
Class iii chitinase |
48,21 |
| 0U34 |
SGN-E990795 |
7e-16 |
Microsomal glutathione s- |
47,84 |
| 0U35 |
SGN-E668445 |
0 |
Serine-type endopeptidase inhibitor |
47,81 |
| 0U36 |
NM |
|
Metallothionein-like protein |
47,79 |
| 0U37 |
NM |
|
Polyphenol oxidase |
46,30 |
| 0U38 |
SGN-E640935 |
0 |
Class iii chitinase |
45,63 |
| 0U39 |
SGN-E657601 |
0 |
Protein |
44,71 |
| 0U40 |
SGN-E835025 |
|
4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
44,21 |
| 0U41 |
SGN-E1320197 |
0 |
Peroxisomal membrane |
43,89 |
| 0U42 |
SGN-E636199 |
0 |
---NA--- |
43,72 |
| 0U43 |
SGN-E1333755 |
0 |
Type ii proteinase inhibitor family protein |
41,88 |
| 0U44 |
SGN-E1337775 |
0 |
Organ-specific protein |
41,68 |
| 0U45 |
SGN-E912118 |
1e-23 |
Tartrate-resistant acid phosphatase type 5 |
41,04 |
| 0U46 |
SGN-E628829 |
8e-94 |
Cell wall protein |
40,77 |
| 0U47 |
SGN-E449176 |
|
Phospholipid glycerol acyltransferase family protein 7 |
39,90 |
| 0U48 |
SGN-E830846 |
0 |
r3h domain containing |
39,64 |
| 0U49 |
SGN-E628829 |
8e-94 |
Oxygen-evolving enhancer protein chloroplast |
39,55 |
| 0U50 |
NM |
|
Auxin-independent growth promoter protein |
39,13 |
| 0U51 |
SGN-E1349312 |
0 |
Lipid transfer protein |
39,09 |
| 0U52 |
SGN-E639273 |
1e-160 |
Protein kinase domain containing expressed |
38,61 |
| 0U53 |
SGN-E1352095 |
0 |
Protein |
35,83 |
| 0U54 |
SGN-E838896 |
0 |
Cytochrome p450 |
34,98 |
| 0U55 |
NM |
|
nadh dehydrogenase subunit f |
34,83 |
| 0U56 |
SGN-E838821 |
0 |
mta sah |
34,62 |
| 0U57 |
SGN-E669832 |
0 |
Cytokinin oxidase |
33,83 |
| 0U58 |
SGN-E643214 |
0 |
vesicle-associated membrane protein 714 |
33,06 |
| 0U59 |
SGN-E1346029 |
0 |
Protein |
32,83 |
| 0U60 |
SGN-E1348577 |
0 |
Protein |
32,50 |
| 0U61 |
SGN-E674849 |
1e-15 |
Mitochondrial chaperonin hsp60 |
32,40 |
| 0U62 |
SGN-E788243 |
6e-20 |
Tartrate-resistant acid phosphatase type 5 |
32,40 |
| 0U63 |
SGN-E109388 |
3e-31 |
Protein |
31,93 |
| 0U64 |
SGN-E1326628 |
0 |
Formate dehydrogenase |
31,83 |
| 0U65 |
SGN-E1333903 |
0 |
Elongation factor-1 alpha |
31,16 |
| 0U66 |
NM |
|
Protein |
30,56 |
| 0U67 |
SGN-E1321812 |
0 |
Peptidylprolyl isomerase |
30,27 |
| 0U68 |
SGN-E1310338 |
0 |
60s ribosomal protein |
30,00 |
| 0U69 |
NM |
|
Conserved hypothetical protein [Ricinus communis] |
29,87 |
| 0U70 |
SGN-E681444 |
0 |
Homeobox-leucine zipper protein |
29,71 |
| 0U71 |
SGN-E1168096 |
2e-20 |
Protein |
29,31 |
| 0U72 |
SGN-E818914 |
2e-36 |
Gibberellin 20 |
29,19 |
| 0U73 |
SGN-E431715 |
7e-11 |
Cellulose synthase |
28,99 |
| 0U74 |
SGN-E650445 |
0 |
Poly -binding protein |
28,29 |
| 0U75 |
SGN-E631106 |
3e-19 |
orf i polyprotein |
28,09 |
| 0U76 |
SGN-E1314273 |
0 |
Ankyrin repeat domain |
27,84 |
| 0U77 |
SGN-E1315499 |
9e-23 |
Atapy2 atpase nucleotide diphosphatase |
27,54 |
| 0U78 |
SGN-E1315958 |
0 |
nadh ubiquinone oxidoreductase b14 subunit |
26,38 |
| 0U79 |
SGN-E830665 |
0 |
sec61 transport protein |
25,52 |
| 0U80 |
SGN-E1316141 |
0 |
Flavanone 3-hydroxylase-like protein |
24,98 |
| 0U81 |
NM |
|
cbl-interacting serine threonine-protein |
24,92 |
| 0U82 |
SGN-E1342733 |
0 |
Transcription factor lim |
24,70 |
| 0U83 |
SGN-E837532 |
8e-30 |
Class iii chitinase |
23,94 |
| 0U84 |
SGN-E1322208 |
5e-90 |
Cysteine proteinase |
23,93 |
| 0U85 |
SGN-E1322866 |
1e-126 |
Protein |
23,85 |
| 0U86 |
SGN-E1315443 |
0 |
Acid phosphatase |
23,69 |
| 0U87 |
SGN-E662706 |
0 |
gdsl-motif lipase hydrolase family protein |
23,27 |
| 0U88 |
SGN-E266690 |
7e-11 |
Protein |
23,16 |
| 0U89 |
SGN-E1319812 |
0 |
Protein |
22,87 |
| 0U90 |
SGN-E1334410 |
0 |
Protein |
22,82 |
| 0U91 |
SGN-E1321222 |
0 |
Triosephosphate isomerase |
22,62 |
| 0U92 |
SGN-E648331 |
1e-68 |
Serine-threonine protein plant- |
22,61 |
| 0U93 |
SGN-E1351186 |
0 |
Class iii chitinase |
22,36 |
| 0U94 |
SGN-E951741 |
2e-22 |
Late embryogenesis abundant protein lea14- |
22,06 |
| 0U95 |
SGN-E686943 |
0 |
Dehydrin |
21,99 |
| 0U96 |
SGN-E674268 |
1e-90 |
mta sah |
21,79 |
| 0U97 |
SGN-E1352075 |
0 |
7-methylxanthine n-methyltransferase |
21,73 |
| 0U98 |
SGN-E1323598 |
0 |
Transcription initiation factor iib |
21,29 |
| 0U99 |
SGN-E1348381 |
1e-151 |
Translation factor |
21,07 |
| 0U100 | SGN-E1322408 | 0 | Beta-glucosidase-like protein | 20,89 |
*The annotation of each sequence was established on the Coffee Genome Database [31].
**No match with any sequence on the Solanaceae Genomic Database.
Table 4.
List of down-regulated genes observed for the T0R_T0S interaction with respective annotation and fold-change values
| Name | Blast Sol network genomics | e-value | Annotation* | Fold change (T0R X T0S) |
|---|---|---|---|---|
| 0D1 |
SGN-E1320843 |
0 |
Protein |
-445,87 |
| 0D2 |
SGN-E676870 |
0 |
Protein |
-235,45 |
| 0D3 |
SGN-E1320843 |
0 |
Zinc finger |
-126,65 |
| 0D4 |
SGN-E1325444 |
0 |
Hypothetical protein VITISV_000181 [Vitis vinifera] |
-62,54 |
| 0D5 |
SGN-E835732 |
0 |
Tapetum-specific protein lla-115 |
-49,63 |
| 0D6 |
SGN-E661762 |
0 |
---NA--- |
-35,43 |
| 0D7 |
NM** |
|
PREDICTED: hypothetical protein [Vitis vinifera] |
-24,73 |
| 0D8 |
NM |
|
Glycerol-3-phosphate acyltransferase 6 |
-17,93 |
| 0D9 |
SGN-E1321887 |
0 |
gdsl-motif lipase hydrolase-like |
-16,98 |
| 0D10 |
SGN-E1033676 |
5e-19 |
er glycerol-phosphate acyltransferase |
-16,50 |
| 0D11 |
SGN-E1322050 |
0 |
Extensin-like protein |
-14,88 |
| 0D12 |
SGN-E791894 |
1e-118 |
Cytochrome p450 |
-12,96 |
| 0D13 |
SGN-E837009 |
0 |
Cytochrome p450 |
-11,39 |
| 0D14 |
NM |
|
Cytochrome b |
-11,12 |
| 0D15 |
SGN-E1349845 |
1e-137 |
gdsl-motif lipase hydrolase family protein |
-10,27 |
| 0D16 |
SGN-E1318049 |
1e-153 |
Gibberellin-regulated protein 1 |
-9,89 |
| 0D17 |
SGN-E660879 |
0 |
Zinc finger |
-9,88 |
| 0D18 |
SGN-E835266 |
0 |
Isocitrate lyase |
-9,22 |
| 0D19 |
SGN-E673783 |
0 |
Isocitrate lyase |
-8,84 |
| 0D20 |
NM |
|
---NA--- |
-8,83 |
| 0D21 |
SGN-E660879 |
0 |
Zinc finger |
-8,73 |
| 0D22 |
SGN-E680272 |
1e-128 |
Protein |
-8,498 |
| 0D23 |
SGN-E898278 |
4e-13 |
Protein |
-8,43 |
| 0D24 |
SGN-E1328871 |
0 |
---NA--- |
-8,16 |
| 0D25 |
NM |
|
Cytochrome p450 |
-8,02 |
| 0D26 |
SGN-E678498 |
0 |
Protein |
-7,36 |
| 0D27 |
SGN-E1319644 |
0 |
Serine-threonine protein plant- |
-7,35 |
| 0D28 |
SGN-E838812 |
1e-40 |
---NA--- |
-7,34 |
| 0D29 |
NM |
|
Pathogenesis-related protein 1 |
-7,27 |
| 0D30 |
SGN-E1312882 |
0 |
Heat shock protein |
-7,17 |
| 0D31 |
NM |
|
---NA--- |
-7,12 |
| 0D32 |
SGN-E830806 |
0 |
Cytochrome p450 |
-7,06 |
| 0D33 |
SGN-E1334002 |
0 |
abc transporter |
-7,00 |
| 0D34 |
SGN-E659349 |
0 |
Glutathione s-transferase gstu6 |
-6,85 |
| 0D35 |
SGN-E1317104 |
0 |
Aspartyl protease family protein |
-6,74 |
| 0D36 |
SGN-E1322588 |
1e-180 |
at1g72120 f28p5_2 |
-6,53 |
| 0D37 |
NM |
|
---NA--- |
-6,46 |
| 0D38 |
SGN-E1350292 |
0 |
Lactoylglutathione lyase family protein |
-6,46 |
| 0D39 |
NM |
|
Achain crystal structure of a cell-wall invertase from Arabidopsis thaliana in complex with sucrose |
-6,36 |
| 0D40 |
SGN-E1335955 |
3e-56 |
Retroelement pol polyprotein |
-6,24 |
| 0D41 |
SGN-E820310 |
2e-64 |
Xyloglucan endotransglucosylase hydrolase protein 22 |
-6,23 |
| 0D42 |
SGN-E836814 |
1e-169 |
Leucine-rich plant specific |
-6,20 |
| 0D43 |
SGN-E686810 |
0 |
Zinc finger |
-6,09 |
| 0D44 |
SGN-E839045 |
0 |
Glucose-methanol-choline oxidoreductase family protein |
-6,06 |
| 0D45 |
SGN-E531670 |
1e-12 |
Hydrolyzing o-glycosyl |
-5,97 |
| 0D46 |
SGN-E1216540 |
2e-27 |
Aminotransferase family protein |
-5,91 |
| 0D47 |
SGN-E747084 |
4e-68 |
Alkaline alpha-galactosidase seed imbibition protein |
-5,81 |
| 0D48 |
SGN-E836814 |
1e-179 |
Leucine-rich plant specific |
-5,71 |
| 0D49 |
SGN-E1345225 |
0 |
---NA--- |
-5,71 |
| 0D50 |
SGN-E1325272 |
0 |
Protein |
-5,69 |
| 0D51 |
SGN-E626178 |
8e-17 |
|
-5,62 |
| 0D52 |
NM |
|
Alkaline alpha-galactosidase seed imbibition protein |
-5,60 |
| 0D53 |
NM |
|
Cinnamoyl reductase-like protein |
-5,57 |
| 0D54 |
SGN-E1349228 |
0 |
Proline dehydrogenase |
-5,54 |
| 0D55 |
SGN-E775239 |
8e-14 |
Kinesin like protein |
-5,46 |
| 0D56 |
NM |
|
Methyl-accepting chemotaxis sensory transducer |
-5,45 |
| 0D57 |
NM |
|
Transcription factor |
-5,42 |
| 0D58 |
NM |
|
3-hydroxyisobutyrate dehydrogenase family protein |
-5,39 |
| 0D59 |
NM |
|
Outer membrane porin protein |
-5,36 |
| 0D60 |
SGN-E1317853 |
0 |
Transcription activator |
-5,31 |
| 0D61 |
SGN-E666413 |
0 |
bahd family clade v |
-5,20 |
| 0D62 |
SGN-E658983 |
0 |
Glycerol-3-phosphate dehydrogenase |
-5,18 |
| 0D63 |
SGN-E1327315 |
0 |
Tonoplast intrinsic |
-5,17 |
| 0D64 |
SGN-1331462 |
0 |
Achain crystal structure of a cell-wall invertase from arabidopsis thaliana in complex with sucrose |
-5,08 |
| 0D65 |
NM |
|
Inner-membrane translocator |
-5,00 |
| 0D66 |
NM |
|
Beta-ig-h3 fasciclin |
-4,94 |
| 0D67 |
SGN-E653486 |
1e-52 |
bahd family clade v |
-4,90 |
| 0D68 |
NM |
|
Stachyose synthase |
-4,86 |
| 0D69 |
SGN-E1349101 |
3e-46 |
60s acidic ribosomal protein p1 |
-4,85 |
| 0D70 |
NM |
|
Disease resistance |
-4,79 |
| 0D71 |
SGN-E666413 |
0 |
bahd family clade v |
-4,75 |
| 0D72 |
SGN-E1313854 |
0 |
Cytosolic aldehyde dehydrogenase |
-4,75 |
| 0D73 |
SGN-E1320568 |
0 |
Protein |
-4,72 |
| 0D74 |
SGN-E748200 |
1e-22 |
Anthranilate synthase alpha subunit |
-4,71 |
| 0D75 |
SGN-E1316428 |
0 |
Heat shock protein |
-4,68 |
| 0D76 |
NM |
|
Protein |
-4,67 |
| 0D77 |
SGN-E700960 |
8e-56 |
Magnesium transporter |
-4,67 |
| 0D78 |
SGN-E1321133 |
0 |
Glutathione s-transferase |
-4,66 |
| 0D79 |
SGN-E1309644 |
1e-33 |
---NA--- |
-4,66 |
| 0D80 |
NM |
|
nac domain ipr003441 |
-4,63 |
| 0D81 |
SGN-E791702 |
0 |
Zinc finger |
-4,57 |
| 0D82 |
SGN-E524668 |
0 |
Protein |
-4,49 |
| 0D83 |
SGN-E667829 |
0 |
gdsl-motif lipase hydrolase family protein |
-4,46 |
| 0D84 |
SGN-E955597 |
4e-15 |
Proline dehydrogenase |
-4,44 |
| 0D85 |
NM |
|
---NA--- |
-4,43 |
| 0D86 |
SGN-E1312314 |
0 |
PREDICTED: hypothetical protein [Vitis vinifera] |
-4,39 |
| 0D87 |
NM |
|
Undecaprenyl pyrophosphate phosphatase |
-4,37 |
| 0D88 |
SGN-E1350610 |
0 |
Cytosolic class i small heat-shock protein |
-4,35 |
| 0D89 |
NM |
|
---NA--- |
-4,34 |
| 0D90 |
SGN-E747084 |
2e-89 |
Alkaline alpha-galactosidase seed imbibition protein |
-4,34 |
| 0D91 |
NM |
|
Protein |
-4,32 |
| 0D92 |
SGN-E528554 |
|
pili assembly chaperone |
-4,31 |
| 0D93 |
SGN-E528554 |
7e-30 |
Cell-wall invertase |
-4,28 |
| 0D94 |
NM |
|
nbs-lrr resistance protein |
-4,26 |
| 0D95 |
NM |
|
Transcriptional family |
-4,26 |
| 0D96 |
NM |
|
---NA--- |
-4,24 |
| 0D97 |
NM |
|
Oxysterol binding protein |
-4,21 |
| 0D98 |
SGN-E1196563 |
2e-53 |
ap2 domain-containing transcription factor |
-4,20 |
| 0D99 |
SGN-E834183 |
0 |
nac domain protein nac2 |
-4,19 |
| 0D100 | SGN-E678677 | 0 | ---NA--- | -4,19 |
*The annotation of each sequence was established on the Coffee Genome Database (Vieira et al., 2006).
**No match with any sequence on the Solanacea Genomic Database.
Figure 3.
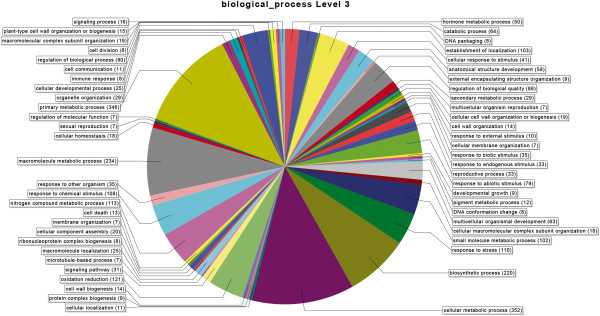
Biological process for up-regulated genes. Functional categorization of up regulated genes (%) in coffee genotypes (susceptible and resistant to leaf miner) without insect.
Figure 4.
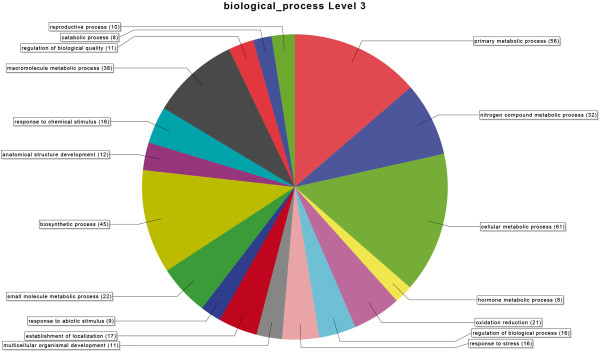
Biological process for down regulated genes. Functional categorization of down-regulated genes (%) in coffee genotypes (susceptible and resistant to leaf miner) without insect.
Metabolic pathways
The categorization of annotated genes using Kegg database was performed with the first 100 up and down regulated genes (Figures 1 and 2). Most of these genes are from starch and purine metabolism, and several others are involved in primary metabolism. Three main metabolic pathways are highlighted here: citrate metabolism, linoleic acid metabolism and phenylpropanoids metabolism.
We choose the citric acid cycle for further characterization as previous analyses using NMR indicated that lower levels of malate, a metabolite resulting from the citrate metabolism, are observed in resistant coffee leaves [33]. Several genes encoding citric acid cycle enzymes exhibit differential-expression (Figure 5). Expression of isocitrate lyase gene is repressed in resistant genotypes at T0 (fold change value -8,84), suggesting that synthesis of malate may be deficient, and therefore low levels of malate may accumulate in those leaves. However, this gene is up-regulated upon oviposition and egg ecclosion (fold change value 2,33).
Figure 5.
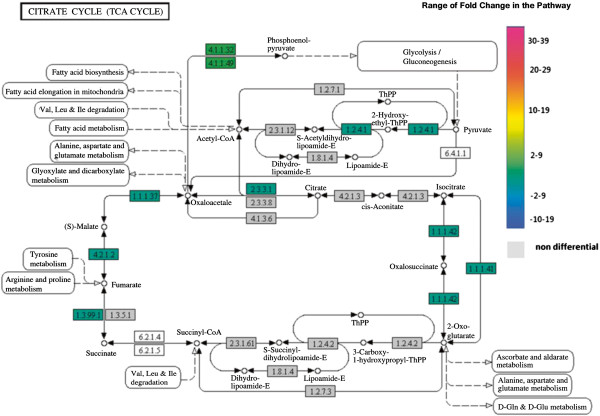
Citric acid pathway (KEGG database). Enzymes are identified by E.C. number and those corresponding to differentially expressed genes are highlighted according to the color scale. Color codes for each gene at T0 are as follows: red for up-regulated, green for down-regulated and grey for no differential expression. E.C number and correspondent enzyme: 1.1.1.37 - Malate dehydrogenase; 1.1.1.41 - Isocitrate dehydrogenase (NAD(+); 1.1.1.42 -Isocitrate dehydrogenase (NADP(+); 1.2.4.1 - Pyruvate dehydrogenase (acetyl-transferring) (Pyruvate dehydrogenase) 1.3.99.1 - Succinate dehydrogenase (Fumarate reductase); 2.3.3.1 - Citrate (Si)-synthase, 4.1.1.32 – Phosphopyruvate carboxylase; 4.2.1.2 – Fumarate hydratase (Fumarase).
The linoleic acid is the first substrate of the Jasmonic acid (JA) pathway, a major signaling pathway during herbivore-defense responses. Control resistant plants (T0) show up-regulation of jasmonate O-methyltransferase and lipoxygenase while differential expression for these genes was not observed at any stage in susceptible genotypes (Figure 6). Also, 13 genes from the alpha-linoleic acid metabolism and 57 genes from jasmonate biosynthesis were regulated in resistant plants (Figure 6). For instance, transcripts of enoyl-CoA hydratase and phospholipase A2 were four times more expressed in T0 than in T1 in resistant genotypes (Figure 6), but increased only at later stages in susceptible leaves.
Figure 6.
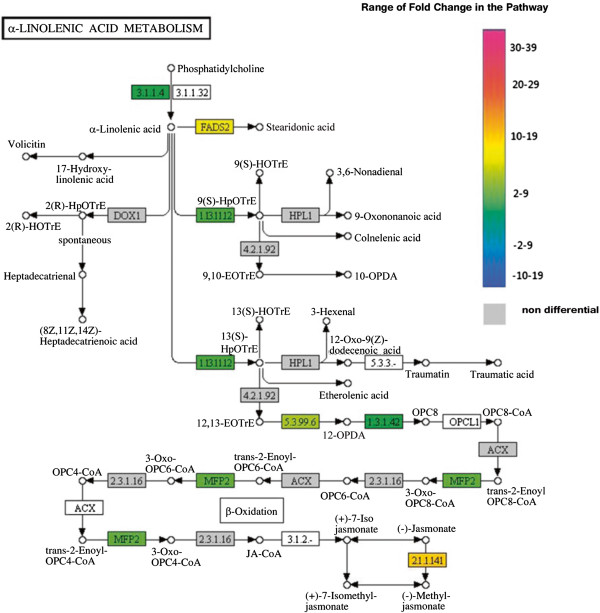
Alfa-Linolenic acid metabolism pathway (KEGG database). Enzymes are identified by E.C. number and those corresponding to differentially expressed genes are highlighted according to the color scale. Color codes for each gene at T0 are as follows: red for up-regulated, green for down-regulated and grey for no differential expression. E.C number and correspondent enzyme: 1.3.1.42 – 12-oxophytodienoate reductase; 3.1.1.4 – Phospholipase A2; 1.13.11.12 – Linoleate 13S-lipoxygenase; 5.3.99.6 – Allene oxide cyclase; 2.1.1.141 - Jasmonate O-methyltransferase; FADS2 – fatty acid desaturase 2 ; MFP2 – 3-hydroxyacyl-CoA dehydrogenase/ enoyl-CoA hydratase.
Phenylpropanoids are major plant phtytoalexins, part of the secondary metabolism (Figure 7). Twenty-seven genes from phenylpropanoids synthesis exhibited differential expression at T0, with foldchange values ranging from 9 to -5. Transcript levels of phenylalanine ammonia lyase (PAL), the enzyme that catalyzes the first step of the pathway, is up-regulated only at T0 in resistant plants (2,05), and this level decreases along insect development. In susceptible plants, PAL transcript levels increase at final steps of insect infection, T2 (2,55). However, genes fromlignin and isoflavones synthesis, downstream metabolites, such as cynnamyl alcoholdehydrogenase (-2,79) and isoflavone reductase (-1,20), are down-regulated in resistant plants. On the other hand, genes from biosynthesis of anthocyanins and tannins, such as flavonoide 3’-hydroxylase (T0 = 24; T1 = 5; T2 = 8) and leucoanthocyanidin dioxygenase (T0 = 2; T1 = 9; T2 = 3) are up-regulated at all times in resistant plants. This expression profile suggests that synthesis of anthocyanins and tannins is favored over synthesis of flavones. Also, activation of upstream genes such as phenylalanine ammonia lyase, chalcone synthase and flavonone dehydrogenase is observed at final stages in susceptible plants, indicating that phenylpropanoid biosynthesis is delayed.
Figure 7.
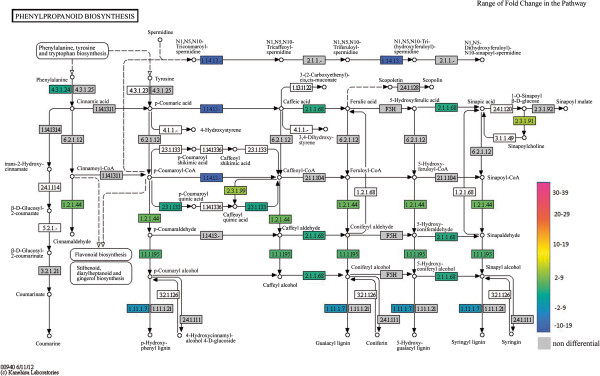
Phenylpropanoid biosysthesis pathway (KEGG database). Enzymes are identified by E.C. number and those corresponding to differentially expressed genes are highlighted according to the color scale. Color codes for each gene at T0 are as follows: red for up-regulated, green for down-regulated and grey for no differential expression. E.C number and correspondent enzyme: 1.1.1.195 - Cinnamyl-alcohol dehydrogenase; 1.2.1.44 - Cinnamoyl-CoA reductase; 1.11.1.7 - Peroxidase; 1.14.13 monoxygenase; 2.1.1.68 Caffeate O-methyltransferase; 2.3.1.91 - Sinapoylglucose--choline O-sinapoyltransferase 2.3.1.99 - Quinate hydroxycinnamoyltransferase, 2.3.1.133 - Shikimate O-hydroxycinnamoyltransferase; 4.3.1.24 - Phenylalanine ammonia lyase.
Several other defense-related genes are also positively regulated in resistant plants, including herbivore-response related genes glutathione-S-transferase and cysteine proteinase inhibitor. Apoptosis-related genes have a variable expression profile: catalase is up-regulated throughout insect development but citocrome c oxidase, superoxide dismutase and a senescence-associated protein have no differential expression, and polygalacturonase is up-regulated only at T0.
Validation of expression profile for selected candidate-genes
We selected 18 genes for validation, listed with corresponding expression levels on Table 5. Genes exhibited a consistent expression pattern when quantified by either microarray or qPCR, and the Pearson coefficient for this comparison is 0.92
Table 5.
Expression Pattern of eighteen genes selected for validation
| Name | Blast SOL network genomics | Annotation | Fold-change value * (microarray) | ΔΔCt value ** (relative expression) | Coef. Pearson.*** |
|---|---|---|---|---|---|
| 0U17 |
NM |
Protein |
94 |
39 |
0,9242 |
| 0U6 |
SGN E1316291 |
Acid phosphatase |
236 |
15 |
|
| 0U14 |
SGN E667484 |
Protein |
115 |
36 |
|
| 0U2 |
SGN E1352064 |
Caffeine synthase |
642 |
179 |
|
| 0U4 |
SGN E1310344 |
Metallothionein-like protein |
280 |
8,45 |
|
| 0U322 |
SGN E450221 |
nadp-dependent d-sorbitol-6-phosphate dehydrogenase |
6,56 |
3 |
|
| 0U1 |
SGN E628893 |
No hits |
1000 |
186,9 |
|
| 0U42 |
SGN E636199 |
---NA--- |
43 |
8,16 |
|
| 0D6 |
SGN E661762 |
---NA--- |
-35 |
-4,3 |
|
| 0D10 |
SGN E1033676 |
Glycerol-phosphate acyltransferase |
-16 |
-1,8 |
|
| 0D2 |
SGN E676870 |
Protein |
-235 |
-120 |
|
| 0D1 |
SGN E1320843 |
Protein |
-445 |
-200 |
|
| 0D19 |
SGN E673783 |
Isocitrate lyase |
-8 |
-6 |
|
| 0D3 |
SGN E1320843 |
Zinc finger |
-126 |
-17 |
|
| 0D4 |
SGN E1325444 |
Hypothetical protein |
-62 |
-36 |
|
| 0D8 |
NM |
Glycerol-3-phosphate acyltransferase 6 |
-17 |
-2,3 |
|
| 0D9 |
SGN E1321887 |
gdsl-motif lipase hydrolase-like |
-16 |
-2,2 |
|
| 0D7 | NM | Hypothetical protein | -24 | -5,8 |
*Microarray analysis value.
**qRT-PCR analysis value.
***Pearson coefficient correlation value.
Further qPCR analyses were performed to validate expression of selected leaf-miner resistance-associated candidate-genes (Figure 8). These included genes from pathways described above and genes with either no significant hits or similarity to unknown proteins, which may represent coffee specific genes, not yet identified or characterized.
Figure 8.
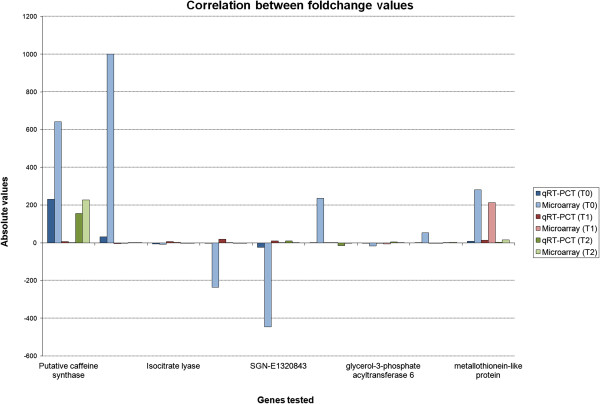
Correlation between foldchange values obtained from microarray analysis and qRT-PCR. Blue bars indicate values for T0 interaction (T0R_T0S), red bars for T1 interaction (T1R_T1S), green bars for T2 interaction (T2R_T2S). Dark colors of each interaction indicate fold change values from microarray analysis and soft color from qRT-PCR.
The putative caffeine synthase gene exhibited the greatest response to leaf-miner infection (Figure 8), as its expression was significantly higher (Relative Quantification value – RQv 230.45) in control resistant leaves, and also in later stages (RQv 155.24). The expression of gene SGN-E628893, encoding an unknown protein, is significantly higher (RQv 1000) in control resistant leaves than in susceptible ones (Figure 8). However, transcript levels dropped in resistant plants at first stages of infection, such as oviposition (RQv -1.53) and egg ecclosion (RQv 1.22). This gene is a good candidate for differentiation of resistant and susceptible plants, although possibly is not related to resistance.
The isocitrate lyase gene is down-regulated (RQv -6.38) in resistant leaves at T0. At initial steps of infection its transcript levels increased rapidly (RQv 5.49), but decreased during feeding stages. Other evaluated genes also exhibited a regulated expression. The glycerol-3-phosphate acyltransferase 6 gene, a trans-membrane protein associated with synthesis of cutin, is up regulated (RQv 11.12). A gene encoding metallothionein-like protein, a class of metal-chelators proteins with possible anti-oxidant role, is also up-regulated (RQv 12.88) at initial stages of insect development.
Genes encoding unknown proteins with lipase protein domain, such as SGN-U585128, SL2.40ch08 and SGN-U585128 were activated in resistant plants, being up-regulated at oviposition and/or egg ecclosion with RQ values of 19.72, 19.03 and 10.30 respectively. As these genes are down-regulated on leaves at T0, this profile during insect development suggests a role during defense response (Figure 8).
The expression profile of polygalacturonase gene was not confirmed by real-time PCR. According to microarray in silico analysis this gene is activated at T0 (fold change value 236,370), and afterwards transcript levels drop along insect development (fold change value 1,628). Yet by real-time PCR analysis transcript levels are similar in susceptible and resistant plants at all times. This result may reflect differences on genetic background of evaluated plants, once they are part of a population still segregating for some characteristics. However, as this was the only observed discrepancy between all performed analyses, these genetic differences may not be associated with the resistance trait.
Discussion
The use of resistant or tolerant cultivars represents an alternative for reducing the use of chemical defensives, the costs of production, and the negative impact over environment. In coffee, breeding for herbivore resistance is important once Coffea arabica, the main commercial Coffea species, is susceptible to almost all known coffee pathogens. Then resistance must be transferred from other compatible species, which is not always possible due to limited efficiency of inter-specific crosses.
At the Agronomic Institute (Campinas, Brazil) leaf-miner resistance genes have been transferred from C. racemosa to C. arabica through traditional breeding strategies, and although the program is currently at advanced generations, no resistant commercial cultivar is available yet. The lack of knowledge of molecular aspects controlling the resistance response, and the restricted genetic variability of breeding populations [34] limit the development of genomic-based selection tools. In this context, we aimed to provide information on molecular aspects of leaf-miner defense mechanisms and identification of reliable candidate-marker genes for assisted-selection. Those genomic tools associated with traditional breeding strategies guarantee that agronomical traits such as productivity and cup quality will be selected in advanced generations. Also, once novel genomic-markers associated with other desirable traits are developed for coffee genotypes, a genome-wide selection strategy will be possible to develop multiple-pathogen resistant cultivars. We chose the microarray analysis approach to assess the co-expression of a large amount of genes, including those that are not looked at in common analyses. Nevertheless, the results described in this work indicated that pathways regularly activated in response to herbivory, such as linoleic acid cycle, phenylpropanoids synthesis and apoptosis, are also activated during coffee-response to leaf-miner. Genes associated with jasmonate (JA) synthesis, such as lipoxygenase and enoyl-CoA hydratase, and with flavonoids synthesis, such as chalcone synthase and flavanone 3-hydroxylase-like, are up-regulated in resistant plants. Also, pathways from the primary metabolism, such as the citric acid cycle are down-regulated during leaf-miner defense response in resistant plants, a profile observed also in conifers [35].
Results of this study provide evidence that most genes encoding enzymes from the citric acid cycle are down-regulated in resistant plants (Figure 5). In a parallel analysis, a metabolite profile was established for resistant and susceptible genotypes using an NMR-based technique [33] and indicated that malate levels on resistant leaves are lower than in susceptible ones. Malate results from conversion of either fumarate or glyoxylate. Expression of fumarase, that converts fumarate into malate, is down-regulated at T0 in resistant genotypes (-2), what could explain the low malate levels. Production of malate from glyoxylate may also be deficient in resistant plants, once genes encoding for malate synthase, that converts glyoxylate into malate, and for isocitrate liase, the upstream enzyme that converts isocitrate into glyoxylate and succinate, are both down-regulated (-2,51 and -9, respectively). In contrast to this profile, susceptible plants exhibit a regular expression levels for these genes at T0. Therefore, both metabolic and transcriptional profiles support the affirmations that citrate cycle is down regulated in leaf-miner resistant coffee plants, and the model of down-regulation of primary metabolism in herbivore-resistant plants [36].
Biosynthesis of JA starts with alpha-linoleic acid release in non-injured tissues, triggered by systemin and phospholipase A2. Alpha-linoleic acid is then converted to JA after enzymatic steps performed by 13-lipoxygenase (LOX), allene oxide synthase (AOS), jasmonate o-methyltransferase and others [37,38]. Several genes from the JA biosynthesis pathway are up-regulated in resistant plants at T0, including those from downstream steps such as jasmonate o-methyltransferase which expression is 10-fold higher than in susceptible plants. All genes of the JA biosynthesis are either down-regulated or up-regulated at later stages in susceptible plants, as for instance expression of LOX (T0 -8.66; T1 1; T2 2) increases only at T1. These observations suggest that the JA signalling pathway, including intermediate signaling-molecules such as oxo-pentenyl-cyclopentane (OPC), may be impaired in susceptible plants. Down-regulation of genes from later steps of JA biosynthesis, such as allene oxide cyclase, allene oxide synthase, carboxyl methyltransferase, the enzyme that converts jasmonic acid into methyl jasmonate, is observed at T1 and T2 in resistant plants. Therefore, a feedback regulation may be activated, with a re-programming of transcriptional response upon leaf-miner infection.
Genes associated with biosynthesis of secondary compounds are shown here to be regulated. Expression profile of genes from phenylpropanoids biosynthesis, both up-stream genes such as PAL and CHS, and downstream genes such as flavonoide 3’-hydroxylase, leucoanthocyanidin dioxygenase, reveals a preferential synthesis of tannins and anthocyanins instead of ligninin, flavones and isoflavones in resistant plants. This profile indicates a direct defense strategy against leaf-miner, once among anthocyanins and tannins are found toxic compounds with antifeedants effects over insects [39,40].
Another gene linked to secondary metabolism is a putative caffeine synthase, which encodes one enzyme from caffeine biosynthetic pathway. Caffeine is an alkaloid distributed in coffee plant tissues and organs. The fact that expression of a gene from its biosynthesis is significantly increased upon leaf-miner infection suggests that caffeine may have a role in defense response. However, several studies regarding caffeine and leaf-miner development indicated that this compound has no effect on insect survival rates [41,42].
Once the ultimate goal of this study is to identify potential candidate for markers, several genes were selected for validation using real-time PCR. Potential candidates include: isocitrate lyase, which increased expression during initial steps of leaf-miner infection may be co-related with reduction of primary carbon metabolism; putative caffeine synthase, part of an important pathway of coffee plants; glycerol-3-phosphate acyltransferase 6, a gene associated with lipid metabolism and part of cutin biosynthetic pathway, a secondary metabolite [43]; and finally metallothionein-like protein gene, a metal-transporter protein family with an uncertain role in plant metabolism but previously associated with redox responses [44]. Future analyses include cloning and re-sequencing genomic regions of target genes from different genotypes in order to identify suitable polymorphisms.
Among selected genes are those that have no similarity with any known reported gene or protein. Although they could not yet be associated with a biological process, their expression profile was very specific and related to defense response. For instance, genes SGN-E676870, SGN-E1128614 SGN-E1320843 were activated upon leaf-miner infection in resistant plants, and therefore represent good candidates for further investigation. Another interesting unknown gene is SGN-E628893, which is highly activated in resistant plants at T0 but is repressed upon infection. The expression profile indicates that this gene is useful for early differentiation between resistant and susceptible plants.
In summary, differential expression profiles between resistant and susceptible genotypes are observed even in the absence of leaf-miner, indicating that defense is already build up in resistant plants, as a priming mechanism. Then, a systemic defense response may be more rapidly activated in resistant plants, once basic compounds such as nitrogen and sugars are readily available as a result of repression of primary metabolism. This shift in plant metabolism is common after a pathogen attack, where defense-related pathways are activated, resulting in reduction of growth and reproduction, and in changes on link-source relationship [45]. During herbivore-defense transcript levels of genes involved in photosynthesis are also down-regulated [21,46], probably as a strategy to liberate nitrogen compounds for the secondary metabolism. Maintenance of these physiological and metabolic states has a high energetic cost, and could represent a survival limitation if nutritional conditions on the field are depleted. Actually, field observations in cultivated areas demonstrated that leaf-miner resistant coffee plants, under a severe nutritional deficit, are attacked by the leaf-miner at the same intensity as susceptible plants [27].
Conclusions
As a basal defense state is decisive for triggering a rapid resistance response, genes associated with priming validated here, represent key genes for assisted-selection. Future studies will focus on comparisons of selected genes genomic sequences, from both resistant and susceptible parental lines, to identify suitable marker polymorphisms.
Methods
Plant materials
Resistant and susceptible coffee progenies were developed by the Coffee Breeding Program from the Agronomic Institute (IAC), Campinas, São Paulo, Brazil. The evaluated population (H14954-46), with 136 plants, is a F2BC5 generation of the inter-specific cross (C. racemosa X C. arabica) X C. arabica.
Plants were evaluated regarding the defense-response to Leucoptera coffeella using infestation methodology described by Guerreiro-Filho et al. [28]. After egg hatching, 1.8 cm leaf discs were taken from the leaves using a cork bore. Discs were placed on damp plastic foam and maintained in a plastic box for two weeks. Resistance/susceptibility response was visually scored according to the evaluation scale defined by Ramiro et al. [26].
Coffee leaf miner infestation of selected plants
Fifteen resistant (R) and fifteen susceptible (S) coffee plants previously selected were used for leaf-miner infestation. Plants/seedlings of each group (R and S) were challenged with L. coffeella in rearing cages and following the same procedures described above. Control non-infestaded plants of each group were also evaluated. Three independent infestation experiments were used for further analyses.
Leaves were collected from the third and fourth pair from plants during different stages of insect development. Stages corresponded to egg hatching, after 1 to 5 days after infestation with L. coffeella (T1), and egg eclosion and tissue injury, after 6 to 10 days after infestation (T2). Control non-infested leaves (T0) were also collected for each genotype. Three leaves of each plant and each stage of insect development were collected (totalizing nine leaf per time of sampling/genotype) and immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. Experimental design was completely randomized including three replicates for each sample.
RNA isolation and preparations
Total RNA for both NimbleGen microarray hybridization and real-time qPCR experiments was isolated using protocol described by Chang et al. [47]. RNA extractions were performed using 2 g of tissue of pooled samples. All RNA samples were analyzed by formaldehyde-agarose gel electrophoresis and by spectrophotometry to assess physical and chemical integrity. To avoid contamination by polyphenols, carbohydrates and proteins, only RNA samples with OD 260/280 and 260/230 > 1.8 were selected for further analysis. For microarray hybridizations, extracted RNA was also checked for purity and degradation using an Agilent Bioanalyzer 1000 (Agilent Technologies). Samples were stored at -80°C until further use.
cDNA double strand synthesis, labeling and hybridization
Ten thousands nanograms (10.000 ng) of each RNA sample were pooled and treated with DNAse- RNAase free for cDNA synthesis and labelling. Three biological replicates of each treatment were used for hybridization with the cDNA microarray chip. Equal amounts of each replicate from resistant and susceptible plants were pooled respectively to minimize variation between individual RNA samples. All RNA samples were sent to Roche NimbleGen Systems, where cDNA synthesis and Cy3 labeling were performed following the manufacturer’s procedures (Nimblegen Gene Expression Analysis protocol, Nimblegen Systems, Inc., Madison, WI, USA). Equal quantities of total RNA of each sample were converted to double strand cDNA (cDNA Synthesis System, Roche Applied Science). All the required equipments, reagents and procedures were provided and executed by Roche/NimbleGen.
Design and production of the Coffea ssp. Nimblegen® custom array
Arrays were designed using sequence information available at the Brazilian Coffee Genome Project, which contains sequences of around 33 K genes identified in EST libraries prepared from different physiological and metabolic situations [31]. The Coffea dataset was composed by quality-filtered contigs from different non-normalized ESTs/cDNA libraries of two coffee species Coffea arabica, Coffea canephora and Coffea racemosa, and by singlets of this assembly. Only sequences with at least one blast hit against NR database (e-value <1e-10) were used as source sequences to generate probes for the 12 coffee microarray. The probes were designed by Roche-NimbleGen software, which selected unique sequences regions for each gene to avoid multiple hybridization with gene family members. Each microarrays consisted of 135.000 probes with length of 48 nucleotides and Tm average from 68°C to 76°C, representing 22,000 genes, with a minimum of 6 probes/gene. The final probe list was submitted to Roche-NimbleGen, Inc. (Madison, WI, USA) for quality control and subsequent probe array layout. Additional probes were also included on the microarray by Roche-NimbleGen, Inc. for quality control of the hybridization process. Microarray manufacture was synthesized in situ by photolithography on glass slides using a random positional pattern by NimbleGen (http://www.nimblegen.com/).
Normalization and statistical analysis
Hybridized-microarray slides were imaged with a high resolution array scanner (GenePix 4000B Microarray Scanner, Molecular Devices Corp., Sunnyvale, CA, USA) and fluorescent signal intensities from each spot were quantified using NimbleScan Software (NimbleGen Systems Inc.). The intensity values were normalized using the oligo package from R statistics software [48]. The workflow used to normalize our data was followed as explained by the package provided for Nimblegen® expression microarrays. Fold change values were calculated comparing resistant and susceptible genotypes with and without infestation. All clusters were annotated using the blast2go software [48] in order to label them with their probable molecular function, biological process and cellular component. An automatic pipeline using Perl scripts was created to map each probe to its corresponding gene and annotation.
Differentially expressed genes (fold change values between 2 and -2) were identified using linear models and by taking into account technical and biological replicates. When individual probes met the criteria that average signals from resistant versus susceptible genotypes differed significantly by at least two fold, probes were selected for final analysis.
Functional characterization of differentially expressed genes was performed using Blast2GO [49] and also through directed searches on Gene Ontology (http://www.geneontology.org), KEGG (http://www.genome.jp/kegg) e InterPro (http://www.ebi.ac.uk/interpro/) databases.
Microarray validation
Validation of selected differentially expressed genes was performed by real-time PCR. Gene-specific primers were designed using Primer Express 3.0 (Applied Biosystems) and Premier Primer 5.0 (Premier Biosoft International, Palo Alto, CA, USA). Gene sequences were aligned with GeneBank reference sequences using the tBLASTx tool. Possible ORFs and functional and conserved domains were identified using the Open Reading Frame Finder (ORF FINDER) and CDD tools from the NCBI database. In order to guarantee gene-specificity and avoid amplification of multigene families, primers were designed upon target regions which included the conserved domain and/or motif and anchoring outside the conserved region. A list of designed primers is shown on Table 6.
Table 6.
Primer sequences used for validation in qRT-PCR analysis
| Name | Blast SOL network genomics | Annotation | Foward sequence | Reverse sequence |
|---|---|---|---|---|
| 0U17 |
NM |
Protein |
ACTACCAACATTCACAGCAGCTC |
TTAACCCTGTTGAAGGTTAGTGC |
| 0U6 |
SGN E1316291 |
Acid phosphatase |
CTAATTAACCCTCTCCGCATGAT |
GCCAACTCAGGCAATTATATACG |
| 0U14 |
SGN E667484 |
Protein |
TAGTCAAGAATATGGGCATGGAC |
ATACCTTCTTGATTCACGCCTTC |
| 0U2 |
SGN E1352064 |
Caffeine synthase |
AAAGGGAGCATTTACTCTTCCAAAG |
AGCATGCATCCTGAGAAATGTGGTA |
| 0U4 |
SGN E1310344 |
Metallothionein-like protein |
ATTCGTCTGCTCTGTGAAGATGT |
ATACATGTTTCCGCAGTTTCCT |
| 0U322 |
SGN E450221 |
nadp-dependent d-sorbitol-6-phosphate dehydrogenase |
CCTTTGTGGCTTCTAAGCAAAT |
GGAAAGCAGAGATTGACAAACAG |
| 0U1 |
SGN E628893 |
No hits |
CAAGGAAGATGCTTTTGACGAT |
TGTAATTATGCTGCTGGTGCTAC |
| 0U1 |
SGN E628893 |
No hits |
CATTTAGTTTGGAAGGGGACAA |
GGATACAGCCGGTAGGACTAACT |
| 0U42 |
SGN E636199 |
---NA--- |
ACCCGCCGGGAAACC |
GATGCACAGACAGGAATCACAAC |
| 0D6 |
SGN E661762 |
---NA--- |
TTGGTAATGATGGAAGTGTCCTC |
GCCAGTAATGGGATTGTAGTTGA |
| 0D10 |
SGN E1033676 |
Glycerol-phosphate acyltransferase |
CTAGCTTGACCAGGAAAGACAAG |
GACTCAGGACTGCTCATTTCATT |
| 0D2 |
SGN E676870 |
Protein |
GGCAACTACTGCATTCTATCAGC |
AAATGGATGAGCTGAAGGAGAAC |
| 0D1 |
SGN E1320843 |
Protein |
ACTAGTACTGGGTGTTGCCTCAA |
GGTGAGCAAATAGTTGTTGTTGC |
| 0D19 |
SGN E673783 |
Isocitrate lyase |
GGCCAGGAGCAACAGACATT |
ATTCTCTCACAATCTTGACTTTGCA |
| 0D3 |
SGN E1320843 |
Zinc finger |
CTGATTACGTCCGCTATCTCATT |
AACCTATCGGACCTGTACCTGTT |
| 0D4 |
SGN E1325444 |
Hypothetical protein |
TACTGGCACTAATGGAGGAAATG |
AATGGGGACAGATGTATCATCAC |
| 0D8 |
NM |
Glycerol-3-phosphate acyltransferase 6 |
TTCAAGAGTTTGGTACTGACGTG |
ATCATGGTCTGTCTCTCGATCTC |
| 0D9 |
SGN E1321887 |
gdsl-motif lipase hydrolase-like |
TCTTAACTGGACTTCCTCCAATG |
CCATTGAAGTTTAGAGCCACAAC |
| 0D7 |
NM |
Hypothetical protein |
CCAAGACAGTTGATCTCCCTCTA |
CGTAGTAGCTAGATGGTGCCAGT |
| 0U9 |
SGN-E1337775 |
Organ-specific protein |
GGTTTCTTTAGGGTTTCCTTCCT |
CACAGTGTGTGTGTTTTGTTCCT |
| 0U11 |
SGN-E1352070 |
Caffeine synthase |
CCTAGCAAGCCATTTTGGAG |
ATTCTTGGCAAACCTGTGGA |
| 0U6 |
SGN-E1316291 |
Acid phosphatase |
ATTACGGCTATGGCAGAATTAGC |
CACCATGTTTCCTTGTTTGAGA |
| 0U3 |
SGN-E1326397 |
Acidic endochitinase se2 |
CAGCAAATTCTTCCCTATGTCC |
CAGCGTTTCAGGGTTAACATAAG |
| 0U8 | SGN-E1327615 | Kunitz trypsin inhibitor | CTCTTCCTTTCATTTCTGCTCTTC | GACGTAGTACTCGACACCAGGAC |
Corresponding cDNAs were synthesized from 250 ng of total RNA using the RevertAidTM Minus First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer protocol. PCR products were amplified using primers designed with Primer Express 3.0 (Applied Biosystems) and analyzed by Premier Primer 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). In order to confirm primer specificity and presence of single amplicons, all PCR products were analyzed through a dissociation curve, with temperature varying from 60°C to 95°C.
Thermocycling and fluorescence detection were performed using ABI Prism 7300 Sequence Detection System (Perkin-Elmer Applied Biosystem). Real-time PCR amplification was carried out in a final volume of 15 μl by reaction using equal amounts of cDNAs (2 μl - 200 ng/μl) as template, 0,2 μM of each primer and 7,5 μl of Maxima SYBR Green/ROX qPCR master Mix (fermentas, USA) at the following conditions: 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 2 min, 62°C for 30 seg, 72°C for 30 seg. Data was collected during extension fase. Three independent qPCR reactions were performed for final quantification.
Expression levels of GAPDH were used as endogenous control. Relative gene expression was calculated using the 2-ΔΔCT method (where CT is threshold cycle) [50]. The Pearson correlation coefficient of linear regression from 18 pairs of microarray/qPCR expression ratios was calculated to validate the qPCR analysis.
Competing interests
The authors declare that they do not have any non-financial competing interests political, personal, religious, ideological, academic, intellectual, commercial or any other competing interests.
Authors’ contributions
JCM and DCC carried out experiments of infestation and the molecular genetic analysis by qRT-PCR. Both authors contributed equally for the experiments. PFG carried out the metabolic pathways analyses. ROV and MFC participated in the microarray analyses and performed the statistical analysis. MPM, LP, OGF performed the design and coordination of the study. JCM, DCC and MPM wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Danielle C Cardoso, Email: danielleccard@yahoo.com.br.
Juliana C Martinati, Email: julianamartinati@gmail.com.
Poliana F Giachetto, Email: poliana.giachetto@embrapa.br.
Ramon O Vidal, Email: ramon.vidal@gmail.com.
Marcelo F Carazzolle, Email: mcarazzo@lge.ibi.unicamp.br.
Lilian Padilha, Email: lilian.padilha@embrapa.br.
Oliveiro Guerreiro-Filho, Email: oliveiro@iac.sp.gov.br.
Mirian P Maluf, Email: mirian.maluf@embrapa.br.
References
- Herms DA, Mattson WJ. The dilemma of plants: to grow or defend. Q Rev Biol. 1992;15:283–335. doi: 10.1086/417659. [DOI] [Google Scholar]
- Giron D, Frago E, Glevarec G, Pieterse CMJ, Dicke M. Cytokinins as key regulators in plant–microbe–insect interactions: connecting plant growth and defence. Funct Ecol. 2012. doi:10.1111/1365-2435.12042.
- Zangerl AR. Evolution of induced plant responses to herbivores. Basic Appl Ecol. 2003;15:91–103. doi: 10.1078/1439-1791-00135. [DOI] [Google Scholar]
- Berger S, Sinha AK, Roitsch T. Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J Exp Bot. 2007;15:4019–4026. doi: 10.1093/jxb/erm298. [DOI] [PubMed] [Google Scholar]
- Rensink WA. et al. Gene expression profiling of potato responses to cold, heat, and salt stress. Funct Integr Genomics. 2005;15:201–207. doi: 10.1007/s10142-005-0141-6. [DOI] [PubMed] [Google Scholar]
- Restrepo S. et al. Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Plant–Microbe Interact. 2005;15:913–922. doi: 10.1094/MPMI-18-0913. [DOI] [PubMed] [Google Scholar]
- Baxter CJ. et al. Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J Exp Bot. 2005;15:1591–1604. doi: 10.1093/jxb/eri154. [DOI] [PubMed] [Google Scholar]
- Eriksson EM. et al. Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004;15:4184–4197. doi: 10.1104/pp.104.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Matthews BFSGMD. The soybean genomics and microarray database. Nucleic Acids Res. 2004;15:D398–D400. doi: 10.1093/nar/gkh126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy P. et al. Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant–Microbe Interact. 2004;15:1051–1062. doi: 10.1094/MPMI.2004.17.10.1051. [DOI] [PubMed] [Google Scholar]
- Clarke B, Rahman S. A microarray analysis of wheat grain hardness. Theor Appl Genet. 2005;15:1259–1267. doi: 10.1007/s00122-005-1962-3. [DOI] [PubMed] [Google Scholar]
- Close TJ. et al. A new resource for cereal genomics: 22 K barley GeneChip comes of age. Plant Physiol. 2004;15:960–968. doi: 10.1104/pp.103.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oztur ZN. et al. Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol. 2002;15:551–573. doi: 10.1023/A:1014875215580. [DOI] [PubMed] [Google Scholar]
- Cho Y, Fernandes J, Kim S-H, Walbot V. Gene-expression profile comparisons distinguish seven organs of maize. Genome Biol. 2002;15:1–16. doi: 10.1186/gb-2002-3-9-research0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, Elumalai R, Galbraith DW, Pierson E, Walbot V. Comparison of RNA expression profiles based on maize EST frequency analysis and microarray hybridization. Plant Physiol. 2002;15:896–910. doi: 10.1104/pp.010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters DLE, Holton TA, Ablett EM, Lee LS, Henry RJ. cDNA microarrays analysis of developing grape (Vitis vinifera cv Shiraz) berryskin. Funct Integr Genomics. 2005;15:40–58. doi: 10.1007/s10142-004-0124-z. [DOI] [PubMed] [Google Scholar]
- Watkinson JI. et al. Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed loblolly pine. Plant Physiol. 2003;15:1702–1716. doi: 10.1104/pp.103.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N, Theologis A, Yang Wei H, Hubbell E, Au M, Chung EY, Lashkari D, Lemieux B, Dean C, Lipshutz RJ, Ausubel FM, Davis RW, Oefner PJ. Genome wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet. 1999;15:203–207. doi: 10.1038/13833. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;15:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;15:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D, Iqbal J, Lehmann K, Gase K, Saluz HP, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: V. microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol. 2003;15:1877–1893. doi: 10.1104/pp.102.018176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell. 2004;15:3132–3147. doi: 10.1105/tpc.104.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Weisser WW, Baldwin IT. An analysis of plant–aphid interactions by different microarray hybridization strategies. Mol Ecol. 2004;15:3187–3195. doi: 10.1111/j.1365-294X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Baldwin IT. Microarray analysis of salicylic acid- and jasmonic acid signalling in responses of Nicotiana attenuate to attack by insects from multiple feeding guilds. Plant Cell Environ. 2004;15:1362–1373. doi: 10.1111/j.1365-3040.2004.01228.x. [DOI] [Google Scholar]
- Rodriguez-Saona CR, Musser RO, Vogel H, Hum-Musser SM, Thaler JS. Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds. J Chem Ecol. 2010;15:1043–1057. doi: 10.1007/s10886-010-9854-7. [DOI] [PubMed] [Google Scholar]
- Ramiro DA, Guerreiro-Filho O, Queiroz-Voltan RB, Matthiesen SC. Anatomical characterization of leaves from coffee plants resistant and susceptible to leaf miner. Bragantia. 2004;15:363–372. doi: 10.1590/S0006-87052004000300006. [DOI] [Google Scholar]
- Guerreiro-Filho O. Coffee leaf miner resistance. Bra J Plant Physiol. 2006;15(1):109–117. doi: 10.1590/S1677-04202006000100009. [DOI] [Google Scholar]
- Guerreiro-Filho O, Silvarolla MB, Eskes AB. Expression and mode of inheritance of resistance in coffee to leaf miner Perileucoptera coffeella. Euphytica. 1999;15:7–15. doi: 10.1023/A:1003427613071. [DOI] [Google Scholar]
- Mondego JMC, Guerreiro-Filho O, Bengtson MH, Drummond RD, Felix JM, Duarte MP, Ramiro D, Maluf MP, Sogayar MC, Menossi M. Isolation and characterization of Coffea genes induced during coffee leaf miner (Leucoptera coffeella) infestation. Plant Sci. 2005;15:351–360. doi: 10.1016/j.plantsci.2005.03.020. [DOI] [Google Scholar]
- Silvestrini M, Maluf MP, Silvarolla MB, Guerreiro-Filho O, Medina-Filho HP, Vanini MMT, Oliveira AS, Gaspari-Pezzopane C, Fazuoli LC. Genetic diversity of a Coffea germoplasm collection assessed by RAPD markers. Gen Resour Crop Evol. 2008;15:901–910. doi: 10.1007/s10722-007-9295-5. [DOI] [Google Scholar]
- Vieira LGE. et al. Brazilian coffee genome project: an EST-based genomic resource. Braz J Plant Physiol. 2006;15(1):95–108. doi: 10.1590/S1677-04202006000100008. [DOI] [Google Scholar]
- Vidal RO, Mondego JM, Pot D, Ambrosio AB, Andrade AC, Pereira LF, Colombo CA, Vieira LG, Carazzolle MF, Pereira GA. A high-throughput data mining of SNPs in Coffea spp ESTs suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol. 2010;15(3):1053–1066. doi: 10.1104/pp.110.162438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbello DM. Standardization of extraction technique for metabolites identification of resistant coffee to leaf miner through 1H NMR. 2009. Master degree in Tropical and Subtropical Agriculture of the Agronomy Institute of Campinas, Campinas, SP. Dissertation.
- Pinto FO, Maluf MP, Guerreiro-Filho O. Study of simple sequence repeat markers from coffee expressed sequences associated to leaf miner resistance. Pesq Agropec Bras. 2007;15(3):377–384. doi: 10.1590/S0100-204X2007000300011. [DOI] [Google Scholar]
- Ralph SG, Yueh H, Friedmann M, Aeschliman D, Zeznik JA. et al. Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell Environ. 2006;15:1545–1570. doi: 10.1111/j.1365-3040.2006.01532.x. [DOI] [PubMed] [Google Scholar]
- Scwachtje J, Baldwin IT. Why does herbivore attack reconfigure primary metabolism? Plant Physiol. 2008;15:845–851. doi: 10.1104/pp.107.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol. 1997;15:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot. 2007;15:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;15:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Lampert E. Influences of plant traits on immune responses of specialist and generalist herbivores. Insects. 2012;15:573–592. doi: 10.3390/insects3020573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzafera P, Guerreiro-Filho O. Caffeine does not protect coffee against the leaf miner Perileucoptera coffeella. J Chem Ecol. 2000;15(6) [Google Scholar]
- Magalhães STV, Fernandes FL, Demuner Y, Picanço MC, Guedes RNC. Leaf alkaloids, phenolics, and coffee resistance to the leaf miner Leucoptera coffeella (Lepidoptera: Lyonetiidae) J Econ Entomol. 2010;15(4):1438–1443. doi: 10.1603/EC09362. [DOI] [PubMed] [Google Scholar]
- Blee E, Schuber F. Biosynthesis of cutin monomers: involvement of a lipoxygenase peroxygenase pathway. Plant J. 1993;15:113–123. doi: 10.1046/j.1365-313X.1993.04010113.x. [DOI] [Google Scholar]
- Blindauer CA, Leszczyszyn OI. Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat Prod Rep. 2010;15:720–741. doi: 10.1039/b906685n. [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;15:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- Tang JY, Zielinski RE, Zangerl AR, Crofts AR, Berenbaum MR, DeLucia EH. The differential effects of herbivory by first and fourth instars of Trichoplusia ni (Lepidoptera: Noctuidae) on photosynthesis in Arabidopsis thaliana. J Exp Bot. 2006;15:527–536. doi: 10.1093/jxb/erj032. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A single and efficient method for isolating RNA from pine trees. Plant Mol Biol. 1993;15:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high density oligonucleotide SNP array data. Biostatistics. 2007;15(2):485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gomes JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;15:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods. 2001;15:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]